Summary
Initiation of infection by herpes simplex virus (HSV-1) involves a step in which the parental virus capsid docks at a nuclear pore and injects its DNA into the nucleus. Once “uncoated” in this way, the virus DNA can be transcribed and replicated. In an effort to clarify the mechanism of DNA injection, we examined DNA release as it occurs in purified capsids incubated in vitro. DNA ejection was observed following two different treatments, trypsin digestion of capsids in solution, and heating of capsids after attachment to a solid surface. In both cases, electron microscopic analysis revealed that DNA was ejected as a single double helix with ejection occurring at one vertex presumed to be the portal. In the case of trypsin-treated capsids, DNA release was found to correlate with cleavage of a small proportion of the portal protein, UL6, suggesting UL6 cleavage may be involved in making the capsid permissive for DNA ejection. In capsids bound to a solid surface, DNA ejection was observed only when capsids were warmed above 4°C. The proportion of capsids releasing their DNA increased as a function of incubation temperature with nearly all capsids ejecting their DNA when incubation was at 37°C. The results demonstrate heterogeneity among HSV-1 capsids with respect to their sensitivity to heat-induced DNA ejection. Such heterogeneity may indicate a similar heterogeneity in the ease with which capsids are able to deliver DNA to the infected cell nucleus.
Keywords: herpes simplex virus, DNA uncoating, portal complex, virus capsid, DNA ends
Introduction
Replication of herpes simplex virus (HSV-1) involves a step in which the virus DNA is uncoated and delivered into the infected cell nucleus. Uncoating occurs when a parental capsid binds to a nuclear pore and injects its DNA through the pore into the nucleoplasm. Once inside the nucleus, the DNA is transcribed and replicated to propagate the infection. In contrast to the DNA, the parental capsid shell does not enter the nucleus or participate further in the infectious process.1–5
Previous studies of HSV-1 DNA uncoating have led to the identification of both cellular and viral proteins involved in the process. For example, analysis of capsid binding to purified nuclei in vitro demonstrated a requirement for both importin β and Ran-GTP.1 This observation suggests that attachment of the HSV-1 capsid to a nuclear pore may involve components of the importin β-dependent nuclear import pathway. A role for the virus-encoded UL36 protein is suggested by studies with a temperature-sensitive mutant, tsB7, in its gene. When cells are infected with tsB7 at the non-permissive temperature, capsids bind normally to nuclear pores, but no DNA release takes place.4 Release is observed, however, when cells are shifted to the permissive temperature. A similar mutant in pseudorabies virus has been described.6
HSV-1 DNA uncoating is thought to resemble uncoating as it occurs in infections by dsDNA bacteriophage such as T4 and λ.7–12 After the phage tail recognizes a specific receptor on the host cell surface, DNA exits the capsid as a single double helix that is extruded beginning at one genome end. As DNA is released, it traverses the channel of the portal, a structure found at a unique capsid vertex and specialized for function in DNA entry and exit. DNA uncoating in HSV-1 is considered to resemble that in dsDNA bacteriophage because of similarities in the portal structure and in the arrangement of DNA inside the capsid. For instance, in both HSV-1 and dsDNA bacteriophage the portal is located at a single capsid vertex and is cylindrical in shape with twelve–fold rotational symmetry.13–19 In both, the packaged DNA is arranged in close-packed strands, and DNA molecules contain no bound protein.20–22
We have been testing the idea that release of DNA from the HSV-1 capsid in vitro may serve as a useful model system to clarify the way egress occurs in infected cells. Experiments are carried out with DNA-containing capsids (C capsids) that are isolated from the nuclei of HSV-1-infected cells. C capsids are closely similar in structure to capsids found in infectious HSV-1 virions, and in infected cells they have the potential to exit the nucleus and mature into live virus. In contrast, two other capsid types, A capsids and B capsids, also accumulate in infected cell nuclei, but do not mature into virions. A capsids and B capsids have the same shell structure as C capsids, but they differ in the contents of the capsid cavity. Whereas C capsids contain the virus DNA, B capsids contain only the scaffolding protein while the A capsid cavity lacks substantial amounts of either DNA or protein. 39,40
In the experiments described here, purified C capsids were subjected to test treatments in vitro and examined for DNA release through a single capsid vertex. The results show that phage-like DNA release can be induced by: (i) treatment of capsids with trypsin; and (ii) attachment of C capsids to a solid surface by followed by warming.
Results
Trypsin treatment of C capsids
Purified C capsids were treated with trypsin in solution, and then examined for DNA release by sucrose density gradient centrifugation. On such gradients C capsids migrate more rapidly than capsids lacking DNA (A and B capsids). Examination of the untreated capsid population indicated it consisted predominately of C capsids (~85%) with small amounts of A and B capsids also present (Fig. 1, gradient 1). In contrast, C capsids were not detected when capsids were treated with 20 μg/ml trypsin (Fig. 1, gradient 4). Loss of C capsids was accompanied by a corresponding increase in a band migrating coincidently with A capsids (Fig. 1), which we interpret as resulting from loss of DNA from C capsids. Disappearance of the C capsid band indicates that DNA was lost from all C capsids treated with 20 μg/ml trypsin (Fig. 1, gradient 4). Intermediate levels of C capsid loss were observed after treatment of capsids with 0.2 μg/ml and 2 μg/ml trypsin (Fig. 1, gradients 2 and 3). Electron microscopic examination of trypsin-treated C capsids confirmed that DNA had been lost and that capsids had not suffered gross morphological damage at any of the trypsin doses tested (micrographs not shown). Control experiments demonstrated that DNA loss from C capsids was promoted by treatment with chymotrypsin at concentrations of 2 μg/ml and 20 μg/ml (data not shown).
Figure 1.
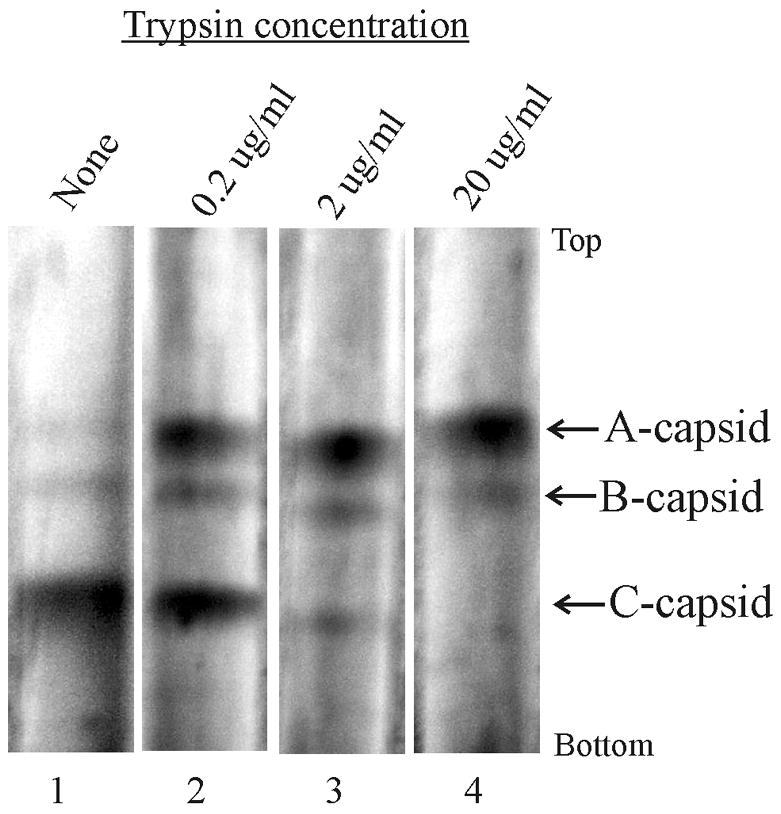
Sucrose density gradient analysis of control HSV-1 C capsids (gradient 1) and C capsids after treatment with the indicated dose of trypsin (gradients 2–4). Sedimentation is from top to bottom and the positions of A, B and C capsids are indicated. Note that with increasing trypsin dose the amount of C capsids declines while that of A capsids increases as expected as DNA is lost from C capsids.
Capsids treated with 0.2 μg/ml trypsin were examined further by: (a) electron microscopic analysis of shadowed preparations; and (b) SDS-polyacrylamide gel electrophoresis and immunoblotting to identify protein cleavages that may correlate with DNA release.
Electron microscopic analysis was performed with capsids that were treated with trypsin, adsorbed to electron microscope grids and rotary shadowed. A dose of 0.2 μg/ml trypsin was chosen with the goal of maximizing the possibility that DNA would be observed in the process of exiting the capsid. In practice, ~3% of capsids were seen to be actively releasing thir DNA (Fig. 2) although many more eventually did so (see Fig. 1, gradient 2). Capsids in which extruded DNA was not observed were assumed to be those that: (i) had not yet released DNA; (ii) had already released all their DNA; or (iii) were enmeshed in DNA tangles in which the capsid source could not be definitively assigned (micrographs not shown).
Figure 2.
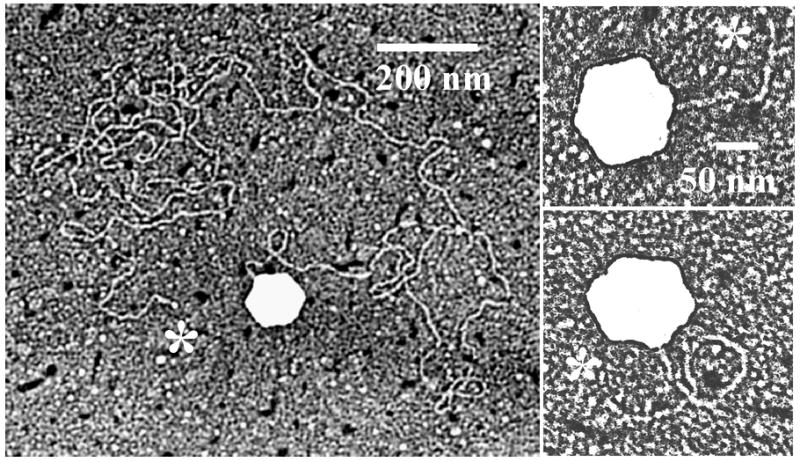
Electron micrographs showing HSV-1 C capsids in the process of releasing their DNA. Micrographs show DNA release following trypsin treatment of capsids in solution. DNA ends are indicated with asterisks. Note that in each case the DNA originates at a capsid vertex.
More than 100 images of capsids in the process of ejecting their DNA were examined. In all cases, DNA was found to be extruded only as a single thread originating at a single capsid vertex. Extrusion at multiple sites was not observed nor was unambiguous evidence for extrusion of DNA loops or release at a non-vertex site. The amount of extruded DNA varied from a few capsid diameters to much longer segments. In some images the distal DNA end was seen suggesting DNA was extruded beginning at an end (see sites labeled with an asterisk in Fig. 2).
SDS-polyacrylamide gel analysis was carried out with trypsin-treated capsids after separation on a sucrose gradient. Results shown in Fig. 1 (lane 2) demonstrated that a trypsin concentration of 0.2 μg/ml caused approximately half of the C capsids to lose their DNA. This can be seen as a reduction in the C capsid band and a corresponding increase in A capsids. The different capsid species from this gradient were isolated, subjected to immunoblot analysis and compared to determine if there was a unique peptide cleavage that correlated with DNA release.
The results showed no evidence for cleavage of the major capsid (UL19) or the large triplex protein (UL38) in A, B or C capsids (Fig. 3a, 3b). In contrast, little intact UL25 was observed after trypsin treatment in any of the three capsid types (Fig. 3b). A doublet of digestion products replaced the intact protein in all cases. A small amount of UL6 cleavage was observed in B and C capsids, and a larger amount (~10%) in A capsids. In control capsids not exposed to trypsin, UL25 cleavage was minimal and cleavage of UL6 was not observed (Fig 3b).
Figure 3.
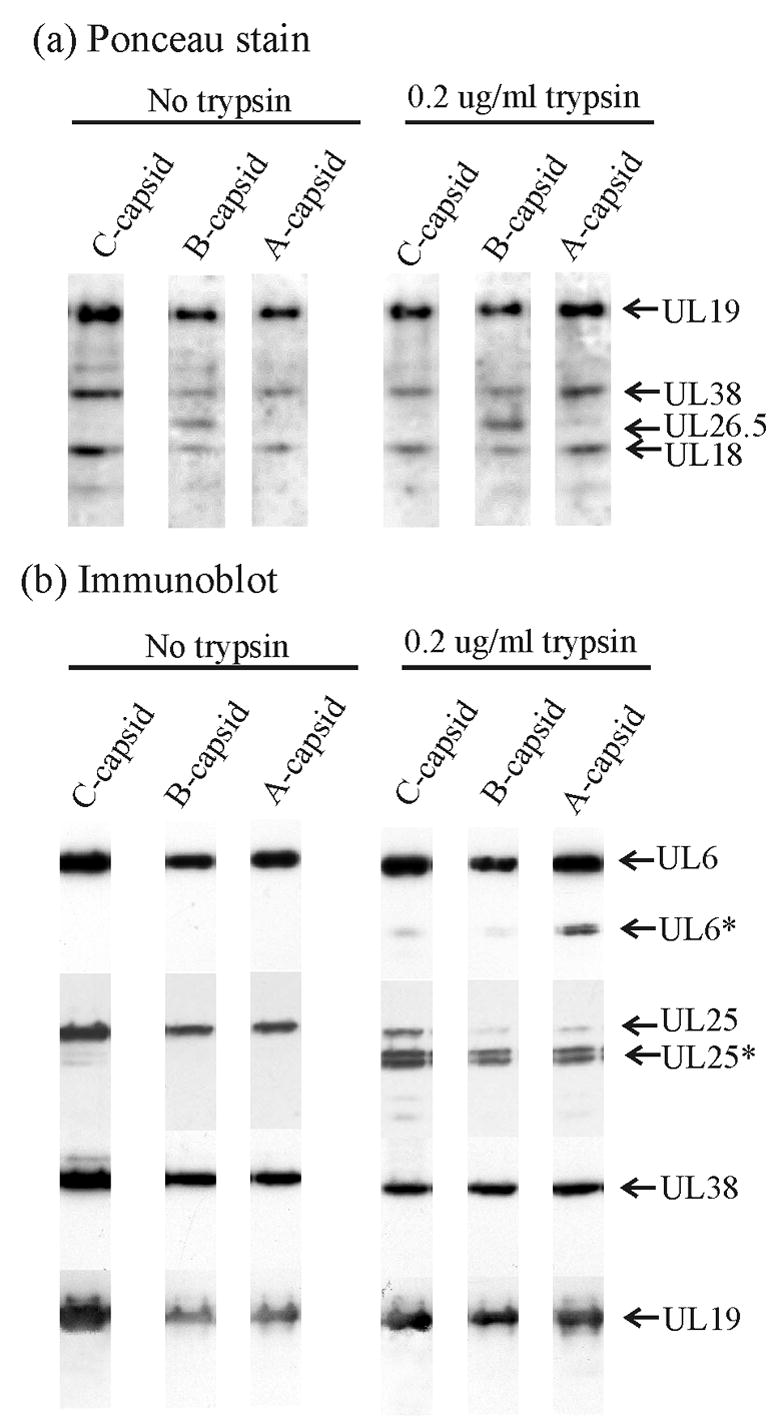
SDS-polyacrylamide gel and immunoblot analyses of control HSV-1 capsids (left three lanes) and HSV-1 capsids after treatment with 0.2 μg/ml trypsin (right three lanes). Procedures described in Materials and Methods were used to treat capsids in solution with trypsin and to separate capsids by sucrose density gradient centrifugation. Capsid-containing fractions were then examined by SDS-polyacrylamide gel electrophoresis (a) and immunoblot analysis for UL6, UL25, UL38 and UL19 (b). Bands corresponding to individual proteins are identified on the right where an asterisk indicates the cleaved form of a protein. Note that cleavage of UL25 is observed in all capsid types while appreciable cleavage of UL6 is found only in A capsids.
Capsids attached to a solid surface
DNA release from C capsids was also examined with capsids after they were adsorbed to the surface of an electron microscope grid. Preliminary studies with grid-associated capsids demonstrated that DNA release did not depend on treatment of capsids with trypsin as it did with capsids in solution. DNA release was observed after grid-associated capsids were simply warmed above 4°C. In a representative experiment, capsids were bound to an electron microscope grid, warmed to 37°C in the absence of trypsin, stained to allow DNA-containing capsids to be distinguished from empty ones, and examined by electron microscopy (Fig. 4).
Figure 4.
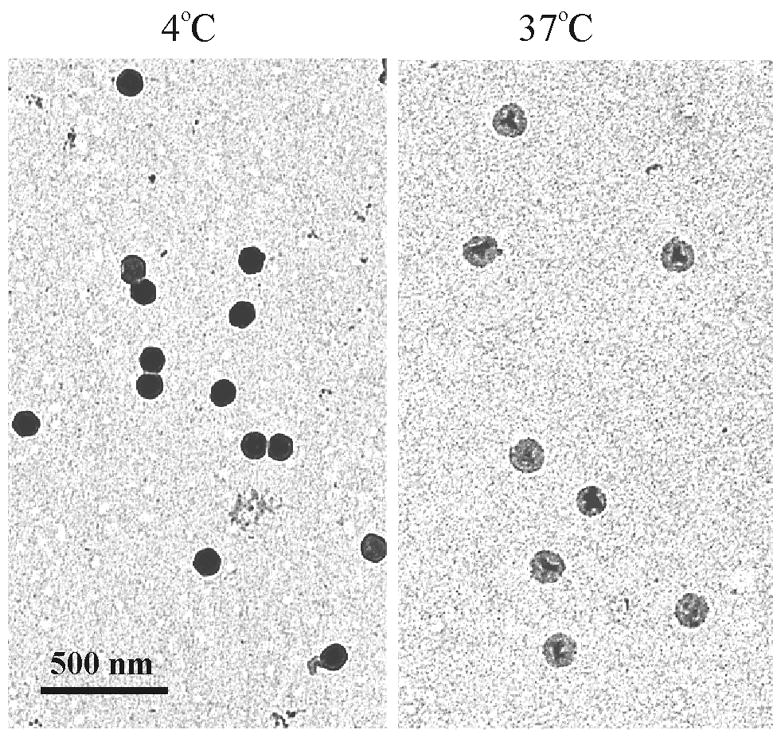
Electron micrographs of HSV-1 C capsids bound to Formvar/carbon-coated electron microscope grids. Capsids were maintained at 4°C (left) or warmed to 37°C (right) for 2 hrs and stained with uranyl acetate. Note that DNA loss, as evidenced by a lighter image, is observed only in capsids heated at 37°C.
Results with this system showed that when grid-associated capsids were incubated for 2 hrs at 37°C, 94% lost their DNA compared to 15% when incubation was at 4°C (Table 1). Observations with C capsids contrasted markedly with similar studies carried out with mature phage λ. Warming of grid associated λ had little effect on the initial proportion of phage with DNA-filled heads (Table 1). We interpret this finding to indicate that the effect of heat to cause DNA release from C capsids was not due to a severe effect that would cause release of any packaged DNA. When C capsids were incubated at 34°C, nearly 80% released their DNA over a period of approximately 130 min as shown in Fig. 5a. DNA was lost from half the capsids in approximately 30 min. Only a background level of DNA ejection (10%–15%) was observed when incubation was at 4°C.
Table 1.
Effect of incubation at 37°C on DNA uncoating in HSV-1 C-capsids and bacteriophage lambdaa
| Specimen | Incubation Temperature | Treatment | Full (%) | Empty (%) |
|---|---|---|---|---|
| HSV-1 C-capsids | 4°C | None | 252 (85%) | 44 (15%) |
| HSV-1 C-capsids | 37°C | None | 26 (6%) | 407 (94%) |
| HSV-1 C-capsids | 37°C | 1% formaldehyde | 330 (97%) | 10 (3%) |
| HSV-1 C-capsids | 37°C | Protease inhibitorsb | 176 (29%) | 431 (71%) |
| HSV-1 C-capsids | 37°C | 10 mM DFPc | 39 (14%) | 240 (86%) |
| Bacteriophage λ | 4°C | None | 172 (86%) | 28 (14%) |
| Bacteriophage λ | 37°C | None | 369 (88%) | 50 (12%) |
HSV-1 C capsids were adsorbed to electron microscope grids, heated in TNE for 2 hrs at 37°C, and stained as described in Materials and Methods. The same procedure was used for bacteriophage λ. Capsids or λ virions where then examined in the electron microscope and scored visually for the presence of DNA inside the capsid.
Roche Diagnostics Complete diluted as described in Materials and Methods.
Diisopropyl-fluorophosphate
Figure 5.
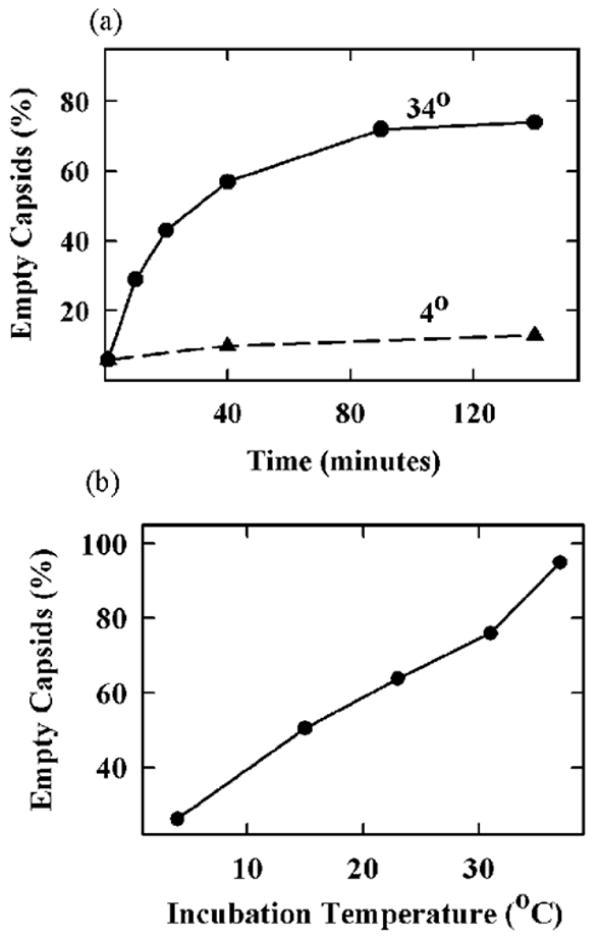
Effect of time and incubation temperature on extrusion of DNA from grid-associated HSV-1 C capsids. (a) Capsids were adsorbed to grids, incubated under the indicated conditions, stained and examined in the electron microscope to determine whether DNA was released. Note that most capsids had released their DNA after 120 minutes of incubation at 34°C. (b) Capsids were absorbed to grids and incubated for 2 hrs at the temperatures indicated. They were then stained and examined for DNA release. Note that the proportion of capsids that released DNA increased with increasing incubation temperature.
Incubation temperature was found to affect the proportion of C capsids that released their DNA. For instance, when capsids were incubated for 2 hrs, 95% of capsids were found to release their DNA when incubation was at 37°C. Lower proportions were observed when the incubation temperature was lower as shown in Fig. 5b. DNA release was almost completely suppressed when grid-associated capsids were treated with 1% formaldehyde prior to incubation at elevated temperature (Table 1). This effect is suggested to result from cross-linking of virus DNA to the capsid shell preventing its ejection.23 Similar suppression of DNA release by 1% formaldehyde was observed with trypsin-treated capsids in solution (data not shown).
In view of the role of trypsin in promoting DNA release from C capsids in solution, we tested the idea that action of an endogenous protease may contribute to DNA release from grid-associated capsids. Cellular protease(s) and the HSV-1-encoded proteases (UL26 and UL36)24, 25 were considered as possible sources of activity. Tests involved heating grid-associated C capsids in the presence of protease inhibitors, which are expected to suppress DNA ejection if protease activity is involved. Tests were performed with a cocktail of protease inhibitors (Roche Complete) and with diisopropyl-fluorophosphate (DFP), an inhibitor of the HSV-1-encoded protease.26 The results demonstrated only a modest effect of either inhibitor (Table 1). For example, whereas DNA release from 94% of capsids was observed in uninhibited reactions, release was 71% in the case of the inhibitor cocktail and 86% in the case of 10 mM DFP. The modest effect of protease inhibitors suggests either that: (i) inhibitors did not have access to the protease as would be the case, for instance, if they did not enter the capsid; or (ii) DNA release is promoted to a greater extent by capsid binding to the carbon-Formvar surface than by the effect of proteases that may be present.
Electron microscopy
Release of DNA from grid-associated capsids was examined by electron microscopy in two ways: (i) by shadowing to identify the site from which DNA is released; and (ii) by examination of frozen hydrated specimens to determine the proportion of DNA released from individual capsids. Both studies were carried out with C capsids that were adsorbed to carbon-Formvar coated grids and warmed for 30 min at 37°C to initiate DNA extrusion.
Of more than 150 capsids examined by shadowing, DNA was found to arise from 30%–35%. As in the case of trypsin-treated capsids, a single DNA thread appeared to extend from a capsid vertex in every case (Fig. 6). DNA loops were not observed nor were capsids with multiple extruded DNA strands. The length of released DNA strands was found to vary from one or a few capsid diameters in some images to tens or perhaps 100 in others (Fig. 6). A DNA end was identified in many images as shown in Fig. 6 (asterisks) suggesting that DNA extrusion was initiated at an end.
Figure 6.
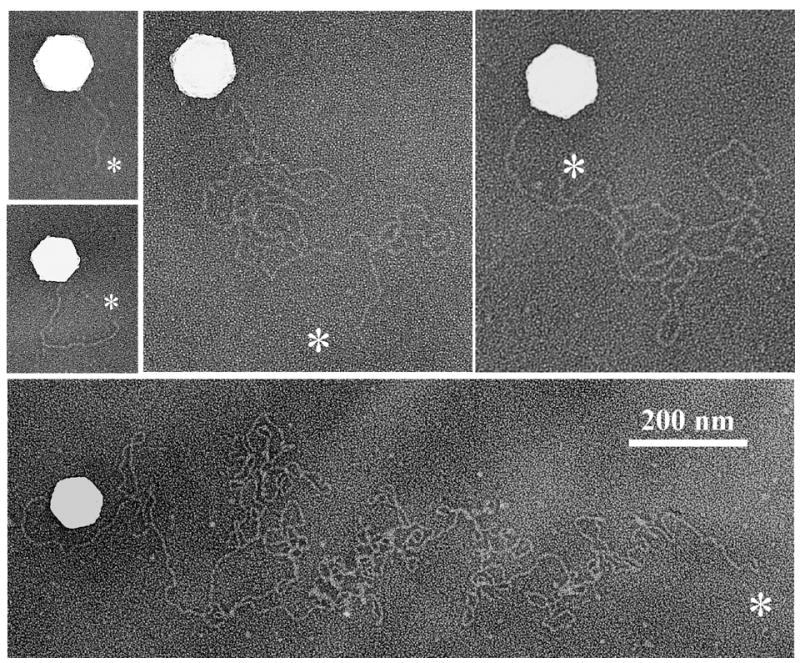
Electron micrographs showing grid-associated HSV-1 C capsids in the process of releasing their DNA. Capsids were warmed to 37°C for 30 min to promote DNA release, and prepared for electron microscopy by shadowing as described in Materials and Methods. White asterisks indicate DNA ends. Note that in each case the DNA originates at a capsid vertex.
Images of frozen-hydrated capsids were recorded with the idea that, since specimens are not stained, the total image density ought to correspond to the DNA remaining inside the capsid plus an invariant density arising from the capsid shell. By subtracting the contribution of the shell, it should be possible to estimate the proportion of DNA remaining in the capsid. Micrographs were therefore recorded of C capsids that had been warmed to 37°C for 30 min and then preserved in the frozen-hydrated state. The integrated density of individual capsid images was then determined by scanning and corrected for the contribution of the protein shell to yield a value proportional to the amount of DNA present.
Representative micrographs are shown in Fig. 7 at low and higher magnification (upper and lower panels, respectively). Compared to control, unheated capsids (4°C), most capsid images in heated specimens were found to be lighter in density as expected from previous results showing that DNA is released from heated capsids. Images showed no evidence of damage to the integrity of the capsid shell. Heated capsids were indistinguishable from controls except for differences due to DNA loss. Among the capsids in the heated specimens it was possible to distinguish: (i) filled capsids whose density is comparable to unheated ones; (ii) empty capsids with a density comparable to the lightest capsids in the field; and (iii) capsids with an intermediate density assumed to be partially filled capsids. Representative filled (F), empty (E) and intermediate (I) capsid images are indicated in Fig. 7 (bottom right).
Figure 7.
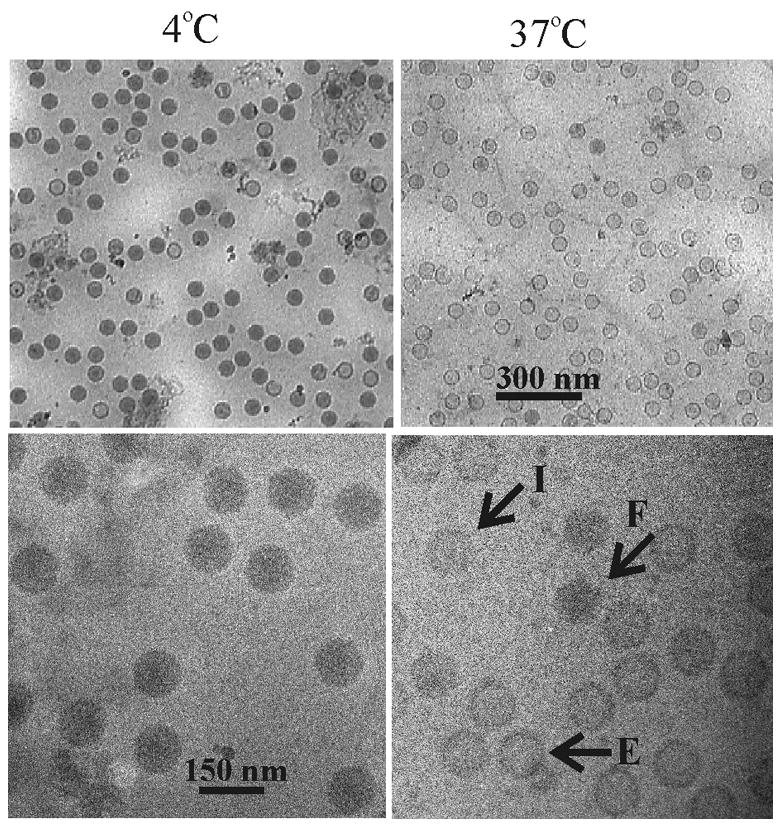
Electron micrographs showing grid-associated capsids preserved in the frozen-hydrated state. C capsids were bound to grids, incubated for 30 min at 4°C (left) or 37°C (right) and examined after freezing at liquid nitrogen temperature (−170°C). Upper and lower panels show capsids at lower and higher magnification, respectively. Note that DNA loss, as evidenced by a lighter image, is observed only in capsids heated at 37°C and that DNA loss is not accompanied by morphological damage to the capsid shell. Indicated in the lower right panel are capsids filled with DNA (F), empty capsids (E) and capsids with an intermediate amount of DNA (I).
The fraction of DNA remaining in 97 randomly selected capsids was determined and the results are shown in Fig. 8. Most capsids (88 of 97) were found to have lost 30% or more of their DNA after 30 min of incubation. Among these capsids, a wide variation of DNA loss was observed. In some, DNA loss was complete or nearly so while in others 60% or more of the DNA was retained. The variability in the amount of DNA remaining is suggested to indicate that there is a difference among capsids in the time at which DNA ejection begins or perhaps in the rate at which DNA is released. The existence of a capsid population from which DNA loss is minimal (30% or less in 9 of 97 capsids; Fig. 8) indicates there may be a small population of capsids in which DNA is resistant to extrusion at 37°C.
Figure 8.
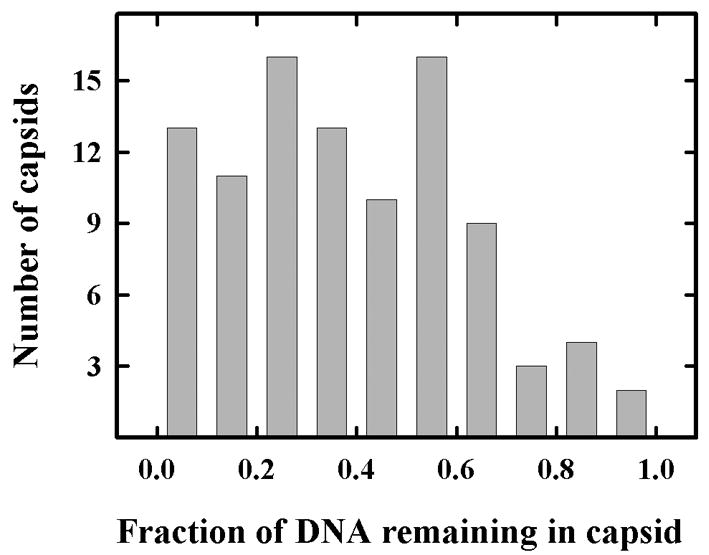
Histogram showing the fraction of DNA remaining in C capsids after incubation at 37°C for 30 min. Capsids were adsorbed to grids, incubated and images were recoded by electron microscopy of frozen-hydrated specimens. The integrated density of individual capsid images (after correction for the contribution of the capsid shell) was then determined as a measure of the DNA remaining inside the capsid. A total of 97 images were measured and the values are shown. Note that most capsids (88 of 97) have lost 30% or more of their DNA.
Discussion
DNA uncoating in vitro
Electron microscopic analysis of capsids extruding their DNA supported the view that release in vitro resembles DNA uncoating as it is thought to occur in infected cells. Images of both trypsin treated and grid-associated capsids showed that DNA was extruded as a single double helix with ejection occurring at a single capsid vertex assumed to be the portal. Tests of the suggested portal involvement are now in progress. No evidence was obtained for release of DNA at multiple vertices or as loops or multi-strand bundles as observed in C capsids treated in vitro with guanidine-HCl.27 Similarly, we did not observe release of DNA as a condensed mass following large-scale damage to the capsid shell as observed by Shahin and colleagues.28 In many images, a single DNA double helix could be traced from its origin at the capsid to a distal end suggesting that DNA extrusion begins at a genome end as expected of DNA uncoating in vivo (Figs. 2 and 6).
In studies of trypsin-treated capsids, Western immunoblotting was employed to test for capsid protein cleavages that might correlate with DNA release. Four capsid proteins were examined, the major capsid protein (UL19), the large triplex protein (UL38), UL25, and the portal protein (UL6). The results showed that of the four only UL6 and UL25 were cleaved and only UL6 cleavage correlated with DNA loss from the capsid (Fig. 3). Since DNA is thought to exit the capsid by way of the portal, it is easy to imagine that digestion of the portal protein might promote DNA release. The small amount of UL6 affected (~10%), however, indicates that DNA release must be able to be initiated by cleavage of no more than one UL6 molecule per capsid. Proteases potentially involved in UL6 cleavage include VP24 (UL26 gene product) and UL36. VP24 is located inside the capsid where it functions to cleave the scaffolding protein.24, 39 UL36 is found outside the capsid shell. As described above, it is required for DNA uncoating in infected cells making it attractive as a candidate for a function in initiation of DNA release.25,4
Grid-associated capsids
In order to examine DNA release from single capsids rather than from capsid populations, experiments were carried out with C capsids bound to a solid surface, a coated electron microscope grid. Experiments with such preparations showed that DNA was extruded when capsids were heated above 4°C. A study of the effect of temperature on the proportion of capsids releasing their DNA demonstrated that DNA was extruded from nearly all capsids incubated for 2 hrs at 37°C (see Fig. 5b). When incubation was at a lower temperature, however, a lower fraction of capsids lost their DNA. We interpret the effects of temperature to indicate that the population of C capsids is heterogeneous with respect to the activation energy required to induce DNA extrusion from grid-associated specimens. We believe our observations are the first to demonstrate a functional heterogeneity among HSV-1 C capsids. This heterogeneity may reflect a related difference in the ease with which capsids are able to deliver their DNA to the infected cell nucleus. In the future, it would be of interest to determine whether the observed functional heterogeneity is correlated with a feature of the capsid structure.
Heterogeneity among C capsids was also observed when cryo-electron microscopy was employed to measure the amount of DNA remaining in capsids incubated for 30 min at 37°C (see Figs. 7–8). Although nearly all capsids had lost some DNA after 30 min of incubation, capsids exhibited a wide variation in the amount of DNA remaining encapsidated (Fig. 8). As described above, this variation may be due to heterogeneity in the time at which DNA release is initiated or in the rate of DNA egress.
In contrast to the situation with capsids in solution, DNA release from grid-associated capsids did not depend on proteolytic digestion of the capsid. Added protease was not required, and ejection was not strongly suppressed by the presence of protease inhibitors (Table 1). We interpret these observations to indicate that attachment to the carbon surface of the grid makes the capsid permissive for DNA egress. For instance, a strong capsid-grid attachment could perturb or compress the capsid in such a way that DNA release would be favored. Alternatively, attachment could produce a perturbation that would be transmitted to the portal to potentiate portal opening and DNA release. Such compression of the capsid or subtle change to the portal could function like trypsin treatment to lower the energy necessary to initiate DNA extrusion.
Cryo-electron microscopic analysis of heated capsids supports the view that the DNA in grid-associated capsids is subject to a degree of internal pressure. DNA exits spontaneously from capsids after warming above 4°C. The amount of DNA released, however, suggests the level of internal pressure must be relatively modest. Even after 30 min of incubation at 37°C, most capsids still contained a substantial amount of DNA (Fig. 8).
DNA uncoating in vivo
It is difficult to define the role internal capsid pressure may play in HSV-1 DNA uncoating in vivo. In view of the relatively modest amount of internal pressure observed here (i.e. enough to cause ejection of ≥30% of the DNA from most capsids in 30 min; see Fig. 8), it would be unreasonable to assume that pressure is a major force propelling the majority of DNA into the nucleus. Some other factor is likely to be involved. Candidates would include diffusion or possibly a property of the nuclear pore complex (NPC). A role for the NPC is suggested by the results of Salman et al.29 who observed in vitro uptake of free DNA into reconstituted nuclei. Such observations suggest a minimal role for the capsid in HSV-1 DNA transport into the nucleus. The role of the capsid may be to deliver the virus DNA to the vicinity of the NPC and release it. Pore proteins would then complete the transport process. Uncoating would therefore resemble the “push-pull” mechanism suggested for ejection of phage ϕ29 DNA.37, 38 The role of the capsid would be to maintain the DNA in a condensed state before it reaches the NPC. The capsid would thereby minimize the resistance to diffusion in the cytoplasm that would apply to an extended DNA molecule.
In vitro uncoating of the HSV-1 genome as described here has important features in common with DNA uncoating in dsDNA bacteriophage. In both, DNA is extruded as a single double helix through the portal vertex, with release initiated at a genome end.12 The major difference has to do with participation of a receptor. Extrusion of DNA from phage such as λ, T5 and SPP1 occurs only after the phage tail has interacted with a specific receptor on the host cell surface.11, 30, 31 The requirement for a receptor is observed for DNA extrusion both in vitro and in vivo. In contrast, no requirement for a receptor was observed in the studies described here. Two possible explanations suggest themselves: (i) A receptor may be involved in HSV-1 DNA uncoating in infected cells, but in vitro the role of the receptor may be substituted by trypsin treatment or capsid binding to a solid surface as described here. (ii) Alternatively, DNA release in vivo may involve a non-specific feature of the capsid-nuclear pore complex environment (e.g. phenylalanine-glycine-containing nucleoporins36) and not a specific receptor. Studies of uncoating in infected cells will be required to resolve this and other issues regarding how DNA is delivered to the host cell nucleus.
Materials and Methods
Cells, viruses and capsids
Experiments were performed with C capsids prepared from the nuclei of BHK-21 cells infected for 20 hrs with the 17MP strain of HSV-1. A recent publication describes the methods used for cell growth, virus infection and capsid purification by sucrose density gradient sedimentation of nuclear lysates.32 Preparations yielded 100 μg–200 μg of C capsids in TNE buffer (0.01 M Tris-HCl, 0.5 M NaCl, 1 mM EDTA, pH 7.5). Contamination with A and B capsids was approximately 10%. For most studies, capsids were adjusted to a concentration of 0.25 mg/ml and stored at 4°C. Purified λ phage (1 × 1010 pfu/ml) was the gift of Dr. Dwight Anderson (University of Minnesota).
Trypsin treatment of C capsids
C capsids (50 μl) were added to 5 μl TNE containing trypsin adjusted to yield a final concentration of 20 μg/ml, 2 μg/ml or 0.2 μg/ml, and the mixtures were incubated for 45 min at 37°C. Capsids were then isolated by centrifugation on a 600-μl gradient of 20%–50% sucrose prepared in TNE containing protease inhibitors (0.1 volume of a solution prepared by dissolving one tablet of Roche Diagnostics Complete in 5 ml phosphate-buffered saline [PBS]).
Centrifugation was for 40 min at 23,000 rpm in a Beckman SW55Ti rotor operated at 4°C. DNA release was not observed if capsids contained protease inhibitors prior to trypsin treatment. Previously described procedures were employed for SDS-polyacrylamide gel electrophoresis, dot blotting to identify capsid-containing fractions, immunoblotting and staining with specific antibodies.32,33 Antibodies used for immunoblotting were: UL25, monoclonal antibody (MAb) 2D9 (1:2000 dilution34; UL6, MAb 1C9 (1:10,000 dilution14); UL38, MAb 4A11 (culture supernatant diluted 1:5,00032); UL19, MAb 3E8 (culture supernatant; diluted 1:50035). Reprobing of blots was performed as described previously.32
DNA extrusion from trypsin-treated C capsids was examined by electron microscopy beginning with 20 μl aliquots of capsids, which were adjusted to 0.2 μg/ml trypsin and incubated for 10 min at 37°C. A 1 μl aliquot of capsids was then mixed with 50 μl 1% formaldehyde at room temperature for 10 seconds, and a sample was loaded onto a carbon-Formvar-coated electron microscope grid. The grid was washed in a drop of TNE (four times) to remove the formaldehyde, stained, rotary shadowed and examined by electron microscopy as described below.
DNA ejection from grid-associated C capsids
An 8 μl aliquot of C capsids (5 μg/ml) was pipetted onto a glow discharged, Formvar-carbon coated electron microscope grid and allowed to attach for 10 seconds at room temperature. Unbound capsids were then removed by floating the grid for one minute face down on a drop (100 μl) of TNE. For warming, as necessary, grids were placed in a 0.5 ml microcentrifuge tube containing 0.4 ml TNE. The tube was laid on its side with the grid face up and incubated. After incubation, the grid was removed from the tube and submerged sequentially at room temperature in: (1) a drop of PBS containing 1% formaldehyde (10 seconds); (2) a drop of TNE containing a 1:500 dilution of Photo-flo (Kodak Photo-flo 200; twice; 30 seconds each); (3) 1% uranyl acetate in 1:500 Photo-flo; and (4) 1:500 Photo-flo (five times). The grid was then placed face up on filter paper and allowed to dry for 5 min prior to examination in the electron microscope or rotary shadowing followed by electron microscopy (see below). In particular experiments, incubation mixtures also contained protease inhibitors (see above), 10 mM diisopropyl-fluorophosphate (DFP), or 1% formaldehyde as described in the text.
Electron microscopy
All electron microscopy was performed on a Philips EM400T transmission instrument operated at 80 keV. Images were recorded on film and converted to digital form by scanning in a flatbed scanner. UN-SCAN-IT v5.1 (Silk Industries) was used to make quantitative measurements of capsid images. Rotary shadowing of specimens was carried out with Pt-carbon at a 15° angle in a Bal-Tec Med-020 instrument operated at 1.90keV and 70 mA. For examination of frozen-hydrated specimens, grids were quick frozen in liquid ethane and maintained at −170°C with liquid nitrogen in a Gatan model 626 cold holder. Images were recorded in low-dose mode on Kodak SO-163 film.
Acknowledgments
Special thanks to Fred Homa, Frank Booy, Anna Maria Copeland and Rebecca Mingo for their comments and criticisms during the course of this investigation. This work was supported by NIH award AI041644.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ojala PM, Sodeik B, Ebersold MW, Kutay U, Helenius A. Herpes simplex virus type 1 entry into host cells: reconstitution of capsid binding and uncoating at the nuclear pore complex in vitro. Mol Cell Biol. 2000;20:4922–4931. doi: 10.1128/mcb.20.13.4922-4931.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sodeik B, Ebersold MW, Helenius A. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J Cell Biol. 1997;136:1007–1021. doi: 10.1083/jcb.136.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roizman B, Knipe DM. Herpes simplex viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. Lippincott Williams and Wilkins; Philadelphia: 2001. pp. 2399–2459. [Google Scholar]
- 4.Batterson W, Furlong D, Roizman B. Molecular genetics of herpes simplex virus VIII. Further characterization of a temperature-sensitive mutant defective in release of viral DNA and in other stages of the viral reproductive cycle. J Virol. 1983;45:397–407. doi: 10.1128/jvi.45.1.397-407.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tognon M, Furlong D, Conley AJ, Roizman B. Molecular genetics of herpes simplex virus. V Characterization of a mutant defective in ability to form plaques at low temperatures and in a viral fraction which prevents accumulation of coreless capsids at nuclear pores late in infection. J Virol. 1981;40:870–880. doi: 10.1128/jvi.40.3.870-880.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feldman L, Blankenship ML, Ben Porat T. Isolation and characterization of a temperature-sensitive uncoating mutant of pseudorabies virus. J Gen Virol. 1981;54:333–342. doi: 10.1099/0022-1317-54-2-333. [DOI] [PubMed] [Google Scholar]
- 7.Grayson P, Evilevitch A, Inamdar MM, Purohit PK, Gelbart WM, Knobler CM, Phillips R. The effect of genome length on ejection forces in bacteriophage lambda. Virol. 2006;348:430–436. doi: 10.1016/j.virol.2006.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evilevitch A, Gober JW, Phillips M, Knobler CM, Gelbart WM. Measurements of DNA lengths remaining in a viral capsid after osmotically suppressed partial ejection. Biophys J. 2005;88:751–756. doi: 10.1529/biophysj.104.045088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evilevitch A, Lavelle L, Knobler CM, Raspaud E, Gelbart WM. Osmotic pressure inhibition of DNA ejection from phage. Proc Natl Acad Sci U S A. 2003;100:9292–9295. doi: 10.1073/pnas.1233721100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rossmann MG, Mesyanzhinov VV, Arisaka F, Leiman PG. The bacteriophage T4 DNA injection machine. Curr Opin Struct Biol. 2004;14:171–180. doi: 10.1016/j.sbi.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 11.Roessner CA, Struck DK, Ihler GM. Injection of DNA into liposomes by bacteriophage lambda. J Biol Chem. 1983;258:643–648. [PubMed] [Google Scholar]
- 12.Molineux IJ. Fifty-three years since Hershey and Chase; much ado about pressure but which pressure is it? Virol. 2006;344:221–229. doi: 10.1016/j.virol.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 13.Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige P, Poliakov A, Potter CS, Carragher B, Johnson JE. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006;312:1791–1795. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- 14.Newcomb WW, Juhas RM, Thomsen DR, Homa FL, Burch AD, Weller SK, Brown JC. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J Virol. 2001;75:10923–10932. doi: 10.1128/JVI.75.22.10923-10932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simpson AA, Tao Y, Leiman PG, Badasso MO, He Y, Jardine PJ, Olson NH, Morais MC, Grimes S, Anderson DL, Baker TS, Rossmann MG. Structure of the bacteriophage phi29 DNA packaging motor. Nature. 2000;408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orlova EV, Gowen B, Droge A, Stiege A, Weise F, Lurz R, van Heel M, Tavares P. Structure of a viral DNA gatekeeper at 10 A resolution by cryo-electron microscopy. EMBO J. 2003;22:1255–1262. doi: 10.1093/emboj/cdg123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trus BL, Cheng N, Newcomb WW, Homa FL, Brown JC, Steven AC. Structure and polymorphism of the UL6 portal protein of herpes simplex virus type 1. J Virol. 2004;2004;78:12668–12671. doi: 10.1128/JVI.78.22.12668-12671.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agirrezabala X, Martin-Benito J, Valle M, Gonzalez JM, Valencia A, Valpuesta JM, Carrascosa JL. Structure of the connector of bacteriophage T7 at 8A resolution: structural homologies of a basic component of a DNA translocating machinery. J Mol Biol. 2005;347:895–902. doi: 10.1016/j.jmb.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 19.Valpuesta JM, Carrascosa JL. Structure of viral connectors and their function in bacteriophage assembly and DNA packaging. Quart Rev Biophys. 1994;27:107–155. doi: 10.1017/s0033583500004510. [DOI] [PubMed] [Google Scholar]
- 20.Cerritelli ME, Cheng N, Rosenberg AH, McPherson CE, Booy FP, Steven AC. Encapsidated conformation of bacteriophage T7 DNA. Cell. 1997;91:271–280. doi: 10.1016/s0092-8674(00)80409-2. [DOI] [PubMed] [Google Scholar]
- 21.Booy FP, Newcomb WW, Trus BL, Brown JC, Baker TS, Steven AC. Liquid-crystalline, phage-like packing of encapsidated DNA in herpes simplex virus. Cell. 1991;64:1007–1015. doi: 10.1016/0092-8674(91)90324-r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lepault J, Dubochet J, Baschong W, Kellenberger E. Organization of double-stranded DNA in bacteriophages: a study by cryo- electron microscopy of vitrified samples. EMBO J. 1987;6:1507–1512. doi: 10.1002/j.1460-2075.1987.tb02393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Solomon MJ, Varshavsky A. Formaldehyde-mediated DNA-protein crosslinking: a probe for in vivo chromatin structures. Proc Natl Acad Sci U S A. 1985;82:6470–6474. doi: 10.1073/pnas.82.19.6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu FY, Roizman B. The herpes simplex virus 1 gene encoding a protease also contains within its coding domain the gene encoding the more abundant substrate. J Virol. 1991;65:5149–5156. doi: 10.1128/jvi.65.10.5149-5156.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kattenhorn LM, Korbel GA, Kessler BM, Spooner E, Ploegh HL. A Deubiquitinating Enzyme Encoded by HSV-1 Belongs to a Family of Cysteine Proteases that Is Conserved across the Family Herpesviridae. Mol Cell. 2005;19:547–557. doi: 10.1016/j.molcel.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 26.DiIanni CL, Stevens JT, Bolgar M, O’Boyle DR, Weinheimer SP, Colonno RJ. Identification of the serine residue at the active site of the herpes simplex virus type 1 protease. J Biol Chem. 1994;269:12672–12676. [PubMed] [Google Scholar]
- 27.Newcomb WW, Brown JC. Induced extrusion of DNA from the capsid of herpes simplex virus type 1. J Virol. 1994;68:433–440. doi: 10.1128/jvi.68.1.433-440.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shahin V, Hafezi W, Oberleithner H, Ludwig Y, Windoffer B, Schillers H, Kuhn JE. The genome of HSV-1 translocates through the nuclear pore as a condensed rod-like structure. J Cell Sci. 2006;119:23–30. doi: 10.1242/jcs.02705. [DOI] [PubMed] [Google Scholar]
- 29.Salman H, Zbaida D, Rabin Y, Chatenay D, Elbaum M. Kinetics and mechanism of DNA uptake into the cell nucleus. Proc Natl Acad Sci U S A. 2001;98:7247–7252. doi: 10.1073/pnas.121067698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Frutos M, Letellier L, Raspaud E. DNA ejection from bacteriophage T5: analysis of the kinetics and energetics. Biophys J. 2005;88:1364–1370. doi: 10.1529/biophysj.104.048785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sao-Jose C, Lhuillier S, Lurz R, Melki R, Lepault J, Santos MA, Tavares P. The ectodomain of the viral receptor YueB forms a fiber that triggers ejection of bacteriophage SPP1 DNA. J Biol Chem. 2006;281:11464–11470. doi: 10.1074/jbc.M513625200. [DOI] [PubMed] [Google Scholar]
- 32.Newcomb WW, Homa FL, Brown JC. Herpes simplex virus capsid structure: DNA packaging protein UL25 is located on the external surface of the capsid near the vertices. J Virol. 2006;80:6286–6294. doi: 10.1128/JVI.02648-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newcomb WW, Trus BL, Cheng N, Steven AC, Sheaffer AK, Tenney DJ, Weller SK, Brown JC. Isolation of herpes simplex virus procapsids from cells infected with a protease-deficient mutant virus. J Virol. 2000;74:1663–1673. doi: 10.1128/jvi.74.4.1663-1673.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McNab AR, Desai P, Person S, Roof LL, Thomsen DR, Newcomb WW, Brown JC, Homa FL. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J Virol. 1998;72:1060–1070. doi: 10.1128/jvi.72.2.1060-1070.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spencer JV, Newcomb WW, Thomsen DR, Homa FL, Brown JC. Assembly of the herpes simplex virus capsid: pre-formed triplexes bind to the nascent capsid. J Virol. 1998;72:3944–3951. doi: 10.1128/jvi.72.5.3944-3951.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim RYH, Huang NP, Köser J, Deng J, Lau KH, Aaron Schwarz-Herion K, Fahrenkrog B, Aebi U. Flexible phenylalanine-glycine nucleoporins as entropic barriers to nucleocytoplasmic transport. Proc Nat Acad Sci USA. 2006;103:9512–9517. doi: 10.1073/pnas.0603521103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gonzalez-Huici V, Salas M, Hermoso JM. The push-pull mechanism of bacteriophage ϕ29 DNA injection. Molec Microbiol. 2004;52:529–540. doi: 10.1111/j.1365-2958.2004.03993.x. [DOI] [PubMed] [Google Scholar]
- 38.Gonzalez-Huici V, Salas M, Hermoso JM. Requirements for Bacillus Subtilis bacteriophage ϕ29 DNA ejection. Gene. 2006;374:19–25. doi: 10.1016/j.gene.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 39.Homa FL, Brown JC. Capsid assembly and DNA packaging in herpes simplex virus. Rev Med Virol. 1997;7:107–122. doi: 10.1002/(sici)1099-1654(199707)7:2<107::aid-rmv191>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 40.Rixon FJ. Structure and assembly of herpesviruses. Semin Virol. 1993;4:135–144. [Google Scholar]


