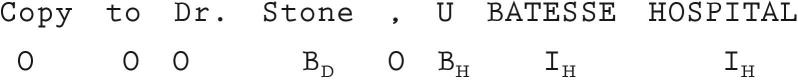Abstract
Objective
This paper describes a successful approach to de-identification that was developed to participate in a recent AMIA-sponsored challenge evaluation.
Method
Our approach focused on rapid adaptation of existing toolkits for named entity recognition using two existing toolkits, Carafe and LingPipe.
Results
The “out of the box” Carafe system achieved a very good score (phrase F-measure of 0.9664) with only four hours of work to adapt it to the de-identification task. With further tuning, we were able to reduce the token-level error term by over 36% through task-specific feature engineering and the introduction of a lexicon, achieving a phrase F-measure of 0.9736.
Conclusions
We were able to achieve good performance on the de-identification task by the rapid retargeting of existing toolkits. For the Carafe system, we developed a method for tuning the balance of recall vs. precision, as well as a confidence score that correlated well with the measured F-score.
Introduction
De-identification is the process of selecting and redacting all of the Protected Health Information (PHI) present in a medical (or other) record so that the record may be shared outside the limited audience authorized to know the identity of the record’s subject. De-identification is a key step toward making clinical data available for many medically important applications, such as epidemiological investigations or collection of data on drug interactions or side-effects. Corpora of de-identified clinical data are also important for research and development of natural language processing systems that capture and map information in clinical records into standardized categories or ontologies. De-identification is a first step towards identification and extraction of other important information, such as medications or diagnoses. A long-term goal is to extract multiple types of information from clinical records and map them onto standard medical terminologies or ontologies for further information processing.
The Team
This work is the result of collaboration between teams at MITRE Bedford and the Harvard Center for Biomedical Informatics, working together on the capture and management of free text information for translational medicine. We were motivated by a desire to participate in the de-identification task of the AMIA Challenges in Natural Language Processing for Clinical Data, 1 which represents the first shared task for the application of natural language processing technology to clinical data. Our approach grows out of previous work on information extraction and text mining in the biomedical area, 2,3 as well as work on information extraction in other domains.
The Approach
Our approach to de-identification focused on the rapid adaptation of two existing toolkits for named entity recognition. One is based on the Carafe toolkit developed at MITRE, and the second used LingPipe, a commercial product from Alias-I. Specifically, our experiments focused on what needed to be done to train the systems, how well they worked “out of the box,” whether there was adequate training data, and how much work was needed for additional performance gains. Our previous experiences with a wide range of information extraction tasks had led us to expect that it would require significant work to adapt a toolkit to a new task such as de-identification. However, we were pleasantly surprised to find that the adaptation process was quite rapid. We were able to achieve good performance with only a few hours of work. We attributed this to several factors, including adequate training data and the structured nature of the task.
The Task and Metrics
This paper reports on systems that were evaluated on a specific, artificially constructed de-identification task, described in Sibanda et al. 4 The data for this task were 910 medical discharge summaries (690 used for training, 220 used for the official evaluation) that had first been de-identified through a combination of machine and human processing, and then repopulated with synthesized identifiers to support the challenge de-identification task. System performance on the de-identification task was evaluated by calculating precision, recall, and balanced F-measure per word (defined below in the Results section) as well as per PHI. Overall, the systems that we fielded for this application performed very well, with token balanced F-measures of well over 0.99 (see ▶). However, we do not know how these measures relate to the real costs of false positives vs. false negatives in an application setting. In such a setting, we expect that false negatives (failures to locate and redact critical information) are more serious than false positives. Overzealous tagging of words in phrases (e.g., including “M.D.” as part of a “doctor name”) may be less harmful, and easier to fix, than failing to tag parts of a proper name. For these reasons, we provided a “high recall” output as one of our submissions. For future evaluations, it would be useful to determine the best trade-off between a thorough “smothering” of protected information and restoration of information that has been over-zealously removed. We also expect that the real value of automated de-identification systems can only be realized in an interactive environment, where a person can quickly review and correct the automatically generated tags.
Table 1.
Table 1 Recall, Precision and F-measure Evaluated Per Phrase and Per Token F-measure
| Phrase Recall | Phrase Precision | Phrase F-measure | Token F-measure | |
|---|---|---|---|---|
| Official Runs | ||||
| Carafe-ALL (b = −1.0) | 0.9597 | 0.9839 | 0.9716 | 0.99714 |
| LingPipe | 0.9212 | 0.9430 | 0.9320 | 0.99400 |
| Carafe-ALL+PP (b = −1.5) | 0.9694 | 0.9778 | 0.9736 | 0.99745 |
| Unofficial Runs | ||||
| Carafe-I (b = −1.0) | 0.9610 | 0.9719 | 0.9664 | 0.99627 |
| Carafe-I+L (b = −1.0) | 0.9672 | 0.9822 | 0.9746 | 0.99758 |
| Carafe-I+RC (b = −1.0) | 0.9574 | 0.9810 | 0.9690 | 0.99682 |
| Carafe-ALL (b = −1.75) | 0.9693 | 0.9818 | 0.9756 | 0.99763 |
Of the official runs, Carafe-ALL+PP achieved our best performance. Unofficial development runs are listed below the official runs.
The following sections of this paper discuss the relationship of this task to previous evaluations of named entity identification; the corpus and our experimental methodology; the use and specialization of the Carafe and LingPipe based systems; the results on the AMIA de-identification challenge evaluation data; and future directions. We include a discussion of possible refinements to the method of scoring a de-identified record as well as the way in which the task and systems might be configured in a complete application for interactive de-identification.
Background
The de-identification task shares many attributes with established named entity extraction tasks. Both involve automatically finding particular types of noun phrases in texts. Traditional named entity tasks such as the MUC evaluations 5 have mainly focused on finding persons, organizations, and locations as well as times and dates in newspaper-style electronic texts such as the Wall Street Journal. However, de-identification in general, and the AMIA de-identification task in particular, differs from earlier named entity extraction tasks in important ways.
In contrast to general named entity tasks, de-identification of medical records involves finding and masking Protected Health Information, including types of persons (doctors and patients), organizations (hospitals), and locations, as well as IDs, ages, dates (but not years), and phone numbers. In addition to these type differences, there are significant source text differences to overcome. In journalistic prose, many named entities may be identified using reliable cues based on case and titles. By contrast, patient medical records consist of semi-structured data with headers and shorter free text fields.
There are further differences that are particular to the present AMIA de-identification challenge. Many approaches to traditional named entity tasks include leveraging databases of known person names, organization names, and location names. Some medical records contain names that are misspelled or simply are not found in such lexical resources. 4 For this reason, the data for the AMIA challenge have been re-identified in a way that discourages reliance on lexical resources, forcing developers to rely on context for de-identification. Such a data preparation strategy may force an over-reliance on context; many cases might be well covered by a lexical approach coupled with a contextual approach to handle the harder cases.
Methods
This section provides a description of the systems we used for the task of identifying phrases of personal health information in medical records.
Identifying Phrases as Sequence Labeling
The problem of identifying phrases can be viewed as a sequence labeling problem in which phrases are denoted by assigning labels to individual words indicating whether the word is part of a phrase of a particular PHI type or whether it is not part of any phrase. A common encoding is to use two labels for each type of phrase: one indicating a word begins a phrase and another indicating a word is within or ending a phrase. ▶ provides an example of the labels assigned to a short excerpt from a medical record. In the figure, O indicates a word outside a phrase, Bτ indicates that the word is the beginning of a phrase of type τ and Iτ indicates the word is inside or at the end of a phrase of type τ. Types of phrases shown include D (doctor) and H (hospital).
Figure 1.
Identifying PHI phrases as a sequence labeling problem.
Our experiments in this paper include two different sequence labeling systems: Carafe, based on an implementation of Conditional Random Fields (CRFs); and LingPipe, a system utilizing hierarchical hidden Markov models. We describe these systems in the following sections.
Conditional Random Fields
Conditional Random Fields (CRFs) are conditionally trained, probabilistic finite-state machines that have been applied successfully to a variety of problems in natural language, including part-of-speech tagging, 6,7 shallow parsing, 8 entity tagging in newswire 9 and tagging genes and proteins in the biomedical domain. 10 CRFs define the conditional probability of a tag (i.e., label) sequence, given an observed sequence of tokens, as follows:
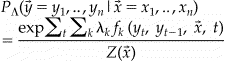 |
where
The probability of a particular label sequence given an observation sequence is computed by summing over each position, t, in the sequence, , and computing the sum over a set of feature functions, each of whose value is multiplied by that feature’s associated weight, . The normalization term, , is determined by computing the above sum for all possible label sequences which can, fortunately, be computed efficiently via dynamic programming.
Each feature function can be viewed most naturally as a predicate over a particular configuration of the observation, , relative to the current position, t, for a particular label pair: yt, yt−1. The feature weights indicate how strongly that predicate over the observations correlates with a particular label pair. For example, the feature below captures a contextual cue for “Dr.” which often indicates that the following token is the beginning of a DOCTOR phrase:
Note also that a feature may ignore the previous label and only correlate the observation predicate with the label at the current position. We call such features node features as they only pay attention to the current “node” (in a graphical representation of the model) whereas features associating observation predicates with the current and previous state are termed edge features.
The weights for each feature in the model are learned by maximizing the conditional log-likelihood of the training data. That is, the weights are set to assign high (log) probabilities to the label sequences found in the training data and low (log) probabilities to all other label sequences over the observations in the training data. The log-likelihood is computed by summing the log-probabilities for a fixed set of weights, , over all the training instances in a data set D:
where the second term, , is a penalty term that biases the model weights towards zero to prevent overfitting. The degree to which the weights are biased is determined by a zero-mean Gaussian distribution with variance σ. Lower variances apply a higher penalty to weights further from zero providing the model with fewer degrees of freedom and thus preventing overfitting to a greater degree than higher variance values. The weights that maximize the above expression can be found using a variety of numerical optimization methods. These methods are iterative, requiring at each step: 1) , the current set of parameters at iteration T; 2) the value of the function to optimize, ; and 3) the value of the gradient of the function . a The result returned from the optimization procedure is a new set of parameters, . This process is repeated until convergence.
Given a trained model, the most likely label sequence for a given observation sequence can be found efficiently with dynamic programming using a slight variation on the Viterbi algorithm. 11
Carafe
Carafe is a toolkit implementing CRFs targeted especially to phrase identification tasks. It includes light-weight, flexible methods for easily introducing new features, an implementation of the L-BFGS numeric optimization routine 12 for weight learning and mechanisms for handling SGML data robustly. This section discusses the steps we took with Carafe to get an initial de-identification system up and running, as well as additional, task-specific feature engineering we carried out to improve performance on this task.
Two questions need to be answered before applying a sequence labeling approach to any tagging problem: 1) what constitute the elements (i.e., words) of a sequence, and 2) what constitutes the beginning and end of a sequence?
The first question is one of tokenization. Crucially, tokens must align properly with the phrases of interest. The system will not be able to properly identify phrases that do not align with the tokens. For the de-identification task and the medical record data provided, the primary tokenization issue arose with dates. Only the month and day portion of date expressions were to be tagged, but not the year. This required modifying our existing tokenizer to split potential date strings into multiple tokens.
In many tasks, sequences consist of natural language sentences. In the challenge task medical records, however, there were no obvious sentence boundaries for much of the record, since it was derived from a set of database fields. As such, we decided to consider each entire medical record as a single sequence (sentence) in our model.
For our initial system, we performed the adjustments to tokenization described above (about four hours of work) and then applied Carafe, out-of-the-box, to the PHI identification task using an existing set of features designed for a different task. Specifically, this other task was Named Entity recognition over newswire texts requiring identification of PERSON, ORGANIZATION and LOCATION phrase types. This constituted the system labeled Carafe-I in ▶.
Trading Off Precision and Recall
For this application, we believed that recall should be more important than precision. However, we also believed that this balance could vary, depending on the specific application or end-user. Therefore we wanted to develop a method to change the balance of recall vs. precision. To achieve this, we introduced a bias parameter, following Minkov et al., 13 that adjusts the model weight associated with the feature capturing the “prior probability” of a token being labeled as O. Specifically, this is the feature —i.e., the feature that returns a value of 1 only when the current label is O and which ignores the previous label and the observations, . Increasing this feature’s weight (positive bias) makes a token more likely to be labeled as O (i.e., the label corresponding to other, thus improving precision). Decreasing the weight (negative bias) makes a token less likely to be labeled O and more likely to be labeled as something besides O (i.e., one of labels corresponding to a phrase type), thus improving recall.
In addition to providing a mechanism to improve recall, we found in general that biasing the resulting system towards recall had a tendency to improve overall F-measure.
Despite introducing the bias parameter above, we noticed a tendency for the system to continue to make certain recall errors. To address this we developed and applied a number of regular expressions (detailed in the section “Task-driven feature customizations” below) as a post-process. This constituted the system labeled Carafe-ALL+PP in ▶.
LingPipe
LingPipe is a software developer’s toolkit of Java classes and sample implementations for performing a variety of natural language processing tasks. For the de-identification task, we chose to use the named-entity tagging features introduced with LingPipe version 2.3.0. Named entity tagging is implemented in LingPipe through the use of chunkers that operate on n-gram based character language models. 14 Given an unlabeled sentence or fragment of text, a properly trained LingPipe Chunker instance will use its hidden Markov model with the statistics it gathered during training to output the most likely “chunking” for the text, which labels each chunk with the best-determined tag. Similarly, an instance of the NBestChunker can output a list of the n most likely chunkings instead of just the first best. A rescoring chunker, implemented by the CharLmRescoringChunker class, uses the output of the NBestChunker and statistics gathered from the tag transitions in the training data to re-score each of the n best chunkings. This rescoring process produces a better top result than the original first-best Chunker because it incorporates information from longer-range relationships in the text.
In order to make predictions about the most likely chunking for an unlabeled segment of text, the hidden Markov model relies on statistics about the tagged text that are gathered during a training phase. Training was accomplished by converting the XML format provided for the task into a MUC-like format that was readily digested by the “TrainMuc6.java” class provided in the LingPipe distribution. We used the default parameters as supplied except for the number of chunkings rescored, which we doubled from 256 to 512 to yield a modest improvement in precision. Each tagged sentence in the MUC-labeled input is taken by LingPipe’s Muc6ChunkParser class as an independent training observation. In order to allow the model to use contextual cues from the semi-structured format of the medical records, we simply labeled the entire text of each medical record as a single “sentence” as was done with the Carafe systems described above.
Tagging was performed by invoking the “chunk” method of the CharLmRescoringChunker for each section of text to be tagged and writing out each chunk as tagged text in the proper format. Our implementation followed the “NamedEntityDemo.java” example supplied with LingPipe and is embedded in a SAX-based XML parser adapted for this task. No post-processing was performed. This constituted the system labeled LingPipe in ▶.
Results
Evaluation Measures
Phrase extraction tasks can be evaluated at either the phrase-level or token-level. Phrase-level measures assign credit to systems based on whether entire phrases of multiple words (the PHI in this task) appropriately match the answer key. Token-level evaluation assigns credit based on whether each individual word is assigned a type matching the answer key phrase type of the phrase to which they belong, independent of the other words that constitute the phrase. For each of these two evaluation paradigms, systems are evaluated based on precision, recall and F-measure.
Precision and recall are computed as:
where tp is the number of true positives, fp is the number of false positives, and fn is the number of false negatives (i.e., misses) as determined from the system output. F-measure is a metric that balances precision and recall and, for the case where precision and recall are weighted evenly, is computed as:
For the token-based evaluation, tp (true positives) are simply the number of tokens assigned the correct type, fp (false positives) are the number of tokens assigned an incorrect type by the system, and fn (false negative or misses) are the number of tokens that the system fails to assign the appropriate type to. Tokens that have a type according to the answer key, but are mistyped will thus count as both a precision and a recall error. The official score reported for the AMIA challenge task was a weighted token balanced F-score that included all true negatives (unlabelled tokens); this accounts for the extremely high scores reported for this measure (see ▶).
For the phrase-based evaluation, tp is the number of phrases correctly typed and whose extent matches the answer key; fp is the number of phrases proposed by the system that do not match (in both type and extent) a phrase in the answer key; and fn is the number of phrases in the answer key that do not match a phrase in the system response. Thus, a system response phrase overlapping with a phrase in the answer key but with a mismatch in either extent or type will count as both a precision and a recall error. Such a metric effectively doubly penalizes extent and type-mismatch errors. For our results in this paper, we use the MUC phrase scorer 5 which separately penalizes type errors and extent errors. This measure provides partial credit for response phrases matching in extent but having the wrong type or for phrases that have the correct type but only partially overlap the key phrase.
Official Results
▶ shows the results for our three submitted systems on the evaluation data as well as four unofficial runs that were part of our development process. Our first submission, Carafe-ALL, consisted of all available features with a precision-recall bias (described below) tuned to favor entity F-measure (bias of −1.0) based on five-fold cross-validation over the training data. Our second system was the LingPipe system. Our final official system, Carafe-ALL+PP, used Carafe tuned for high recall (bias of −1.5), plus the manually-derived post-processing approach described above to further improve recall.
Our highest official F-measure was attained with Carafe-ALL+PP, achieving a phrase-based F-measure of 0.9736. This system achieved a weighted-F-measure of 0.99745 using the official AMIA Challenge scoring metric, a weighted token F-measure which counted both true positives and true negatives as the basis of the score. Counting only true positives, the F-measure was 0.9755. The token-level metric is useful since even partial matches of entities may be useful in practice. Moreover, token-level precision errors from extent mismatches may not be problematic—this metric effectively gives partial credit for such cases, compared to the phrase level metric. A number of unofficial development system results are also reported in ▶ for comparison against our official runs. These systems are discussed in more detail below.
Carafe Development Experiments
This section describes the development we did for Carafe prior to submitting our official runs. We begin with the performance of the Carafe-I system (see ▶), which was a quick port to this task from a standard Named Entity task, and then we examine the contribution of various task specific features on the performance of Carafe. Note that for all experiments in this paper with Carafe, we set the Gaussian prior, σ, to 10.0 b and all results are reported on the evaluation data for comparison with our official results.
The contextual features used for the Carafe-I system are shown in ▶. The left column indicates the formal description of the feature types and the right column provides example feature instantiations for a fragment of text, “A call to Dr. Smith at 1-5555” where the current position in the sequence is at the word Smith. Regular expression features over words are shown in ▶. These features were developed for a separate Named Entity recognition task, as described above.
Table 2.
Table 2 Contextual Features (Word and Character) Used for Baseline PHI Identification System
| Feature Predicate Type | Feature Instantiations at “Smith” in: A Call to Dr. Smith at 1-5555 | |||
|---|---|---|---|---|
| Word Unigrams | <to> <Dr.> | <Smith> | <at> | <1-5555> |
| W−2w−1 | w0 | w1 | w2 | |
| Word Bi-grams | <Dr., Smith> | <to, Dr.> | <at, 1-5555> | <Dr., at> |
| w−1, w0 | w−2, w−1 | w1, w2 | w−1, w1 | |
| Character affixes up to length 2 at w0 | <S> | <Sm> | <th> | <h> |
| pre(1,w0) | pre(2,w0) | suf(2,w0) | suf(1,w0) | |
Table 3.
Table 3 Regular Expression Features Used for Baseline PHI Identification System
| Reg. Exp. Feature | Reg. Exp. |
|---|---|
| INITCAP | [A-Z].*$ |
| ALLCAPS | [A-Z]+$ |
| CAPS_MIX | [A-Za-z]+$ |
| HAS_DIGIT | .*[0-9].*$ |
| SINGLE_DIGIT | [0-9]$ |
| DOUBLE_DIGIT | [0-9][0-9]$ |
| FOUR_DIGIT | [0-9][0-9][0-9][0-9]$ |
| NAT_NUM | [0-9]+$ |
| REAL_NUM | [0-9]+.[0-9]+$ |
| ALPHA_NUM | [0-9A-Za-z]+$ |
| HAS_DASH | .*-.*$ |
| PUNCTUATION | [∧A-Za-z0-9]+$ |
Task-driven Feature Customizations
Analysis of the errors made by the Carafe-I system revealed two trends: 1) precision was higher than recall by a considerable margin, and 2) performance was poor for LOCATION and HOSPITAL, and only fair for DATE, DOCTOR and PHONE types.
We therefore customized specific features to capture some of the characteristics of this task. We added features consisting of regular expressions that matched particular tokens or token sequences. For example, we introduced regular expressions to match phone numbers, tokens consisting of exactly n digits (for phone numbers or zip codes), tokens with a mix of numbers and letters (often IDs), etc. For capturing larger PHI phrases, such as addresses or certain hospital names, we created patterns that matched larger phrases, such as “<CapWord> Medical Center”. A feature then indicated whether a token was part of a particular pattern. We also added features to capture context including the words (and affixes of words) three to the left and two to the right as well as additional word bi-grams and uni-grams in the surrounding context. Finally, we obtained substantial improvement using a lexicon that included US state names, months, and a large list of English words. The latter list appeared to help with words not seen in the training data that were in fact common English terms. This avoided confusion with unseen words that might be entity names. A summary of the additional regular expressions and contextual features are shown in ▶ and ▶.
Table 4.
Table 4 Task-Specific Regular Expressions Features
| Feature Name | Regular Expression |
|---|---|
| PHONE_REG1 | [0-9][0-9][0-9]-[0-9][0-9][0-9][0-9]$ |
| PHONE_REG2 | [0-9][0-9][0-9]-[0-9][0-9][0-9]-[0-9][0-9][0-9][0-9]$ |
| FIVE_DIGIT | [0-9][0-9][0-9][0-9][0-9] |
| NO_VOWELS | [∧AaEeIiOoUu]+$ |
| HAS_DASH_NUM_ALPHA | .*[A-z].*-.*[0-9].*$ | *.[0-9].*-.*[0-9].*$ |
| DATE_SEP | [-/]$ |
Table 5.
Table 5 Contextual Pattern Features for Capturing Hospitals and Locations
| Example Contextual Patterns |
|---|
| <INITCAP> <Hospital|Health|Care Center|Medical Center|Morgue> |
| <INITCAP> <INITCAP> <Hospital|Health|Care Center|Medical Center|Morgue> |
| <INITCAP> <INITCAP> <INITCAP> <Hospital|Health|Care Center|Medical Center|Morgue> |
| <NAT_NUM> <INITCAP> <Dr.|Blvd.|Ln.|etc.>, <INITCAP>, <INITCAP>, <FIVE_DIGIT> |
To ascertain the utility of these features, we added the lexicon features (called Carafe-I+L) and the additional contextual features, including regular expressions, (called Carafe-I+RC) separately to the Carafe-I system to determine the improvement in performance provided by each of these feature sets. The results of adding these feature groups are shown in ▶. Early runs on the development data did not indicate a strong contribution from the lexicon features, but the lexicon clearly provided a very large improvement on the evaluation data for both precision and recall. There was considerable, but lesser, improvement from the additional regular expressions and contextual features.
Effect of Bias and Summary of Performance
After receiving our official evaluation results, we investigated whether or not we selected the optimal bias for our systems. Using the Carafe-ALL system, we plotted the precision-recall curve obtained by setting the bias to different values. This curve is shown in ▶ together with precision/recall scores for all of the official and unofficial Carafe runs shown in ▶. The Carafe-ALL system using an optimal bias (of −1.75) achieved a balanced phrase F-measure score of 0.9756 which is slightly better than our official best run, Carafe-ALL+PP (with a balanced phrase F-measure of 0.9736). Note that the Carafe-ALL+PP results are slightly below this curve indicating that although post-processing rules do improve recall, this comes at a cost in overall F-measure potential. The Carafe-ALL official run clearly did not have the optimal bias with respect to the evaluation data, showing considerably lower recall.
Figure 2.
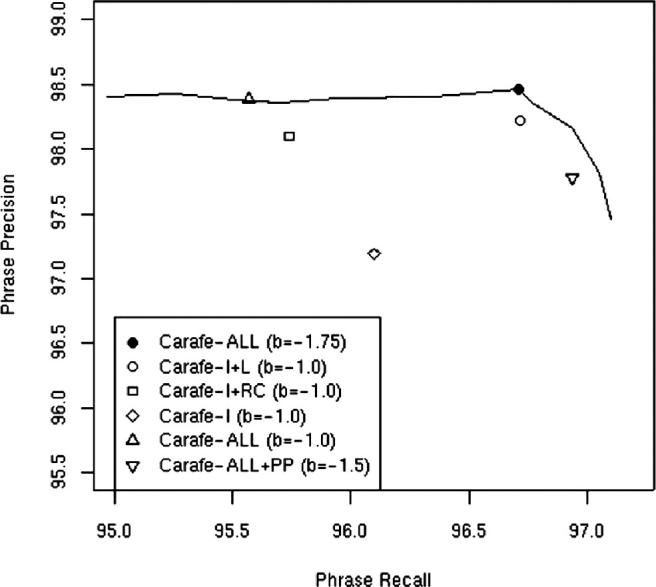
Phrase precision and recall for the official and unofficial Carafe systems and precision-recall curve for Carafe-ALL.
Learning Curves
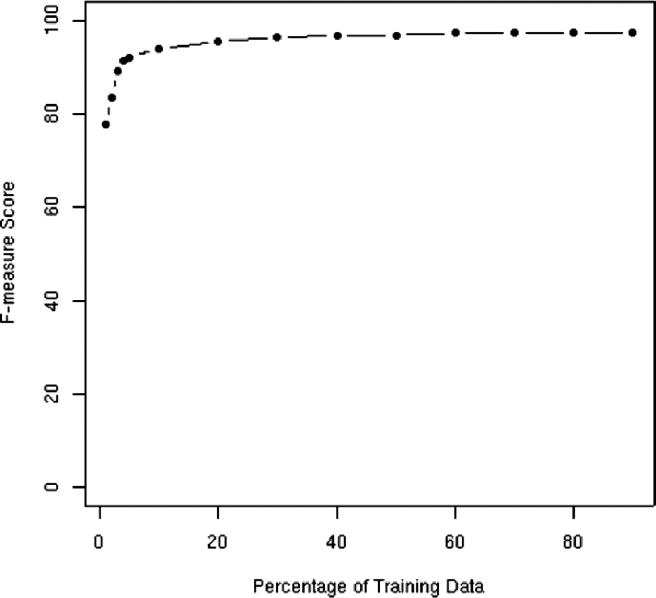
Typically, manual annotation of training data is expensive and time consuming. An important question with any machine learning-based approach is: How much training data is required to achieve a certain level of performance? We address this question in this section by examining the learning curve on the test data. ▶ shows the number of training documents (on the x-axis) and the corresponding F-measure results (on the y-axis) for the evaluation data. The subsets of training documents were selected randomly from the entire training set. These results use the Carafe-ALL system with a bias value of −1.75.
Figure 3.
Learning curve showing the Phrase F-measure score on the evaluation data for systems trained with varying percentages of the entire set of training data.
The results here are somewhat surprising. With only 2 percent of the training data (only 13 medical records), the performance is over 0.80 F-measure. Additionally, various adjacent points on the curve are, in fact, statistically insignificant from each other. For example, the system trained with 60 percent of the training data is statistically indistinguishable from the system trained with 70 percent of the data.
Error Analysis of Carafe System
We performed an error analysis of the output of the Carafe-ALL system. Errors fall into several broad classes. Instances where the system assigned the wrong label to an entity are type errors. An example is “<DOCTOR>Veteran’s Day</DOCTOR>”: the extent of tagged material is correct, but the label should be DATE. Instances of the wrong amount of text (either too much or too little) are extent errors. For example, “<HOSPITAL>Tarcet Health</HOSPITAL> Systems” is an extent error because in the key the full string “Tarcet Health Systems” is tagged HOSPITAL. The other major classes of errors are missing (tag present in key but not in system output), and spurious (tag present in system output but not in key). ▶ shows a summary of all errors produced by Carafe.
Table 6.
Table 6 Errors Produced by Carafe System
| Count | % of Errors | |
|---|---|---|
| INCORRECT | 8 | 3.1% |
| MISSING | 131 | 50.0% |
| SPURIOUS | 40 | 15.3% |
| EXTENT | 83 | 31.7% |
Although the total number of errors produced by Carafe is small, 50% of the errors are missing tags. From the standpoint of protecting PHI, missing tags can result in the disclosure of PHI and are therefore more serious than spurious tags. ▶ shows the breakdown by type of correct, incorrect, missing, and spurious tags produced by the Carafe system (extent errors appear later in ▶).
Table 7.
Table 7 Breakdown of Carafe Correct, Incorrect (Type Errors), Missing (Right Column), and Spurious (Bottom Row) Tags
| Output |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Key | AGE | DATE | DOC | HOSP | ID | LOC | PT | PHONE | Missing | Total |
| AGE | 2 | 1 (33.3%) | 3 | |||||||
| DATE | 1906 | 1 | 24 (1.2%) | 1931 | ||||||
| DOC | 1050 | 20 (1.9%) | 1070 | |||||||
| HOSP | 1 | 645 | 30 (4.4%) | 676 | ||||||
| ID | 1140 | 3 (0.3%) | 1143 | |||||||
| LOC | 5 | 74 | 40 (33.6%) | 119 | ||||||
| PT | 1 | 236 | 8 (3.3%) | 245 | ||||||
| PHONE | 53 | 5 (8.6%) | 58 | |||||||
| spurious | 0 | 22 (1.1%) | 6 (0.6%) | 5 (0.8%) | 3 (0.3%) | 3 (3.9%) | 1 (0.4%) | 0 | 40 | |
| Total | 2 | 1929 | 1058 | 655 | 1143 | 77 | 237 | 53 | 131 | 5285 |
Table 8.
Table 8 Counts of Extent Errors in Carafe Output
| AGE | DATE | DOCTOR | HOSP | ID | LOC | PATIENT | PHONE | Total | |
|---|---|---|---|---|---|---|---|---|---|
| SHORT | 0 | 1 | 0 | 33 | 2 | 3 | 0 | 4 | 43 |
| LONG | 0 | 3 | 16 | 17 | 0 | 2 | 1 | 0 | 40 |
| S&L | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |
| Total | 0 | 4 | 16 | 51 | 2 | 5 | 1 | 4 | 83 |
Correctly tagged items can be seen reading down the diagonal in ▶. Type errors appear in bold text, missing tags appear down the right in italic text, and spurious tags appear along the bottom in italic text. Extent errors are not visible in ▶. Type errors are rare, with only eight occurrences, five of those being LOCATIONs mis-tagged as HOSPITALs (e.g., “…transfer back to Balau for care…” or “returns back to Grovecu And…”).
Spurious tags are also rare (indicating high precision) with the exceptions of DATE (22 spurious tags of of 1929), and LOCATION (3.9% or 3 out of LOCATION tags are spurious). Missing tags occur most often for LOCATION (40 out of 119 tags or 33.6% missed) and HOSPITAL (30 of out of 676 or 4.4% missed), but also affect DATE, DOCTOR, and PHONE). All missed tags are harmful, but the percentages and the percentages for PATIENT (3.3% or 8 out of 245), and ID (0.3% or 3 out of 1143) should be particular cause for concern in a real application setting.
▶ shows a breakdown of extent errors. Numbers in the short row indicate instances where the tag produced by Carafe did not span the entire text of the corresponding tag in the key, while numbers in the long row indicate instances where the tag produced by Carafe spanned more text than the corresponding tag in the key.
More than half of the extent errors occurred on HOSPITAL, with many instances of both long and short errors. For example, the key has an instance of “Box Memorial Hospital Nursing Home” tagged as HOSPITAL, while Carafe tagged just “Box Memorial Hospital” as HOSPITAL (short extent error). An example long extent error on HOSPITAL is “Kentnaalvet Medical Center” (key), versus “Burn Unit on Kentnaalvet Medical Center” (Carafe output). Note that there was one extent error that was both short and long—the Carafe output was offset compared to the key, failing to capture the full key string, but was also longer than the key string. Extent errors for some tag types are skewed in one direction. All of the sixteen extent errors on DOCTOR are long (e.g., “NISTE MARHALT”—key; “SURG AG NISTE MARHALT”—output). There were only 4 extent errors for PHONE, but they were all short (failure to include either area code or extension). A breakdown of precision, recall, and F-measure for each tag type, accounting for all error types, appears in ▶. In much the same way that missing tags are more important that spurious tags for de-identification, short tags are more potentially harmful than long tags, because they expose a portion of PHI. It should be noted however that not all tokens missed because of a short tag are equally problematic. When the missed word is very common (such as “hospital”, “clinic”, or even “John”), there is a very low chance that any real exposure of the patient’s identity will occur. In the final discussion, we consider ways to factor this into the scoring.
Table 9.
Table 9 Phrase Precision, Recall, and f-measure for Carafe-ALL (b = −1.0) Output
| AGE | DATE | DOCTOR | HOSP | ID | LOC | PATIENT | PHONE | Overall | |
|---|---|---|---|---|---|---|---|---|---|
| Recall | 66.67 | 98.71 | 98.13 | 95.41 | 99.74 | 62.18 | 96.33 | 91.38 | 97.50 |
| Precision | 100.00 | 98.81 | 99.43 | 99.08 | 99.74 | 90.24 | 99.16 | 100.00 | 99.22 |
| F-measure | 80.00 | 98.76 | 98.78 | 97.21 | 99.74 | 73.63 | 97.72 | 95.50 | 98.35 |
Discussion
For medical record anonymization, perhaps more than most other tasks, perfect or near-perfect performance is critical. Given the privacy protections required by the Health Insurance Portability and Accountability Act (HIPAA), perfect recall is most essential to prepare the release of de-identified records: false negatives represent unauthorized disclosure of PHI. Systems can, of course, be adjusted to provide extremely high recall, at the expense of precision. The intended application of the materials will dictate whether the loss of precision is tolerable at the required recall.
For the foreseeable future, the best solution for most applications may be an interactive system that combines automated de-identification with human review. It is important to understand that even a fully human process will result in occasional errors and may result in unintentional disclosure of PHI. The aim of a partially automated system would be to make the process much more efficient with negligible danger of revealing sensitive information. One approach to improving the efficiency of human review is to have the person review only those records (or portions thereof) that cannot be confidently marked by automatic means. To do this, it is important for systems to provide well-calibrated confidence estimates along with their output. Records with low confidence estimates will be subject to thorough review; those with high confidence can be reviewed quickly or not at all. This approach is used, for example, in Pakhomov et al., 2006 15 for partially automated assignment of diagnostic codes to medical records.
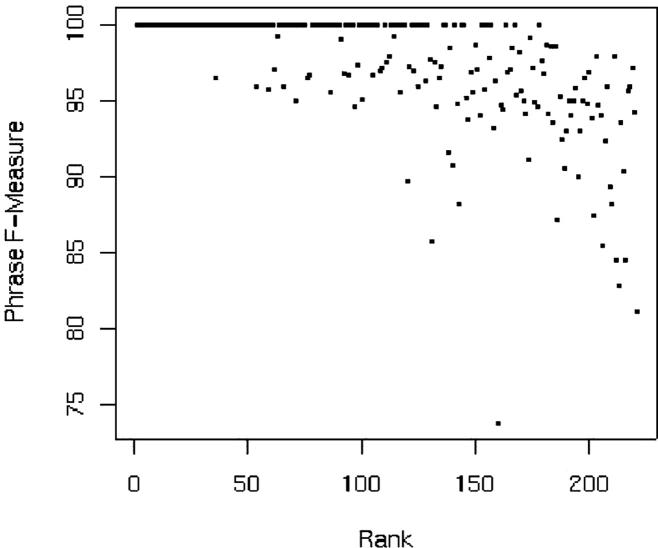
In our system, since we treat entire medical records as single sequences, we were able to obtain the (negative) log-probabilities for each processed record using the forward-backward algorithm in Carafe. ▶ shows how these tagged records, ranked by conditional log-probability, correlate with the F-measure from the Carafe-ALL system. The x-axis indicates the rank for a particular record based on the probability assigned to it. Importantly, the top 35 documents ranked by probability were tagged perfectly. Note also that a record’s length correlates with F-measure (shorter documents tend to having higher scores), but this correlation isn’t as strong as with Carafe’s probability estimates: using Spearman’s rank correlation statistic (rs), we have rs = 0.7267 for rank correlation with log-probability and rs = 0.5392 for rank correlation with record length.
Figure 4.
Scatter plot showing F-measure correlating with rank as determined by negative log-probability.
There are a number of refinements in the task scoring function that might be used to close the gap between system performance and the requirements of a deployed application for de-identification. We hinted at some of these earlier when discussing the types of errors made by our system. Consider first that type errors (e.g., identifying a hospital as a location) would not result in unintentional disclosure of PHI. If the type were used to replace the PHI with synthetic data, the effect of misclassified types would be potentially confusing but it is otherwise innocuous. Next consider extent errors. When the marked extent of some PHI is over-long, no leakage of PHI occurs. In such a case the token recall metric with respect to PHI tokens does correctly reflect the fact that no leakage has occurred, though its effects are largely obscured because that metric is dominated by all of the non-PHI tokens that are correctly “not-tagged”.
A closer examination of the extent errors that are too short suggest several approaches for refining the scoring metric. The bulk (77%) of our short-tag errors occurred with hospitals. The untagged words for these 33 cases occur in two groups: 1) common nouns and phrases which disclose virtually no information about a patient:
Systems (as in “Tarcet Health Systems”)
Services (as in “Lympnanier Health Services”)
Nursing Home (as in “Box Memorial Hospital Nursing Home”);
or 2) proper names that might allow one to deduce the relevant PHI:
Fairm of (as in “Fairm of Ijordcompmac Hospital”)
Ingree and Ot (as in “Ingree and Ot of Weamanshy Medical Center”).
A refinement of the scoring function that distinguished these cases would better represent the system’s true performance at protecting PHI. A more accurate metric, used during system training, might yield improved system performance as well. From these examples, we can infer that we might better judge the cost of not tagging these words by understanding the likelihood that revealing these words would allow one to specifically identify a patient’s PHI. We might model these likelihoods using the “information content” of the words, using, for example, the a priori probabilities of the occurrence of these words in relevant corpora. Words such as “systems,” “services,” and “nursing” are common in most corpora whereas words such as “Fairm” and “Ingree” are not. It should be possible then, to more accurately predict the probability of leaking PHI using information content measures in the scoring metric. There may be a way to use these unigram probabilities during training to improve the recall error rate.
A further consideration of the information content approach could be applied during human review to identify missed tags. For categories of Doctor, Hospital, and Patient, many of our misses include low-probability (in fact, previously unseen) words. In order to catch our misses we could favor showing a reviewer phrases with these high-content terms.
In an interactive system, we would also have an opportunity to explore a machine learning paradigm known as active learning. In active learning, iterative feedback from a human review is used to retrain the automated methods. Examples are selected for review primarily on the basis of their present ambiguity. That is, it is usually most advantageous to seek additional judgments on records (or portion thereof) that will provide the greatest additional discriminatory power to the algorithm. Records whose taggings have low-confidence estimates are often used as review candidates in these systems. Given that our learning curves demonstrate that we quickly reached a region of diminishing returns, active learning might be a key to boosting performance beyond the current level without tagging inordinate amounts of additional data.
Conclusion
De-identification is an important task in medical informatics. Its successful application has the potential to vastly improve research in many areas of medicine by making large amounts of clinical data available to researchers while protecting patient privacy. We see the primary goals of de-identification systems as being: 1) High performance—systems must perform at very high levels of overall accuracy to protect privacy and to avoid removing important medical data; 2) Rapid Retargeting—systems should require limited task-specific feature engineering and as little annotated training data as possible to perform well; 3) Adjustability—systems should be adjustable, to accommodate different evaluation metrics, since, for example, recall may be more important than precision by some degree; 4) Introspectivity—systems should be able to reliably characterize the certainty of their own output to allow for more intelligent review, and corrective annotation of automatically de-identified data; and 5) Interactivity—even very high-performing systems will need to be subject to human review. This requires a system able to interact with users to ensure properly de-identified data. Additionally, interactive annotation would allow the system to improve faster by selecting data for user annotation that, when annotated, offers the best chance of improving the system.
In this paper, we have addressed goals 1–4 in significant ways. We have demonstrated a high performing de-identification system that achieved the best overall results at the AMIA De-identification challenge workshop. Our methodology was specifically focused on rapidly tailoring existing systems for this task with great success: both Carafe and Lingpipe worked well out of the box with little tailoring. Additionally, the learning curve for Carafe indicated that high-performance is achievable on this task with a fraction of the total training data. We introduced a bias parameter in this work that begins to address the notion of cost sensitivity by allowing the system to trade off precision and recall at runtime. Finally, we have demonstrated an introspective system by establishing that Carafe’s confidence scores for its own output correlate well with performance (F-measure). This positions us well to pursue the development of interactive de-identification systems in future work.
Footnotes
The gradient of the log-likelihood is a vector where each component, corresponding to a particular feature, is the difference between the observed frequency of that feature in the training data and the expected frequency of that feature according to the current model (i.e., the current set of weights). These feature expectations can be computed efficiently using the forward-backward algorithm. See Sha and Pereira, 2003 8 for details.
We carried out a number of experiments with different Gaussian prior values and noticed remarkably little difference in the results with different values on these data.
References
- 1.Uzuner O, Luo Y, Szolovits P. Evaluating the State-of-theArt in De-identification J Am Med Inform Assoc 2007:14. [DOI] [PMC free article] [PubMed]
- 2.Wellner B. Weakly Supervised Learning Methods for Improving the Quality of Gene Name Normalization Data ACL-ISMB joint workshop on linking literature, information and knowledge for biology BioLINK. 2005. Detroit, MI, June.
- 3.Morgan AA, Hirschman L, Colosimo M, Yeh AS, Colombe JB. Gene name identification and normalization using a model organism database J Biomed Inform 2004;37(6):396-410. [DOI] [PubMed] [Google Scholar]
- 4.Sibanda T, Uzuner O. Role of Local Context in De-identification of Ungrammatical, Fragmented Text North American Chapter of the Association for Computational Linguistics (NAACL-HLT 2006). 2006. New York, NY.
- 5.Sundheim B. Overview of the Results of the MUC-6 Evalution. Proc. of the 6th Mess Understand Conf (MUC-6). Defense Advanced Research Projects Agency; 1995.
- 6.Lafferty J, McCallum A, Pereira F. Conditional Random Fields: Probabilistic Models for Segmenting and Labeling Sequence DataSan Francisco, CA: Morgan Kaufmann; 2001. pp. 282-28918th Int Conf Mach Learn 2001.
- 7.Wellner B, Vilain M. Leveraging Machine-Readable Dictionaries in Discriminative Sequence Models Lang Res Eval Conf (LREC 2006). 2006. Genoa, Italy.
- 8.Sha F, Pereira F. Shallow parsing with conditional random fields Proc HLT-NAACL. 2003.
- 9.Krishnan V, Manning CD. An Effective Two-Stage Model for Exploiting Non-Local Dependencies in Named Entity Recognition Conf Assoc Comp Ling. 2006. Sydney, Australia.
- 10.McDonald R, Pereira F. Identifying Gene and Protein Mentions in Text Using Conditional Random Fields BMC Bioinform 2005;6(Suppl 1):S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lafferty JD, McCallum A, Pereira FCN. Conditional Random Fields: Probabilistic Models for Segmenting and Labeling Sequences ICML 01: Proc 18th Int Conf Mach Learn. San Francisco CA, USA: Morgan Kaufmann; 2001. pp. 282-289.
- 12.Byrd RH, Lu P, Nocedal J. A Limited Memory Algorithm for Bound Constrained Optimization SIAM J Sci Stat Comp 1995;16:1190-1208. [Google Scholar]
- 13.Minkov E, Wang RC, Cohen WW. NER Systems That Suit User’s Preferences: Adjusting the Recall-Precision Trade-off for Entity Extraction 2006. HLT/NAACL.
- 14.Carpenter B, 5th ACL Chinese Special Interest Group (SIGHAN) Character Language Models for Chinese Word Segmentation and Named Entity Recognition 2006. Sydney, Australia.
- 15.Pakhomov SVS, Buntrock JD, Chute CG. Automating the Assignment of Diagnosis Codes to Patient Encounters Using Example-based and Machine Learning Techniques J Am Med Inform Assoc 2006;13(5):516-525. [DOI] [PMC free article] [PubMed] [Google Scholar]