Abstract
Tipin and its interacting partner Tim1 (Timeless) form a complex at replication forks that plays an important role in the DNA damage checkpoint response. Here we identify Xenopus laevis Tipin as a substrate for cyclin E/cyclin-dependent kinases 2 that is phosphorylated in interphase and undergoes further phosphorylation upon entry into mitosis. During unperturbed DNA replication, the Tipin/Tim1 complex is bound to chromatin, and we were able to detect interactions between Tipin and the MCM helicase. Depletion of Tipin from Xenopus extracts did not significantly impair normal replication but substantially blocked the ability of stalled replication forks to recover after removal of a block imposed by aphidicolin. Tipin-depleted extracts also showed defects in the activation of Chk1 in response to aphidicolin, probably because of a failure to load the checkpoint mediator protein Claspin onto chromatin.
Keywords: cell cycle, checkpoint, cyclin, DNA damage
Eukaryotic cells have evolved multiple mechanisms that promote the fidelity of DNA replication. Stalled replication forks arising at sites of DNA damage or after nucleotide depletion are sensed and repaired by surveillance mechanisms called checkpoints. Once activated, the checkpoint machinery triggers a number of events that activate DNA repair pathways to coordinate a delay in cell cycle progression with repair of damage, ensuring the integrity of the genome (1–4). The importance of the checkpoint pathways is underlined by the conservation of the proteins involved from yeast to humans (5, 6) and by the fact that mutations in genes encoding checkpoint proteins tend to predispose affected individuals to cancer (7).
Different types of DNA damage trigger specific checkpoint pathways; UV light and stalled replication forks activate the ATR-Chk1 pathway, whereas DNA double-strand breaks activate the ATM-Chk2 pathway. The key function of the ATR-Chk1 pathway is thought to be to stabilize and preserve the stalled replication forks until the damage is repaired, permitting replication to resume (8–12).
Recent studies in yeast and mammalian cells have identified three proteins, Timeless (Tim1), Tipin, and Claspin, important for the repair of stalled DNA replication forks. The budding and fission yeast homolog of Tim1 are, respectively, Tof1 and Swi1. Tof1 (13, 14) is part of the Mrc1 (Claspin) pathway that is required for replication forks to pause when barriers to replication are encountered (13, 15–17). Swi1, a functional homolog of Tof1 (18), is essential for full activation of Cds1 and stabilization of replication forks (17, 19, 20). Recently, human Tim1 was reported to be a mediator of the intra S-phase checkpoint (21).
Tipin was originally identified as a Tim1 interacting protein by a yeast two-hybrid screen (22). Tipin homologs have been identified in budding (Csm3) and in fission yeast (Swi3) (18, 23). Csm3 and Swi3 interact with Tof1 and Swi1, respectively, and are required for checkpoint responses to replication stress (24–29). Recent data in mammalian cells showed that Tipin is required for efficient cell cycle arrest in response to DNA damage (30). Reduction in the levels of Tipin or Tim1 by siRNA renders cells sensitive to ionizing radiation (31) and results in reduced level of Chk1 phosphorylation upon hydroxyurea or ultraviolet radiation (100–280 nm) treatment. These findings indicate that the Tipin/Tim1 complex is a mediator of the UV-induced intra S-phase checkpoint (30–33). Tim1 is also important for replication fork progression in the absence of damage whereas Tipin slows DNA chain elongation in active replicons in UV-damaged cells (32).
In this work we focused our attention on the Xenopus laevis homolog of Tipin and studied its role during S phase progression. We demonstrate that depletion of Tipin from Xenopus extracts does not alter the overall efficiency of DNA replication. However, Tipin is necessary for the intra S-phase checkpoint response to replication block, which may result from a failure to load Claspin onto chromatin when Tipin is absent. Furthermore, we find that Tipin associates with replication forks and ensures fork stability by coupling the replicative polymerase and helicase activities, thereby preventing the helicase from excessively unwinding the DNA when the polymerase has stalled. Our data support a model in which Tipin mediates replication fork pausing in response to replication stress and thus participates in the DNA damage checkpoint response.
Results
Tipin Is a Cyclin E/Cyclin-Dependent Kinase (CDK) 2 Substrate.
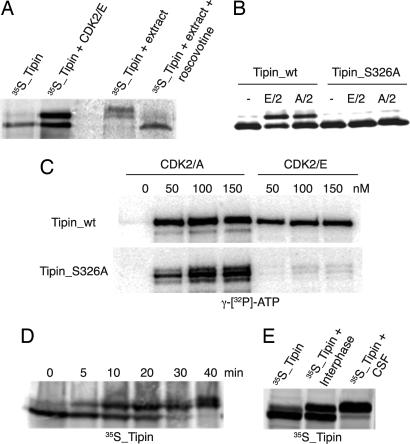
In a screen for cyclin E/CDK2 substrates (unpublished data), we identified Tipin as a protein that undergoes a mobility shift upon phosphorylation by CDK2 (Fig. 1A). To confirm that Tipin is phosphorylated by CDK in vivo we added35S-labeled protein to an interphase extract and observed retardation in electrophoretic mobility, which was abolished if the extract was pretreated with roscovitine to inhibit CDK activity (Fig. 1A) (34).
Fig. 1.
Tipin is a substrate for cyclin E/CDK2. (A) In vitro-translated Tipin was supplemented or not with recombinant cyclin E/CDK2 and ATP. Alternatively, 35S-labeled protein was added to a Xenopus extract in the presence or absence of roscovitine. (B) Cyclin E- or cyclin A-CDK2 and ATP were added to in vitro-translated Tipin (wild type or S326A mutant). (C) Recombinant GST-Tipin (wild type or S326A) was incubated with increasing concentrations of cyclin E- or cyclin A-CDK2 and [γ-32P]ATP. (D) 35S-labeled Tipin was added to a Xenopus cycling extract, and samples were collected from 0 to 40 min. (E) Wild-type 35S-labeled Tipin was added to an interphase or metaphase arrested extract (CSF, cytostatic factor-mediated metaphase II arrested extract). Samples were analyzed by SDS/PAGE and autoradiography.
Tipin is a 386-residue protein that contains six S/T-P motifs and one complete CDK consensus site, (S/T)PXK/R (35, 36), at Ser-326. We generated a S326A mutant and tested the ability of this mutant to be phosphorylated by cyclin E/CDK2 or cyclin A/CDK2. For both kinases the mobility shift was abolished by the S326A mutation (Fig. 1B). However, when the phosphorylation status of the S326A mutant was assessed by 32PO4 incorporation, cyclin A/CDK2 was still able to phosphorylate the S326A mutant, whereas cyclin E/CDK2 did not (Fig. 1C). This suggests that there are other CDK phosphorylation sites on the protein. We therefore analyzed the behavior of Tipin in different stages of the cell cycle. Addition of 35S-labeled Tipin to an extract in interphase induced a rapid mobility shift, which presumably reflects the phosphorylation by cyclin E/CDK2. A further mobility shift is induced at 40 min, when the extract enters mitosis (data not shown), suggesting the existence of additional phosphorylation sites in Tipin (Fig. 1D). To further confirm this, the mobility of Tipin was compared between interphase extracts and a metaphase arrested egg extract, revealing an additional retardation in the mobility of Tipin in the M-phase environment (Fig. 1E).
Tipin Is a Phosphoprotein in Xenopus Extracts.
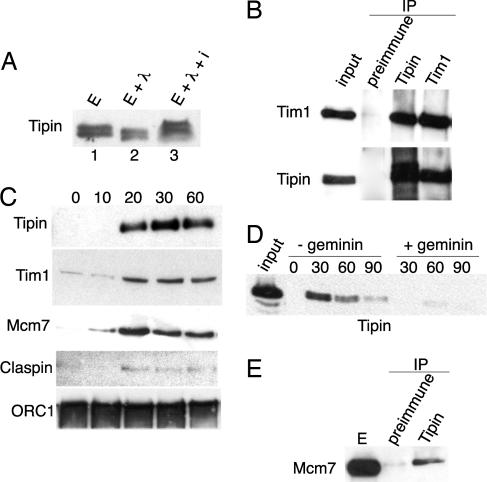
To study the function of Tipin in Xenopus extracts, we generated polyclonal antibodies against the full-length recombinant protein expressed in bacteria. Although Tipin has a calculated molecular mass of 42 kDa, the antibody recognizes a specific band ≈60 kDa in Xenopus extract, which disappeared after Tipin depletion. Because our data indicate that Tipin is a substrate for cyclin E/CDK2 and this kinase is always present in early Xenopus embryos (37), we treated the extract with λ phosphatase in the presence or absence of sodium vanadate, a phosphatase inhibitor. The band corresponding to endogenous Tipin is shifted down (Fig. 2A) upon phosphatase treatment showing that Tipin is constitutively phosphorylated in an interphase extract.
Fig. 2.
Endogenous Tipin is a chromatin-associated phosphoprotein that interacts with Tim1 and MCM7. (A) Xenopus extract was treated (lane 2) or not (lane 1) with λ-phosphatase in the presence (lane 3) or absence of sodium vanadate. Endogenous Tipin was detected by immunoblotting. (B) Equal amounts of extract were immunoprecipitated with anti-Tipin and anti-Tim1 antibodies or preimmune serum. Purified proteins were immunoblotted as indicated. (C) Sperm nuclei (3,000 nuclei per microliter) were added to Xenopus extract, and chromatin was harvested at different times. Chromatin-bound proteins were analyzed by immunoblotting. (D) Sperm nuclei were added to Xenopus extract, and chromatin was harvested in the presence or absence of geminin (5 ng/μl). Samples were probed with anti-Tipin antibody. (E) Xenopus extract was immunoprecipitated with either anti-Tipin or preimmune serum. Samples were probed with anti-MCM7 antibody.
Tipin Associates with Chromatin and Forms a Stable Complex with Tim1.
To determine whether Tipin interacts with Tim1 in Xenopus, we performed coimmunoprecipitation experiments. Tipin and Tim1 coimmunoprecipitated from Xenopus extracts, demonstrating that these two proteins form a complex in vivo (Fig. 2B). Because Tipin is phosphorylated in Xenopus extracts, we asked whether the Tipin/Tim1 interaction depended on phosphorylation, but treatment with λ-phosphatase had no effect on the interaction of Tipin and Tim1 (data not shown).
Previous work in both yeast and human cells demonstrated that the Tipin/Tim1 complex is associated with the replication fork (33), is required for efficient S-phase progression (30), and is important for the intra S-phase checkpoint (30–33). We therefore investigated the role of Tipin in DNA replication in Xenopus extracts. To determine whether Tipin associated with chromatin in Xenopus extracts, we added demembranated sperm nuclei to an interphase extract and purified chromatin at different times. Members of the prereplication complex, such as ORC1, are rapidly loaded on to the chromatin when nuclei are added to the extract (Fig. 2C); other proteins required for replication initiation are found in the chromatin at later time points. MCM7 is loaded onto the chromatin 20 min after nuclei addition. Fig. 2C shows that Tipin, Tim1, and Claspin, a mediator of the intra S-phase checkpoint response, appeared in the chromatin fraction with the same kinetics as MCM7 (Fig. 2C). In the absence of deliberate DNA damage, very little Claspin loads onto chromatin.
To determine whether the association of Tipin with chromatin required DNA replication, we added geminin to prevent loading of the MCM proteins onto chromatin (38–40). Fig. 2D shows that addition of geminin inhibited the loading of Tipin onto chromatin. We next asked whether Tipin was directly associated with the replication machinery. Even without adding DNA to the extract, Tipin immunoprecipitates were found to contain MCM7, a member of the putative replicative helicase (Fig. 2E). The idea that Tipin and Tim1 are members of a multiprotein complex at the replication fork is strengthened by the fact that they migrate as a high-molecular-mass complex of ≈600–850 kDa and coelute with Claspin and MCM7 upon gel filtration (data not shown).
Tipin Is Not Essential for Replication.
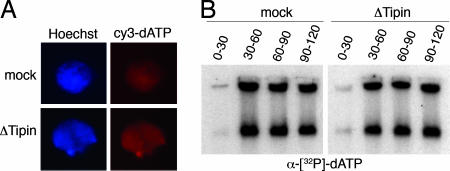
Although Tipin is associated with the replication fork, its role during DNA replication is controversial. Reduction of Tipin levels in human cells by siRNA resulted in a delay in S-phase progression (30–32). To gain further insight into the role of Tipin during DNA replication, we assessed the impact of Tipin depletion from Xenopus extracts on DNA replication. Perhaps surprisingly, both mock-depleted and Tipin-depleted extracts formed nuclei that incorporated cy3-dATP at the same rate (Fig. 3A). Similarly, there was no reproducible difference in the labeling of the newly replicated DNA with [α-32P]dATP between extracts containing or lacking Tipin (Fig. 3B).
Fig. 3.
Tipin is not essential for replication. (A) Mock- or Tipin-depleted extracts were supplemented with sperm nuclei (3,000 nuclei per microliter) and cy3-dATP (Amersham). After 30 min, 3 μl was spotted on a microscope slide and mixed with a fixative containing Hoechst 33258 (blue fluorescence). Red fluorescence due to cy3-dATP incorporation indicates a replicating nucleus. (B) Sperm nuclei (1,000 nuclei per microliter) were added to an interphase Xenopus extract (mock- or Tipin-depleted). Replication was labeled with [α-32P]dATP from 0 to 30 min, from 30 to 60 min, from 60 to 90 min, or from 90 to 120 min from nuclei addition. Replicated DNA was purified, loaded on agarose gel, and analyzed by autoradiography.
Tipin Is Important for the Checkpoint Response to Replication Block.
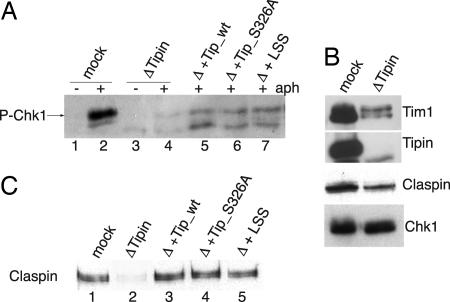
Budding and fission yeast orthologues of Tipin and Tim1 are involved in DNA replication checkpoint responses (13, 17, 28). Recent data in mammalian cells showed that down-regulation of Tipin or Tim1 by siRNA resulted in reduced levels of Chk1 phosphorylation (21, 30–33). Therefore, we examined the role of Tipin in Xenopus S-phase checkpoint responses by monitoring the phosphorylation of Chk1 in response to a replication block. Sperm nuclei were added to an interphase Xenopus extract treated or not with aphidicolin, a DNA replication inhibitor that induces replication fork stalling and triggers the activation of Chk1 (41, 42). After 70 min of aphidicolin treatment, nuclei were isolated and the level of Chk1 phosphorylation assessed by using a phosphoantibody that recognizes P-Ser-344 in Chk1 (Fig. 4A). In Tipin-depleted extracts, the phosphorylation of Chk1 was barely detectable compared with the control, although the level of Chk1 in the depleted extract was unaffected (Fig. 4B). Phosphorylation of Chk1 was partially rescued by adding back recombinant Tipin to the depleted extract (Fig. 4A), but, compared with the mock-depleted extract, the level of Chk1 phosphorylation was considerably reduced. In all Tipin depletion experiments, however, Tim1 levels were also greatly reduced. Thus, by adding back Tipin alone, rather than the Tipin/Tim1 complex, the residual Tim1 remaining in the extract may be limiting and not sufficient to completely restore Chk1 phosphorylation. On the other hand, we observed a similarly low level of rescue when 1 μl of mock-depleted extract [low-speed supernatant (LSS)] was added back to the depleted extract, suggesting that the process of depletion has compromised something else, which can only be partially rescued even when complete extract is added. There does seem to be an excess of Tim1 over Tipin in the extract, so that when Tipin is added back, it can find its partner and form an active complex, but this may be at too low a concentration to perform a complete rescue.
Fig. 4.
Tipin is required for Chk1 phosphorylation and Claspin loading on chromatin. (A) Sperm nuclei were added to mock-depleted (lanes 1 and 2) or Tipin-depleted (lanes 3 and 4) extract in the presence (lanes 2 and 4) or absence (lanes 1 and 3) of aphidicolin and incubated at 23°C for 70 min. Nuclei were purified, and proteins were analyzed by immunoblot with anti-phospho-Chk1 antibody. Recombinant Tipin (wild type, lane 5; S326A, lane 6) or 1 μl of mock-depleted extract (LSS, lane 7) were added to the depleted extract, and the experiment was performed as described above. (B) Mock- or Tipin-depleted extract (1 μl) was analyzed by immunoblotting as indicated. (C) Chromatin was purified from mock-depleted (lane1) or Tipin-depleted (lanes 2–5) extract supplemented with aphidicolin and recombinant wild-type Tipin (lane 3) or S326A Tipin (lane 4) or 1 μl of extract (LSS, lane 5). Samples were probed with anti-Claspin antibody.
When replication is blocked, the Xenopus homologue of ATR is responsible for the activation of Chk1 (43, 44). This process requires the mediator protein Claspin (45–47), which also associates with the replication fork (48, 49). Because Claspin is an essential mediator of the checkpoint response, we analyzed its behavior in Tipin-depleted extracts. Although Claspin levels were not significantly reduced in Tipin-depleted extracts (Fig. 4B), Claspin was not loaded onto the chromatin in the absence of Tipin (Fig. 4C). The loading of Claspin onto the chromatin was rescued by the addition of wild-type recombinant Tipin to the depleted extract (Fig. 4C). The defect in Claspin loading onto the chromatin and the phosphorylation of Chk1 was also rescued by adding back recombinant Tipin with the S326 mutated to either Ala or Glu (Fig. 4A and data not shown), indicating that Tipin phosphorylation is not important for its checkpoint function.
Tipin Is Required for Stabilizing the Replication Fork During DNA Damage.
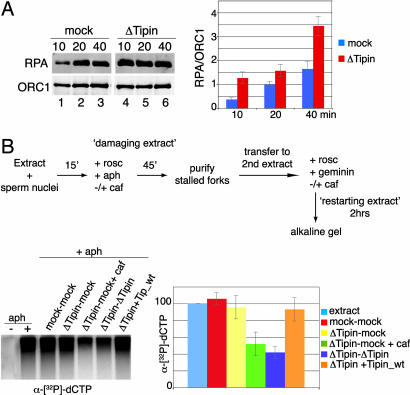
The Human Tipin/Tim1 complex associates with replication protein A (RPA)34, Claspin, MCM helicase, and DNA polymerase, and it has been suggested that a possible role for the Tipin/Tim1 complex at the DNA replication fork is to couple the MCM-mediated unwinding activity with the polymerase-mediated DNA synthesis, stabilizing the replisome (33). To address the role of Tipin at the replication fork, we depleted Tipin from the extract and monitored the accumulation of RPA on chromatin at different times after blocking replication with aphidicolin. Fig. 5A shows that RPA rapidly accumulated to high levels on chromatin in Tipin-depleted extract compared with the mock-depleted control, suggesting that single-stranded DNA accumulates at blocked replication forks in the absence of Tipin. The quantitation shown in the accompanying bar graph (Fig. 5A Right) was performed in three independent experiments using the Li-Cor system (Lincoln, NE) for the analysis of the immunoblots.
Fig. 5.
Tipin depletion induces accumulation of ssDNA and impairs recovery from stalled replication forks. (A) Mock-depleted (lanes 1–3) or Tipin-depleted (lanes 4–6) extract was incubated with 20 μM aphidicolin. Chromatin was harvested at 10, 20, and 40 min and analyzed by immunoblotting with anti-RPA and anti-ORC1 antibodies. The amount of RPA loaded on the chromatin was determined with respect to ORC1. Three independent experiments are averaged in the bar graphs; mock-depleted extract is represented by blue bars, and Tipin-depleted extracts are represented by red bars. (B) Chromatin replication was initiated in an interphase extract (damaging extract). Sperm nuclei were added to the extract and incubated at 23°C to allow origins to fire and replication to start. After 15 min, fork progression was blocked by adding 40 μM aphidicolin and 0.5 mM roscovitine. Damaged forks were isolated at 60 min and added to a second extract (restarting extract) made incompetent for origin firing and origin assembly by addition of 0.5 mM roscovitine and 5 ng/μl geminin. Different damaging and restarting (mock- or Tipin-depleted) extracts were used as stated in the different lanes. Lanes 1 and 2, Xenopus extract either with (lane 2) or without (lane 1) aphidicolin in the first incubation; lanes 3–7, all had aphidicolin in the first incubation; lane 3, both damaging and restarting extract were mock-depleted; lanes 4 and 5, the damaging extract was Tipin-depleted, and forks were restarted in a mock-depleted extract; lane 5, caffeine was added to the second incubation; lane 6, both damaging and restarting extract were Tipin-depleted; lane 7, the same as lane 6 except that recombinant Tipin was added in the second incubation. In all cases, [α-32P]dCTP was added in the second incubation to monitor replication. Incorporation was analyzed by alkaline agarose gel electrophoresis and autoradiography. Three independent experiments are averaged in the bar graphs, taking the amount of replication of the positive control (e.g., lane 2) as 100%. The error bars represent standard deviations from the mean values.
We next examined the role of Tipin during the recovery of stalled replication forks using a chromatin transfer assay (Fig. 5B) (50). First, sperm nuclei were added to an extract (“damaging extract”) to allow origin firing and the initiation of DNA replication. After 15 min, aphidicolin was added to stall replication forks. Roscovitine was added at the same time to inhibit CDK activity and block further origin firing (where indicated, caffeine was added to inhibit ATM and ATR activity). After a further 45 min, chromatin containing the stalled replication forks was isolated and transferred to a second extract (“restarting extract”) rendered incompetent for origin firing or assembly of new origins by addition of both roscovitine and geminin. Aphidicolin treatment led to replication arrest in the damaging extracts. Fig. 5B shows that, when the chromatin from the first incubation was added to the restarting extract, significant incorporation of [α-32P]dCTP into DNA occurred if the damaging extract contained aphidicolin (Fig. 5B, lane 2), but not if aphidicolin was omitted and no damage was inflicted (Fig. 5B, lane 1). When both the damaging extract and the restarting extract were depleted of Tipin, DNA synthesis was reduced to ≈50% of the control (Fig. 5B, lane 6). Tipin must be present in the restarting extract (Fig. 5B, lane 4). To test whether Tipin was directly involved in restart of damaged replication forks, Tipin-depleted damaging and restarting extracts were supplemented with recombinant Tipin. This restored normal DNA replication (Fig. 5B, lane 7). Trenz et al. (50) have shown that ATR-ATM-dependent checkpoint is required for restarting of collapsed fork, but not of stalled forks, and we found that addition of caffeine to both damaging and restarting extracts prevented restart (Fig. 5B, lanes 4 and 5), suggesting that restart of forks impaired by Tipin depletion requires ATM-ATR activity. In a Tipin-depleted extract, loss of Chk1 activation in response to DNA damage indicates that the ATR checkpoint is defective, but ATM is presumably still active, which helps to prevent collapse of replication forks. When both ATR and ATM are inhibited by caffeine, however, the checkpoint is completely inactive, and forks collapse in the absence of Tipin. Once collapsed, replication forks are not able to restart.
Discussion
The DNA damage checkpoint network is extremely important for the faithful completion of DNA replication during S-phase. When a replication fork is stalled, checkpoint signaling pathways are activated and DNA replication is temporarily inhibited to prevent the irreversible breakdown of replication forks (51). A replication-pausing complex is thought to assemble at arrested forks to activate the checkpoint, allowing repair of the damaged DNA and resumption of replication (16–18, 28). In budding yeast, the Mec1/Rad53 (ATR/Chk2) checkpoint response prevents the collapse of stalled replication forks and allows the restart of DNA replication after recovery (52). This response requires both Tof1 (Tim1) and Mrc1 (Claspin) proteins (16, 48). Nedelcheva et al. (27) suggested a mechanism where Tof1-Csm3-Mrc1 (Tim1-Tipin-Claspin) contributes to a “pausing complex” that prevents uncoupling of the replication machinery from DNA unwinding upon DNA damage. After pausing, there is limited accumulation of ssDNA to which RPA binds and promotes the recruitment of the Mec1-Ddc2 (ATR-ATRIP) complex, which activates the checkpoint cascade (53). A similar model has been suggested for the human S-phase checkpoint, where the Tim/Tipin/Claspin complex is believed to be present at the replisome (6).
In mammals, Tim1, Tipin, and Claspin are considered checkpoint mediators, because loss of any one of these proteins results in reduced phosphorylation of Chk1 and a somewhat inefficient progression through S-phase (30). In Xenopus, Claspin associates directly with Chk1, an interaction that strongly enhances the ability of ATR to phosphorylate Chk1 (54, 55). We have shown that immunodepletion of Tipin from Xenopus extract also results in a substantial removal of Tim1, whereas Claspin and Chk1 levels are unaffected. In these Tipin (and Tim1)-depleted extracts Chk1 phosphorylation in response to aphidicolin treatment is considerably reduced, consistent with the postulated mediator functions of Tipin. We also found that Claspin is not loaded onto chromatin in Tipin-depleted extracts, in agreement with recent experiments in human cells (30). It is likely that the failure to load Claspin is responsible for the failure to phosphorylate Chk1 in the absence of Tipin after replication stress. Both the loading of Claspin onto the chromatin and Chk1 phosphorylation are rescued by adding back recombinant Tipin. Truncated forms of Claspin lacking several hundred residues from the N terminus show reduced ability to bind chromatin but can still activate Chk1 to some extent, indicating that stable association of Claspin with chromatin is not absolutely essential for its Chk1-activating function, although full-length Claspin was much more effective at activating Chk1 (56). It would be interesting to know whether the N terminus of Claspin contained domains that interacted with Tim1 and/or Tipin.
Taken together, the homology of Tipin with Csm3 and the recent results in human cells strongly support a role for Tipin/Tim1 as part of a pausing complex at the replication fork. Gotter et al. (33) suggested a model in which Tipin/Tim1 complexes are closely associated with the replisome and are important for coupling the DNA unwinding activity of the MCM helicase with DNA synthesis catalyzed by the polymerases. According to this model, when DNA synthesis is inhibited by aphidicolin treatment, the MCM helicase continues to unwind DNA, which generates a signal that leads to Chk1 activation (57). Consistent with this model, aphidicolin treatment generated regions of ssDNA, which were considerably increased when Tipin/Tim1 complexes were depleted. This supports the idea that at least one role of Tipin and Tim1 is to help couple the helicase to the replisome to minimize excessive unwinding of the DNA. In support of this, we were able to detect an interaction between Tipin and MCM7.
We also assessed the importance of Tipin in maintaining fork stability and promoting fork restart by monitoring the recovery of stalled replication forks (50). We found that, in the absence of Tipin, the ability of stalled replication forks to restart was seriously impaired. Importantly, this phenotype could be rescued by the addition of recombinant wild-type Tipin in the restarting extract. To understand whether this restart failure was due to a checkpoint signaling defect we treated extracts with caffeine before adding back the recombinant Tipin. When ATR and ATM activities were inhibited by caffeine, the damaged forks were unable to restart. It has been shown that ATM and ATR are not necessary to resolve replication forks that have been stalled by aphidicolin treatment but are required for the recovery of replication forks collapsed by addition of camptothecin or mitomycin C (50, 58). This implies that the absence of Tipin leads to the collapse of replication forks, which are therefore unable to resume replication when aphidicolin is removed. Tipin may thus be considered as a component of the replication machinery that helps to stabilize the fork when DNA replication is impaired. At present, there is no quantitative information concerning the stoichiometry of Tim1 and Tipin in relation to other components of the replisome. Consistent with the idea that loss of Tipin leads to the accumulation of aberrant structures during DNA replication, caused by failure to stabilize stalled replication forks, it has recently been shown that loss of Tipin results in spontaneous γ-H2AX foci, a marker for DNA double-strand breaks (31).
Our data provide clear and direct experimental evidence in favor of the proposed model for checkpoint activation and fork stabilization upon DNA damage, strongly suggesting a role for the Tim1/Tipin complex in stabilizing the replication fork and helping to bring the highly diffusible Chk1 to sites of stalled replication forks where it can be activated (via Claspin interaction) by ATR and ATRIP.
We originally identified Tipin as a substrate for cyclin E-CDK2 and found that Tipin is a phosphoprotein in Xenopus extracts. So far we have not found a functional role for this phosphorylation. Phosphorylation at S326 does not appear to be required for Tim1 interaction, binding to chromatin, rescue of stalled replication forks, or activation of Chk1.
The Xenopus extract provides an excellent system to further investigate the role of Tipin and Tim1 in DNA replication using biochemical approaches. Previous results in human cells and yeast (19, 31), as well as our results, suggest that depletion of the Tipin/Tim1 complex may contribute to genomic instability. Tim1 is an essential gene in mice and Caenorhabditis elegans (59, 60); one would guess the same would be true for Tipin, although this has not been tested. Further studies of these proteins should prove most illuminating to understanding replication fork collapse and recovery as well as the checkpoint signaling pathways responsible for reporting and reacting to DNA damage.
Materials and Methods
Plasmids.
Expression plasmids for Tipin were generated by inserting PCR-amplified Tipin cDNA in pDEST15 and pDEST17. Mutagenesis was performed by using the QuikChange site-directed mutagenesis kit (Qiagen, Crawley, U.K.).
Antibodies.
Rabbit Tipin antiserum was generated against a 6His-Tipin protein by Biogenes (Berlin, Germany). Anti-Tim1 was a gift of Jan-Michael Peters (Research Institute of Molecular Pathology, Vienna, Austria). Anti-Claspin and anti-Chk1 were a gift from Howard Lindsay (University of Lancaster, Lancaster, U.K.). Additional antibodies used included anti-Mcm7 (Santa Cruz Biotechnology, Santa Cruz, CA), anti-phospho-Chk1 (New England Biolabs, Hitchin, U.K.). Anti-ORC1 TK15 was generated previously (61).
Xenopus Egg Extracts and Nuclear Fraction.
S-phase extracts and demembranated sperm nuclei were prepared as described (62). Aphidicolin was added to the extracts at a concentration of 40 μg/ml to inhibit DNA replication. Roscovitine (Sigma, Poole, Dorset, U.K.) was added to the extracts at 0.5 mM. Caffeine (Sigma) was added at 5 mM. Nuclear fractions to detect endogenous Chk1 phosphorylation were prepared as previously described (41).
Chromatin Isolation and Chromatin Transfer.
To isolate chromatin, sperm nuclei (3,000 nuclei per microliter) were added to 30 μl of egg extracts for appropriate times (see figure legends). For immunoblotting, samples were diluted with 10 volumes of EB buffer (100 mM KCl/2.5 mM MgCl2/50 mM Hepes·KOH, pH 7.5) containing 0.25% Nonidet P-40 and centrifuged through a 30% sucrose layer at 10,000 × g at 4°C for 5 min. Pellets were suspended in sample buffer. For transfer experiments, samples from damaging extracts were diluted with 10 volumes of EB buffer containing 15 mM MgCl2 and 0.08% Triton X-100 and centrifuged through a 15% sucrose layer at 6,000 × g at 4°C for 5 min; chromatin pellets were washed and resuspended in 25 μl of egg extract.
Immunodepletion and Immunoprecipitation.
Immunodepletion was carried out as described (63). To deplete 1 ml of extract, 30 μg of Tipin antibody was used. For immunoprecipitations, antibodies (5 μg) were conjugated with 30 μl of protein A-Sepharose FF (GE Healthcare, Little Chalfont, U.K.) and added to 100 μl of Xenopus extract. After 1 h of incubation, beads were washed and harvested.
In Vitro Transcription–Translation and Kinase Assays.
In vitro transcription–translation was performed with a Quick Coupled SP6 kit according to the manufacturer's instructions (Promega, Southhampton, U.K.). Kinase assays were performed by incubating the substrate (5 μl of in vitro transcription–translation or 5 μg of recombinant protein) in 80 mM β-glycerolphosphate, 0.5 mM EGTA, 100 μM ATP, and 2.5 mM MgCl2, in the presence or absence of recombinant cyclin E/CDK2 or cyclin A/CDK2 at 23°C for 30 min.
Acknowledgments
We thank Dr. Simon Boulton for helpful reading of the manuscript and Dr. Haruhiko Takisawa for hospitality and practical advice. This work was supported by the European Union Mitocheck Consortium and Cancer Research UK.
Abbreviations
- CDK
cyclin-dependent kinase
- RPA
replication protein A.
Footnotes
The authors declare no conflict of interest.
References
- 1.Osborn AJ, Elledge SJ, Zou L. Trends Cell Biol. 2002;12:509–516. doi: 10.1016/s0962-8924(02)02380-2. [DOI] [PubMed] [Google Scholar]
- 2.McGowan CH, Russell P. Curr Opin Cell Biol. 2004;16:629–633. doi: 10.1016/j.ceb.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 3.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 4.O'Connell MJ, Cimprich KA. J Cell Sci. 2005;118:1–6. doi: 10.1242/jcs.01626. [DOI] [PubMed] [Google Scholar]
- 5.Zhou BB, Elledge SJ. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 6.Melo J, Toczyski D. Curr Opin Cell Biol. 2002;14:237–245. doi: 10.1016/s0955-0674(02)00312-5. [DOI] [PubMed] [Google Scholar]
- 7.Sherr CJ. Cell. 2004;116:235–246. doi: 10.1016/s0092-8674(03)01075-4. [DOI] [PubMed] [Google Scholar]
- 8.Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP. Nat Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 9.Zhao H, Piwnica-Worms H. Mol Cell Biol. 2001;21:4129–4139. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takai H, Tominaga K, Motoyama N, Minamishima YA, Nagahama H, Tsukiyama T, Ikeda K, Nakayama K, Nakanishi M, Nakayama K. Genes Dev. 2000;14:1439–1447. [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K, Luo G, Carattini-Rivera S, DeMayo F, Bradley A, et al. Genes Dev. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuoka S, Huang M, Elledge SJ. Science. 1998;282:1893–1897. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- 13.Foss EJ. Genetics. 2001;157:567–577. doi: 10.1093/genetics/157.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park H, Sternglanz R. Yeast. 1999;15:35–41. doi: 10.1002/(SICI)1097-0061(19990115)15:1<35::AID-YEA340>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 15.Alcasabas AA, Osborn AJ, Bachant J, Hu F, Werler PJ, Bousset K, Furuya K, Diffley JF, Carr AM, Elledge SJ. Nat Cell Biol. 2001;3:958–965. doi: 10.1038/ncb1101-958. [DOI] [PubMed] [Google Scholar]
- 16.Katou Y, Kanoh Y, Bando M, Noguchi H, Tanaka H, Ashikari T, Sugimoto K, Shirahige K. Nature. 2003;424:1078–1083. doi: 10.1038/nature01900. [DOI] [PubMed] [Google Scholar]
- 17.Noguchi E, Noguchi C, Du LL, Russell P. Mol Cell Biol. 2003;23:7861–7874. doi: 10.1128/MCB.23.21.7861-7874.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dalgaard JZ, Klar AJ. Cell. 2000;102:745–751. doi: 10.1016/s0092-8674(00)00063-5. [DOI] [PubMed] [Google Scholar]
- 19.Sommariva E, Pellny TK, Karahan N, Kumar S, Huberman JA, Dalgaard JZ. Mol Cell Biol. 2005;25:2770–2784. doi: 10.1128/MCB.25.7.2770-2784.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsumoto S, Ogino K, Noguchi E, Russell P, Masai H. J Biol Chem. 2005;280:42536–42542. doi: 10.1074/jbc.M510575200. [DOI] [PubMed] [Google Scholar]
- 21.Unsal-Kacmaz K, Mullen TE, Kaufmann WK, Sancar A. Mol Cell Biol. 2005;25:3109–3116. doi: 10.1128/MCB.25.8.3109-3116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gotter AL. J Mol Biol. 2003;331:167–176. doi: 10.1016/s0022-2836(03)00633-8. [DOI] [PubMed] [Google Scholar]
- 23.Rabitsch KP, Toth A, Galova M, Schleiffer A, Schaffner G, Aigner E, Rupp C, Penkner AM, Moreno-Borchart AC, Primig M, et al. Curr Biol. 2001;11:1001–1009. doi: 10.1016/s0960-9822(01)00274-3. [DOI] [PubMed] [Google Scholar]
- 24.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. Proc Natl Acad Sci USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 26.Mayer ML, Pot I, Chang M, Xu H, Aneliunas V, Kwok T, Newitt R, Aebersold R, Boone C, Brown GW, et al. Mol Biol Cell. 2004;15:1736–1745. doi: 10.1091/mbc.E03-08-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nedelcheva MN, Roguev A, Dolapchiev LB, Shevchenko A, Taskov HB, Shevchenko A, Stewart AF, Stoynov SS. J Mol Biol. 2005;347:509–521. doi: 10.1016/j.jmb.2005.01.041. [DOI] [PubMed] [Google Scholar]
- 28.Noguchi E, Noguchi C, McDonald WH, Yates JR, III, Russell P. Mol Cell Biol. 2004;24:8342–8355. doi: 10.1128/MCB.24.19.8342-8355.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohanty BK, Bairwa NK, Bastia D. Proc Natl Acad Sci USA. 2006;103:897–902. doi: 10.1073/pnas.0506540103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshizawa-Sugata N, Masai H. J Biol Chem. 2007;282:2729–2740. doi: 10.1074/jbc.M605596200. [DOI] [PubMed] [Google Scholar]
- 31.Chou DM, Elledge SJ. Proc Natl Acad Sci USA. 2006;103:18143–18147. doi: 10.1073/pnas.0609251103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Unsal-Kacmaz K, Chastain PD, Qu PP, Minoo P, Cordeiro-Stone M, Sancar A, Kaufmann WK. Mol Cell Biol. 2007;27:3131–3142. doi: 10.1128/MCB.02190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gotter AL, Suppa C, Emanuel BS. J Mol Biol. 2007;366:36–52. doi: 10.1016/j.jmb.2006.10.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Azevedo WF, Leclerc S, Meijer L, Havlicek L, Strnad M, Kim SH. Eur J Biochem. 1997;243:518–526. doi: 10.1111/j.1432-1033.1997.0518a.x. [DOI] [PubMed] [Google Scholar]
- 35.Songyang Z, Blechner S, Hoagland N, Hoekstra MF, Piwnica-Worms H, Cantley LC. Curr Biol. 1994;4:973–982. doi: 10.1016/s0960-9822(00)00221-9. [DOI] [PubMed] [Google Scholar]
- 36.Holmes JK, Solomon MJ. J Biol Chem. 1996;271:25240–25246. doi: 10.1074/jbc.271.41.25240. [DOI] [PubMed] [Google Scholar]
- 37.Hartley RS, Rempel RE, Maller JL. Dev Biol. 1996;173:408–419. doi: 10.1006/dbio.1996.0036. [DOI] [PubMed] [Google Scholar]
- 38.McGarry TJ, Kirschner MW. Cell. 1998;93:1043–1053. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- 39.Wohlschlegel JA, Dwyer BT, Dhar SK, Cvetic C, Walter JC, Dutta A. Science. 2000;290:2309–2312. doi: 10.1126/science.290.5500.2309. [DOI] [PubMed] [Google Scholar]
- 40.Lutzmann M, Maiorano D, Mechali M. EMBO J. 2006;25:5764–5774. doi: 10.1038/sj.emboj.7601436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumagai A, Guo Z, Emami KH, Wang SX, Dunphy WG. J Cell Biol. 1998;142:1559–1569. doi: 10.1083/jcb.142.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Michael WM, Ott R, Fanning E, Newport J. Science. 2000;289:2133–2137. doi: 10.1126/science.289.5487.2133. [DOI] [PubMed] [Google Scholar]
- 43.Guo Z, Kumagai A, Wang SX, Dunphy WG. Genes Dev. 2000;14:2745–2756. doi: 10.1101/gad.842500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hekmat-Nejad M, You Z, Yee MC, Newport JW, Cimprich KA. Curr Biol. 2000;10:1565–1573. doi: 10.1016/s0960-9822(00)00855-1. [DOI] [PubMed] [Google Scholar]
- 45.Kumagai A, Dunphy WG. Mol Cell. 2000;6:839–849. doi: 10.1016/s1097-2765(05)00092-4. [DOI] [PubMed] [Google Scholar]
- 46.Chini CC, Chen J. J Biol Chem. 2003;278:30057–30062. doi: 10.1074/jbc.M301136200. [DOI] [PubMed] [Google Scholar]
- 47.Lin SY, Li K, Stewart GS, Elledge SJ. Proc Natl Acad Sci USA. 2004;101:6484–6489. doi: 10.1073/pnas.0401847101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Osborn AJ, Elledge SJ. Genes Dev. 2003;17:1755–1767. doi: 10.1101/gad.1098303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sar F, Lindsey-Boltz LA, Subramanian D, Croteau DL, Hutsell SQ, Griffith JD, Sancar A. J Biol Chem. 2004;279:39289–39295. doi: 10.1074/jbc.M405793200. [DOI] [PubMed] [Google Scholar]
- 50.Trenz K, Smith E, Smith S, Costanzo V. EMBO J. 2006;25:1764–1774. doi: 10.1038/sj.emboj.7601045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tercero JA, Longhese MP, Diffley JF. Mol Cell. 2003;11:1323–1336. doi: 10.1016/s1097-2765(03)00169-2. [DOI] [PubMed] [Google Scholar]
- 52.Harrison JC, Haber JE. Annu Rev Genet. 2006;40:209–235. doi: 10.1146/annurev.genet.40.051206.105231. [DOI] [PubMed] [Google Scholar]
- 53.Zou L, Elledge SJ. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 54.Kumagai A, Dunphy WG. Nat Cell Biol. 2003;5:161–165. doi: 10.1038/ncb921. [DOI] [PubMed] [Google Scholar]
- 55.Kumagai A, Kim SM, Dunphy WG. J Biol Chem. 2004;279:49599–49608. doi: 10.1074/jbc.M408353200. [DOI] [PubMed] [Google Scholar]
- 56.Lee J, Gold DA, Shevchenko A, Shevchenko A, Dunphy WG. Mol Biol Cell. 2005;16:5269–5282. doi: 10.1091/mbc.E05-07-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luciani MG, Oehlmann M, Blow JJ. J Cell Sci. 2004;117:6019–6030. doi: 10.1242/jcs.01400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gotter AL, Manganaro T, Weaver DR, Kolakowski LF, Jr, Possidente B, Sriram S, MacLaughlin DT, Reppert SM. Nat Neurosci. 2000;3:755–756. doi: 10.1038/77653. [DOI] [PubMed] [Google Scholar]
- 60.Chan RC, Chan A, Jeon M, Wu TF, Pasqualone D, Rougvie AE, Meyer BJ. Nature. 2003;423:1002–1009. doi: 10.1038/nature01697. [DOI] [PubMed] [Google Scholar]
- 61.Tugal T, Zou-Yang XH, Gavin K, Pappin D, Canas B, Kobayashi R, Hunt T, Stillman B. J Biol Chem. 1998;273:32421–32429. doi: 10.1074/jbc.273.49.32421. [DOI] [PubMed] [Google Scholar]
- 62.Kubota Y, Takisawa H. J Cell Biol. 1993;123:1321–1331. doi: 10.1083/jcb.123.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hashimoto Y, Takisawa H. EMBO J. 2003;22:2526–2535. doi: 10.1093/emboj/cdg238. [DOI] [PMC free article] [PubMed] [Google Scholar]