Introduction
Chromatin and gene regulatory molecules tend to operate in multisubunit complexes in the process of controlling gene expression. Accumulating evidence suggests that varying the amount of any one member of such complexes will affect the function of the whole via the kinetics of assembly and other actions (1). In effect, they exhibit a “balance” among themselves in terms of the activity of the whole (Figure 1). When this fact is coupled with genetic and biological observations stretching back a century, a synthesis emerges that helps explain at least some aspects of a variety of phenomena including aneuploid syndromes, dosage compensation, quantitative trait genetics, regulatory gene evolution following polyploidization, the emergence of complexity in multicellular organisms, the genetic basis of evolutionary gradualism and potential implications for heterosis and co-evolving genes complexes involved with speciation. In this article we will summarize the evidence for this potential synthesis.
Figure 1. Model showing effect of stoichiometry of gene regulatory complexes and its consequences in generating phenotypes.
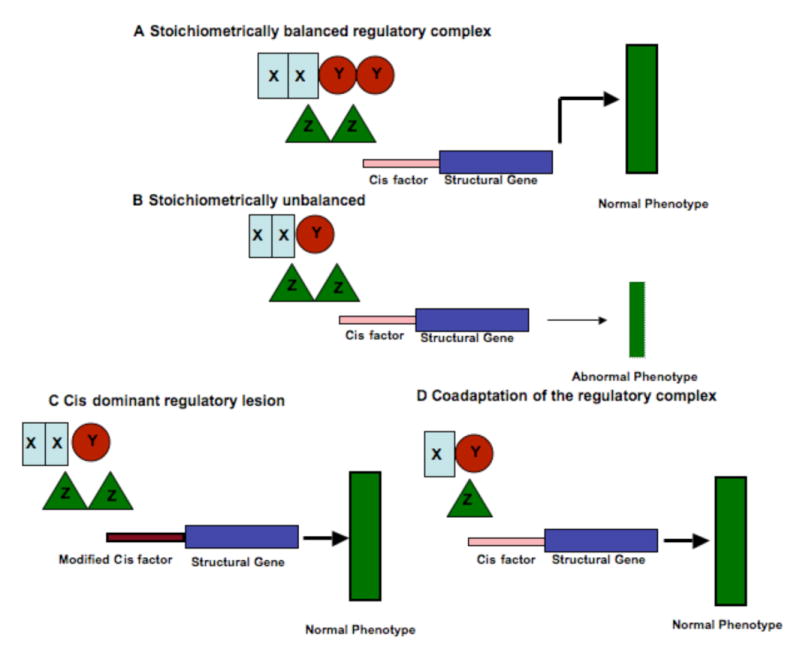
Panel A shows a stoichiometrically balanced regulatory complex depicted by sets of rectangles, circles and triangles (XX:YY:ZZ::1:1:1), which is able to properly communicate/interact with the cis factor of a target structural gene, which leads to a (normal) phenotype depicted by a thick green rectangle. In panel B the stoichiometry of the regulatory complex is altered and thus unbalanced (XX:Y:ZZ::2:1:2) so interaction with the cis elements of target loci is modified such that an abnormal phenotype (depicted by a thinner green rectangle) is produced. This phenotype might be selected against. Panel C shows modification in a cis factor (depicted by brown rectangle) such that a shift from the balanced regulatory complex (XX:YY:ZZ::1:1:1) to the unbalanced state (XX:Y:ZZ::2:1:2) is tolerated. Panel D depicts co-adaptation of the unbalanced regulatory complex over time to attain a new balance (X:Y:Z::1:1:1) leading to the normal phenotype.
Aneuploidy versus ploidy on the phenotype level
The concept of “balance” in genetics traces back to the early days of genetics in studies of chromosomal duplication in Datura (2) and of sex determination in the moth, Lymantria (3), and the fruitfly, Drosophila (4). In Datura, the variation in copy number of individual chromosomes (aneuploidy) was compared to variation for the whole genome (ploidy). In insects, the determination of sex was influenced by a changing dose of the sex chromosome in relation to the whole genome. The most extensive studies in the plant kingdom were performed by Blakeslee and colleagues. They produced a whole set of extra chromosomes (trisomics) for all of the twelve members of the karyotype of Datura, each of which produced a characteristic change in phenotype from the normal type (5). In these studies, they also found extra chromosomes that contain two copies of the same chromosome arm (isochromosomes, secondary trisomics). These lines exhibited greater extremes of a subset of the altered phenotypic characteristics found with the trisomics from which they were derived. Such aneuploid syndromes typically involve reduced stature, vigor and in some cases reduced fertility.
In contrast, the production of a ploidy series with increasing doses of the whole genome, while generating specific changes in phenotype, did not result in as extreme effects as found with varying individual chromosomes or chromosome arms (5). Moreover, haploid plants were found for Datura and the addition of an extra chromosome to this genotype produced a very extreme deviant phenotype (6). These types of observations have been recapitulated in many other plant species over the decades. The results indicate that there is a “balance” within the genome that, if upset, will alter the phenotype (Figure 1). This balance has been interpreted for many years as resulting from an altered level of expression of genes on the varied chromosome relative to the remainder of the genome. Typically, the imbalance was considered to involve gene products of housekeeping or metabolic processes. However, recent evidence from studies of gene expression in aneuploidy and ploidy series and the identification of the genes responsible for these effects (7), together with studies of the retention of regulatory genes during the process of diploidization following multiple rounds of polyploidization in the evolutionary lineage of the plant kingdom (8-10), would suggest that the balance is primarily one involving regulatory molecules. This balance would affect the expression of the target genes, which might cause the aneuploid syndromes by reductions in expression of critical products regardless of whether there is an addition or subtraction of a chromosome from the genotype (11-12).
Gene expression in aneuploidy and ploidy series
Studies of the expression of individual genes in aneuploidy and ploidy series, primarily conducted in maize, indicate that changes in dosage of individual chromosomes or chromosomal segments have global effects on gene expression and that these effects are more numerous and extreme than what is generally found in studies on gene expression in a ploidy series (11-12). In early studies, enzyme activities encoded by various genes located around the genome were assayed in a dosage series of 1-4 copies of the long arm of chromosome 1 in maize (11). Further, protein profiles were generated from dosage series of several chromosome arm dosage series and compared to those from a ploidy series (12). The results from these studies can be summarized as follows. Any specific chromosome arm dosage series will modulate the expression of many genes located at various positions in the genome--in other words the effects operate in trans. Second, any one gene could be modulated by several different dosage series. Third, the modulations could be both positive or negative correlations with the dosage of the varied chromosome arm, but the negative effects predominated. Finally, the modulations of the same genes in a ploidy series were minimal in comparison. Together, these results suggested a balance of regulatory factors controlling gene expression levels.
Dosage compensation
The trans-acting dosage effects that operate in aneuploid series not only affect the expression of genes located elsewhere in the genome, but also modulate the expression of the genes present on the same chromosome arm. Because the major type of effect is a negative correlation with dosage, referred to as an “inverse dosage effect”, many cases of dosage compensation result in an aneuploid dosage series (11-13). This compensation results because the target locus is being varied in dosage as well. In other words, in a dosage series the target gene is increasing as does the copy number of the chromosome arm, but the inverse dosage effect counteracts the amount of expression from each allele present by an inverse increment. These two effects cancel to produce about the same total amount of gene product from each chromosomal dose in the series (Figure 2). This situation was first demonstrated for the Alcohol dehydrogenase (Adh) gene in maize, which resides in the long arm of chromosome 1 (13). In the aforementioned dosage series, the total Adh expression is approximately equal for 1 to 4 copies of the chromosome arm. However, if the chromosome arm is assayed for dosage effects in smaller segments using some genetic tricks, the Adh structural gene dosage series produced a positive correlation between the total expression and the copy number of the gene present. When a separate portion of 1L was varied in 1-3 copies, but holding the dosage of Adh constant, an inverse dosage effect was found on the level of ADH present. Thus, the two effects occurring simultaneously cancel to produce dosage compensation. This type of mechanism has been recapitulated for cases of dosage compensation in Drosophila (14-17) and recent evidence suggests an involvement of balance in cases of dosage compensation in plants and for sex chromosomes in Drosophila, C. elegans and mammals (18).
Figure 2. Dosage Compensation.

The copy number of a gene located on a chromosome varies with each change in chromosomal dose but the total amount of gene expression remains the same due to transacting regulatory dosage effects on the gene. In normal diploids there are two doses of each chromosome and each allele of the gene contributes 50% to the total gene expression. However, when there is only a single dose of the chromosome, as in monoploids, the transacting regulators interact such that the remaining allele contributes 100% to gene expression (complete green rectangle). On the other hand when chromosome dose is tripled as in trisomics, each of the three alleles contributes to only a third of the total gene expression (green red and blue rectangles).
The changes in gene expression found in aneuploidy and ploidy series have also been documented on the RNA level. The same types of effects, i.e. positive and negative with a prevalence of the latter have been documented in embryo tissues of developing kernels of maize (19). Cases of dosage compensation were also found. In a ploidy series, there is also less modulation of RNA levels than expected from a cumulative effect from the aneuploidy results (20). In the triploid endosperm tissues, there are also aneuploid effects, but of lesser magnitude as might be expected from a balance relationship because the addition or subtraction of a chromosome in a triploid genotype creates less imbalance.
What are the types of genes responsible for the dosage effects?
Over the course of two decades, single gene mutations were sought in Drosophila that would mimic the types of effects that occur in the aneuploid dosage series in order to gain insight into the molecular nature of the phenomenon (7, 21). The approach was to mutagenize flies and screen for heterozygous mutations that would produce a semidominant effect on the expression of a leaky allele of the white eye color gene as a reporter. Such semidominant mutations, if they result from a loss of function, would be equivalent to a one dose situation of the affected gene relative to the normal two dose diploid. To date, there are 47 known genes that produce such a dosage effect on the white reporter (7). As with the aneuploid dosage effects, there is a mix of positive and negative effects with a prevalence of the latter. The molecular nature of many of these modifiers is now known. The group is comprised of chromatin components, transcription factors and members of signal transduction cascades (7).
An obvious question that arises is how a single target reporter could be affected by so many regulatory genes. It has been proposed that an explanation for this result might lie in the fact that regulatory hierarchies are known to occur and if each is composed of dosage dependent members, then varying the dosage of any member within the hierarchy will pass along the dosage effect at some magnitude to most of the targets (7). In other words, a dosage effect of a regulatory gene at the top of the hierarchy, which might modulate a subsequent dosage dependent regulator which in turn affects the expression of a target gene, will pass along the effect to ultimately alter the expression of that target. Clearly, of course, each regulator modulates the expression of multiple targets, which creates a complicated web of interactions.
Haplo-insufficiency
Recent studies from clinical medicine have indicated that many genetic syndromes in humans result from a heterozygosity for a loss of function allele of transcription factors, oncogenes and signal transduction components (1). Theoretical modeling has provided an underpinning of how these effects can result from interactions and assembly of different subunits of macromolecular complexes (1). From studies in yeast (22) and humans (23), genes that exhibit a haploinsufficiency are strongly skewed toward those that are involved in molecular complexes, suggesting the importance of stoichiometry of the various subunits. Thus, there are parallels to the above-mentioned studies of dosage balance in aneuploidy/ploidy studies of regulatory genes. While other classes of genes than regulatory loci are involved with molecular complexes and exhibit haploinsufficiency, we will focus on the fact that regulatory genes fall within this category.
Parallels with quantitative trait loci
Quantitative traits are usually controlled by multiple loci each of which modulates the phenotype in increments (24). They encompass a wide variety of aspects of the phenotype, so generalizations are difficult to formulate, but some principles do emerge. Typically, quantitative trait loci (QTL) show some level of semidominance between the two alleles being examined at any one locus, namely the heterozygote is intermediate between the two parents for the phenotype rather than identical to one of them (dominant/recessive) as often occurs with qualitative traits. Multiple loci contribute to any one phenotype. Thus, there are parallels with the impact of aneuploidy on the phenotype in that the effects are dosage sensitive and there are several factors influencing any one characteristic (7, 19).
The molecular definition of quantitative trait loci is still in its infancy, but some trends have emerged from those examples that are now known, as previously summarized (7, 25). For the most part, QTL appear to have some type of regulatory function, particularly transcription factors and members of signal transduction cascades. Some have a positive impact on the phenotype, while others act in a negative dosage sensitive manner. It is also likely that genes encoding the rate limiting step in biochemical pathways would contribute to quantitative phenotypes and act in a dosage dependent manner (25).
Thus, from what is known to date, there appears to be a connection between the behavior of aneuploid effects on the phenotype and the control of quantitative traits. This parallel not only involves the multigenic nature of control and a dosage dependence, but also the molecular nature of the genes involved.
Retention of regulatory genes during diploidization
The availability of genomic sequences for Arabidopsis and rice has revealed that there have been repeated cycles of polyploidization and diploidization in the evolution of the angiosperms (8-10, 26-28). Allotetraploids have occurred by the hybridization of related species followed by chromosomal doubling. Because of the ploidy hybridization barrier in angiosperms (29), the new polyploids usually become reproductively isolated from the diploid progenitor species. Being tetraploid, they can tolerate the deletion of genes--at least certain classes of genes. As this process continues over evolutionary time, there is a progression toward a diploid state again. However, recent evidence indicates that there is a preferential retention of transcription factors and signal transduction components as duplicates from the most recent respective events of polyploidization and diploidization in both Arabidopsis and rice (8-10, 28). Indeed, there is evidence of purifying selection for the genes that are preferentially retained (10). By comparison, studies of the types of genes present in segmental duplications, which increase the copy number of only a small portion of the genome, provide a complementary picture, namely, a relative dearth of transcription factors and signal transduction components (30). These observations suggest there is usually a selection against creating a regulatory imbalance by segmental duplication.
The sum of these results suggests that transcription factors and signal transduction components that are duplicated during the tetraploidization events are resistant to deletion. In other words, if one copy of a duplicate pair is removed from the genotype by random deletion processes, this circumstance could mimic aneuploid effects and result in reduced reproductive fitness. Thus, such events would be subject to purifying selection and cause the retention of the duplicate pairs. For other types of genes, deletion of one pair might not have any consequences for fitness and would be reduced to the diploid level. As noted above, segmental duplications that alter the balance of regulators would likely be selected against (30). Thus, duplications of small parts of the genome are preferentially represented by housekeeping genes (30).
These results suggest that the concept of balance of regulatory genes extends to the finding of evolutionary retention following polyploidization. The same general classes of genes are involved in the processes of retention from whole genome duplication events and exclusion from segmental duplication. Thus, regulatory genes, namely transcription factors and signal transduction components, exhibit parallel and consistent behavior in studies of aneuploidy, quantitative trait specification and their evolutionary fates following duplication.
Complexity drive?
Freeling and Thomas (28) have suggested that the duplication and retention of duplicate regulatory factors following polyploidization could contribute to an increase in complexity during the evolution of multicellular organisms. Whether such increase in complexity has occurred during evolution has been a topic of debate for some time. The resistance to this idea comes from the idea of “progress” in evolution, which is unlikely to occur. It is important to realize however that “progress” and “complexity” are not necessarily synonymous. The new suggestion is that complexity is driven by degenerative processes, such as mutation and deletion, following polyploidization events within the context of regulatory balance and selection. This proliferation of regulatory functions can foster greater complexity.
New variation in life comes from mutations and new genes from duplications. Numerous examples are known of proliferation of genes by tandem duplication and divergence. Accumulation of mutations in one copy of the duplicate results in divergence to a new function, which is referred to as neofunctionalization. Alternatively, the two copies could diverge such that each member of the pair performs separate aspects of the original function, which is called subfunctionalization (31).
One explanation for the retention of duplicate regulatory genes during diploidization might be that sub or neo functionalization (i. e. altered tissue specific expression or enzymatic action) has occurred for one member of the duplicate pair so that deletion of either copy removes a required function from the genotype which would be detrimental and thus selected against. To some degree, this almost certainly occurs, but the evidence for purifying selection against mutation accumulation for the retained classes of genes suggests otherwise for many genes (10). Sub and neo functionalization of duplicates is apparently a slower process than once thought. As pointed out by Freeling and Thomas (28), maintenance of balance of the regulators is required immediately upon polyploidization and deletion of one member of a pair is predicted to have detrimental consequences.
Therefore, one might envision the following scenario. The duplication of the genome would increase the copy number of all genes. Regulatory factors are present in a balanced stoichiometry that makes them resistant to loss. The retention over long evolutionary time makes them available for potential mutational change that could lead to sub and neo functionalization. However, in order for this to happen, they must escape from the balanced stoichiometry. One might imagine that one way in which this could happen might be by the deletion of specific target genes. This must occur in such a manner that only some targets of a balanced regulatory system are removed at a time so that a detrimental situation is not created. As a gradual deletion of multiple targets accumulate, selection pressure might occur to shift a regulatory balance. Another possibility might be the occurrence of cis-dominant regulatory lesions in critical target loci that would alter their expression in such a manner to allow a shift in the balance of regulators (Figure 1). It is certain, however, that, in order for whole genome duplication to contribute to an increase in complexity, divergence of duplicate regulators must occur at some point following the various rounds of polyploidization and diploidization during plant evolution.
The principle proposed by Freeling and Thomas (28) is that with multiple whole genome duplication events in the history of angiosperms, there has been a continual proliferation of regulatory genes. They are held in the evolutionary lineage against deletion because of their balance property. As a new balance is eventually found and divergence occurs, developmental and metabolic processes will become more complex. Thus, on this hypothesis, complexity is driven at least in part by the cycle of polyploidization and diploidization followed by random degenerative mechanisms (deletion and mutations) that do not invoke “progress”.
Is there an involvement in hybrid vigor?
Hybrid vigor or heterosis is the phenomenon that hybrid individuals often show superior phenotypes compared to their inbred parents for such characteristics as fertility, biomass and flowering time. The genetic and molecular basis of heterosis has eluded a consensus (32). However, recently an exhaustive study of introgression lines of overlapping portions of a wild tomato genome into domesticated tomato has suggested that the heterotic response results from so-called overdominant QTL (33). In other words, heterozygosity for the introgressed segment of the genome produced a response that was superior to homozygous lines of the respective introgressed line or of domesticated tomato itself. QTL for other types of traits not involved with reproductive success did not exhibit overdominance. The implication of this result is that an allelic interaction causes the superior response as opposed to a complementation of partially detrimental alleles in the heterozygote.
The authors propose that the superior performance of the heterozygous state would be selected for due to the increased reproductive success of the hybrids. An analogy is made to self incompatibility genes that foster outcrossing. This scenario would suggest that the selection must occur in the heterozygous state and the question arises whether dosage dependent regulators are also involved with this type of quantitative response. Examples of single gene heterosis were reported many years ago (34) and now that the molecular basis of these loci has been defined, an example of a transcription factor and a signal transduction component are represented (35,36). This sample of course is too small to form any generalizations, but it is consistent with other studies of the molecular basis of QTL and dosage dependent regulators.
Could regulatory complexes represent coevolving gene incompatibilities for speciation?
From studies of hybrids of different Drosophila species, Herman Muller and Theodosius Dobzhansky recognized that gene incompatibilities occurred (37-39). While two related species are obviously individually viable and fertile, hybrids between them can show lethality of one of the two sexes or sterility of various sorts. To the extent that genetics can be applied to the problem, it is obvious that multiple genes are involved that are incompatible in the hybrid. In at least one study of the determination of the quantitative trait loci involved in differences between species, the multiple genes showed semidominant effects (40). Thus, it appears that interacting genes are evolving in concert in the diverging lineages.
Of the very few speciation genes identified molecularly, dosage dependent transcription factors are represented (41-43). Thus, an interesting pursuit in the future would be to explore the possibility that hybrid incompatibility genes represent members of interacting macromolecular complexes that are co-evolving. One might imagine that a change in one member in a balanced stoichiometry might present conditions that favor selective changes in other members of the same complex. In one sense, different members of such complexes might be in conflict for evolving characteristics that foster their perpetuation. If a selection occurs on one member of a complex, selection pressure might occur for coevolution of other members of a stoichiometric group to change as well (Figure 1). Because each member is dosage sensitive and new mutations can produce a subtle change in the phenotype, such changes can rapidly be selected because the mutations have an effect in the heterozygote and are thus available for selection.
Gradualism in evolution
An unresolved issue in biology is whether evolution proceeds by few changes of large effect or by many changes of small effect. The realization of the fact that gene regulatory processes are dosage dependent and exist in hierarchies, which provides numerous modifiers of a single phenotype, provides a potential explanation for evolutionary gradualism for large changes in phenotype by continual selection for many changes of small effect. Changes in members of multi-subunit complexes can all have the potential to produce a subtle change on the phenotype. If selection on a particular trait is strong, the phenotype can shift in one direction to great extreme by many subtle steps via sequential selection on different members of a dosage dependent regulatory hierarchy. It is also likely that cis-dominant changes in the regulation of target genes could contribute to the evolutionary shift as well. Of course, pleiotropy of individual regulators and epistasis among interacting genes will no doubt occur and affect the process. A challenging future research direction would be to examine these issues and determine the contribution of these various factors to evolutionary trends.
Concluding remarks
In this article we have summarized the evidence which suggests that regulatory genes in multicellular eukaryotes are dosage dependent as a result of their membership in macromolecular complexes for which the contributing subunits produce stoichiometric effects on the function of the whole. This evidence comes from the biology of aneuploidy and ploidy series, gene expression studies in the same, the molecular basis of dosage dependent modifiers, the behavior of quantitative trait loci, the molecular basis of QTLs and the maintenance of balance of these classes of genes over evolutionary time following polyploidization events. The implication of this system is that new mutations in regulatory genes are immediately available for selection in the heterozygote, be that purifying or adaptive. It has been known since the first mutagenesis experiments that most mutations are completely recessive (44) and most of these are present in housekeeping genes. New mutations that are completely recessive will be unavailable for selection in the heterozygote and are only subject to evolutionary forces when made homozygous. Thus, evolution for these genes may be largely neutral, while adaptive changes might be more common with dosage dependent regulatory genes.
Natural selection allows for adaptive change, but also for maintenance of the status quo. The early emergence of a regulatory balance in the evolution of multicellular eukaryotes was conducive for both. A regulatory system that can produce subtle changes in the diploid state allows selective change in diploid heterozygous organisms and provides a means by which many new mutations can be available for selection. Any new mutation that fosters greater reproductive success over the alternative will sweep through the population assuming no detrimental homozygous effect, and those mutations that are detrimental will be eliminated quickly. Such a hypothesis produces a synthesis of many genetic and evolutionary observations.
Acknowledgments
Funding on this topic in our laboratory is provided by a grant from the NSF Plant Genome Program, DBI 0501712 and NIH R01 GM068042.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Veitia RA. Exploring the etiology of haploinsufficiency. Bioessays. 2002;24:175–184. doi: 10.1002/bies.10023. [DOI] [PubMed] [Google Scholar]
- 2.Blakeslee AF, Belling J, Farnham ME. Chromosomal duplication and Mendelian phenomena in Datura mutants. Science. 1920;52:388–390. doi: 10.1126/science.52.1347.388. [DOI] [PubMed] [Google Scholar]
- 3.Goldschmidt RB. Untersuchngen uber Inter-sexualitat. Zeit Abst Vererb. 1920;23:1–199. [Google Scholar]
- 4.Bridges CB. Sex in relation to chromosomes and genes. Am Naturalist. 1925;59:127–137. [Google Scholar]
- 5.Blakeslee AF. New Jimson weeds from old chromosomes. J Hered. 1934;25:80–108. [Google Scholar]
- 6.Satina S, Blakeslee AF, Avery AG. Balanced and unbalanced haploids in Datura. J Hered. 1937;28:192–202. [Google Scholar]
- 7.Birchler JA, Bhadra U, Pal-Bhadra M, Auger DL. Dosage dependent gene regulation in higher eukaryotes: implications for dosage compensation, aneuploid syndromes and quantitative traits. Dev Biol. 2001;234:275–288. doi: 10.1006/dbio.2001.0262. [DOI] [PubMed] [Google Scholar]
- 8.Blanc G, Wolfe KH. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. The Plant Cell. 2004;16:1679–1691. doi: 10.1105/tpc.021410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maere S, DeBodt S, Raes J, Casneuf T, Van Montagu M, Kuiper M, Van de Peer Y. Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci USA. 2005;102:5454–5459. doi: 10.1073/pnas.0501102102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chapman BA, Bowers JE, Feltus FA, Paterson AH. Buffering of crucial functions by paleologous duplicated genes may contribute cyclicality to angiosperm genome duplication. Proc Natl Acad Sci USA. 2006;103:2730–2735. doi: 10.1073/pnas.0507782103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Birchler JA. A study of enzyme activities in a dosage series of the long arm of chromosome one in maize. Genetics. 1979;92:1211–1229. doi: 10.1093/genetics/92.4.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Birchler JA, Newton KJ. Modulation of protein levels in chromosomal dosage series of maize: The biochemical basis of aneuploid syndromes. Genetics. 1981;99:247–266. doi: 10.1093/genetics/99.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Birchler JA. The genetic basis of dosage compensation of alcohol dehydrogenase-1 in maize. Genetics. 1981;97:625–637. doi: 10.1093/genetics/97.3-4.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Devlin RH, Holm DG, Grigliatti TA. Autosomal dosage compensation in Drosophila melanogaster strains trisomic for the left arm of chromosome 2. Proc Natl Acad Sci USA. 1982;79:1200–1204. doi: 10.1073/pnas.79.4.1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Birchler JA, Hiebert JC, Paigen K. Analysis of autosomal dosage compensation involving the Alcohol dehydrogenase locus in Drosophila melanogaster. Genetics. 1990;124:677–686. [PMC free article] [PubMed] [Google Scholar]
- 16.Birchler JA, Hiebert JC, Krietzman M. Gene expression in adult metafemales of Drosophila melanogaster. Genetics. 1989;122:869–879. doi: 10.1093/genetics/122.4.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Birchler JA. Expression of cis-regulatory mutants of the white locus in metafemales of Drosophila melanogaster. Genetical Research. 1992;59:11–18. doi: 10.1017/s0016672300030123. [DOI] [PubMed] [Google Scholar]
- 18.Birchler JA, Fernandez H, Kavi H. Commonalities in Compensation. Bioessays. 2006;28:565–568. doi: 10.1002/bies.20408. [DOI] [PubMed] [Google Scholar]
- 19.Guo M, Birchler JA. Trans-acting dosage effects on the expression of model gene systems in maize aneuploids. Science. 1994;266:1999–2002. doi: 10.1126/science.266.5193.1999. [DOI] [PubMed] [Google Scholar]
- 20.Guo M, Davis D, Birchler JA. Dosage effects on gene expression in a maize ploidy series. Genetics. 1996;142:1349–1355. doi: 10.1093/genetics/142.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rabinow L, Nguyen-Huynh AT, Birchler JA. A trans-acting regulatory gene that inversely affects the expression of the white, brown and scarlet loci in Drosophila melanogaster. Genetics. 1991;129:463–480. doi: 10.1093/genetics/129.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papp B, Pal C, Hurst LD. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424:194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- 23.Kondrashov FA, Koonin EV. A common framework for understanding the origin of genetic dominance and evolutionary fates of gene duplications. Trends Genet. 2004;20:287–290. doi: 10.1016/j.tig.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 24.Tanksley SD. Mapping polygenes. Annu Rev Genetics. 1993;27:205–233. doi: 10.1146/annurev.ge.27.120193.001225. [DOI] [PubMed] [Google Scholar]
- 25.Birchler JA, Riddle NC, Auger DL, Veitia RA. Dosage balance in gene regulation: biological implications. Trends in Genetics. 2005;21:219–226. doi: 10.1016/j.tig.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 26.Bowers JE, Chapman BA, Rong J, Paterson AH. Unraveling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;2003;422:433–438. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- 27.Simillion C, Vandepoele K, Montagu MC, Zabeau M, Van de Peer Y. The hidden duplication past of Arabidopsis thaliana. Proc Natl Acad Sci USA. 2002;99:13627–13632. doi: 10.1073/pnas.212522399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freeling M, Thomas BC. Gene-balanced duplications, like tetraploid, provide predictable drive to increase morphological complexity. Genome Research. 2006;16:805–14. doi: 10.1101/gr.3681406. [DOI] [PubMed] [Google Scholar]
- 29.Birchler JA. Dosage analysis of maize endosperm development. Annual Review of Genetics. 1993;27:181–204. doi: 10.1146/annurev.ge.27.120193.001145. [DOI] [PubMed] [Google Scholar]
- 30.Davis JC, Petrov DA. Do disparate mechanisms of duplication add similar genes to the genome? Trends in Genetics. 2005;21:548–551. doi: 10.1016/j.tig.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 31.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 32.Birchler JA, Auger DL, Riddle NC. In search of a molecular basis of heterosis. The Plant Cell. 2003;15:2236–2239. doi: 10.1105/tpc.151030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Semel Y, Nissenbaum J, Menda N, Zinder M, Krieger U, Issman N, Pleban T, Lippman Z, Gur A, Zamir D. Overdominant QTL for yield and fitness in tomato. Proc Natl Acad Sci USA. 2006 doi: 10.1073/pnas.0604635103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Redei GP. Single locus heterosis. Zeitschrift fur Verebungslehre. 1962;93:164–170. [Google Scholar]
- 35.Shpak ED, Berthiaume CT, Hill EJ, Torii KU. Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development. 2004;131:1491–1501. doi: 10.1242/dev.01028. [DOI] [PubMed] [Google Scholar]
- 36.Kim GT, Shoda K, Tsuge T, Cho KH, Uchimiya H, Yokoyama R, Nishitani K, Tsukaya H. The ANGUSTIFOLIA gene of Arabidopsis, a plant CtBP gene, regulates leaf-cell expansion, the arrangement of cortical microtubules in lead cells and expression of a gene involved in cell-wall formation. EMBO J. 2002;21:1267–1279. doi: 10.1093/emboj/21.6.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dobzhansky T. Genetics and the Origin of Species. Columbia University Press; New York: 1937. [Google Scholar]
- 38.Muller HJ. Isolating mechanisms, evolution and temperature. Biol Symp. 1942;6:71–125. [Google Scholar]
- 39.Orr HA, Turelli M. The evolution of postzygotic isolation: accumulating Dobzhansky-Muller incompatibilities. Evolution Int J Org Evolution. 2002;55:1085–94. doi: 10.1111/j.0014-3820.2001.tb00628.x. [DOI] [PubMed] [Google Scholar]
- 40.Laurie CC, True JR, Liu J, Mercer JM. An introgression analysis of quantitative trait loci that contribute to a morphological difference between Drosophila simulans and D. mauritiana. Genetics. 1997;145:339–348. doi: 10.1093/genetics/145.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peres DE, Wu CI. Further characterization of the Odysseus locus of hybrid sterility in Drosophila: one gene is not enough. Genetics. 1993;140:201–206. doi: 10.1093/genetics/140.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ting CT, Tsaur SC, Wu ML, Wu CI. A rapidly evolving homeobox at the site of a hybrid sterility gene. Science. 1998;282:1501–1504. doi: 10.1126/science.282.5393.1501. [DOI] [PubMed] [Google Scholar]
- 43.Barbash DA, Siino DF, Tarone AM, Roote J. A rapidly evolving MYB-related protein causes species isolation in Drosophila. Proc Natl Acad Sci USA. 2003;100:5302–5307. doi: 10.1073/pnas.0836927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stadler LJ. Mutations in barley induced by X-rays and radium. Science. 1928;68:186–187. doi: 10.1126/science.68.1756.186. [DOI] [PubMed] [Google Scholar]


