Abstract
Single and multiple mutations at residues 16, 51, 59, 108, and 164 of Plasmodium falciparum dihydrofolate reductase (pfDHFR) have been linked to antifolate resistance in malaria. We prepared and characterized all seven of the pfDHFR mutants found in nature, as well as six mutants not observed in nature. Mutations involving residues 51, 59, 108, or 164 conferred cross resistance to both the antifolates pyrimethamine and cycloguanil, whereas mutation of residue 16 specifically conferred resistance to cycloguanil. The antifolate resistance of enzyme mutants found in nature correlated with in vivo antifolate resistance; however, mutants not found in nature were either poorly resistant or had insufficient catalytic activity to support DNA synthesis. Thus, specific combinations of multiple mutations at target residues were selected in nature to optimize resistance. Further, the resistance of multiple mutants was more than the sum of the component single mutations, indicating that residues were selected for their synergistic as well as intrinsic effects on resistance. Pathways inferred for the evolution of pyrimethamine-resistant mutants suggested that all multiple mutants emerged from stepwise selection of the single mutant, S108N. Thus, we propose that drugs targeted to both the wild-type pfDHFR and S108N mutant would have a low propensity for developing resistance, and hence could provide effective antimalarial agents.
Keywords: malaria, synthetic gene, mutagenesis, pyrimethamine, cycloguanil
Dihydrofolate reductase (DHFR) catalyzes the NADPH-dependent reduction of 7,8-dihydrofolate (H2folate) to 5,6,7,8-tetrahydrofolate (1). In protozoa and plants, DHFR resides on the same polypeptide chain as thymidylate synthase (2–6).
Pyrimethamine (Pyr) and cycloguanil (Cyc) are potent inhibitors of Plasmodium falciparum DHFR (pfDHFR) that were effectively used for the treatment of P. falciparum malaria until the widespread appearance of resistant parasites reduced their utility. Recently, sequencing of the DHFR gene from Pyr- and Cyc-resistant P. falciparum isolates revealed linkages between point mutations of the gene and the degree of drug resistance of the parasites (7–11), which have been supported by transfection experiments (12–15).
We have undertaken an investigation of pfDHFR to assess the correlation between antifolate inhibition of the enzyme and resistance of the parasite and to obtain an understanding of the molecular basis of antifolate resistance. Although the sparse amounts of enzyme obtainable from malaria parasites or cloned genes precluded meaningful enzyme studies (16–18), the chemically synthesized pfDHFR domain was highly expressed in Escherichia coli as inactive inclusion bodies. Homogeneous, active enzyme with appropriate kinetic and inhibition characteristics could be obtained by unfolding and refolding, and purification by methotrexate (MTX) affinity chromatography (19). Because the synthetic gene contains numerous unique restriction sites in the coding sequence, it was ideally suited for cassette mutagenesis.
Here, we report the preparation and characterization of the pfDHFR mutants at residues 16, 51, 59, 108, and 164 that have been linked to Pyr and Cyc resistance. Analysis of the kinetics of mutant enzymes and their inhibition by Pyr and Cyc aided in the elucidation of the contribution of mutated residues to antifolate resistance, the generation of an evolutionary tree describing the genesis of antifolate resistance, and the generation of ideas that could lead to antifolate regimens with low propensity for development of resistance.
MATERIALS AND METHODS
Materials.
The expression vector pET-17b and the host strain E. coli BL21(DE3)pLysS were from Novagen. Cyc was a gift from Burroughs Wellcome. MTX-Sepharose CL-6B (≈1 μmol MTX/ml resin) (20) was prepared as described. Oligonucleotides longer than 50 bases were purified by denaturing gel electrophoresis (21). DNA sequences were verified by the dideoxy chain-termination method (22). Methods for DNA manipulation were performed as described (21). Other materials were the purest grades commercially available.
Mutation of the Synthetic pfDHFR Gene.
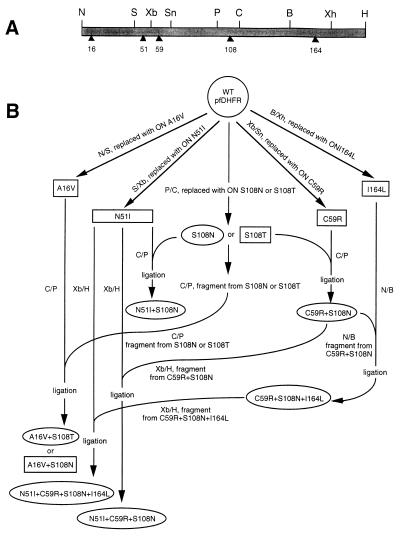
pUC-pfDHFR (19) containing the wild-type synthetic pfDHFR gene was employed for the construction of all DHFR mutants. Single and multiple mutations at amino acid residues 16, 51, 59, 108, and 164 of the domain were accomplished by oligonucleotide cassettes (Table 1). Relevant unique restriction sites and the residues mutated in the pfDHFR synthetic gene are shown in Fig. 1A. The recombinant plasmids were transformed into E. coli DH5α, and the cells were plated on Luria–Bertani (LB) agar containing 100 μg/ml of ampicillin. Plasmid DNA was characterized by restriction analysis, and the nucleotide sequences of mutated regions were verified.
Table 1.
Oligonucleotide cassettes for the construction of pfDHFR mutants
| Residue no. | Restriction site | Length of oligonucleotide cassette,* bases | Mutation (codon) |
|---|---|---|---|
| 16 | NdeI–SacII | 115/111 | Ala (GCA) to Val (GTA) |
| 51 | SacII–XbaI | 42/48 | Asn (AAC) to Ile (ATC) |
| 59 | XbaI–SnaBI | 36/32 | Cys (TGC) to Arg (CGC) |
| 108 | PstI–ClaI | 35/41 | Ser (TCG) to Asn (AAC) |
| Ser (TCG) to Thr (ACC) | |||
| 164 | BglII–XhoI | 67/67 | Ile (ATC) to Leu (CTC) |
Except for the indicated codon change, oligonucleotide cassettes had the same sequences as in the synthetic gene (19).
Figure 1.
Cassette mutagenesis of pfDHFR. (A) Coding sequence of the synthetic pfDHFR domain; restriction sites are shown above the gene whereas the amino acid residues mutated are shown as solid triangles below the gene. (B) Strategy for the construction of mutant DHFRs from the wild-type synthetic DHFR template. B, BglII; C, ClaI; H, HindIII; N, NdeI; P, PstI; S, SacII; Sn, SnaBI; Xb, XbaI; Xh, XhoI.
The single point mutants, namely A16V, N51I, C59R, S108N, S108T, and I164L, were constructed first and used as parent plasmids to make the multiple mutants (Fig. 1B). Single mutations at residues 16, 51, 59, 108, and 164 were performed by cassette replacement of specific oligonucleotide duplexes at NdeI–SacII, SacII–XbaI, XbaI–SnaBI, PstI–ClaI, and BglII–XhoI sites of pUC-pfDHFR, respectively. For the construction of double mutants, the PstI–ClaI fragments were excised from pUC-pfDHFR(S108T) and pUC-pfDHFR(S108N), purified, and cloned into the corresponding sites of pUC-pfDHFR(A16V) to give pUC-pfDHFR(A16V+S108T) and pUC-pfDHFR(A16V+S108N), respectively. Likewise, the PstI–ClaI fragment from pUC-pfDHFR(S108N) was cloned into the corresponding sites of pUC-pfDHFR(N51I) and pUC-pfDHFR(C59R) to give pUC-pfDHFR(N51I+S108N) and pUC-pfDHFR(C59R+S108N), respectively. The triple mutant pUC-pfDHFR(N51I+C59R+S108N) was constructed by cloning a 550-bp XbaI–HindIII fragment from pUC-pfDHFR(C59R+S108N) into the corresponding sites of pUC-pfDHFR(N51I), whereas cloning of a 440-bp NdeI–BglII fragment from pUC-pfDHFR(C59R+S108N) into the corresponding sites of pUC-pfDHFR(I164L) yielded the triple mutant pUC-pfDHFR(C59R+S108N+I164L). For the quadruple mutant pUC-pfDHFR(N51I+C59R+S108N+I164L), a 550-bp XbaI–HindIII fragment from pUC-pfDHFR (C59R+S108N+I164L) was cloned into the corresponding sites of pUC-pfDHFR (N51I). Using the above strategy, a total of 13 mutants were obtained (Table 2).
Table 2.
List of mutants generated by cassette mutagenesis of wild-type synthetic pfDHFR domain
| Construct | Field isolates with equivalent mutation* | Drug susceptibility†
|
Amino acid residue of DHFR domain‡
|
|||||
|---|---|---|---|---|---|---|---|---|
| Pyr | Cyc | 16 | 51 | 59 | 108 | 164 | ||
| Wild type | 3D7, SL/D6 | S | S | Ala | Asn | Cys | Ser | Ile |
| A16V§ | Val | Asn | Cys | Ser | Ile | |||
| N51I§ | Ala | Ile | Cys | Ser | Ile | |||
| C59R§ | Ala | Asn | Arg | Ser | Ile | |||
| S108N | HB3 | MR | LR | Ala | Asn | Cys | Asn | Ile |
| S108T§ | Ala | Asn | Cys | Thr | Ile | |||
| I164L§ | Ala | Asn | Cys | Ser | Leu | |||
| A16V + S108T | FAC8, FAC3, UPA, FCB, It.G2.F6 | S | HR | Val | Asn | Cys | Thr | Ile |
| A16V + S108N§ | Val | Asn | Cys | Asn | Ile | |||
| N51I + S108N | 7G8, It.D12 | MR | MR | Ala | Ile | Cys | Asn | Ile |
| C59R + S108N | K1, V1 | MR | MR | Ala | Asn | Arg | Asn | Ile |
| N51I + C59R + S108N | W2 | HR | MR | Ala | Ile | Arg | Asn | Ile |
| C59R + S108N + I164L | Cs1-2 | HR | HR | Ala | Asn | Arg | Asn | Leu |
| N51I + C59R + S108N + I164L | V1/S | HR | HR | Ala | Ile | Arg | Asn | Leu |
Expression and Preparation of Mutant DHFRs.
The recombinant pUC plasmids containing mutant DHFRs were digested with NdeI and HindIII. The DHFR fragments (≈0.7 kb) were gel-purified, cloned into the corresponding sites of the pET-17b expression vector, and transformed into E. coli BL21(DE3)pLysS, and expression was induced by isopropyl β-d-thiogalactoside (19). The expressed inclusion bodies were partially purified, unfolded, refolded, and purified by affinity chromatography on a MTX-Sepharose CL-6B column as described (19). The DHFR quadruple mutant (N51I+C59R+S108N+I164L) showed low affinity for MTX-Sepharose and was purified by hydroxyapatite chromatography. Briefly, the refolded enzyme was adsorbed on a column of hydroxyapatite (1.5 × 5.0 cm) that was pre-equilibrated with buffer A (20 mM potassium phosphate, pH 7.0/0.1 mM EDTA/10 mM DTT/20% glycerol) at a flow rate of ≈0.8 ml/min. The column was washed with buffer B (50 mM potassium phosphate, pH 7.0/0.1 mM EDTA/10 mM DTT/20% glycerol) until no protein was detected in the eluate (≈100 ml). Buffer C (400 mM potassium phosphate, pH 7.0/0.1 mM EDTA/10 mM DTT/20% glycerol) was then applied. Fractions (4 ml) containing DHFR activity were pooled and concentrated.
Enzyme Assay and Kinetic Analysis.
The standard assay for DHFR activity and kinetic analysis of the mutant DHFRs were as described (19). The in vitro resistance indices for Pyr and Cyc were calculated from the ratio of IC50 values of the mutant DHFRs versus IC50 values of the wild-type DHFR. The in vivo resistance indices were determined using the IC50 values for Pyr and Cyc reported previously for the drug-sensitive and drug-resistant parasite isolates (9, 11). Free energies of binding to Pyr and Cyc of mutant DHFRs were calculated from the Ki values, based on the equation described by Wilkinson et al. (23).
RESULTS
Construction of DHFR Mutants.
The wild-type synthetic pfDHFR gene and synthetic oligonucleotide cassettes were used for the construction of single and multiple mutants at amino acid residues 16, 51, 59, 108, and 164. A total of 13 mutants were obtained, comprising 6 single point mutants, 4 double mutants, 2 triple mutants, and 1 quadruple mutant. Of the 13 mutants prepared, 6 (A16V, N51I, C59R, S108T, I164L, and A16V+S108N) have not been found in nature (Table 2).
Expression and Purification of Mutant DHFRs.
The fragments encoding DHFR mutants were excised from pUC construction vectors, inserted into pET expression vectors and expressed in E. coli BL21(DE3)pLysS. As with wild-type DHFR (19), SDS/PAGE of the particulate fractions showed that the mutant DHFRs were expressed as insoluble inclusion bodies. With the exception of the A16V+S108N mutant, all mutants yielded DHFR activity upon unfolding and refolding of the inclusion bodies (19). All mutants, except the N51I+C59R+S108N+I164L quadruple mutant, bound to MTX-Sepharose and could be eluted by H2folate. The quadruple mutant was purified to homogeneity by hydroxyapatite chromatography. With the exception of the quadruple mutant purified mutants were titratable with MTX (data not shown). We assumed the homogeneous quadruple mutant was fully active.
Kinetics of the Mutant DHFRs.
The kinetic parameters of the purified mutant DHFRs are summarized in Table 3. The C59R mutant showed an elevated kcat, while the S108N and N51I+S108N mutants exhibited kcat values comparable to wild-type DHFR. The remaining mutants showed decreased kcat values ranging from ≈2% to 40% of wild type. Two single mutants (S108T and I164L), one triple mutant (C59R+S108N+I164L), and the quadruple mutant (N51I+C59R+S108N+I164L) had Km values for H2folate that were within experimental error of that for the wild-type enzyme, but exhibited Km values for NADPH that were 3- to 7-fold higher than the wild-type enzyme. Unchanged or moderately increased (up to 5-fold) Km values for both substrates were observed for S108N, C59R+S108N, and A16V+S108T mutants. The remaining mutants showed 2- to 8-fold reductions in Km for H2folate, and Km values for NADPH within a factor of 3 of that for wild-type DHFR. Except for C59R and N51I+S108N, the kcat/Km values for the mutant DHFRs were decreased by 2- to 50-fold.
Table 3.
Kinetic parameters of synthetic mutant DHFRs and inhibition by antifolates Pyr and Cyc
| Construct | Kinetic parameter
|
|||||
|---|---|---|---|---|---|---|
| kcat, sec−1 | Km H2folate, μM | Km NADPH, μM | kcat/Km,* (M−1·sec−1) × 106 | Inhibition by antifolate, Ki, nM
|
||
| Pyr | Cyc | |||||
| Wild type† | 88 | 13 ± 5 | 5 ± 1 | 6.8 | 1.5 ± 0.2 | 2.6 ± 0.3 |
| A16V‡ | 1.6 | 168 ± 14 | 57 ± 7 | 0.01 | 6 ± 1 | 565 ± 40 |
| N51I‡ | 2.6 | 3.2 ± 0.2 | 4.0 ± 0.2 | 0.81 | 0.4 ± 0.03 | 4.7 ± 0.2 |
| C59R‡ | 122 | 1.6 ± 0.1 | 4.4 ± 0.3 | 76 | 1.1 ± 0.1 | 4.7 ± 0.2 |
| S108N† | 92 | 25 ± 9 | 7 ± 2 | 3.7 | 13 ± 4 | 15 ± 2 |
| S108T‡ | 36 | 14 ± 2 | 17 ± 3 | 2.6 | 1.4 ± 0.2 | 1.6 ± 0.2 |
| I164L‡ | 33 | 14 ± 1 | 15 ± 1 | 2.4 | 0.83 ± 0.05 | 9.0 ± 0.5 |
| A16V + S108T | 19 | 29 ± 2 | 24 ± 1 | 0.64 | 3.6 ± 0.3 | 2129 ± 100 |
| A16V + S108N§ | ||||||
| N51I + S108N† | 77 | 6 ± 1 | 1.9 ± 0.4 | 13 | 37 ± 6 | 24 ± 4 |
| C59R + S108N | 4.2 | 24 ± 1 | 15 ± 1 | 0.18 | 72 ± 3 | 82 ± 4 |
| N51I + C59R + S108N | 3.2 | 6.2 ± 0.3 | 10 ± 1 | 0.53 | 120 ± 5 | 103 ± 8 |
| C59R + S108N + I164L | 3.0 | 18 ± 1 | 36 ± 3 | 0.17 | 383 ± 33 | 1141 ± 98 |
| N51I + C59R + S108N + I164L | 15 | 14 ± 1 | 25 ± 6 | 1.1 | 859 ± 117 | 730 ± 19 |
Calculated from Km values for H2folate.
Data from ref. 19.
Not found in nature.
No DHFR activity detected and not found in nature.
Inhibition of Mutant DHFRs by Pyr and Cyc.
As reported previously (19), a single mutation at residue 108 from Ser to Asn resulted in 10- and 6-fold higher Ki values for Pyr and Cyc, respectively, compared with wild-type enzyme. The Ki values for inhibition of the other mutants by Pyr and Cyc are given in Table 3. The double mutants N51I+S108N and C59R+S108N exhibited 10- to 50-fold increases in the Ki values for both antifolates relative to the wild-type enzyme. The triple mutants N51I+C59R+S108N and C59R+S108N+I164L exhibited Ki values for Pyr and Cyc 40- to 600-fold higher than the wild-type enzyme. The quadruple mutant exhibited 300- to 600-fold higher Ki values for both inhibitors compared with the wild-type enzyme. The single mutant A16V showed a more than 200-fold increase in the Ki for Cyc, but only a 4-fold increase in the Ki value for Pyr. Similarly, the A16V+S108T double mutant exhibited a Ki value for Cyc about 800-fold higher than wild type, but only a 2-fold increase in the Ki for Pyr.
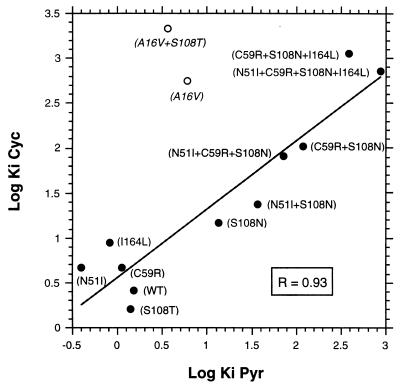
The cross resistance between Pyr and Cyc inhibition of the mutant DHFRs was evaluated by plotting the log Ki values for Pyr against the log Ki values for Cyc (Fig. 2). A significant correlation (R = 0.93) was observed when A16V and A16V+S108T were excluded.
Figure 2.
Correlation of Pyr and Cyc resistance among DHFR mutants. The log Ki values for Pyr were plotted against the log Ki values for Cyc. Mutants and amino acids mutated were indicated. Data were analyzed by a linear regression fitting program. Mutants indicated by open circles and italicized names were not included in the analysis.
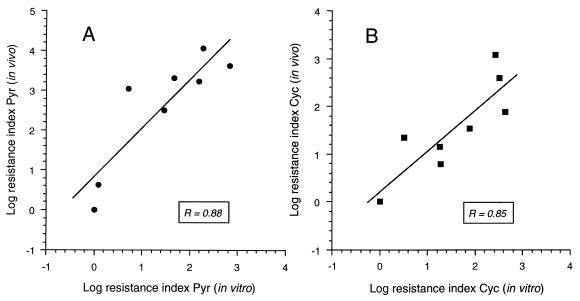
To determine whether there was a correlation between the in vitro inhibition of DHFR mutants and the in vivo inhibition of the parasites reported previously (9, 11), the resistance indices (IC50 mutant/IC50 wild type) were calculated for each mutant DHFR and each parasite isolate containing equivalent mutations (9, 11). There was a significant correlation for the log resistance indices between in vitro and in vivo data for both Pyr (Fig. 3A) and Cyc (Fig. 3B).
Figure 3.
Correlation of antifolate inhibition between mutant DHFRs, in vitro, and parasite isolates, in vivo. The log resistance indices for mutant DHFRs were plotted against the log resistance indices for parasites. (A) Inhibition by Pyr. (B) Inhibition by Cyc. Data were analyzed by a linear regression fitting program.
The free energies for Pyr and Cyc binding to mutants with single and multiple mutations are presented in Table 4. The observed free energies for Pyr binding for all multiple mutants, except A16V+S108T, were lower than the sum of the corresponding single mutations. Moreover, the differences of these values (interaction energies, ΔΔG°) increased as the extent of mutation progressed from double mutants to multiple mutants. For Cyc binding, the A16V+S108T double mutant and the C59R+S108N+I164L triple mutant showed the largest increase in ΔΔG°, while the C59R+S108N, N51I+C59R+S108N, and N51I+C59R+S108N+I164L mutants showed slight to moderate changes in the interaction energies.
Table 4.
Free energies and interaction energies for Pyr and Cyc binding of multiple and the component mutations
| Mutation | Pyr binding, kcal/mol
|
Cyc binding, kcal/mol
|
||||
|---|---|---|---|---|---|---|
| Free energy*
|
Interaction energy, ΔΔG° | Free energy*
|
Interaction energy, ΔΔG° | |||
| Observed ΔG° | Sum ΔG° | Observed ΔG° | Sum ΔG° | |||
| A16V | −0.811 | −3.187 | ||||
| N51I | 0.783 | −0.351 | ||||
| C59R | 0.184 | −0.351 | ||||
| S108N | −1.297 | −1.026 | ||||
| S108T | 0.041 | 0.288 | ||||
| I164L | 0.351 | −0.735 | ||||
| A16V + S108T | −0.519 | −0.770 | −0.252 | −3.973 | −2.900 | 1.073 |
| N51I + S108N | −1.890 | −0.514 | 1.376 | −1.314 | −1.377 | −0.063 |
| C59R + S108N | −2.290 | −1.113 | 1.177 | −2.045 | −1.377 | 0.668 |
| N51I + C59R + S108N | −2.595 | −0.330 | 2.265 | −2.179 | −1.727 | 0.452 |
| C59R + S108N + I164L | −3.283 | −0.763 | 2.520 | −3.603 | −2.112 | 1.491 |
| N51I + C59R + S108N + I164L | −3.761 | 0.020 | 3.781 | −3.339 | −2.463 | 0.876 |
Data within 10% of experimental error.
DISCUSSION
A linkage between certain mutations in the DHFR gene of P. falciparum and resistance of malaria parasites to the antifolates Pyr and Cyc has been established (9, 11). DHFR mutations including certain combinations of N51I, C59R, S108N, and I164L are associated with Pyr-resistant organisms showing cross resistance to Cyc (Table 2) (9, 11, 24). However, organisms with DHFR A16V+S108T are sensitive to Pyr but resistant to Cyc. In the present work, we sought to establish a direct linkage between purified mutant DHFRs and parasite resistance, to assess the importance of each mutant target residue to the degree of resistance, and to develop strategies for overcoming resistance.
We describe here thirteen mutants of the pfDHFR domain which contain one to four of the point mutations at residues 16, 51, 59, 108, and 164 found in nature, and include all mutations reported in Pyr- and Cyc-resistant field isolates of P. falciparum. The seven mutants with direct counterparts in antifolate-resistant malaria included one single mutant, three double mutants, two triple mutants, and one quadruple mutant; the remaining six mutants have not been found in parasites and were produced as intermediates leading to the naturally occurring mutants (Table 2).
The multiple unique restriction sites in the pfDHFR synthetic gene permitted facile cassette mutagenesis by oligonucleotides to produce the mutants. The mutants were expressed as insoluble inclusion bodies and unfolded–refolded to recover activity. With the exception of the quadruple mutant, the active mutants were purified by MTX-Sepharose affinity chromatography.
The mutants found in parasites must possess adequate catalytic efficiency to support DNA synthesis, so it was of interest to determine their catalytic properties (Table 3). The only single mutant found in parasites, S108N, and one of the two double mutants seen in vivo, N51I+S108N, had similar kinetic parameters to the wild-type enzyme. The remaining naturally occurring mutants generally showed either moderate (<6-fold) or large (20- to 30-fold) reductions in kcat compared with wild-type enzyme, similar Km values for H2folate, and 2- to 7-fold greater Km values for NADPH. The kcat/Km values for all naturally occurring mutants except S108N and N51I+S108N were decreased by 2- to 50-fold.
The sensitivity of mutant pfDHFRs toward Pyr and Cyc correlated well with the sensitivity of malaria parasites harboring these mutant enzymes (Fig. 3). DHFR S108N, the only single mutant found in nature, exhibited Ki values for Pyr and Cyc ≈10-fold higher than wild-type enzyme (19); correspondingly, the parasite isolate HB3 carrying this mutation is moderately resistant to Pyr and slightly resistant to Cyc (9–11). Further mutation of DHFR S108N at residue 51 or 59 to give the double mutants N51I+S108N and C59R+S108N conferred 10- to 50-fold increased resistance to Pyr and Cyc compared with wild-type enzyme; in vivo, these double mutants are moderately resistant to both drugs. The other double mutant, A16V+S108T, was as sensitive as wild-type enzyme to Pyr, but showed about 1000-fold greater resistance to Cyc than the wild-type enzyme; parasites with a DHFR carrying this double mutation were likewise sensitive to Pyr but resistant to Cyc (9, 11). Triple mutants with changes at residues 108, 59, and either 51 or 164 were 40- to 200-fold more resistant to the inhibitors than the wild-type enzyme; in vivo, parasites carrying these triple mutant DHFRs were highly resistant to Pyr and moderately to highly resistant to Cyc (9–11). Finally, the quadruple mutant, N51I+C59R+S108N+I164L, was highly resistant to both Pyr and Cyc and conferred extremely high antifolate resistance to parasites in vivo. The correlations between in vitro and in vivo sensitivity of the DHFR mutants supports proposals (9, 11) that point mutations are the primary determinants of antifolate resistance in the parasites. Further, the results agree with epidemiologic studies that show relationships between the prevalence of mutations arising in nature with in vivo resistance and degree of antifolate use.‖
The five single mutants (S108T, N51I, C59R, I164L, and A16V) used as precursors for the multiple mutants were also studied (Table 3). Except for C59R and A16V, the steady-state kinetic parameters of these mutants were generally unremarkable. C59R was highly active, showing a more than 10-fold increase in kcat/Km compared with wild-type enzyme. In contrast, the activity of A16V was very low, with a 150-fold decrease in kcat/Km. Inhibition of the five unnatural single mutants by Pyr and of all except A16V by Cyc was unremarkable, with Ki values falling within about a 4-fold range of the wild-type enzyme. DHFR A16V, which was susceptible to Pyr, was some 200-fold more resistant to Cyc than the wild-type enzyme; it is possible that this mutant did not survive Cyc selection in nature because of its poor handling of substrates as indicated in the dramatic reduction of kcat/Km.
With Pyr-resistant DHFR mutants involving N51I, C59R, S108N, and/or I164L, there was an excellent correlation between the Ki values for Pyr and those for Cyc (Fig. 2). The correlation suggests that residues 51, 59, 108, and 164 are involved in the binding of both drugs, which explains the cross resistance of parasites containing these mutants toward both drugs (9).
DHFR A16V+S108T and parasites harboring this mutant are resistant to Cyc but sensitive to Pyr (9, 11). Unlike the other mutants, which presumably arose in response to Pyr selection, DHFR A16V+S108T would not have survived Pyr, and must have arisen in response to Cyc. Because the S108T mutant contributes little to Cyc resistance whereas A16V is highly resistant, Cyc resistance in the A16V+S108T mutant is attributed almost completely to the A16V mutation. It is interesting that the poor activity of DHFR A16V, which is not found in nature, is restored by a second-site mutation of Ser to Thr at position 108 to give the naturally occurring A16V+S108T (Table 3).
We wished to determine whether the resistance of the multiple mutants was caused by the additive effects of the component mutations or whether the mutations interacted with one another in a cooperative manner. To address this question, we calculated free energies (ΔG°) of binding and compared the sum of the free energies of binding of the single mutants to the observed ΔG° values for the corresponding multiple mutants. The difference in these values, termed the interaction energy (ΔΔG°), reflects the synergistic (positive value) or antagonistic (negative value) effects of the multiple mutations on resistance.
Except for A16V+S108T, the ΔΔG° for multiple mutations in Pyr binding are all positive, indicating that when combined, the mutations contribute more than the sum of the component mutations (Table 4). Further, as the mutations progress from double to triple to quadruple, the synergistic effect increases. Thus, the interaction energy, or synergistic effect, for each additional mutation on Pyr resistance is about 1.2 kcal/mol, and at the stage of the quadruple mutant the effect on resistance contributed by synergism is over 1000-fold. We speculate that the specific point mutations in multiple mutants evolved not only for their individual contributions toward Pyr resistance, but also for their synergistic contributions in the context of the residues previously mutated.
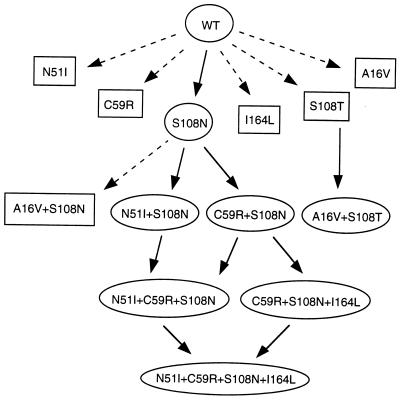
We have proposed a pathway for the evolution of antifolate resistance in pfDHFR (Fig. 4). The premise is that the emergence of drug resistance involved sequential selection of single point mutations, starting from the least resistant single mutant that survived selection, S108N, and proceeding stepwise to the more resistant multiple mutants. If an amino acid mutation expected in a pathway was not observed, it was assumed that the kinetic or inhibition properties of the mutant prevented its survival. Because some mutant enzymes not found in nature were also available, our hypotheses could be tested.
Figure 4.
Proposed evolutionary tree for the development of antifolate resistance in malaria parasites. Solid arrows are the anticipated pathways by which antifolate-resistant mutants were developed. Dashed arrows indicate the pathways that are unlikely to have occurred in nature for reasons given in the text. Mutants surrounded by ellipses are naturally occurring parasites whereas those with square boxes have not been found in nature.
The single mutant S108N would have arisen by point mutation of the AGC codon of the wild-type gene to AAC. S108N would have been a favorable precursor for the double mutants because it has almost unperturbed kinetic parameters but is about 10-fold more resistant to the antifolates (Table 3). The other five mutations possible by single base changes of AGC (C, G, I, R, or T) were inactive or less active than wild-type pfDHFR, and 2- to 10-fold less resistant to Pyr than S108N (unpublished results). Of the possible mutations of S108, the S108N mutant that emerged in nature seems to be the most favorable for Pyr resistance. The five single mutants found at the target sites of multiple mutants (A16V, N51I, C59R, S108T, and I164L) have not been observed as single mutants in nature. Four of these remain highly sensitive to both antifolates (N51I, C59R, S108T, and I164L), with one (N51I) also showing a significantly reduced kcat. A16V is highly resistant to Cyc but retains sensitivity to Pyr; however, the kinetic properties of DHFR A16V are severely impaired, and this mutant may not be competent to support DNA synthesis in vivo. Thus, under pressure of selection by antifolates, the most favorable single mutation appears to be DHFR S108N. The fact that the S108N mutation is found in all of the Pyr-resistant multiple mutants found in nature further supports the idea that it was a common parent.
Except for A16V+S108T, the two double mutants found in vivo (N51I+S108N and C59R+S108N) could have arisen from point mutation of the S108N DHFR single mutant; they both provide a lower increment of resistance toward Pyr which would have led to their selection (Table 3). The N51I+S108N mutant had kinetic parameters similar to wild-type DHFR and was about 2- to 3-fold more resistant to Pyr than its presumed parent S108N. C59R+S108N had a significantly reduced kcat compared with wild-type and to S108N DHFR, but was 5-fold more resistant to Pyr than was S108N. There is no unambiguous pathway leading to the Cyc-resistant double mutant, A16V+S108T, because neither possible single mutant precursor is found in nature. DHFR A16V+S108T may have arisen by mutation of either S108T or A16V; however, neither mutant is a favorable parent because S108T is quite sensitive to Cyc, and A16V has very unfavorable kinetic properties (Table 3). Alternatively, A16V+S108T could have arisen from mutation of A16V+S108N, which in turn could have arisen by Ala to Val mutation at residue 16 of the common precursor S108N. However, the A16V+S108N intermediate has not been found in nature, and we were unable to detect activity for this mutant.
The triple mutants provide a significant synergistic enhancement of resistance over the double mutants and could have all occurred by point mutation of double mutants. The triple mutant C59R+S108N+I164L would have arisen from mutation of I164L in C59R+S108N, whereas the triple mutant C59R+S108N+N51I could have arisen either by C59R mutation of N51I+S108N or by N51I mutation of C59R+S108N. Finally, the highly resistant quadruple mutant could have arisen by I164L mutation of N51I+C59R+S108N or by N51I mutation of C59R+S108N+I164L; in either case, it provides both an enhancement of resistance and a more favorable kcat.
As described above, our data suggest that all of the Pyr-resistant strains were generated from sequential point mutations of the common intermediate S108N; other single mutants at the target positions probably would not have provided resistant or viable organisms. Also, whereas Pyr resistant mutants show cross resistance to Cyc, the Cyc-resistant A16V+S108T is sensitive to Pyr. These observations suggest new approaches to drug therapy that might be useful in antifolate resistant malaria. The first approach would target potential Pyr-resistant mutants. Here, the idea is that if an agent, or combination of agents, could be developed that inhibited both the wild-type and the S108N single mutant, the emergence of the common, essential precursor S108N would be prevented. Drug-resistant mutants would have to proceed through a yet unknown single mutation which was resistant to both drug(s) or through a highly improbable double mutation. A second approach would target the Cyc-resistant A16V+S108T mutant with Pyr after it has emerged and been identified. This double mutant is sensitive to Pyr and could revert to a Pyr-resistant form only after an improbable double mutation—i.e., both single mutants A16V and S108T are sensitive to Pyr. The low probability of such a double mutation may make this a viable chemotherapeutic approach, although it would be impractical to practice on a widespread basis. These proposals assume that we have a reasonably complete knowledge of the molecular basis of resistance, which may not be correct. Nevertheless, with current technology, the hypotheses can be tested experimentally and would at the least provide new insight into antifolate-resistant malaria.
Acknowledgments
We wish to thank Dr. Kathryn Ivanetich for assistance with the manuscript. This work received financial support from Tropical Disease Research Grant ID 940096 (W.S.), U.S. Public Health Service Grant AI 19358 (D.V.S.), and in part by a Rockefeller Biotechnology Career Fellowship to W.S.
Footnotes
Abbreviations: DHFR, dihydrofolate reductase; pfDHFR, Plasmodium falciparum DHFR; H2folate, 7,8-dihydrofolate; Cyc, cycloguanil; Pyr, pyrimethamine; MTX, methotrexate.
Plowe, C., Djimde, A., Cortese, J., Taylor, T., Nwanyanwu, O., Watkins, W., Winstanley, P., Estrada, J., Cespedes, J., Mollinedo, R., Peterson, D. & Doumbo, O. Seventh Molecular Parasitology Meeting, Sept. 15–19, 1996, Woods Hole, MA, p. 98.
References
- 1.Blakley R L. In: Folates and Pteridines. Blakley R L, Benkovics S J, editors. Vol. 1. New York: Wiley; 1984. pp. 191–253. [Google Scholar]
- 2.Cella R, Carbonera D, Orsi R, Ferri G, Iadarola P. Plant Mol Biol. 1991;16:975–982. doi: 10.1007/BF00016070. [DOI] [PubMed] [Google Scholar]
- 3.Ferone R, Roland S. Proc Natl Acad Sci USA. 1980;77:5802–5806. doi: 10.1073/pnas.77.10.5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ivanetich K M, Santi D V. FASEB J. 1990;4:1591–1597. doi: 10.1096/fasebj.4.6.2180768. [DOI] [PubMed] [Google Scholar]
- 5.Lazar G, Zhang H, Goodman H M. Plant J. 1993;3:657–668. doi: 10.1046/j.1365-313x.1993.03050657.x. [DOI] [PubMed] [Google Scholar]
- 6.Garrett C E, Coderre J A, Meek T D, Garvey E P, Claman D M, Beverley S M, Santi D V. Mol Biochem Parasitol. 1984;11:257–265. doi: 10.1016/0166-6851(84)90070-7. [DOI] [PubMed] [Google Scholar]
- 7.Basco L K, de Pecoulas P E, Wilson C M, Le Bras J, Mazabraud A. Mol Biochem Parasitol. 1995;69:135–138. doi: 10.1016/0166-6851(94)00207-4. [DOI] [PubMed] [Google Scholar]
- 8.Cowman A F, Morry M J, Biggs B A, Cross G A M, Foote S J. Proc Natl Acad Sci USA. 1988;85:9109–9113. doi: 10.1073/pnas.85.23.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foote S J, Galatis D, Cowman A F. Proc Natl Acad Sci USA. 1990;87:3014–3017. doi: 10.1073/pnas.87.8.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peterson D S, Walliker D, Wellems T E. Proc Natl Acad Sci USA. 1988;85:9114–9118. doi: 10.1073/pnas.85.23.9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peterson D S, Milhous W K, Wellems T E. Proc Natl Acad Sci USA. 1990;87:3018–3022. doi: 10.1073/pnas.87.8.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Dijk M R, Waters A P, Janse C J. Science. 1995;268:1358–1362. doi: 10.1126/science.7761856. [DOI] [PubMed] [Google Scholar]
- 13.Van Dijk M R, Janse C J, Waters A P. Science. 1996;271:662–665. doi: 10.1126/science.271.5249.662. [DOI] [PubMed] [Google Scholar]
- 14.Wu Y, Sifri C D, Lei H-H, Su X-Z, Wellems T E. Proc Natl Acad Sci USA. 1995;92:973–977. doi: 10.1073/pnas.92.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Y, Kirkman L A, Wellems T E. Proc Natl Acad Sci USA. 1996;93:1130–1134. doi: 10.1073/pnas.93.3.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bzik D J, Li W-B, Horii T, Inselburg J. Proc Natl Acad Sci USA. 1987;84:8360–8364. doi: 10.1073/pnas.84.23.8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall S J, Sim P F G, Hyde J E. Mol Biochem Parasitol. 1991;45:317–330. doi: 10.1016/0166-6851(91)90100-k. [DOI] [PubMed] [Google Scholar]
- 18.Sirawaraporn W, Sirawaraporn R, Cowman A F, Yuthavong Y, Santi D V. Biochemistry. 1990;29:10779–10785. doi: 10.1021/bi00500a009. [DOI] [PubMed] [Google Scholar]
- 19.Sirawaraporn W, Prapunwattana P, Sirawaraporn R, Yuthavong Y, Santi D V. J Biol Chem. 1993;268:21637–21644. [PubMed] [Google Scholar]
- 20.Meek T D, Garvey E P, Santi D V. Biochemistry. 1985;24:678–686. doi: 10.1021/bi00324a021. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 22.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilkinson A J, Fersht A R, Blow D M, Winter G. Biochemistry. 1983;22:3581–3586. doi: 10.1021/bi00284a007. [DOI] [PubMed] [Google Scholar]
- 24.Basco L K, Ramiliarisoa O, Le Bras J. Am J Trop Med Hyg. 1994;45:492–497. doi: 10.4269/ajtmh.1994.50.193. [DOI] [PubMed] [Google Scholar]