Abstract
The SAGE (Standards-Based Active Guideline Environment) project was formed to create a methodology and infrastructure required to demonstrate integration of decision-support technology for guideline-based care in commercial clinical information systems. This paper describes the development and innovative features of the SAGE Guideline Model and reports our experience encoding four guidelines. Innovations include methods for integrating guideline-based decision support with clinical workflow and employment of enterprise order sets. Using SAGE, a clinician informatician can encode computable guideline content as recommendation sets using only standard terminologies and standards-based patient information models. The SAGE Model supports encoding large portions of guideline knowledge as re-usable declarative evidence statements and supports querying external knowledge sources.
Introduction
In recent years, clinical guidelines and protocols have gained support as vehicles for disseminating evidence-based best practices in clinical medicine, with the aim of improving quality of care and reducing costs. Unfortunately, the traditional method for disseminating clinical practice guidelines (CPGs) as educational documents is not effective in improving behavior. 1 Furthermore, the delay in bringing clinical research into daily practice is excessive. 2 To overcome these problems, a number of groups have developed models for marking up or encoding CPGs. 3–10 The expectation is that machine-readable formats will facilitate delivery of context-sensitive guideline content to clinicians. However, the technology for delivering situation-specific decision support to clinical settings has not matched the increased flow of guideline production. Organizations implementing guideline-based decision-support systems either create custom software or use commercial packages that are often little more than if-then rules. Researchers developing sophisticated guideline modeling formalisms and execution software are generally forced by practical and technical limitations to confine the use of their technology to their home institutions.
To demonstrate the feasibility of widespread, interoperable dissemination of guideline-based care in commercial clinical information systems (CIS), the SAGE Project (Standards-Based Active Guideline Environment) formed a consortium consisting of medical informatics groups at GE Healthcare, the University of Nebraska Medical Center (UNMC), Intermountain Health Care, Apelon, Inc., Stanford University, and the Mayo Clinic (Rochester, MN). During a four-year project, we created the methodology and technological infrastructure for standards-based encoding and interoperable deployment of computable guideline content.
There are formidable challenges to sharing computer-interpretable guidelines across multiple institutions and integrating guideline-based clinical decision support (CDS) into vendor CISs. A decision-support application may be designed as a separate system but must be able to query computerized patient data and knowledge sources and deliver decision-support interventions through features available within a vendor’s CIS. To be effective, guideline CDS must be invoked at an opportune moment in the clinical care process and must facilitate clinical workflow non-intrusively. The guidelines must provide rich decision support, yet must be efficient and allow easy inspection of the underlying clinical logic. This paper focuses on innovations in guideline modeling that the SAGE project devised to meet these challenges.
Background
The SAGE project builds upon previous work on guideline modeling (including Asbru, 6 GEM, 9,11 GLIF3, 7,12 EON, 10 PROforma, 4 GUIDE, 8 and PRODIGY 5 ). It advances the state of the art by focusing on requirements that previous models have not met simultaneously: A) incorporation of workflow awareness, B) employment of information and terminology standards, C) incorporation of simple flow-of-control standards, and D) attention to integration with vendor CIS. These requirements characterized and guided our approach in developing the SAGE Guideline Model. a
Workflow
The success of clinical decision-support systems (CDSSs) depends heavily on their integration into the workflow of the care process. 13,14 SAGE DSS does not control the host system’s workflow. Rather, it responds to opportunities for decision support in the care process, as suggested by Osheroff et al. 15 The guideline model must formalize sufficient workflow context to enable clinical and administrative events to trigger SAGE DSS and for it to then deliver appropriate recommendations through CIS facilities. For example, a physician might see an “inbox” notification of guideline-based recommendations or be presented with an order set for a patient. A nurse might receive a guideline flowsheet reminding her to chart against immunization orders.
This approach links knowledge to appropriate clinical contexts by an event-driven system. The system monitors and responds to the care setting with knowledge of provider roles and the CIS capabilities. Instead of merely reformulating a published clinical practice guideline, the SAGE model formalizes guideline knowledge to capitalize on workflow awareness. While the event-driven approach to workflow integration is not new in the clinical decision support literature, 16,17 most previous guideline modeling approaches failed by either (1) leaving the workflow context outside the guideline model (EON, 10 PRODIGY, 5 and Asbru 6 ), (2) considering workflow integration to be a matter of adapting fully encoded guidelines (GLIF3 7 and GEM 11 ), or (3) attempting to detail every element of workflow (the University of Pavia’s Careflow methodology). 8
Information Standards
The Department of Health and Human Services is aggressively promoting information model standards such as Electronic Health Record standards based on Health Level Seven’s Version 3 Reference Information Model (HL7 v3 RIM), 18 and reference terminology standards including the SNOMED Clinical Terms® 19 and LOINC. 16 This has created an opportunity to create a guideline model that could achieve the next step in semantic interoperability for clinical knowledge.
We incorporated the HL7 v3 data types directly into our model and worked with HL7 to define a RIM-based view of patient data appropriate for clinical system modeling. Reference terminologies such as SNOMED CT allowed us to use advanced terminological features including subsumption relationships and post-coordination of guideline concepts. Employing content standards for encoding guidelines is not simply a process of linking guideline concepts with published code sets, as existing standards do not satisfy every requirement of guideline modeling. Thus, examining deployment requirements for standards in guideline modeling and quantifying them are major deliverables of the SAGE project. Previous approaches either did not account for standard terminologies and information models (e.g., EON, 10 Asbru, 6 PROforma, 4 and GEM 9 ) or did not systematically investigate the implications of binding standards to vendor information systems. GLIF3, for example, adopted a version of HL7 v3 RIM as its data model and used controlled terminologies. However, it lacked mechanisms for managing limitations of pre-coordinated content and for defining the precise model of meaning required by the guideline. 7
Flow-of-Control Standard
A distinguishing feature of multi-step guideline modeling languages is their use of a network of tasks to express the flow-of-control specification in guideline recommendations. 20 SAGE’s initial requirements for this task include (1) a formalism for specifying activities required in complex decision making, (2) an expressive process model that allows sequencing, repetition, and concurrency of decisions and activities, and (3) a clear and understandable meaning for control-flow constructs. Precise specification of control-flow can be subtle and difficult. 21 We synthesized the task-network models of previous guideline modeling languages and reflected on the Workflow Management Coalition’s process definition model. 22 One overriding concern was ease of clinical encoding of control-flow in guideline-directed activities.
Vendor Collaboration
With the leadership of GE Healthcare and the collaboration of the University of Nebraska Medical Center and the Mayo Clinic, we integrated guideline-based decision support into an enterprise CIS. The diversity of GE clinical implementations allowed direct assessment of interoperability at multiple sites. We determined that SAGE DSS should augment a CIS using a standard set of CIS actions and capabilities, and should not require modification of the CIS. Further, because SAGE will be deployed in an existing enterprise setting, it should not duplicate existing system capabilities. In particular, it should use existing external knowledge sources as needed and should use CIS resources such as order sets as vehicles for decision support. No other guideline modeling formalism has attempted to develop general principles for resolving these integration problems.
Method
We used Protégé, 23 an open-source knowledge base modeling environment developed at Stanford University, as the vehicle for guideline representation. We initially synthesized previous modeling work to create a prototype of the SAGE Guideline Model. To ensure that SAGE computer-interpretable guidelines addressed decision-support requirements within the clinical workflow, we developed a deployment-driven guideline encoding methodology. 24 For each guideline, clinicians created workflow-aware decision support scenarios to be aided by the guideline via recommendation sets. After many cycles of encoding, testing, and evaluation, the SAGE Guideline Model satisfied the requirements for implementing the following:
• Immunization: based on comprehensive recommendations for immunization issued by the Center for Disease Control25
• Diabetes: based on primary care practices from the Standards of Medical Care in Diabetes—200626
• Diabetic hypertension: based on the Seventh Report of the Joint National Committee on Prevention, Diagnosis and Management of Hypertension27
• Community acquired pneumonia (CAP): based on the Practice Guidelines for the Management of Community-Acquired Pneumonia in Adults.28,29 The SAGE CAP guideline also supports compliance with the JCAHO Pneumonia Core Measure Setb and reminders for key care events.
Model Description and Illustrative Example
As an example scenario, a diagnosis of CAP is made at an outpatient clinic. SAGE detects this event when the diagnosis is added to the patient’s Problem List. The SAGE Engine accesses the patient’s clinical record, queries the clinician for additional data, and then computes and displays the Pneumonia Severity Index (PSI). c The PSI is used to place a patient into an appropriate level of in- or outpatient care. SAGE provides these recommendations, and for inpatients, it provides an order set specific to the patient, with the orders pre-selected and annotated where appropriate.
Workflow Context and Invocation of the DSS
The need for tightly-coupled, real-time interaction between guideline-based CDS and clinical workflow in a host CIS required that we encode specific scenarios of clinical usage. An encoded scenario creates two key requirements for workflow-aware modeling of guideline content. First, it defines the workflow context and trigger events that become opportunities for intervention. Second, it specifies decision-support services and information vehicles to be used. These might include a documentation reminder, test summary, or a patient-specific order set. As we have described, 24 after decision-support scenarios are formulated, clinicians and informaticians analyze guidelines and medical literature to distill and disambiguate knowledge required for decision-support services, identify medical concepts required by the guideline logic, and map them to terms in standard terminology.
Events triggering SAGE-CIS interactions may originate as events in the Patient Care process, be generated by a SAGE guideline action, or be driven by the clock in order to generate periodic population-based surveillance. In the first instance, to identify characteristics of the workflow context in which decision-support interventions occur, Patient Care events are defined by an agent in a role performing an act on an object for a patient in a setting. An example of such an event may be a physician accessing the record of a patient in an emergency room. SAGE encodes the role, act, object, and setting properties using terminological codes. Possible values of the role property, for example, are constrained to be SNOMED CT codes for medical practitioners (concept code 158965000 and its descendants).
Events generated by guideline action specifications may occur within a guideline scenario. In the CAP protocol, for example, if JCAHO requirements have not been met at time of admission, SAGE generates an event that triggers repeat checking in 30 minutes.
Finally, in population-based scenarios, a SAGE guideline may specify periodic checking of all eligible patients to determine if a reminder or report should be generated. For these events, the SAGE Guideline Model uses the HL7 version 3 Periodic Interval of Time data type to specify recurring events that must be generated at specific intervals.
In our CAP example, the SAGE DSS is activated when the pneumonia diagnosis is recorded. SAGE determines the relevance of any guideline to the patient through enrollment criteria that must be met for a guideline to be applied. The SAGE Guideline Model organizes guideline interventions in terms of Recommendation Sets. Each set has Context instances linked to the event triggers and may have preconditions restricting relevant patient and management states. In this case, “Community Acquired Pneumonia” must be on the problem list and the PSI score should not be in the medical record.
Standard Terminology, Patient Information Model, and Clinical Expressions
Once the SAGE DSS is triggered, it must interact with patient data from the CIS. To support localization at multiple institutions, SAGE guidelines are encoded using a normalized representation of patient data. Given the labor-intensive process of encoding a guideline and validating it, it is more cost effective to map an institution’s data source to a standardized patient data model than to tailor guideline encoding to the idiosyncrasies of every institution’s data format. The latter process defeats interoperability, as it requires that a DSS’s execution engine be modified to reflect each local data representation.
Semantic interoperability of clinical data requires standardization of the data types, terminology, information models, and conventions for expressing clinical statements. 30 SAGE uses HL7 version 3 data types as the basis for its modeling. 31
Standard Terminologies and Extensions
Knowledge bases created for SAGE use the core vocabulary resources of SNOMED CT, LOINC, and National Drug File—Reference Terminology (NDF-RT). 32 As we have described elsewhere, 24 the SAGE project delineates several levels of use of standard terminologies, and also identifies a suite of terminological services that are required both for encoding guideline knowledge bases and executing them.
At the simplest level, a concept in a guideline corresponds exactly to a pre-coordinated term in a standard terminology (e.g., alcohol abuse; SNOMED CT 15167005). In this case, the terminology service allows the guideline knowledge base to reference the concept, and, at run time, to identify all concept instances stored in the CIS subsumed by it. For example, if a patient’s medical record indicates persistent alcohol abuse (SNOMED CT 284591009), the terminology service will return the more specific condition when the SAGE DSS queries about alcohol abuse in a patient’s problem list.
Next are guideline concepts that are not pre-coordinated. Compositional terminologies like SNOMED CT allow logical definition and modeling of novel concepts. For example, contaminated wound lesion (finding) can be defined by extending a wound lesion (finding) (SNOMED CT 23915507) with an associated morphology (SNOMED CT 116676008) of contaminated laceration (SNOMED CT 62604006). Extension concepts required to encode a recommendation set are included with the guideline knowledge base as concept definitions. They are meant to be included in the vocabulary services for run-time support. Each is assigned an extension identifier and classified within the concept taxonomy. Concepts not fully definable using the terminology elements are given an extension status of primitive.
Alternatively, some concepts can be expressed as Boolean combinations of existing concepts. For example, one risk factor in the calculation of the PSI is congestive heart failure (CHF). SAGE clinicians determined that CHF-related concepts affecting the CAP risk corresponded to the concept congestive heart failure (SNOMED CT 4234307) and its descendants but did not include pleural effusion due to CHF (SNOMED CT: 90727007). To specify Boolean combinations over terminologies with hierarchically organized content, SAGE defines concept expressions as terminological expressions composed using the set operators AND, OR, and NOT. In this expression language, atomic expressions consist of concept identifiers that denote sub-taxonomies rooted at the identified concept. For instance, in ▶, the root concept congestive heart failure denotes the SNOMED CT concept congestive heart failure and all of its sub-concepts.
Figure 1.
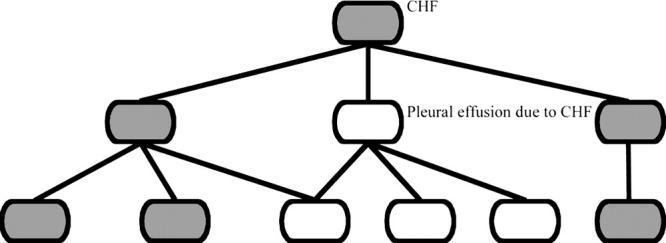
Terminology hierarchy showing terms defined by the expression CHF ∧ (¬Pleural effusion due to CHF).
Composite expressions are formed from atomic expressions via logical operators. AND and OR are interpreted as set intersection and union respectively. Negation is interpreted as difference from root concept(s). In ▶, CHF affecting the CAP risk (shaded nodes) is interpreted as sub-concepts of congestive heart failure that are not part of the pleural effusion due to CHF tree.
At the time of guideline encoding, a terminology server should support post-coordination of new concepts when supported by the terminology or via Boolean concept expressions. When an encoded guideline is used for decision support at the point of care, the SAGE DSS queries a terminology server to test subsumption of concepts in patient data with those encoded in the guideline.
Information Model
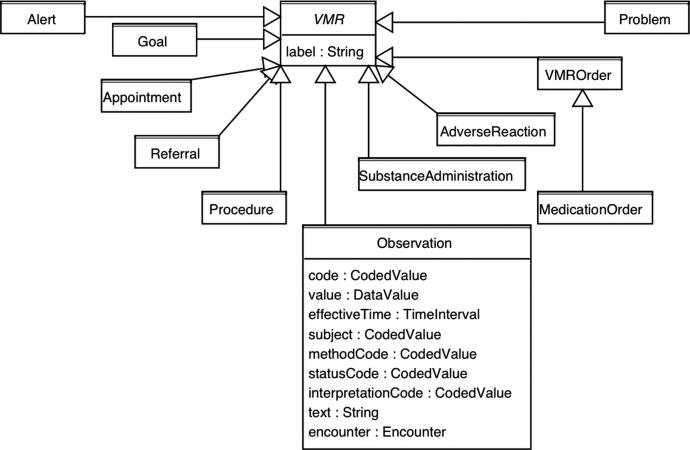
SAGE represents clinical data by adapting the idea of virtual medical records (VMR) from the EON and PRODIGY projects, 33 and extends them to use the HL7 v3 RIM. 18 A VMR is a simplified patient information model using classes and attributes of patient data that pragmatically reflect current clinical information system designs and their relevance to encoding of guidelines for CDS. A patient’s address, for example, is unlikely to factor into guideline recommendations, and is thus omitted from a VMR.
▶ shows the VMR class hierarchy and details of the Observation class. The SAGE Observation class corresponds to a specialization of the Observation class in the HL7 v3 RIM, where the mood code equals EVENT and properties like negationInd and confidentiality code have been omitted. Other VMR classes use similar simplification of RIM classes.
Figure 2.
SAGE VMR classes and details of the Observation class.
The use of standard data types, terminologies, and an information model such as a RIM-based VMR are necessary, but not sufficient for semantic interoperability of patient information. Data can still be represented in multiple equivalent ways. An observation about supine systolic blood pressure, for example, might logically be expressed in two ways (▶). Such multiplicity would make automated clinical decision support unfeasibly complex.
Figure 3.
Alternative representations of Supine Systolic Blood Pressure.
To specify the unique data representation expected in SAGE, we introduced a Clinical Expression Model (CEM) that describes the data element used in the guideline encoding. A CEM is a specialized VMR class whose possible property values are further restricted by concept-specific constraints. For example, a SAGE CEM for a supine blood pressure observation may be a subclass of Observation where the code must be the term lying systolic blood pressure (SNOMED CT: 407556006) or its descendants. Furthermore, the value of the observation must be Physical Quantity instances whose units are mm Hg (SNOMED CT: 259018001). Thus, this CEM makes the representation for observations about supine systolic blood pressure in the SAGE guideline knowledge base explicit and unique. A CEM defines the relationship between information model and terminology similar to the Code Binding Interface proposed by Rector et al., 34 albeit with less formal rigor.
In order to support interoperable data queries, there must be agreement regarding the SAGE VMR’s classes and attributes. The SAGE Guideline Model provides a simple, yet effective, query construct constrained by the VMR. By creating a partially specified instance of a VMR class as part of a query template (e.g., an Observation instance whose code is for a serum creatinine laboratory test result), a domain expert can easily specify a query for data matching the template (e.g., select the most recent value of serum creatinine test results).
Expression Language
By employing HL7 data types, the VMR patient information model, standard terminologies and conventions for their use, SAGE supports unambiguous specification for patient information queries required for guideline encoding. In order to write decision criteria, formulas and other expressions employing CIS data, we use GELLO as the foundation of SAGE’s expression language.
GELLO is the common expression language standardized by HL7. 35 It is a generic language that can be used with any object-oriented data model. However, it is complex and string-based, and difficult for someone without technical training. To allow SAGE guideline encoders to easily author computable expressions, we introduced classes that serve as expression templates. They include variables with derivation expressions, functions which implement a very limited subset of GELLO, and queries and criteria that serve as components of stereotypical expressions. For example, an expression for a PSI score can be built as a function object that holds a GELLO expression referencing variables such as Presence of CHF risk. This variable in turn employs a Presence Criterion that tests for an Observation whose code is a descendant of CHF disease affecting CAP risk—a Boolean concept expression that resolves into a list of applicable CHF diagnoses (▶). d With these templates, encoders can easily specify common queries and decision criteria without concern for expression language syntax.
Figure 4.
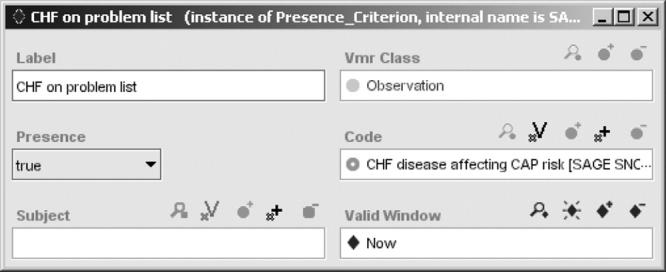
A SAGE expression template specifying a criterion testing for the presence of congestive heart failure observation during an encounter.
Not all institutions use reference standard vocabularies, although one reward for their use includes “plug-and-play” implementation of interoperable guidelines such as those from SAGE. For the run-time evaluation of decision criteria and queries in the setting of legacy vocabulary use, we developed a format for specifying mappings between the standards and an institution’s local terminologies. Queries specified in the standard formalisms are resolved via the mappings into queries on the local information sources. The results are translated into the standard format through a suite of web services.
Recommendation Sets
Standards for expression language, information model, terminologies and event definition are necessary building blocks for encoding guideline recommendations. Recommendation statements are the fundamental assertions of guidelines. We employ the organizing concept of a recommendation set, defined as a collection of related recommendations applicable in one or more shared contexts and organized according to a computable formalism. A context is characterized by a combination of clinical settings (e.g., outpatient encounter in an emergency room), triggering events (e.g., a patient checking into a clinic or entry of a newly diagnosed problem), care providers for whom recommendations are directed, and relevant patient states (e.g., a patient diagnosed with community-acquired pneumonia and not yet having a PSI score). Within each context, a recommendation may describe the preferred choice of a management decision (e.g., whether to manage the patient as an in- or outpatient), or it may specify performance of a series of actions (e.g., recording JCAHO measures such as pneumococcal and influenza vaccinations and previous antibiotics). A recommendation set specifies how computable decisions and actions are related to each other to implement the guideline recommendation.
A single guideline may encompass multiple recommendation sets. For example, the SAGE CAP guideline has three top-level recommendation sets. Each is organized as a use case scenario:
• Upon diagnosis of CAP in an outpatient setting, calculate PSI score and determine CAP therapy disposition based on score.
• Upon hospital admission, monitor critical JCAHO measures, such as timely administration of antibiotics and blood test. Offer an admission order set to the admitting clinician.
• During hospital stay, determine if patient is ready for discharge.
Recommendation sets are modeled as either Activity Graphs that represent guideline-directed processes or Decision Maps that represent recommendations involving decisions at one point in time. The details of the recommendation set representation have been described. 36, 37
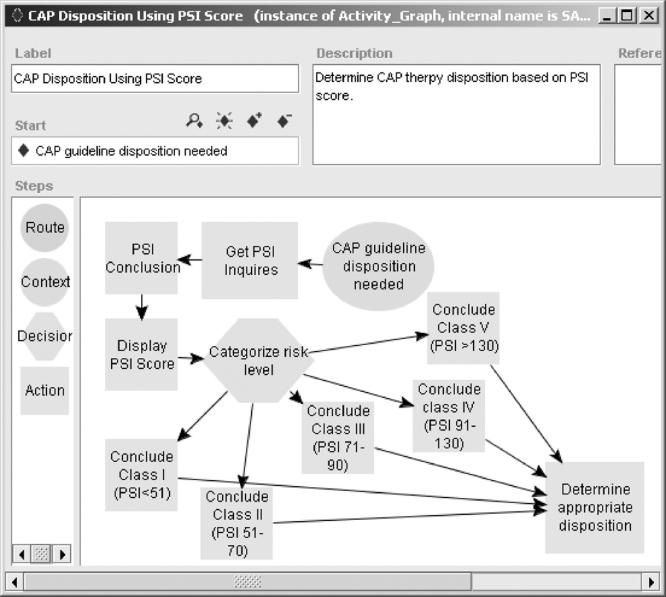
▶ shows a partial Activity Graph specifiying the SAGE DSS’s activities in supporting the management of a patient diagnosed with CAP in an outpatient clinic. The Context node (“CAP guideline disposition needed”) contains a triggering event that activates the SAGE DSS upon diagnosis. The subsequent action node (rectangular icon) initiates retrieval of the data items needed to calculate a PSI score. SAGE queries the clinician if there is no recent data, such as no patient observation for CHF disease affecting CAP risk (see ▶). The clinician’s response is an observation that conforms to the specification of the VMR. Upon data collection, the PSI score is computed using a GELLO expression and the result is displayed. The system then categorizes the patient in terms of CAP risk levels, and calls a subguideline (labeled “Determine appropriate disposition”) to determine the best therapy choice.
Figure 5.
Activity graph specifying SAGE DSS’s response to decision-support opportunities in the clinical workflow.
The subguideline is a Decision Map for determining the disposition of the CAP patient based on the risk class determined via the PSI score, as defined in the original paper guideline. The decision is linked to a set of alternatives: outpatient treatment or ICU/non-ICU admission and orders. A decision model supports the determination of preferences for each alternative.
The current SAGE decision model derives from PROforma, 4 GLIF3, 7 EON, 10 and PRODIGY. 5 It expresses preferences for an alternative as a for-against argumentation structure. For example, the structure for recommending ICU admission contains two strict rule-out criteria (patient’s PSI class is II or III). If any apply to a patient, ICU admission is ruled out. Assignment to PSI class V is the strict rule-in criterion and assignment to PSI class IV is the rule-in criterion. If the number of applicable strict-rule in criteria is greater than the recommendation threshold (in this case, one), the alternative is recommended. If only rule-in criteria are applicable, the alternative is not recommended but remains an option for manual selection by a clinician. In all cases, the resulting recommendation is viewed by the clinician as additional information for consideration.
A guideline-based recommendation includes actions specifying interventions (e.g., alerts and prescription recommendations) to be communicated to the host CIS. In the SAGE Guideline Model, the VMR constrains action specifications that interact with the patient record. The basic set of actions includes:
• Conclude: asserts instances of an Observation, a Problem, or a Goal derived by the system to the CIS medical record or to the SAGE transient data store.
• Retract: retracts instances of concluded Observations, Problems, or Goals.
• Display: shows instances queried from VMR, evaluated expressions, or reference materials to some addressee using a particular communication mode.
• Inquire: requests information, specified by Clinical Expression Models, from some addressee.
• Recommend: proposes specific orders, referrals, appointments, or order sets.
• Notify: sends a text message with some priority to an addressee, using a communication mode.
• Generate event: creates instances of events which may trigger context to spawn future guideline activity.
Because the SAGE DSS does not control the workflow in the clinic, it communicates decision-support actions to the CIS, depending on the CIS to perform the actions for appropriate users or to provide appropriate services.
Integration with Order Sets
In a clinical enterprise, order sets may be integrated into the CIS as a vehicle through which complex patient care plans are coordinated and delivered. To support such an environment, guideline decision support should be able to manipulate order sets hosted by the CIS. The decision support we have tested in SAGE includes the use of order sets that may be dynamically modified and annotated during guideline execution according to specific patient situations.
Order sets are “orders linked in sequences that can be invoked to generate many orders quickly.” 38 To augment order sets with guideline-based and patient-specific decision support, the SAGE Guideline Model assumes that (1) there is an external collection of reference order sets, implemented as eXtensible Markup Language (XML) documents that conform to a standardized order set schema, and (2) SAGE can reference specific order items in an order set by their unique identifiers and offer decision-support services by modifying the content of the order item. Specifically, a SAGE guideline knowledge base may contain logic to:
• Select among alternative pre-defined order sets;
• Pre-select an item or a set of related items within an order set for the physician;
• Compute patient-specific annotations for orders and combinations of orders.
HL7 is developing a standard representation for order sets. 39 In the HL7 standards proposal, an order set includes Boolean combinations of order items that are defined by a RIM-compliant specification of Orders, Medication orders, Observations and Goals. An order item may also be a pointer to an order set, allowing the nesting of order sets.
Because of the need to compute annotations and pre-select order set items based on patient-specific information, the representation of order sets in the SAGE Guideline Model corresponds to, but is not identical with, the proposed standard representation. First, a SAGE guideline knowledge base does not encode the entire order set that is available externally. Instead, a guideline encoder represents solely order items for which SAGE will provide decision support. Second, when representing an item in the order set, the SAGE formalism specifies (1) a Boolean preference condition that, if evaluated to true, pre-selects the item as a default in the order set, and (2) one or more order-alerting or explanation text annotations which may incorporate dynamically generated patient-specific information. The order item in ▶ will be pre-selected if the pre-selection preference criterion (“No CAP antibiotic therapy in past 3 months”) evaluates to true. In addition, the order-alerting (or explanatory) text regarding the need to check recent antibiotic therapy will be added to the order item.
Figure 6.
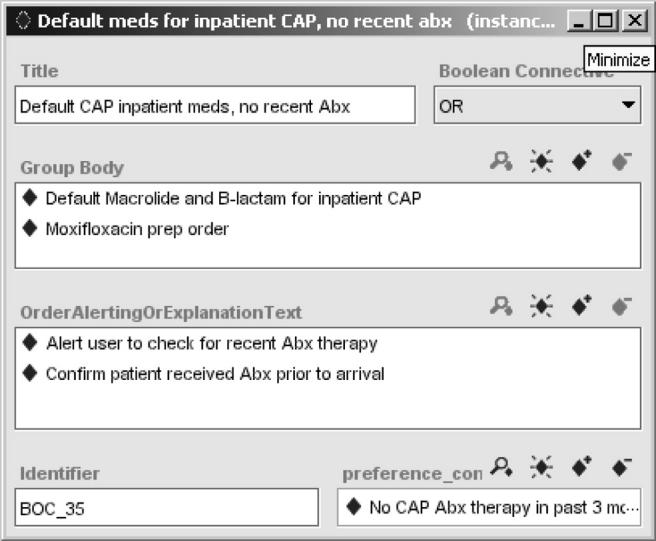
A SAGE order set shown in the Protégé editor. The order set specifies alternative (OR Boolean connective) drug orders for managing community-acquired pneumonia.
In the CAP example, the SAGE DSS may recommend non-ICU admission because of the patient’s PSI score. The SAGE DSS evaluates all the pre-selection preference criteria and order-alerting text, and generates a patient-specific XML file that is merged with the external order set to produce an annotated order set.
▶ shows an example of such an order set. The condition for pre-selecting the default CAP inpatient medication order (no recent antibiotics) holds in the patient, and the system generates a message requesting confirmation of no previous antibiotic therapy. Furthermore, it selects Macrolide plus Beta-lactam as the preferred antibiotic combination, and provides a warning about interactions between its recommendation and the warfarin the patient is already taking. It deselects moxifloxacin due to allergy, and due to a relative contraindication to fluoroquinolones, in this case epilepsy.
Figure 7.
The Medication section of the CAP order set shows checked pre-selection flags and patient-specific annotations.
Evidence Statements
We have shown that effective decision support for medication orders requires detailed information about properties of the medications. Some decision-support services, such as drug-drug interactions and drug-allergy checking, may be provided by capabilities in the enterprise CIS. Much of this specialized medical knowledge may already exist in knowledge bases commonly used in vendor CIS software. A developer of guideline-based DSS faces two challenges: (1) integrating similar knowledge from guideline and non-guideline references, and (2) querying available knowledge in external knowledge sources.
For the SAGE project, we concluded that the DSS needed to assess drug-drug interactions and drug-allergies prior to making recommendations; otherwise the CIS drug-drug/drug allergy checking within its medication module could potentially negate SAGE recommendations. We developed two methods to deal with this challenge: Evidence Statements and Virtual Knowledge Bases.
The SAGE Guideline Model does not mandate a particular division between knowledge that should be encoded in a guideline knowledge base, and that which should be externally supplied. The division is likely guideline- and application environment-specific. Information in a generic drug knowledge base may overlap with that presented in the guideline. The SAGE project defines the concept of Evidence Statements as relationships between clinical conditions and interventions, with associated contextual information and supporting references. 40 An Evidence Statement allows SAGE to encode a statement such as “In managing community-acquired pneumonia, presence of epilepsy is a relative contraindication for Moxifloxacin; reference: Micromedex and Epocrates).” Evidence Statements require that relationships such as relative contraindication be modeled as terminological concepts populating the relationship property of the Evidence Statement. Patient conditions such as presence of epilepsy are encoded as Boolean expressions referencing states that may be in the patient record.
Evidence Statements have these characteristics; they (1) assert that some relatively simple relationships exist between patient conditions and possible interventions, with no flow-of-control or behavioral assumptions, (2) allow statements from multiple sources to be represented in the same format, and (3) can be authored and maintained by clinician-informaticians with minimal training in the modeling tool. For SAGE’s hypertension-in-diabetes guideline, we formulated many of the recommendations extracted from JNC 7 as Evidence Statements. 40 Similarly, for the CAP guideline, we collated drug information from various sources and represented them as Evidence Statements. These statements can be used as knowledge elements in more abstract and maintainable algorithms for determining preferred alternatives in guideline-directed decision-making. In the order set example above, they are used to generate explanatory messages prioritizing antibiotic choices.
The Evidence Statements are declarative in that they include no implied actions or prescribed behaviors. Like records in a database, they can be queried using a structured query language. To specify queries on Evidence Statements, we defined a query template to support the retrieval of instances of Evidence Statements for which the patient condition evaluates to true or false. For example, a query may involve finding all instances of Evidence Statements applying to CAP which are relative contraindications to beta-lactams. The simple query template significantly eases the encoding burden. These queries are incorporated into decision criteria supporting a variety of guideline-directed decisions and actions.
Virtual Knowledge Bases: Medication Knowledge
For accessing knowledge sources external to the guideline knowledge base, we adopted the same approach we used to query patient data. Thus, we defined an information model, called the Virtual Knowledge Base (VKB). VKB is analogous to the VMR, and specifies the structure of external information that can be queried by SAGE. In integrating the SAGE DSS into a CIS, queries to the VKB must be translated into queries understood by the real knowledge source, just as VMR queries must be implemented in terms of the CIS data source.
For managing hypertension in diabetic patients, the SAGE project validated this approach for accessing external knowledge sources by implementing the VKB for a drug knowledge base. We adopted the drug model defined in Veterans Administration NDF-RT. The model contains entities such as ActiveIngredientPreparation, DrugComponent, and ClinicalDrug, where, for example, an ActiveIngredientPreparation like LISINOPROL PREPARATION has roles such as may_treat, and contraindication_with. While external knowledge sources available to local implementers may also represent this knowledge, we modeled ActiveIngredientPreparation, DrugComponent, and ClinicalDrug as extensions of terminology classes in a SAGE knowledge base.
A virtual knowledge-base query (VKB query) has a very simple structure with two main attributes: (1) a partially specified instance of a VKB class constraining the properties that the target instances of VKB classes must satisfy, and (2) a selection attribute that determines if the query should return, when a selection attribute is unspecified, instances of the VKB class or, when the attribute is specified, the attribute values of the VKB instance. For example, a query for the maximum dose of Lisinopril in hypertension requires specifying (1) an instance of ActiveIngredientPreparation with code LISINOPROL PREPARATION FOR HYPERTENSION and (2) the selection attribute maximum dose.
The SAGE DSS implements the VKB query by reformulating it in terms of the actual representation of the external knowledge source. For example, the SAGE DSS transforms the VKB query in the previous paragraph into an SQL query for the LISINOPRIL drug record in the SAGE extraction of the NDF-RT knowledge base. The selection attribute of the VKB Query would be applied to the result queried from the external knowledge base.
Discussion
Status
We analyzed, encoded, tested, and implemented SAGE guidelines in clinical information systems located at GE Healthcare, the Mayo Clinic and the University of Nebraska Medical Center. They were verified in simulated patient encounters on test installations of the Mayo Clinic and UNMC enterprise clinical information systems. The results, not reported here, supported the possibility of using SAGE technology to provide guideline decision support in disparate settings. Each feature of the SAGE Guideline Model described in this paper was tested and incrementally refined while modeling and encoding the guidelines. As a technology development project, SAGE was not funded to test the system in real clinical settings.
Comparison with Earlier Models
To meet the challenges of the SAGE project, we have synthesized prior work and, wherever possible, established mappings between SAGE and other models. In particular, the Activity Graph formalism is a generalization of similar constructs in EON 10 and GLIF3. 7 The Decision Map concept derives from the PRODIGY 5 project, and the decision model used to determine preferences among alternatives is derived from PROforma. 4 The use of events to trigger the execution recommendation set is derived from PROforma and GLIF. The SAGE project emphasizes that what we model in the Activity Graphs is executed by the DSS in response to opportunities for decision support. The top-level activity graphs model the DSS’s interactions with the CIS within the clinical workflow to present guideline-based decision support services. Thus, the processes in the SAGE activity graphs may differ from the workflow that takes place in the care process and the clinical algorithms in guideline documents that are designed for human comprehension.
The SAGE Evidence Statement is a generalization of similar constructs in the EON model. 40 Whereas the EON model hard codes the relationships such as compelling indication, relative indication, relative contraindication, absolute contraindication, and good drug partners as properties in the model, the SAGE Guideline Model’s Evidence Statement specifies these relationships as terminology codes, thus (1) allowing much more flexibility and generality in the relationships that can be encoded and retrieved, and (2) enhancing the ease of updating and maintaining the evidence statements.
Strengths and Limitations
The major innovation of the SAGE Guideline Model is its demonstration that heterogeneous information sources, such as patient data, order sets, and external knowledge sources, can be integrated and used within encoded guideline knowledge bases. For each information source, the SAGE project first defines an information model, such as the Virtual Medical Record for patient data, the order set schema for order sets, the Evidence Statements for relationships in medical knowledge, and the Virtual Knowledge Base for medication information. The information models enable us to formulate statements, using standard terminologies, such as observations on a patient’s laboratory test results or comorbidities as indications for the use of a certain drug class. The SAGE project demonstrates that guideline knowledge bases, enterprise order sets, drug knowledge bases, standard terminologies, and patient data can interoperate once the information model and associated roles for each are clearly defined.
The project exposed difficulties for wide adoption of this technology. The primary bottlenecks are developing and maintaining comprehensive guideline knowledge bases and a lack of comprehensive integrated standards. Analyzing, selecting, refining, encoding, and testing guideline knowledge in computable format is still labor-intensive, requiring collaboration between expert clinicians and computer scientists. Computable encoding of paper-based guidelines requires introducing additional clinical knowledge to disambiguate knowledge in generalized statements. Standard terminologies would reduce this problem, but they are still the exception to the rule in clinical information systems.
The SAGE specification context makes strong assumptions about clinical workflow. The system cannot adapt dynamically to changes in pre-specified workflow patterns, though workflow modifications using the Protégé workbench were easily accomplished. Encoding interventions, such as physician inbox messages and order sets, may be too specific and therefore not as interoperable as expected. The advantage of tightly coupled guideline encoding with clinical workflow is that it optimizes the opportunity for a well-integrated presentation of guideline recommendations to busy clinicians. The disadvantage is that changes in workflow (in time or across institutions) may require more changes to a guideline with workflow-specific encoding.
For years, workers have attempted to find commonalities among various guideline modeling formalisms, for possible convergence and standardization of guideline representation. 20,41 Our experience points to the possibility of new directions in these efforts. Instead of attempting to standardize a guideline model, for which there is little consensus on syntax and semantics, a more attainable approach may be to standardize the relatively simple information models and the operations that can be performed on them. The HL7 Clinical Statement project is already defining a collection of clinical statement patterns that are similar to the SAGE VMR. The HL7 Clinical Decision Support Technical Committee is formulating a standard representation for complex order sets. The Evidence Statement is similarly an information model for a class of declarative “factual” knowledge for which a consensus may be easier to reach. Such consensus would allow these declarative statements, like patient data, to be shared and used by different guideline modeling and execution systems.
Footnotes
This work was partially supported by grant 70NANB1H3049 of the U.S. National Institute of Standards and Technology, Advanced Technology Program. The Protégé resource is supported by Grant P41 LM007885 from the National Library of Medicine. Views expressed are those of the authors and not necessarily those of affiliated organizations. The authors wish to thank Dr. Mor Peleg for detailed comments on the draft version of the paper.
Sections on workflow and information standards draw on previously published materials. 13
http://www.jointcommission.org/PerformanceMeasurement/PerformanceMeasurement/Pneumonia+Core+Measure+Set.htm (Accessed on 4/30/2007).
http://www.ahrq.gov/clinic/pneumonia/pneumonia.htm#pneumonia (Accessed on 4/30/2007.)
The Presence Criterion in ▶ is equivalent to the following GELLO expression: Observation.exists(code.implies(“CHF disease affecting CAP risk code”) and effectiveTime.within(Now), where Observation is a collection of Observation instances, code and effectiveTime are attributes of the Observation class, implies and within are operators associated with HL7 CodedValue and interval of PointInTime data types, and Now is a variable whose value is the current time.
References
- 1.Bero LA, Grilli R, Grimshaw JM, Harvey E, Oxman AD, Thomson MA. Closing the gap between research and practice: an overview of systematic reviews of interventions to promote the implementation of research findings BMJ 1998;317(7156):465-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balas EA, Boren SA. Managing Clinical Knowledge for Health Care Improvement. Yearbook of Medical Informatics 2000: Patient-centered SystemsStuttgart, Germany: Schattauer; 2000. pp. 65-70. [PubMed]
- 3.de Clercq PA, Hasman A, Blom JA, Korsten HHM. Design and Implementation of a Framework to Support the Development of Clinical Guidelines Int J Med Inf 2001;64(2–3):285-318. [DOI] [PubMed] [Google Scholar]
- 4.Fox J, Das SK. Safe and SoundCambridge: MIT Press; 2000.
- 5.Johnson PD, Tu SW, Booth N, Sugden B, Purves IN. Using Scenarios in Chronic Disease Management Guidelines for Primary Care Proc AMIA Symp 2000. 38993. [PMC free article] [PubMed]
- 6.Miksch S, Shahar Y, Johnson P. Asbru: A Task-Specific, Intention-Based, and Time-Oriented Language for Representing Skeletal PlansIn: Motta E, Harmelen Fv, Pierret-Golbreich C, Filby I, Wijngaards N, editors. 7th Workshop on Knowledge Engineering: Methods & Languages (KEML-97). Milton Keynes, UK. 1997. pp 9-1-9-20.
- 7.Boxwala AA, Peleg M, Tu SW, Ogunyemi O, Zeng Q, Wang D, et al. GLIF3: A Representation Format for Sharable Computer-Interpretable Clinical Practice Guidelines J Biomed Inform 2004;37(3):147-161. [DOI] [PubMed] [Google Scholar]
- 8.Quaglini S, Stefanelli M, Cavallini A, Micieli G, Fassino C, Mossa C. Guideline-Based Careflow Systems Artif Intell Med. 2000;5(22):5-22. [DOI] [PubMed] [Google Scholar]
- 9.Shiffman RN, Karras BT, Agrawal A, Chen R, Marenco L, Sujai Nath M. GEM: A Proposal for a More Comprehensive Guideline Document Model Using XML J Am Med Inform Assoc 2000;7:488-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tu SW, Musen MA. A Flexible Approach to Guideline Modeling. Proc AMIA Symp; 1999Washington DC: Hanley & Belfus, Inc; 1999. pp. 420-424. [PMC free article] [PubMed]
- 11.Shiffman RN, Michel G, Essaihi A, Thornquist E. Bridging the Guideline Implementation Gap: A Systematic, Document-Centered Approach to Guideline Implementation J Am Med Inform Assoc 2004;11:418-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peleg M, Boxwala AA, Tu SW, Zeng Q, Ogunyemi O, Wang D, et al. The InterMed Approach to Sharable Computer-Interpretable Guidelines: A Review J Am Med Inform Assoc 2004;11(1):1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fieschi M, Dufour J, Staccini P, Gouvernet J, Bouhaddou O. Medical decision support systems: old dilemmas and new paradigms? Methods Inf Med 2003;42(3):190-198. [PubMed] [Google Scholar]
- 14.Kawamoto K, Houlihan CA, Balas EA, Lobach DF. Improving clinical practice using clinical decision support systems: a systematic review of trials to identify features critical to success BMJ 2005. 765. [DOI] [PMC free article] [PubMed]
- 15.Osheroff JA, Pifer EA, Sittig DF, Jenders RA, Teich JM. Improving Outcomes: A Practical Guide to Clinical Decision Support ImplementationChicago: HIMSS; 2005.
- 16.McDonald CJ, Huff SM, Suico JG, et al. LOINC, a universal standard for identifying laboratory observations: a 5-year update Clin Chem. 2003;49(4):624-633Apr. [DOI] [PubMed] [Google Scholar]
- 17.Hripcsak G, Ludemann P, Pryor TA, Wigertz OB, Clayton PD. Rationale for the Arden Syntax Comput Biomed Res 1994;27(4):291-324. [DOI] [PubMed] [Google Scholar]
- 18.Health Level Seven. HL 7 Reference Information Model. Available at: http://www.hl7.org/library/data-model/RIM/modelpage_non.htm. Accessed on July 7, 2003.
- 19.Wang AY, Sable JH, Spackman KA. The SNOMED clinical terms development process: refinement and analysis of content Proc AMIA Symp 2002. 8459. [PMC free article] [PubMed]
- 20.Peleg M, Tu SW, Bury J, Ciccarese P, Fox J, Greenes RA, et al. Comparing Computer-Interpretable Guideline Models: A Case-Study Approach J Am Med Inform Assoc 2003;10(1):52-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mulyar N, van der Aalst WMP, Peleg M. A Pattern-based Analysis of Clinical Computer-Interpretable Guideline Modelling Languages: BPMcenter.org; 2006. Report No.: BPM Center Report BPM-06-29. [DOI] [PMC free article] [PubMed]
- 22.Workflow Management Coalition. Interface 1: Process Definition Interchange, Process Model. Lighthouse Point, FL: Workflow Management Coalition; 1999 October 29. Report No. WfMC TC-1010-P.
- 23.Gennari JH, Musen MA, Fergerson RW, et al. The Evolution of Protégé: An Environment for Knowledge-Based Systems Development Int J Hum Comput Stud 2003;58(1):89-123. [Google Scholar]
- 24.Tu SW, Musen MA, Shankar R, et al. Modeling Guidelines for Integration into Clinical Workflow 2004. Medinfo; 2004; San Francisco, CA, USA 1748. [PubMed]
- 25.Kroger AT, Atkinson WL, Marcuse EK, Pickering LK. General recommendations on immunization: recommendations of the Advisory Committee on Immunization Practices (ACIP) MMWR Recomm Rep. 2006;55(RR-15):1-48(Dec 1). [PubMed] [Google Scholar]
- 26.Standards of medical care in diabetes—2006. Diabetes Care;(Jan 29)[Suppl 1]:S4–42. [PubMed]
- 27.The Joint National Committee on Prevention Diagnosis and Management of Hypertension The Seventh Report of the Joint National Committee on Prevention Diagnosis and Management of HypertensionU.S. Department of Health and Human Services; 2004.
- 28.Bartlett JG, Dowell SF, Mandell LA, File Jr TM, Musher DM, Fine MJ. Practice guidelines for the management of community-acquired pneumonia in adultsInfectious Diseases Society of America. Clin Infect Dis 2000;31(2):347-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mandell LA, Bartlett JG, Dowell SF, File Jr. TM, Musher DM, Whitney C. Update of practice guidelines for the management of community-acquired pneumonia in immunocompetent adults Clin Infect Dis 2003;37(11):1405-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mead CN. Data interchange standards in healthcare IT—computable semantic interoperability: now possible but still difficult, do we really need a better mousetrap? J Healthc Inf Manag 2006;20(1):71-78. [PubMed] [Google Scholar]
- 31.Health Level Seven. Data Types—Abstract Specification, 2004. Available at: http://www.hl7.org/library/data-model/RIM/modelpage_non.htm. Accessed on July 7, 2006.
- 32.Carter J, Brown S, Erlbaum M, et al. Initializing the VA medication reference terminology using UMLS metathesaurus co-occurrences Proc AMIA Symp 2002. 11620. [PMC free article] [PubMed]
- 33.Johnson PD, Tu SW, Musen MA, Purves I. A Virtual Medical Record for Guideline-Based Decision Support Proc AMIA Symp 2001. 2948. [PMC free article] [PubMed]
- 34.Rector A, Qamar R, Marley T. Binding Ontologies & Coding Systems to Electronic Health Records and Messages Proc 2nd Int Workshop Formal Biomed Knowl Rep (KR-MED 2006) 2006:11-19.
- 35.Sordo M, Boxwala A, Ogunyemi O, Greenes R. Description and Status Update on GELLO: a Proposed Standardized Object-oriented Expression Language for Clinical Decision Support Medinfo 2004:164-168. [PubMed]
- 36.Tu SW, Campbell J, Musen MA. The Structure of Guideline Recommendations: A Synthesis Proc AMIA Symp 2003;679:83. [PMC free article] [PubMed] [Google Scholar]
- 37.Tu SW. The SAGE Guideline Model Technical SpecificationStanford: Stanford Medical Informatics, Stanford University; 2006. Report No. 1243.
- 38.Payne TH, Hoey PJ, Nichol P, Lovis C. Preparation and use of preconstructed orders, order sets, and order menus in a computerized provider order entry system J Am Med Inform Assoc 2003;10(4):322-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McClay JC, Campbell JR, Parker C, Hrabak KM, Tu SW, Abarbanel R. Structuring Order Sets for Interoperable Distribution Proc AMIA Symp 2006. 54953. [PMC free article] [PubMed]
- 40.Tu SW, Hrabak KM, Campbell JR, et al. Use of Declarative Statements in Creating and Maintaining Computer-Interpretable Knowledge Bases for Guideline-Based Care Proc AMIA Symp 2006. 7848. [PMC free article] [PubMed]
- 41.Wang D, Tu SW, Peleg M, et al. Representation Primitives, Process Models and Patient Data in Computer-Interpretable Clinical Practice Guidelines: A Literature Review of Guideline Representation Models Int J Med Inf 2002;68(1–3):59-70. [DOI] [PubMed] [Google Scholar]