Abstract
In animals, fatty acid desaturases catalyze key reactions in the synthesis of arachidonic acid and other polyunsaturated fatty acids. A search of the Caenorhabditis elegans DNA databases, using the sequences of Arabidopsis genes, identified several putative desaturases. Here we describe the characterization of the first of these genes, fat-1. The predicted protein encoded by a fat-1 cDNA showed 32–35% identity with both FAD2 and FAD3 of Arabidopsis. When expressed in transgenic plants, fat-1 resulted in a 90% increase in the proportion of α-linolenic acid in root lipids. Wild-type Arabidopsis incorporated ω-6 fatty acids (Δ8,11,14–20:3 and Δ5,8,11,14–20:4) into membrane lipids but did not desaturate them. By contrast, fat-1 transgenic plants efficiently desaturated both of these fatty acids to the corresponding ω-3 products. These findings indicate that the C. elegans fat-1 gene encodes the first animal representative of a class of glycerolipid desaturases that have previously been characterized in plants and cyanobacteria. The FAT-1 protein is an ω-3 fatty acyl desaturase that recognizes a range of 18- and 20-carbon ω-6 substrates.
Keywords: fatty acid desaturation, expressed sequence tag, fat-1, arachidonic acid, polyunsaturated fatty acids
Polyunsaturated fatty acids are important as structural components of membrane glycerolipids and as precursors of families of signaling molecules including prostaglandins, thromboxanes, and leukotrienes (1, 2). The principal fatty acid precursors of these signaling compounds are arachidonic acid (Δ5,8,11,14–20:4), providing an ω-6 substrate that is responsible for the major synthesis of these compounds, and eicosapentaenoic acid (Δ5,8,11,14,17–20:5), an ω-3 substrate that is responsible for the parallel synthesis of many eicosanoids with an additional double bond. An important class of enzymes involved in the synthesis of polyunsaturated fatty acids are the fatty acid desaturases that catalyze the introduction of double bonds into the hydrocarbon chain. In vertebrates, desaturases are known to act at the Δ4, -5, -6, -8, and -9 positions (3). The 18:0-CoA Δ9 desaturase from rat liver has been characterized biochemically (4, 5), and the corresponding gene has been cloned (6). However, the remaining four enzymes have remained recalcitrant to purification and genes that encode them have not been isolated. Based on available information, and by analogy to the 18:0-CoA desaturase, it is likely that these additional enzymes are integral membrane proteins that require other membrane components (cytochrome b5 and NADH:cytochrome b5 reductase) for activity (4), and it is these features that have limited progress in studying the biochemistry and molecular genetics of these important synthetic reactions.
Biochemical studies of membrane-bound fatty acid desaturases in plants have proven equally difficult, and only one enzyme has been purified to homogeneity (7). Higher plants produce many different unsaturated fatty acids (8), but in membrane lipids the major locations for double bonds are at the Δ9, -12, and -15 (ω-3) positions of 18-carbon acyl chains and the corresponding Δ7, -10, and -13 (ω-3) positions of 16-carbon chains (9). In Arabidopsis, the isolation of mutants with altered fatty acid compositions (10) has facilitated studies of the biochemistry and function of the seven membrane desaturases present in this plant (11). Furthermore, the extensive molecular genetic tools available in Arabidopsis allowed genes encoding desaturases to be cloned without the need to first solubilize and purify the enzymes themselves (12, 13). Interestingly, all the plant membrane-bound desaturases use complex glycerolipids rather than acyl-CoAs as substrates. This finding and data indicating that the Δ5 desaturase from rat liver acts on glycerolipids (14) suggest the possibility that the majority of animal desaturases also catalyze glycerolipid-linked desaturation. However, the lack of progress in characterizing other animal desaturases has precluded further discussion of this issue.
We set out to use the Arabidopsis desaturase genes and the extensive information now available from the Caenorhabditis elegans genome project (15) to identify and clone genes encoding fatty acid desaturases in animals. C. elegans is a good model in this respect because it elaborates a wide range of polyunsaturated fatty acids including arachidonic and eicosapentaenoic acids, from very simple precursors available in its diet (16, 17). In this communication, we report the isolation and characterization of a cDNA from C. elegans that, when expressed in Arabidopsis, encodes a novel fatty acid desaturase which can catalyze the introduction of an ω-3 double bond into a range of 18- and 20-carbon fatty acids.
MATERIALS AND METHODS
Database Searches.
The National Center for Biotechnology Information’s (NCBI) peptide sequence database was searched using a blast server, with the peptide sequences of the Arabidopsis thaliana FAD2, FAD6 and FAD7 fatty acid desaturases as queries. The GenBank accession numbers for the corresponding Arabidopsis cDNAs are L26296L26296, U09503U09503, and L22931L22931, respectively.
Cloning and Sequencing of a fat-1 cDNA.
The partial cDNA identified by a database search, CEL10e11, was obtained from the C. elegans Genome Sequencing Center (Washington University School of Medicine, St. Louis). The cDNA insert was released by double digestion with HindIII and SacI, gel purified, and labeled with [32P]dCTP using a random prime kit (Promega). The denatured probe was used to screen a mixed stage C. elegans cDNA λ phage (Uni-Zap XR) library (Stratagene). Nucleic acid hybridizations and high stringency washes were performed as described (13). Hybridizing plaques were visualized by autoradiography. Positive clones were isolated and excised from the phage vector according to the manufacturer’s protocol to yield pBluescript plasmids. The plasmid clone with the longest insert, pCE8, was completely sequenced in both directions. Sequence analysis was carried out using the programs available in the Genetics Computer Group package (18) using default settings for parameters unless indicated. The DNA sequence corresponding to the ORF of the fat-1 cDNA was used to search the database of the C. elegans genome sequencing project using the blast server accessed through the World Wide Web interface (15). No homologous sequence was found (highest blast score, 145; P = 0.034), indicating that the fat-1 genomic sequence had not yet been included by this project.
Gene Construct for Plant Expression.
The cauliflower mosaic virus 35S promoter/nopaline synthase terminator cassette of Baulcombe et al. (19) was cloned into the XbaI/EcoRI sites of pBIN400 (20) to make the binary transformation vector pBIN420. The cDNA insert of pCE8 was released with a EcoRI/KpnI double digest, end-filled with Klenow fragment, and blunt-ligated into the SmaI site of pBIN420 to make pBIN420-CE8. These vectors contain the NPTII gene within their T-DNA, thus conferring kanamycin resistance to transgenic plants.
Plant Material, Transformation, and Growth.
The Columbia ecotype of the wild-type line of A. thaliana (L.) Heynh. was used for these experiments. The binary vector pBIN420-CE8 was introduced into the Agrobacterium strain PC2760 by the freeze–thaw method (21). Agrobacterium-mediated transformation was accomplished with the in planta vacuum infiltration method (22). Primary generation transformed seeds were selected on plates containing Murashige and Skoog basal salts (4.3 g/liter), 1% (wt/vol) sucrose, 0.8% (wt/vol) Bacto-Agar, 200 mg/liter carbenecillin, 50 mg/liter kanamycin, and adjusted to pH 5.8 with KOH. In vitro roots were grown from sterilized seeds placed on vertical plates at 23°C under continuous illumination (50–100 micromol quanta m−2·s−1). The media for in vitro roots contained Gamborg B5 salts (3.1 g/liter), 2% (wt/vol) glucose, 0.2% phytagel (Sigma), and adjusted to pH 5.8 with KOH.
Exogenous Fatty Acid Treatments.
Plants were grown in a growth chamber at 20°C on a 12-h day/night cycle. Fatty acid treatments began when the plant rosettes reached ≈2 cm in diameter. Sodium soaps of homogamma-linolenic acid (Δ8,11,14–20:3) and arachidonic acid (Δ5,8,11,14–20:4) (Nu Check Prep, Elysian, MN) were made to a 0.1% aqueous solution and frozen in 5 ml aliquots. Plants were sprayed daily at the beginning of the dark period using a perfume atomizer. Groups of 15 plants were sprayed with 5 ml of soap solution for 20 consecutive days.
Lipid Analyses.
Methods for extraction and separation of lipids, and for the preparation of fatty acid methyl esters, have been described (23). Analysis of fatty acid methyl esters by gas chromatography was carried out using a 15 m × 0.53 mm Supelcowax column (Supelco) with flame ionization detection. The initial column temperature of 160°C was held for 1 min then raised at 20°C/min to 190°C followed by a ramp of 5°C/min to 230°C. The final temperature was held for 5 min. When wild-type and fat-1 transgenic plants were sprayed with exogenous fatty acids, the peaks for the ω-6 substrates Δ8,11,14–20:3 and Δ5,8,11,14–20:4, and of the ω-3 desaturation products Δ8,11,14,17–20:4 and Δ5,8,11,14,17–20:5, were identified based on their coelution with authentic standards (Nu Check Prep) and on the results of gas chromatography–mass spectrometry analysis. For this analysis, fatty acid methyl esters derived from phosphatidylcholine were separated on a 30 m × 0.2 mm AT1000 column (Alltech Associates) in a HP6890 Instrument (Hewlett–Packard). Oven temperature at injection was 150°C, and this was increased at 5°C/min to 230°C then held at 230°C for 10 min. Criteria for identification of Δ5,8,11,14,17–20:5 in phosphatidylcholine from fat-1 transgenic plants where the identification of a mass peak at m/z = 316, which corresponds to the expected molecular ion, and a retention time (36.11 min) and fragmentation pattern that were identical to those of the authentic Δ5,8,11,14,17–20:5 standard. No commercial standard was available for Δ8,11,14,17–20:4. A fatty acid methyl ester present in fat-1 transgenic plants sprayed with Δ8,11,14–20:3 but not in wild-type control plants had a retention time of 35.74 min during gas chromatography–mass spectrometry. This compound showed a mass peak at m/z = 318 (the expected molecular ion for 20:4) and a fragmentation pattern very similar to that of the authentic Δ5,8,11,14–20:4 standard. The retention time of Δ5,8,11,14–20:4 was 35.04 min for both the authentic standard and for the methyl esters recovered from plants sprayed with soaps of this isomer. Therefore it was concluded that the new compound detected only in fat-1 transgenic plants sprayed with Δ8,11,14–20:3 was an isomer of 20:4 and most probably Δ8,11,14,17–20:4.
Nucleic Acid Analysis.
Total RNA was extracted from leaves according to the method of Verwoerd et al. (24). Twenty-five micrograms of total RNA was separated on 1.2% agarose–formaldehyde gels and blot transferred to nylon membranes. Genomic DNA was isolated according to the method of Dellaporta et al. (25), restricted with BamHI, separated by agarose gel electrophoresis, and alkaline blotted to nylon membranes. The HindIII/SacI fragment of pCE8 was used as a probe on the RNA and DNA blots. Probe labeling, hybridizations, and washings were as described above for cDNA library screens.
RESULTS
Cloning and Sequencing of a fat-1 cDNA.
In database searches using FAD2, FAD6, or FAD7 as queries, the highest scoring C. elegans expressed sequence tag (EST) clones were NCBI-5443, NCBI-a5881, and NCBI-5049. The corresponding GenBank accession numbers were Z14935Z14935, M88884M88884, and Z14543Z14543, respectively. An alignment of cDNA sequences revealed a common identity of 301 bp among all three clones, indicating that the three ESTs originated from a single gene. The greatest amount of sequence data (486 bp) was available from EST clone NCBI-5881. This clone was requested from its origin at the C. elegans Genome Sequencing Center, with its original source identifier (CEL10e11). Upon receipt of CEL10e11, its identity was confirmed by partial sequencing. To obtain a full-length cDNA for the corresponding gene, the insert from CEL10e11 was 32P-labeled and used to probe ≈30,000 plaques from a C. elegans cDNA library. This screen yielded 13 positive clones. The longest eight clones were all ≈1.4 kb in length as judged by agarose gel electrophoresis. One clone, pCE8, was fully sequenced in both directions and found to contain a 1410-bp cDNA insert. This sequence was deposited in GenBank under accession no. L41807L41807.
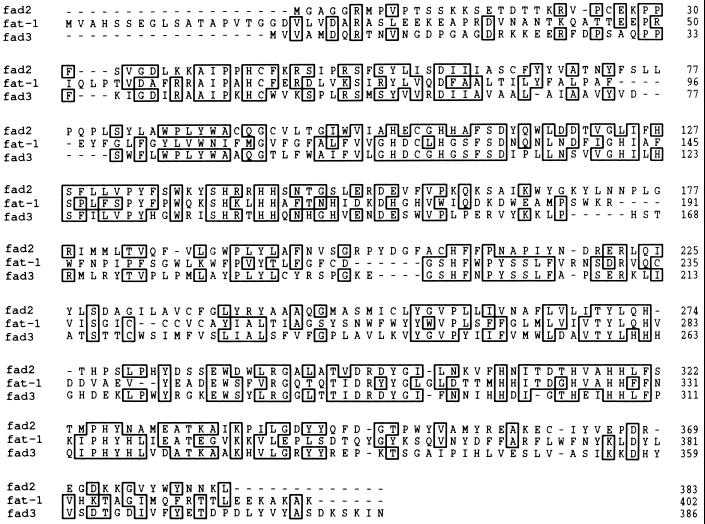
The cDNA insert in pCE8 contained an ORF that would be expected to encode a protein of 402 amino acids, with a molecular mass of 46.4 kDa. The predicted amino acid sequence of the protein showed several regions of common identity with the predicted sequences of the FAD2 and FAD3 desaturases of Arabidopsis. Alignment with FAD2 revealed 35% sequence identity and 61% similarity. Alignment with FAD3 indicated 32% sequence identity and 54% similarity. The FAD2 and FAD3 genes are known to encode enzymes that desaturate oleate (FAD2) or linoleate (FAD3) esterified to phosphatidylcholine of the endoplasmic reticulum (12, 13). Within a tripartite alignment of the three sequences (Fig. 1), there are 69 residues common to all three sequences including eight histidines (amino acids 123, 127, 159, 162, 163, 324, 327, and 328 in the C. elegans sequence) whose presence and locations are highly conserved among all the membrane desaturases (13). These findings strongly indicate that the gene represented by the pCE8 cDNA encodes a fatty acid desaturase or a related enzyme function. We have designated this gene fat-1 (Fatty acid metabolism 1). It is somewhat surprising that the C. elegans gene shows a similar amino acid sequence homology to each of the two Arabidopsis desaturases, especially in view of the fact that the FAD2 and FAD3 sequences are relatively divergent with only 37% common amino acid identity (13). These observations mean that it is difficult to deduce—from these comparisons alone—whether the fat-1 gene is likely to represent a Δ12 desaturase (like FAD2), an ω-3 desaturase (like FAD3), or a more distantly related enzyme.
Figure 1.
Deduced amino acid sequences and shared identities of the fat-1 gene of C. elegans (fat-1) and the FAD2 and FAD3 genes of A. thaliana (fad2 and fad3).
Expression in Arabidopsis Reveals the Function of the fat-1 Gene.
In higher plants, desaturases have been characterized from two cellular compartments. Enzymes localized to the chloroplast (or plastid) use soluble ferredoxin as the electron donor for the reaction (7, 26). Enzymes localized to the endoplasmic reticulum (including the FAD2 and FAD3 gene products) are similar to known yeast and animal desaturases in as much as they rely on cytochrome b5 and cytochrome b5 reductase to supply electrons from NAD(P)H (26). Mutants deficient in each of the major desaturases are available in Arabidopsis (11). Genes that encode the 18:0-CoA desaturases from yeast and mammals have been expressed in plants and shown to alter the fatty acid compositions of the plant tissues (27, 28). These considerations recommended Arabidopsis as a suitable heterologous system to study the expression and function of the fat-1 gene. We therefore cut the cDNA insert from pCE8, ligated it into the plant expression vector pBIN420 to yield pBIN420-CE8, and used this construct to transform Arabidopsis.
Five individual transformants were obtained and allowed to set seed. Lines 9.7 and 10.5 were selected for further analysis. A Southern blot of genomic DNA from these two lines probed with the insert from pCE8 confirmed the presence of at least one copy of the transgene in line 9.7 and at least two copies in line 10.5 (data not shown). When RNA from plants of the two lines and wild-type Arabidopsis was subject to gel blot analysis (Fig. 2), the appropriate fat-1 transcript was shown to accumulate in both transgenic lines. Plants from line 9.7 consistently produced higher transcript levels than line 10.5.
Figure 2.
Gel blot analysis of fat-1 transcript levels in wild-type and transgenic Arabidopsis. Total RNA (25 μg) isolated from leaves was separated by electrophoresis on a 1.2% agarose–formaldehyde gel, transferred to a nylon membrane, and probed with a HindIII/SacI DNA fragment of pCE8.
Characterization of the Arabidopsis lipid mutants has indicated that lesions in the fad2 and fad3 genes are partly masked in leaf tissue by action of the chloroplast desaturases (encoded by FAD6, FAD7, and FAD8). For this reason, a first attempt to determine the function of the fat-1 gene product was made by analyzing the overall fatty acid composition of root tissues from wild-type and fat-1 transgenic plants. The data in Table 1 show very large increases in the proportion of 18:3 in both transgenic lines compared with wild-type Arabidopsis. These increases were accompanied by concomitant decreases in the proportion of 18:2 but no significant changes in the levels of any other fatty acid. The alterations in root fatty acid composition induced by expression of the C. elegans fat-1 gene are comparable to those observed by overexpression of the plant FAD3 gene (12). The FAT1 protein is thus operating as an efficient ω-3 desaturase in Arabidopsis.
Table 1.
Compositions of total fatty acids from in vitro grown roots of the wild-type (WT) and two transgenic lines (9.7 and 10.5) of Arabidopsis expressing a C. elegans fat-1 cDNA
| Genotype | Mol % of total fatty acids
|
|||||
|---|---|---|---|---|---|---|
| 16:0 | 16:1(c) | 18:0 | 18:1 | 18:2 | 18:3 | |
| WT | 16.8a | 1.2a | 1.4a | 22.7a | 39.3a | 18.0b |
| 9.7 | 17.8a | 1.2a | 2.2a | 21.7a | 22.7b | 33.7a |
| 10.5 | 17.5a | 1.0a | 1.6a | 20.9a | 23.7b | 34.5a |
Values are means of quadruplicate measurements. Values for each fatty acid with the same letter do not differ significantly (P < 0.01).
fat-1 Transgenic Arabidopsis Desaturate 20-Carbon Fatty Acids.
In Arabidopsis, linolenic acid (Δ9,12,15–18:3) is the only significant product resulting from fat-1 expression. However, C. elegans contains a wider range of polyunsaturated fatty acids than does Arabidopsis. Three ω-3 fatty acids are present in the membrane lipids of the worm and of these linolenic acid is the least abundant (0.15% of total fatty acids). The 20-carbon fatty acids Δ8,11,14,17-eicosatetraenoic acid (an isomer of arachidonic acid) and Δ5,8,11,14,17-eicosapentaenoic acid (the expected product of ω-3 desaturation of arachidonic acid) account for 7.7% and 8.7% of total fatty acids, respectively (16).
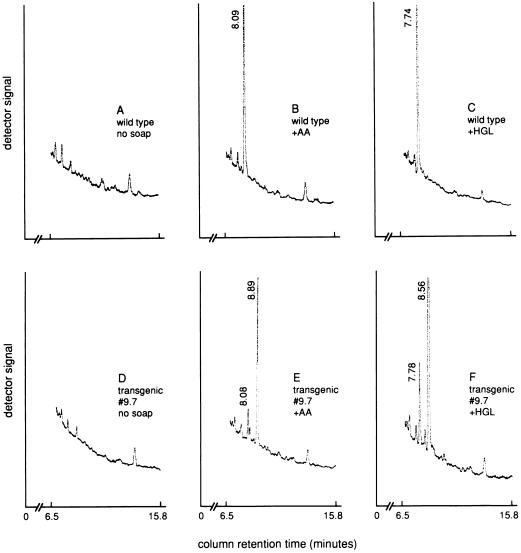
We have previously found that exogenous fatty acids, when applied to Arabidopsis leaves as sodium soaps, are readily taken up and incorporated into membrane glycerolipids to levels that correspond to 2–5% of the total leaf lipids (29). To test whether the fat-1-encoded desaturase is likely to be involved in synthesis of the 20-carbon ω-3 fatty acids in C. elegans, wild-type and transgenic Arabidopsis plants were sprayed once a day for 20 days with solutions of arachidonic acid (Δ5,8,11,14–20:4) or a homogamma-linolenic acid (Δ8,11,14–20:3) as the sodium salts. In this experiment, analyses of total leaf lipids indicated that the exogenously supplied fatty acids were incorporated at levels of 1–3% of the total fatty acids (data not shown). Extensive incorporation occurred into phosphatidylcholine, which is the major lipid of the endoplasmic reticulum and the major substrate for the plant 18:1 and 18:2 desaturases (23, 30). In wild-type leaves, the peak corresponding to Δ5,8,11,14–20:4 or Δ8,11,14–20:3 accounted for ≈3–5% of the total fatty acids in phosphatidylcholine but there was no detectable conversion of either of these fatty acids to their ω-3 unsaturated derivatives (Fig. 3). By contrast, in leaves from plants expressing the fat-1 gene, the peaks corresponding to the exogenously supplied ω-6 fatty acids were substantially replaced by peaks that correspond to the expected ω-3 desaturated products, Δ5,8,11,14,17–20:5 and Δ8,11,14,17–20:4. Thus, in contrast to the Arabidopsis FAD3 gene product, the C. elegans FAT-1 protein is a desaturase that acts on a range of ω-6 fatty acid substrates.
Figure 3.
Partial gas chromatograph traces of the component fatty acids of phosphatidylcholine extracted from leaves of (A) wild-type Arabidopsis, (B) wild-type Arabidopsis sprayed with the sodium soap of arachidonic acid (Δ5,8,11,14–20:4; AA), (C) wild-type Arabidopsis sprayed with the sodium soap of homogamma-linolenic acid (Δ8,11,14–20:3; HGL), (D) transgenic Arabidopsis line 9.7 expressing a fat-1 cDNA, (E) transgenic Arabidopsis line 9.7 sprayed with the sodium soap arachidonic acid, and (F) transgenic Arabidopsis line 9.7 sprayed with the sodium soap of homogamma-linolenic acid.
DISCUSSION
The ω-3 fatty acids (Δ9,12,15–18:3; Δ8,11,14,17–20:4, and Δ5,8,11,14,17–20:5) account for 17% of the total fatty acids in C. elegans (16) and 20:5 ω-3 is the major fatty acid in phosphatidylcholine from this organism (17). These lipids are produced even when the worms are grown exclusively on Escherichia coli which provides only saturated and monounsaturated fatty acids (17). Evidently, C. elegans must contain all the enzymes required for the synthesis of these highly unsaturated acyl groups. The fat-1 gene described here encodes an ω-3 desaturase that is able to carry out the final step in the synthesis of all these fatty acids. The fat-1 gene was identified through its homology with the FAD2 and FAD3 genes of Arabidopsis. In pairwise homology comparisons, the deduced FAT-1 sequence exhibited 32–35% identity to the products of these two genes. By contrast, homology to the yeast and rat genes that encode 18:0-CoA desaturases was at the level of 17–23% identity—only slightly above the level from comparisons with entirely unrelated genes. For this reason, it is unlikely that a database search using an 18:0-CoA desaturase as the query could have identified the fat-1 sequence.
The predicated protein sequence of the fat-1 gene product includes the three histidine-rich sequences that are highly conserved among all the membrane-bound fatty acid desaturases and that are believed to be the residues that coordinate the diiron-oxo structure at the active site of these enzymes (31, 32). Furthermore, two long stretches (>40 residues each) of hydrophobic residues are present (80–124 and 229–284). The length of these stretches and their positions relative to the conserved histidine sequences are similar to other desaturases so that the FAT-1 protein could conform with the model proposed by Stukey et al. (32) in which the bulk of the protein is exposed on the cytosolic face of the endoplasmic reticulum while two membrane-traversing loops (each comprised of two membrane-spanning, α-helical segments) lock the protein into the bilayer. In common with many, though not all, of the proposed endoplasmic reticulum desaturases, the FAT-1 protein contains a carboxyl-terminal motif (KAKAK) which conforms to a consensus retention signal for transmembrane proteins in the endoplasmic reticulum (33).
These features are consistent with FAT-1 being a member of the membrane-bound desaturase/hydroxylase family of diiron-oxo proteins (31). However, the FAT-1 sequence shows equal homology to both the Δ12 glycerolipid desaturase encoded by FAD2 and the ω-3 glycerolipid desaturase encoded by FAD3. To determine which class of reaction is catalyzed by FAT-1 and to explore the substrate chain length and regiochemical specificities of the enzyme it was necessary use heterologous expression of a fat-1 cDNA in a host that contained potential fatty acid substrates. Both E. coli and Saccharomyces cerevisiae, which are two common laboratory hosts for heterologous expression, possess a very limited range of endogenous desaturation activities and hence fatty acid compositions. By contrast, plants possess both a wider range of desaturase activities and fatty acids which are potential substrates for a desaturase of unknown function. In addition, the lipid and fatty acid metabolism of plants, especially Arabidopsis, have been well characterized. These features make Arabidopsis a more attractive host for transgenic studies of putative eukaryotic fatty acid desaturases than either E. coli or S. cerevisiae. Such considerations were important in allowing us to identify the C. elegans fat-1 gene product as a functional homolog of the Arabidopsis FAD3 protein. The ability of Arabidopsis plants to take up exogenous fatty acids provided us with a means to extend the biochemical characterization of the FAT-1 desaturase by showing that all the 18- and 20-carbon ω-6 fatty acids normally present in C. elegans are recognized as its substrates.
The FAD2 and FAD3 desaturases are known to use membrane glycerolipids, not acyl-CoAs, as substrates (23, 30). The high efficiency with which the FAT-1 enzyme desaturates the 18:2 of Arabidopsis membrane lipids, and the high homology of FAT-1 to FAD2 and FAD3 strongly suggest that FAT-1 is also a glycerolipid desaturase and in this respect the enzyme is similar to the Δ5 desaturase activity described in rat liver (14). These findings raise the possibility that other desaturases required for the synthesis of arachidonic acid in mammals also use membrane phospholipids as their substrates.
The results reported here demonstrate the power of well resourced models such as Arabidopsis and molecular–genetic techniques to solve recalcitrant problems in the biochemistry and cell biology of very unrelated organisms. The identification of the fat-1 gene will allow the generation and isolation of stable mutations at fat-1. Such C. elegans mutants will provide new opportunities to elucidate the roles of polyunsaturated fatty acids and compounds derived from them in the development and cell biology of this model organism.
Acknowledgments
We are grateful to Tom Savage for help and advice on gas chromatography–mass spectrometry identification of fatty acid methyl esters. This work was supported in part by Research Grant 9500759 from the U.S. Department of Agriculture–National Research Initiative Competitive Grants Program and by the Agricultural Research Center, Washington State University.
Footnotes
References
- 1.Needleman P, Turk J, Jachshik B A, Morrison A R, Lefkowith J B. Annu Rev Biochem. 1986;55:69–102. doi: 10.1146/annurev.bi.55.070186.000441. [DOI] [PubMed] [Google Scholar]
- 2.Smith W L, Borgeat P. In: Biochemistry of Lipids and Membranes. Vance D E, Vance J E, editors. Menlo Park, CA: Benjamin/Cummings; 1986. pp. 325–360. [Google Scholar]
- 3.Holloway P W. In: The Enzymes. Boyer P D, editor. Vol. 16. New York: Academic; 1983. pp. 63–83. [Google Scholar]
- 4.Strittmatter P, Spatz L, Corcoran D, Rogers M J, Setlow B, Redline R. Proc Natl Acad Sci USA. 1974;71:4565–4569. doi: 10.1073/pnas.71.11.4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thiede M A, Ozols J, Strittmatter P. J Biol Chem. 1985;260:14459–14463. [PubMed] [Google Scholar]
- 6.Thiede M A, Ozols J, Strittmatter P. J Biol Chem. 1986;261:13230–13235. [PubMed] [Google Scholar]
- 7.Schmidt H, Dresselhaus T, Buck F, Heinz E. Plant Mol Biol. 1994;26:631–642. doi: 10.1007/BF00013749. [DOI] [PubMed] [Google Scholar]
- 8.Hilditch T P, Williams P N. The Chemical Constituents of Natural Fats. 4th Ed. London: Chapman & Hall; 1964. [Google Scholar]
- 9.Browse J, Somerville C. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:467–506. [Google Scholar]
- 10.Browse J A, McCourt P J, Somerville C R. Science. 1985;227:763–765. doi: 10.1126/science.227.4688.763. [DOI] [PubMed] [Google Scholar]
- 11.Browse J, Somerville C. In: Arabidopsis. Meyerowitz E, Somerville C, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. pp. 881–912. [Google Scholar]
- 12.Arondel V, Lemieux B, Hwang I, Gibson S, Goodman H M, Somerville C R. Science. 1992;258:1353–1355. doi: 10.1126/science.1455229. [DOI] [PubMed] [Google Scholar]
- 13.Okuley J, Lightner J, Feldmann K, Yadav N, Lark E, Browse J. Plant Cell. 1994;6:147–158. doi: 10.1105/tpc.6.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pugh E L, Kates M. J Biol Chem. 1997;252:68–73. [PubMed] [Google Scholar]
- 15.Waterston R, Sulston J. Proc Natl Acad Sci USA. 1995;92:10836–10840. doi: 10.1073/pnas.92.24.10836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hutzell P A, Krusberg L R. Comp Biochem Physiol. 1982;73B:517–520. [Google Scholar]
- 17.Satouchi K, Hirano K, Sakaguchi M, Takehara H, Matsuura F. Lipids. 1993;28:837–840. doi: 10.1007/BF02536239. [DOI] [PubMed] [Google Scholar]
- 18.Devereaux J, Haeberli P, Smithies O. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baulcombe D C, Saunders G R, Bevan M W, Mayo M A, Harrison B D. Science. 1986;321:446–449. [Google Scholar]
- 20.Spychalla J P, Bevan M W. In: Plant Tissue Culture Manual: Fundamentals and Applications. Lindsay K, editor. B11. Dordrecht, The Netherlands: Kluwer; 1993. pp. 1–18. [Google Scholar]
- 21.Holsters M, de Waele D, Depicker A, Messens E, Van Montagu M, Schell J. Mol Gen Genet. 1978;163:181–187. doi: 10.1007/BF00267408. [DOI] [PubMed] [Google Scholar]
- 22.Bouchez D, Camilleri C, Caboche M. CR Acad Sci Paris. 1993;316:1188–1193. [Google Scholar]
- 23.Miquel M, Browse J. J Biol Chem. 1992;267:1502–1509. [PubMed] [Google Scholar]
- 24.Verwoerd T C, Dekker B M M, Hoekema A. Nucleic Acids Res. 1989;17:2362. doi: 10.1093/nar/17.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dellaporta S L, Wood J, Hicks J B. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- 26.Heinz E. In: Lipid Metabolism in Plants. Moore T S, editor. Boca Raton, FL: CRC; 1993. pp. 33–89. [Google Scholar]
- 27.Polashok J J, Chin C-K, Martin C E. Plant Physiol. 1992;100:894–901. doi: 10.1104/pp.100.2.894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grayburn W S, Collins G B, Hildebrand D F. Bio/Technology. 1992;10:675–677. doi: 10.1038/nbt0692-675. [DOI] [PubMed] [Google Scholar]
- 29.McConn M, Browse J. Plant Cell. 1996;8:403–416. doi: 10.1105/tpc.8.3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Browse J, McConn M, James D, Miquel M. J Biol Chem. 1993;268:16345–16351. [PubMed] [Google Scholar]
- 31.Shanklin J, Whittle E, Fox B G. Biochemistry. 1994;33:12787–12794. doi: 10.1021/bi00209a009. [DOI] [PubMed] [Google Scholar]
- 32.Stukey J E, McDonough V M, Martin C E. J Biol Chem. 1990;265:20144–20149. [PubMed] [Google Scholar]
- 33.Jackson M R, Nilsson T, Peterson P A. EMBO J. 1990;9:3153–3162. doi: 10.1002/j.1460-2075.1990.tb07513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]