Abstract
Many positive-stranded RNA viruses contain short, single-stranded 3′ ends that are vulnerable to degradation by host cellular RNases. Therefore, the existence of a 3′-end repair mechanism (analogous to cellular telomerases) must be required and/or advantageous for RNA viruses. Accordingly, we provide evidence suggesting that deletions of up to 6 nt from the 3′ end of satellite (sat-) RNA C (a small parasitic RNA associated with turnip crinkle carmovirus) are repaired to the wild-type sequence in vivo and in vitro. The novel 3′-end repair mechanism involves the production of 4–8 nt oligoribonucleotides by abortive synthesis by the viral replicase using the 3′ end of the viral genomic RNA as template. Based on our in vitro results, we postulate that the oligoribonucleotides are able to prime synthesis of wild-type negative-strand sat-RNA C in a reaction that does not require base pairing of the oligoribonucleotides to the mutant, positive-strand RNA template. The discovery of a 3′-end repair mechanism opens up new strategies for interfering with viral infections.
Keywords: viral polymerase, satellite RNAs, telomerase, abortive cycling, priming
Organisms with linear genomes must develop mechanisms for repairing their 3′ ends, which are exposed to nuclease degradation and/or loss during replication. While the 3′-end repair of cellular chromosomes is a function of a specialized and evolutionary conserved enzyme complex known as telomerase (1–4), an analogous mechanism operating for single-stranded RNA viruses has been proposed but not yet defined (5–8). The significance of a 3′-end repair mechanism for RNA viruses is emphasized by the vulnerability of their single-stranded 3′ ends to cellular RNases. RNA viruses likely use “passive” protection mechanisms, such as sequestering replicating RNAs into virus-induced membranous structures (9–11) and binding specific viral proteins to terminal sequences (12). However, RNA viruses that do not have the stabilizing features of esterified amino acids or poly(A) tails covalently linked to their 3′ ends (13–15) may have periods in their life-cycle when single-stranded RNA 3′ ends are exposed to exonucleases (e.g., translation).
We have used a positive (+)-strand RNA virus, turnip crinkle carmovirus (TCV, Fig. 1A) to demonstrate the function and mechanism of “active” 3′-end repair. TCV is associated with several small, subviral RNAs such as sat-RNAs that require products encoded by the viral genomic RNA for accumulation in plants and protoplasts (17, 18). Both the genomic RNA and the sat-RNAs contain short single-stranded 3′-end sequences that terminate with the motif CCUGCCC-3′ (19). Deletions of 6–13 bases at the 3′ end of sat-RNA D (194 bases) were repaired in vivo by additional truncations to the −14 position followed by replacement of the missing sequence with a short segment derived from various regions of TCV or a nonviral template, followed by the motif C1-2UGC1-3 (7, 8).
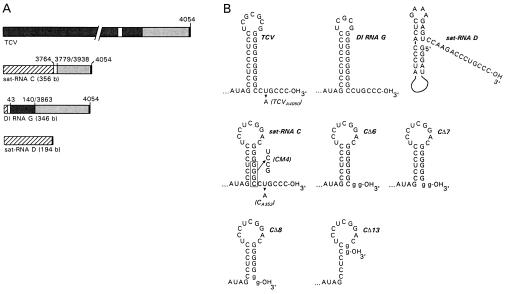
Figure 1.
Sequences and secondary structures of TCV and TCV-associated subviral RNAs. (A) Schematic representations of TCV, satellite RNA (sat-RNA) C, defective interfering (DI) RNA G, and sat-RNA D. Similar sequences are shaded alike. Positions of TCV-related sequence in the subviral RNAs are indicated. (B) Secondary structures of the 3′-terminal sequences of TCV, DI RNA G, sat-RNA D, sat-RNA C (16), and mutants derived from sat-RNA C. Plasmid-derived nucleotides present in the transcripts of sat-RNA C mutants containing 3′-terminal deletions are in lowercase letters. Point mutations that were generated in wild-type TCV and sat-RNA C are indicated, with names of mutants in parentheses.
Analysis of repair of 3′-terminal deletions has now been extended to include a second TCV sat-RNA, sat-RNA C (Fig. 1A). We have found that sat-RNA C containing a deletion of the 3′-terminal 6-base motif was repaired in vivo using the analogous motif of TCV. To determine the possible mechanism of repair, we demonstrate that the TCV replicase produces 4- to 8-base abortive products in vitro and can use such oligoribonucleotides to prime negative-strand synthesis using both wild-type and mutated sat-RNA C templates. Based on these results, a novel, viral replicase-mediated 3′-end repair model is proposed that involves the synthesis of a pool of oligoribonucleotides and their subsequent utilization to prime negative-strand synthesis, thereby correcting deletions at the 3′ ends of TCV-associated RNAs.
MATERIALS AND METHODS
Construction of 3′ End Mutants of TCV and Sat-RNA C.
To generate transcripts of sat-RNA C and TCV with deletions at their 3′ ends, pT7C(+), containing a full-length cDNA of sat-RNA C downstream from a T7 RNA polymerase promoter (20), or pTCV66, containing a full-length cDNA of TCV isolate TCV-M downstream from a T7 RNA polymerase promoter (21), were linearized with SmaI, treated with Bal31 (New England Biolabs), and plasmids were then digested for a range of empirically determined times. Blunt ends were generated with polymerase I large fragment (New England Biolabs), and plasmids were then digested with NcoI [for pT7C(+)] or EcoRI (for pTCV66), which cleave within the cDNA sequence. Fragments containing deletions were recloned into either pT7C(+) or pTCV66 that had been previously digested with SmaI and either NcoI [for pT7C(+)[ or EcoRI (for pTCV66).
To construct TCVA4050 and CA352, pTCV66 and pT7C(+) were used for PCR-based mutagenesis. Primers containing altered bases spanned the 3′ ends of the cloned molecules and adjacent vector sequences that contained restriction enzyme sites. Upstream primers were selected that allowed restriction digestion of the PCR products to facilitate recloning into similarly digested, dephosphorylated, parental clone vectors. To construct C/3′TCV, the SpeI–SmaI fragment of pT7C(+) was replaced with the corresponding fragment of pTCV66.
RNA Synthesis, Plant Inoculations, and Detection of TCV and Sat-RNAs.
KpnI-digested pT7C(+), XbaI-digested pTCV66, or their similarly digested deletion mutants were templates for bacteriophage T7 RNA polymerase transcription reactions (7, 20). This resulted in transcripts containing wild-type 5′ ends, and 3′ ends terminating with the plasmid-derived sequence GG-3′ (for sat-RNA C) or GGGGAUCCUCUAG-3′ (for TCV). For reactions requiring TCV or sat-RNA C without 3′-end deletions, plasmids were digested with SmaI, resulting in transcripts with wild-type 5′ and 3′ ends.
Inoculation of turnip cv. Just Right plants, isolation of total plant RNA, and detection of TCV and sat-RNAs in infected plants was as described (7).
Analysis of 3′-Terminal Sequences.
Sat-RNA C and TCV were gel purified and poly(A) tails added to the 3′ ends using poly(A) polymerase as described (7). cDNAs were synthesized using a primer containing a 19-nt sequence joined to 17 T residues as described (7). PCR was performed using primers identical to positions 57–78 of sat-RNA C, or 3773–3789 of TCV, and a second primer consisting of the 19-nt sequence mentioned above as described (7). PCR products were cloned into the SmaI site of pUC19 and sequenced.
Analysis of Aborted Synthesis Products Generated in TCV RdRp Reactions in Vitro.
Partially purified TCV RNA-dependent RNA polymerase (RdRp) was prepared as described (20). Plus-strand RNA templates (3 μg) were transcribed by the partially purified TCV RdRp using reaction conditions previously described (20) except that [α-32P]ATP was used as the radioactive nucleotide and tRNA was omitted. After the RdRp reactions, RNA samples were extracted with phenol/chloroform (1:1), and the RNAs were precipitated with the addition of 1/10 vol of 3 M sodium acetate and 3 vol of 95% ethanol. The RNA was recovered by centrifugation for 25 min at 13,200 rpm in a microcentrifuge at 4°C and subjected to electrophoresis on 20% polyacrylamide/8 M urea gels. A synthetic double-stranded oligodeoxynucleotide containing a T7 RNA polymerase promoter (template strand, 5′-GCTCGAATTCCCTATAGTGAGTCGTATTA-3′; a gift of A. Ujvari and C. T. Martin, University of Massachusetts, Amherst) and [α-32P]ATP were used in T7 RNA polymerase reactions to obtain labeled 5-, 6-, and 7-mer ribonucleotide size markers (22).
Analysis of Primer-Extension Products Synthesized in TCV RdRp Reactions in Vitro.
Three micrograms of (+)- or minus (−)-strand RNA transcripts were used as templates for RdRp reactions using conditions previously described (20) except that in some of the reactions the mononucleotide label was omitted and replaced with either 1 or 10 μg of the following oligoribonucleotide primers synthesized by T7 RNA polymerase and labeled with [α-32P]ATP (NEN): 6a, GACGGG-5′; 5, AAGGG-5′; 6b, CCCAGG-5′. In addition, the amount of GTP was reduced from 1 mM to 66 μM, because the reduction in GTP levels facilitated the priming reaction by 2-fold. The double-stranded oligodeoxynucleotides used for the synthesis of labeled ribonucleotide primers were (template strand is shown): 6a, 5′-CTGCCCTATAGTGAGTCGTATTA-3′; 5, 5′-TTCCCTATAGTGAGTCGTATTA-3′; 6b, 5′-GGGTCCTATAGTGAGTCGTATTA-3′ (22). Unincorporated nucleotides were removed by two isopropanol/ammonium acetate precipitations. The oligoribonucleotides were recovered by centrifugation and analyzed using 20% polyacrylamide gel/8 M urea gels. The absence of unincorporated [α-32P]ATP in the labeled ribonucleotide primer preparations was determined by the lack of incorporation of trace amounts of labeled nucleotides into products using the highly active (−)-strand sat-RNA C as template in subsequent RdRp reactions in the presence of the primer and nonradioactive nucleotides (1 mM of ATP, CTP and UTP; 66 μM of GTP). After the RdRp reactions, RNA samples were extracted with phenol/chloroform (1:1), and the RNAs recovered by isopropanol/ammonium acetate precipitation and centrifugation. Plasmid pT7C(+) was linearized with SmaI or SpeI and used in T7 transcription reaction (20) in the presence of [α-32P]ATP to produce 356- and 256-nt RNA markers, respectively. Samples were subjected to electrophoresis through 5% polyacrylamide/8 M urea gels and analyzed using a PhosphorImager (Molecular Dynamics).
RESULTS AND DISCUSSION
Sequences Participating in Repair of the 3′ Ends of Sat-RNA C Mutants in Vivo Are Derived from TCV.
The 3′ ends of TCV (the use of “TCV” throughout refers to the genomic RNA) and sat-RNA C fold into stable stem–loop structures followed by the single-stranded motif CUGCCC-3′ (Fig. 1B). This 3′-terminal hairpin, which serves as the promoter for (−)-strand synthesis (16), is analogous in structure and function to other elements found at the 3′ end of many other RNA viruses of plants and animals (23–27). A series of sat-RNA C mutants were constructed with 3′-end deletions to mimic the action of cellular RNases (Fig. 1B). Of the six plants inoculated with transcripts of sat-RNA C with a deletion of the 3′-terminal 6 bases (CΔ6; the number reflects the number of bases deleted), three accumulated sat-RNA C-sized species and three had no detectable sat-RNA C (data not shown). Twenty-three of the 24 full-sized CΔ6-derived clones analyzed from these plants had regained the wild-type termination sequence CUGCCC-3′, while the remaining clone terminated in CGGCCC-3′ (Table 1).
Table 1.
The 3′-end sequences in sat-RNA C generated in vivo from transcripts with 3′-end alterations
| T | Plant | Genomic RNA
|
|
|---|---|---|---|
| TCV [GGGGCCUGCCC] | TCVA4050 [GGGGCCAGCCC] | ||
| C | [GGGGGGCCUGCCCgg] | [GGGGGGCCUGCCCgg] | |
| 1 and 2 | GGGGGGCCUGCCC (7, 4) | GGGGGGCCUGCCC (9, 18) | |
| 1 | UUU (1) | ||
| 1 | 13-base deletion (1) | ||
| 2 | GG (1) | ||
| CA352 | [GGGGGGCCAGCCCgg] | ||
| 1 | GGGGGGCCAGCCC (7) | ||
| 1 | GGGGGGCCUGCCC (2) | ||
| C + CA352 | [GGGGGGCCUGCCCgg] | ||
| [GGGGGGCCAGCCCgg] | |||
| 1 and 2 | GGGGGGCCUGCCC (11, 10) | ||
| CΔ6 | [GGGGGGCgg] | [GGGGGGCgg] | |
| 1–3 | GGGGGGCCUGCCC (3, 6, 7) | GGGGGGCCAGCCC (4, 10) | |
| 1 | GGGGGGCGGCCUGCCC (5) | ||
| 1 | 41-base deletion (1) | ||
| 2 | G (1) | GGGGGGGGGGGCCAGCCCG (1) | |
| 3 | GGGGGGCCGGCCC (1) | ||
| 3 | GGGGGGGGGCCUGCCC(G) (2) | ||
| CΔ7 | [GGGGGGgg] | ||
| 1 | GGGGGGGGGGCCUGCCC (9) | ||
| 2 | GGGGGGCCUGCCC (4) | ||
| CΔ8 | [GGGGGgg] | ||
| 1 and 2 | GGGGGGGGCCUGCCC (6, 3) | ||
| 2 | GGGGGGGGGCCUGCCC (2) | ||
| 2 | GGGGGGGG (1) | ||
| CΔ13 | [gg] | ||
| 1–6 | ND | ||
Six turnip plants were inoculated with transcripts of (+)-strand sat-RNA C, either wild-type or containing the alterations shown, and transcripts of either (+)-strand TCV or TCV with a U to A alteration at position 4050 (TCVA4050). Transcript sequences are enclosed in brackets; only the sequence between positions 4044 or 344 and the 3′ terminus is shown for TCV and sat-RNA C, respectively. Lowercase nucleotides in the transcript sequence are plasmid-derived. Unbracketed sequences are from cloned sat-RNA C generated in planta from the transcripts indicated. Sequences in italics denote differences between sat-RNA C accumulating in plants and the inoculated transcripts. Sequences underlined can be derived from the 3′ end of TCV. Several clones also contained additional G residues upstream from the terminal 7-nt motif that could be derived from either the plasmid-derived nucleotides (GG) at the ends of the transcripts or from TCV. Numbers in parentheses reflect the number of clones with the sequence indicated from one or more plants. All clones terminated in poly(A), which was used in the cloning procedure. T, transcript used in the inoculum; ND, no sat-RNA detected; C, wild-type sat-RNA C; CA352, sat-RNA C with a U to A alteration at position 352. Other sat-RNA C transcripts have the 3′-terminal deletions indicated.
Both CΔ7 and CΔ8 had deletions of the terminal sequence CCUGCCC-3′ and extended the number of upstream G residues to eight and seven, respectfully (due to two plasmid-derived G residues at the ends of the transcripts). As with the CΔ6 transcripts, CΔ7 and CΔ8 transcripts were infectious, with four of six or three of six plants accumulating sat-RNA C-sized species, respectively (data not shown). The sat-RNA C-sized species were cloned from two CΔ7- and CΔ8-positive plants. All sat-RNA clones contained variable numbers of upstream contiguous G residues (from 6 to 10), and 11 of 12 clones had regained the wild-type terminal sequence CCUGCCC-3′. None of the six plants inoculated with TCV and CΔ13 transcripts accumulated sat-RNA C-sized species.
To determine the origin of the repaired 3′ ends in the sat-RNA C deletion mutants, a U to A alteration was introduced into TCV at position 4050 generating TCVA4050 (terminating sequence: CCAGCCC-3′). All six turnip plants inoculated with TCVA4050 had normal TCV symptoms and accumulated wild-type levels of viral RNA as determined by ethidium bromide-stained gel electrophoresis, and 14 of 14 cDNA clones sequenced from two plants contained the U to A alteration (data not shown). TCVA4050 was coinoculated onto six turnip plants with CΔ6 to determine if the marker mutation appeared in the repaired sat-RNA. Fourteen CΔ6-derived clones generated from two plants had precisely repaired 3′ ends with the marker mutation, while 1 clone had the marker mutation and additional upstream G residues (Table 1). This demonstrates that TCV is involved in 3′-end repair of sat-RNA C. In the opposite scenario, six plants inoculated with sat-RNA C terminating in CAGCCC-3′ (CA352) and TCV with a deletion of the 3′-terminal sequence CUGCCC-3′ (TCVΔ6) did not become infected. This suggests either that only TCV can be used for the 3′-end repair function or that nonreplicating TCV generates insufficient RdRp for use in repair.
Abortive Synthesis of Oligoribonucleotides by the TCV RdRp in Vitro.
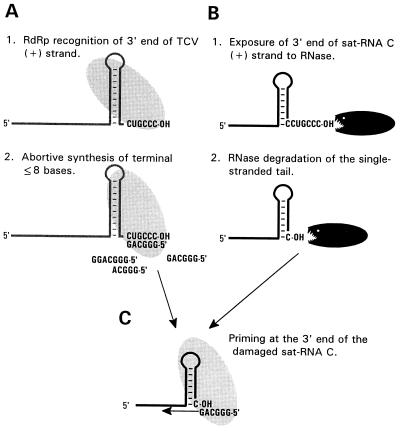
To explain how the 3′ end of TCV could be used for repair of sat-RNA C 3′-end truncations, a model is proposed in which short oligoribonucleotides complementary to the 3′ terminus of (+)-strand TCV are synthesized by the TCV RdRp through abortive cycling (Fig. 2). Abortive cycling is the process by which short, abortive oligoribonucleotides are synthesized by an initial polymerase complex that has not yet undergone the transition to an elongation complex (22, 28). After synthesis of the abortive products, the model suggests that the oligoribonucleotides are then used by the RdRp in a primer-extension reaction at the promoter for (−)-strand synthesis located at the 3′ end of the (+)-strand of sat-RNA C.
Figure 2.
Model for the repair of deletions at the 3′ end of sat-RNA C. (A) Synthesis of abortive products by the RdRp at the 3′ end of (+)-strand TCV. (B) RNase-mediated damage at the single-stranded 3′ end of sat-RNA C (a deletion of six bases is shown). (C) Use of the “abortive” oligoribonucleotides by the RdRp to prime (−)-strand sat-RNA C synthesis, thereby repairing the damaged 3′ end.
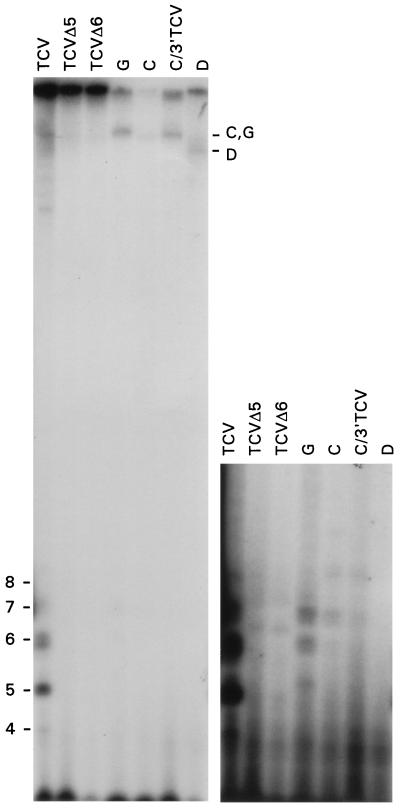
To test for the synthesis of short abortive products, in vitro transcription reactions were performed with wild-type and mutant transcripts of (+)-strand TCV, sat-RNA C, and other TCV subviral RNAs using a partially purified, template-dependent RdRp extract prepared from TCV-infected turnip plants (Fig. 3). RdRp reactions with wild-type TCV template accumulated oligoribonucleotides of 4–8 nt in more than 100-fold molar excess compared with higher molecular weight products, supporting the model (Fig. 2A, step 2). When the TCV 3′-terminal 5 or 6 nt were replaced with a 13 nt, plasmid-derived sequence (TCVΔ5, TCVΔ6), only the high molecular weight RNAs were synthesized in amounts significantly above background, indicating that the oligoribonucleotides were not derived from degradation of high molecular weight RNAs and were not products of internal initiation at cryptic promoter sites. Because both sat-RNA C with the 3′ 100 nt replaced with the corresponding sequence of TCV (C/3′TCV) and DI RNA G, which has high sequence and structural similarity with TCV at the 3′ end (see Fig. 1B) produced substantially less abortive products (Fig. 3), sequence and/or structural features of TCV outside the 3′-terminal 100 nt may be required for high level abortive product synthesis.
Figure 3.
Analysis of aborted synthesis products generated in TCV RdRp reactions in vitro. Plus-strand RNA templates indicated above each lane (3 μg per lane) were added to partially purified TCV RdRp using reaction conditions as described, and using [α-32P]ATP as the radioactive nucleotide. Products were separated through 20% polyacrylamide/8 M urea gels and analyzed by autoradiography. Lanes: TCV, wild-type TCV; TCVΔ5 and TCVΔ6, mutants of TCV with the 3′ terminal 5 (UGCCC-3′) and 6 (CUGCCC-3′) nucleotides, respectively, replaced with the sequence GGGGAUCCUCUAG-3′; G, DI RNA G; C, sat-RNA C; C/3′TCV, sat-RNA C with the 3′ 100 bases replaced with the corresponding region from TCV; D, sat-RNA D. (Left) Low and high molecular weight products. (Right) Overexposure of the lower portion of the gel shown at left to visualize low abundant oligoribonucleotide products. The migration positions of full-length subviral RNAs and oligoribonucleotides of 4–8 bases are shown. The sizes of the oligoribonucleotides products were determined by the use of labeled 5-, 6-, and 7-mer ribonucleotide size-markers. TCV migrates only slightly into these gels.
Use of Oligoribonucleotide Primers by the TCV RdRp in Vitro.
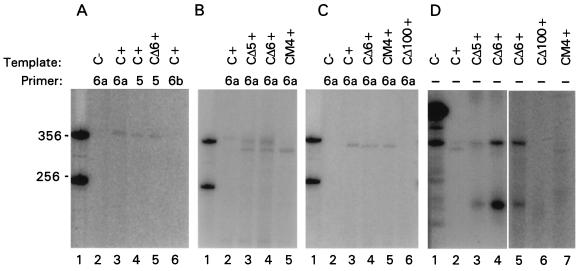
To test if the TCV RdRp can use oligoribonucleotides complementary to the 3′ end of (+)-strand TCV as primers for synthesis of sat-RNA C, a 32P-labeled oligoribonucleotide complementary to the 3′-terminal 6 nt (5′-GGGCAG-3′) was added to the RdRp reaction without labeled mononucleotides. Synthesis of full-length RNAs were observed using (+)-strand sat-RNA C but not (−)-strand sat-RNA C (Fig. 4A, lanes 2 and 3). The RdRp could also extend on the same oligoribonucleotide primer with a similar efficiency using (+)-strands of CΔ5 or CΔ6 (Fig. 4B, lanes 3 and 4). This result indicates that base pairing along the length of the primer with the template was not required and supports the model shown in Fig. 2, which describes the repair of CΔ6. In addition to the full-length products, some shorter than full-length products were also synthesized. The product migrating slightly faster than the full-length product had been previously shown to be synthesized in high amounts when transcripts containing mutations in the stem of the 3′ hairpin promoter were used in primer-independent RdRp reactions (ref. 16; see below).
Figure 4.
Analysis of primer-extension products synthesized in TCV RdRp reactions in vitro. The (+)- or (−)-strand RNA templates indicated above each lane (3 μg per lane) were added to partially purified TCV RdRp reactions with or without the primers indicated, and the products were separated on 5% polyacrylamide/8 M urea gels and analyzed using a PhosphorImager. Lanes: C, sat-RNA C; CΔ5, CΔ6, and CΔ100, sat-RNA C with deletions of the 3′ terminal 5, 6, or 100 nt, respectively; CM4, sat-RNA C with four mismatch mutations that destabilize the base of the 3′ terminal hairpin (Fig. 1B); “+” or “−” indicate (+)-strand or (−)-strand template, respectively. (A–C) The mononucleotide label was omitted and replaced with the following oligoribonucleotide primers produced by T7 RNA polymerase transcription and labeled with [α-32P]ATP: 6a, GACGGG-5′; 5, AAGGG-5′; 6b, CCCAGG-5′. One microgram of primer was used per reaction in A and B, and 10 μg of primer was used in C. Lanes 1 in A–C contain 356- and 256-nt markers. (D) The mononucleotide label was [α-32P]ATP.
To determine the importance of the oligoribonucleotide sequence in the primer-extension reaction, two additional oligoribonucleotides were tested: 5′-GGGAA-3′(primer 5; differences from primer 6a, which contains the wild-type complementary sequence [5′-GGGCAG-3′], are underlined) was used nearly as efficiently as primer 6a (Fig. 4A, compare lanes 3 and 4). In contrast, 5′-GGACCC-3′ (primer 6b) was not efficiently utilized by the RdRp (Fig. 4A, compare lanes 3 and 6). This suggests that the TCV RdRp shows specificity in binding and/or elongating the oligoribonucleotide primers in vitro.
To determine whether the 3′-terminal hairpin that is required for (−)-strand synthesis is also required for the priming reaction, the sat-RNA C mutant CM4 (4-nt mismatch mutation in the hairpin; Fig. 1B) was used in the primer-mediated RdRp reaction. Previous studies had shown that CM4 is not a template for synthesis of full-length products in vitro (16) and is not a viable sat-RNA in vivo (V. Stupina and A.E.S., unpublished results). RdRp reactions with the labeled 6a primer produced no detectable full-length complementary-strand products (Fig. 4B, lane 5). However, the shorter than full-length RNA was synthesized using CM4 as template in reactions with (Fig. 4B, lane 5) and without primer (Fig. 4D, lane 7). This result suggests that the primer was used by the RdRp to initiate (−)-strand synthesis from CM4, but initiation did not begin at the 3′ end. Such shorter products were exclusively synthesized using 10-fold additional primer and sat-RNA C and CΔ6, but not sat-RNA C with a deletion of the 3′-terminal 100 nt (CΔ100) (Fig. 4C, lanes 3, 4, and 6). The species migrating slightly faster than full-length may be derived from internal initiation by the RdRp within the 3′ promoter region that might be folding into two alternative conformations. Together, these results suggest that the hairpin required for initiation of full-length (−)-strand synthesis is also required for priming at the 3′ end.
The lack of RdRp products in the absence of the 3′ promoter sequence and the generation of similar sized full-length and close to full-length products in the primer-dependent and independent reactions (e.g., mutant CM4; Fig. 4 B, lane 5, and D, lane 7) exclude the possibility that the primers were attached to the template RNAs by ligation. Furthermore, the labeling of RNAs was observed with TCV-related primers [primers 5 (one base mis-match) and 6a (no mismatched bases)], but not with the unrelated primer [primer 6b (three mismatched bases)].
Our finding that the TCV RdRp can use primers derived from the 3′ end of TCV for synthesis of (−)-strand sat-RNA C from (+)-strand sat-RNA C template in vitro, and that this mechanism may be responsible for repair of truncated sat-RNA C in vivo, raises the possibility that a similar priming reaction could be used during the normal synthesis of sat-RNA C. To address this possibility, turnip plants were inoculated with wild-type sat-RNA C and TCVA4050. If a percentage of sat-RNA C replication was mediated by extension from the TCV-derived primer, then some sat-RNA C should terminate with CCAGCCC-3′ (the terminal motif of TCVA4050). Twenty-seven sat-RNA C clones were generated from two plants and all contained the sat-RNA C wild-type CCUGCCC-3′ sequence (Table 1). All 14 clones derived from TCV accumulating in the same plants terminated in CCAGCCC-3′ (data not shown). This result indicates either that replication of sat-RNA C does not proceed to any great extent by a primer-mediated mechanism or that sat-RNA C terminating with CCAGCCC-3′ is at a selective disadvantage when accumulating in the presence of wild-type sat-RNA C. This possibility was supported when plants inoculated with transcripts of wild-type TCV and a 1:1 mix of wild-type sat-RNA C and CA352 only accumulated wild-type sat-RNA C at 3 weeks postinoculation (Table 1). This result suggests that low level usage of a primer-mediated mechanism for the synthesis of sat-RNA C when coinoculated with TCVA4050 would not be detectable, and the possibility that such a mechanism is used for a portion of sat-RNA synthesis remains open.
Generation of short abortive RNAs has been shown to occur for T7 bacteriophage, Escherichia coli, and eukaryotic DNA-dependent RNA polymerases (22, 28–30) and may be a general phenomenon among RNA polymerases although not previously reported for viral RdRp. In addition, primases can be regarded as specialized RNA polymerases that only produce short oligoribonucleotides using DNA templates (31, 32). The use of short RNA primers by polymerases for promoter-dependent replication has been shown for the reverse transcriptase encoded by the Mauriceville plasmid (33) and DNA-dependent RNA polymerases of T7 bacteriophage and E. coli (34). In addition, the use of RNA primers by viral polymerases during mRNA transcription has been demonstrated for influenza virus (35, 36) and proposed for coronaviruses (37, 38).
In summary, we have demonstrated that TCV RdRp synthesizes abortive RNA products of up to 8 nt and can use these RNAs as primers in in vitro RdRp reactions. This mechanism explains the in vivo repair of sat-RNA C with 3′-terminal deletions of 6–7 residues and supports the model shown in Fig. 2. The 3′-end repair function uses (+)-strand TCV, the only known template from which the RdRp generates substantial abortive synthesis products. Thus, the parasitic sat-RNA C depends on its helper virus for both replication and 3′-end repair. The ability of the TCV RdRp to mediate primer-directed synthesis may explain the conservation of CCUGCCC-3′ at the 3′ ends of all RNAs associated with TCV. It is possible that the production of a pool of primer RNAs is a mechanism responsible for conservation and maintenance (repair) of 3′-end sequences for other RNA viruses. While the tripartite brome mosaic virus, whose RNAs contains a 3′ tRNA-like structure terminating in -CCA, may repair deletions of the 3′-terminal CCA using cellular nucleotidyltransferase (5, 39), a primer-directed synthesis mechanism may be especially advantageous for maintaining the terminal sequences of multicomponent viruses without tRNA-like structures.
Acknowledgments
This work was supported by grants from the National Science Foundation (MCB-9315948 and MCB-9419303) to A.E.S.
Footnotes
Abbreviations: TCV, genomic RNA of turnip crinkle virus; sat-RNAs, satellite RNAs; RdRp, RNA-dependent RNA polymerase.
References
- 1.Greider C, Blackburn E H. Cell. 1987;51:887–898. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- 2.Greider C, Blackburn E H. Nature (London) 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 3.Yu G-L, Bradley J D, Attardi L D, Blackburn E H. Nature (London) 1990;344:126–132. doi: 10.1038/344126a0. [DOI] [PubMed] [Google Scholar]
- 4.Blackburn E H. Annu Rev Biochem. 1992;61:113–129. doi: 10.1146/annurev.bi.61.070192.000553. [DOI] [PubMed] [Google Scholar]
- 5.Rao A L N, Dreher T W, Marsh L E, Hall T C. Proc Natl Acad Sci USA. 1989;86:5335–5339. doi: 10.1073/pnas.86.14.5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalmay T, Russo M, Burgyan J. Virology. 1993;192:551–555. doi: 10.1006/viro.1993.1071. [DOI] [PubMed] [Google Scholar]
- 7.Carpenter C D, Simon A E. J Virol. 1996;70:478–486. doi: 10.1128/jvi.70.1.478-486.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carpenter C D, Simon A E. Virology. 1996;226:153–160. doi: 10.1006/viro.1996.0641. [DOI] [PubMed] [Google Scholar]
- 9.Garnier M, Candresse T, Bove J M. Virology. 1986;151:100–109. doi: 10.1016/0042-6822(86)90107-8. [DOI] [PubMed] [Google Scholar]
- 10.Froshauer S, Kartenbeck J, Helenius A. J Cell Biol. 1988;107:2075–2086. doi: 10.1083/jcb.107.6.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bienz K, Egger D, Pfister T, Troxler M. J Virol. 1992;66:2740–2747. doi: 10.1128/jvi.66.5.2740-2747.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Kuyl A C, Neeleman L, Bol J F. Virology. 1991;183:687–694. doi: 10.1016/0042-6822(91)90997-p. [DOI] [PubMed] [Google Scholar]
- 13.Joshi R L, Joshi S, Chapeville F, Haenni A L. EMBO J. 1983;2:1123–1127. doi: 10.1002/j.1460-2075.1983.tb01556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Omata T, Kohara M, Sakai Y, Kameda A, Imura N, Nomoto A. Gene. 1984;32:1–10. doi: 10.1016/0378-1119(84)90026-x. [DOI] [PubMed] [Google Scholar]
- 15.Neufeld K L, Galarza M, Richards O C, Summers D F, Ehrenfeld E. J Virol. 1994;68:5811–5818. doi: 10.1128/jvi.68.9.5811-5818.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song C, Simon A E. J Mol Biol. 1995;254:6–14. doi: 10.1006/jmbi.1995.0594. [DOI] [PubMed] [Google Scholar]
- 17.Simon A E, Howell S H. EMBO J. 1986;5:3423–3428. doi: 10.1002/j.1460-2075.1986.tb04664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang C, Simon A E. J Virol. 1994;68:8466–8469. doi: 10.1128/jvi.68.12.8466-8469.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carpenter C D, Oh J-W, Zhang C, Simon A E. J Mol Biol. 1995;245:608–622. doi: 10.1006/jmbi.1994.0050. [DOI] [PubMed] [Google Scholar]
- 20.Song C, Simon A E. Proc Natl Acad Sci USA. 1994;91:8792–8796. doi: 10.1073/pnas.91.19.8792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oh J-W, Kong Q, Song C, Carpenter C D, Simon A E. Mol Plant-Microbe Interact. 1995;8:979–987. doi: 10.1094/mpmi-8-0979. [DOI] [PubMed] [Google Scholar]
- 22.Martin C T, Muller D K, Coleman J E. Biochemistry. 1988;27:3966–3974. doi: 10.1021/bi00411a012. [DOI] [PubMed] [Google Scholar]
- 23.Bujarski J J, Ahlquist P, Hall T C, Dreher T W, Kaesberg P. EMBO J. 1986;5:1769–1774. doi: 10.1002/j.1460-2075.1986.tb04425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takamatsu N, Watanabe Y, Meshi T, Okada Y. J Virol. 1990;64:3686–3693. doi: 10.1128/jvi.64.8.3686-3693.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai C-H, Dreher T W. J Virol. 1992;66:5190–5199. doi: 10.1128/jvi.66.9.5190-5199.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobson S J, Konings D A M, Sarnow P. J Virol. 1993;67:2961–2971. doi: 10.1128/jvi.67.6.2961-2971.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duggal R, Lahser F C, Hall T C. Annu Rev Phytopathol. 1994;32:287–309. [Google Scholar]
- 28.Carpousis A J, Gralla J D. Biochemistry. 1980;19:3245–3253. doi: 10.1021/bi00555a023. [DOI] [PubMed] [Google Scholar]
- 29.Yamakawa M, Furuichi Y, Nakashima K, La Fiandra A J, Shatkin A J. J Biol Chem. 1981;256:6507–6514. [PubMed] [Google Scholar]
- 30.Ackerman S, Bunick D, Zandomeni R, Weinmann R. Nucleic Acids Res. 1983;11:6041–6064. doi: 10.1093/nar/11.17.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.So A G, Downey K M. Crit Rev Biochem Mol Biol. 1992;27:129–155. doi: 10.3109/10409239209082561. [DOI] [PubMed] [Google Scholar]
- 32.Marians K J. Annu Rev Biochem. 1992;61:673–719. doi: 10.1146/annurev.bi.61.070192.003325. [DOI] [PubMed] [Google Scholar]
- 33.Wang H, Lambowitz A M. Cell. 1993;75:1071–1081. doi: 10.1016/0092-8674(93)90317-j. [DOI] [PubMed] [Google Scholar]
- 34.Daube S S, von Hippel P H. Science. 1992;258:1320–1324. doi: 10.1126/science.1280856. [DOI] [PubMed] [Google Scholar]
- 35.Honda A, Mizumoto K, Ishihama A. J Biol Chem. 1986;261:5987–5991. [PubMed] [Google Scholar]
- 36.Chung T D Y, Cianci C, Hagen M, Terry B, Matthews J T, Krystal M, Colonno R. Proc Natl Acad Sci USA. 1994;91:2372–2376. doi: 10.1073/pnas.91.6.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Makino S, Lai M M C. J Virol. 1989;63:5285–5292. doi: 10.1128/jvi.63.12.5285-5292.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X, Lai M M C. J Virol. 1994;68:6626–6633. doi: 10.1128/jvi.68.10.6626-6633.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller W A, Bujarski J J, Dreher T W, Hall T C. J Mol Biol. 1986;187:537–546. doi: 10.1016/0022-2836(86)90332-3. [DOI] [PubMed] [Google Scholar]