Abstract
Background
The mammalian receptor protein tyrosine kinase (RTK), Anaplastic Lymphoma Kinase (ALK), was first described as the product of the t(2;5)chromosomal translocation found in non-Hodgkin’s lymphoma. While the mechanism of ALK activation in non-Hodgkin’s lymphoma has been examined, to date, no in vivo role for this orphan insulin receptorfamily RTK has been described.
Results
We describe here a novel Drosophila melanogaster RTK, DAlk, which we have mapped to band 53on the right arm of the second chromosome. Fulllength DAlk cDNA encodes a phosphoprotein of 200 kDa, which shares homology not only with mammalian ALK but also with the orphan RTK LTK. Analysis of both mammalian and Drosophila ALK reveals that the ALK family of RTKs contains a newly identified MAM domain within their extra cellular domains. Like its mammalian counterpart, Dalk appears to be expressed in the developing CNS by in situ analysis. However, in addition to expression of DAlk in the Drosophila brain, careful analysis reveals anadditional early role for DAlk in the developing visceralmesoderm where its expression is coincident withactivated ERK.
Conclusion
In this paper we describe a Drosophila melanogaster Alk RTK which is expressed in the developing embryonic mesoderm and CNS. Our data provide evidence for the existence of a DAlk RTK pathway in Drosophila. We show that ERK participates in this pathway, and that it is activated by DAlk in vivo. Expression patterns of dALK, together with activated ERK, suggest that DAlk fulfils the criteria of the missing RTK pathway, leading to ERK activation in the developing visceral mesoderm.
Introduction
A wide range of processes involving intercellular communication are mediated by receptor tyrosine kinases (RTKs) and their signalling pathways. The growing list of processes regulated by these receptors across the phylogenetic tree is extremely broad, and includes the induction of cell fates, guidance of cell and axon migration and cell proliferation. A definition of cellular responses such as proliferation, differentiation or survival by a variety of external stimuli involves the regulation of transcriptional events in eukaryotic cells through intracellular signalling cascades, including pathways that activate protein kinases of the mitogen-activated protein kinase (MAPK) family (Treisman 1996; Widmann et al. 1999). Cellular responses, including proliferation and differentiation, are influenced by extracellular factors such as polypeptide growth factors. Most of the effects of these growth factors are mediated by RTKs—high-affinity receptors with protein tyrosine kinase (PTK) activity. RTKs are composed of three domains: an extracellular ligand-binding domain, a single membrane-spanning domain and a cytoplasmic catalytic domain (Yarden & Ullrich 1988). Ligand binding to the extracellular domain induces activation of the kinase on the cytoplasmic side, which initiates the intracellular signalling. The activated RTKs phosphorylate themselves and the cytoplasmic substrates, leading to activation of a number of downstream signaling molecules, and ultimately inducing changes in gene expression and the phenotypic state of the cell (Fantl et al. 1993; van der Geer et al. 1994). RTKs thus play important roles in cellular proliferation and differentiation. In addition, RTKs reveal an oncogenic potential when their kinase activities are constitutively enhanced by point mutation, amplification or rearrangement of the corresponding genes (Robertson et al. 2000).
Mammalian Anaplastic Lymphoma Kinase (ALK) was originally identified as a member of the insulin receptor subfamily of receptor tyrosine kinases (RTKs) which acquire their transforming capability when they are truncated and fused to nucleophosmin (NPM) in the t(2;5) chromosomal rearrangement associated with non-Hodgkin’s lymphoma (Morris et al. 1994; Shiota et al. 1994). To date, several chromosomal rearrangements leading to an activated ALK RTK have been described, including NPM-ALK (Morris et al. 1994; Shiota et al. 1994) and ATIC-ALK (Colleoni et al. 2000; Ma et al. 2000; Trinei et al. 2000) which are constitutively dimerized through the fused domain. However, there are few insights into the normal structure and function of the ALK RTK. Full-length cDNA encoding the mammalian ALK RTK has been identified (Iwahara et al. 1997; Morris et al. 1997) as a first step towards a functional assessment of the receptor. This has shown ALK to be a member of the Insulin Receptor superfamily, most closely related to the orphan RTK leucocyte tyrosine kinase (LTK) (Iwahara et al. 1997; Morris et al. 1997). In situ hybridization studies have revealed ALK expression in the developing nervous system and ALK is currently a novel orphan receptor tyrosine kinase that is suspected to play important role in the normal development and function of the nervous system.
In this paper we describe a Drosophila melanogaster homologue of ALK, which we have named DAlk. This novel RTK was identified using a degenerate PCR approach (Palmer et al. 1999). DAlk is a 200 kDa RTK that has strong homology with both ALK and LTK. Due to the conserved nature of many receptor signalling systems in Drosophila, ALK RTK mediated signalling may also be conserved from Drosophila to vertebrates. Drosophilahas a smaller number of RTK genes than vertebrates, with ≈21 RTKs now predicted to be encoded by the Drosophila melanogaster genome (G. Plowman, personal communication). In addition, since the sequencing of the Drosophila melanogaster genome has now been completed (Adams et al. 2000) we can say that while an Insulin Receptor homologue is present (Fernandez et al. 1995; Ruan et al. 1995), there appears to be no homologue for the ALK relative RTK, LTK in Drosophila melanogaster. We have been able to show, both by in situ hybridization analysis and by immunostaining, that DAlk is expressed during early mesodermal development as well as within the developing nervous system. Interestingly, early expression of DAlk in the mesoderm correlates with ERK activation in the developing embryo mesoderm in vivo (Gabay et al. 1997). Furthermore, using the UAS-GAL4 expression system, together with clonal overexpression techniques, we observe that DAlk can indeed activate ERK in vivo.
Results
Identification of a novel Drosophila melanogaster RTK: DAlk
To identify novel PTKs in Drosophila melanogaster, we have utilized a degenerate PCR-based approach (Palmer et al. 1999). Highly conserved residues within subdomains VIb and IX of known PTKs were targeted for degenerate PCR primer design (Lai & Lemke 1991), leading to the identification of several novel putative Drosophila melanogaster PTKs. Multiple PCR products were obtained and sequenced, identifying novel as well as previously described Drosophila melanogaster PTKs (Palmer et al. 1999). One of the novel PCR products, displayed the greatest similarity to members of the mammalian Insulin Receptor RTK superfamily (Whitehead et al. 2000).
Full length cDNAs were obtained by screening Drosophila melanogaster adult cDNA libraries. Multiple cDNAs were obtained, falling into two classes, based on alternate splicing within the 5′ UTR (see below, Fig. 2). No alternate splicing was observed within theORF of these novel cDNA species. We have named this locus DAlk53 (see below). Figure 1 A shows the complete amino acid sequence of full-length Dalk cDNA. The DAlk open reading frame of predicts a 1701 amino acid, 180 kDa novel protein. Analysis of the predicted amino acid sequence reveals an amino terminal signal sequence, as well as a hydrophobic transmembrane domain. BLAST homology searching of the NCBI database revealed that DAlk does indeed appear to encode a novel RTK in the insulin receptor superfamily (Fig. 1C). A Drosphila insulin receptor already exists (Fernandez et al. 1995; Ruan et al. 1995), but no Drosophila counterpart for the LTK/ALK single pass RTK branch of the INR superfamily has been described. Our novel Drosophila RTK shows the most homology with a previously described mammalian RTK, ALK with 34% identity to ALK (52% in the cytoplasmic region) as well as a conserved overall structure (Fig. 1C,D). DAlk, like mammalian ALK, encodes for several putative domains, an aminoterminal signal sequence, an extracellular domain, a hydrophobic transmembrane region and a cytoplasmic PTK domain. The kinase domain of DAlk is most similar (58% identity; 85% homology with hALK) to those of the Insulin Receptor superfamily (Fig. 1A; shaded) and contains several sequence motifs conserved among PTKs, including the tripeptide motif DFG that is found in most kinases, and a consensus ATP-binding motif GxGxxG followed by an AxK sequence downstream (Fig. 1A; underlined). The cytoplasmic domain of DAlk contains a NPNY putative IRS/Shc-binding consensus sequence at amino acid 1170 (Fig. 1A; boxed), homologous to the NPXY motif in p80–NPM/ ALK, which has been shown to bind to mammalian IRS1 when tyrosine phosphorylated. Within the amino-terminal extracellular domain of DAlk several features are found: (i) an LDLa domain (Daly et al. 1995; Fass et al. 1997), (ii) a MAM domain (named after meprins, A-5 protein and receptor protein tyrosine phosphatase mu) and (iii) a glycine-rich region. Careful analysis of the mammalian ALK sequence revealed that both the LDLa and MAM domains can also be seen in mammalian ALK, although they have not previously been noted. The MAM domain is not found in LTK. MAM domains comprise about 160 amino acids that are present in transmembrane proteins such as the meprins and receptor protein-tyrosine phosphatases, where they appear to function in cell/cell interactions (Beckmann & Bork 1993; Jiang et al. 1993). Thus we propose that the ALK family of RTKs is novel within the RTK family by virtue of a MAM domain within their extracellular domains. Currently the functions of this domain in the regulation of ALK family RTKs are unknown.
Figure 2.
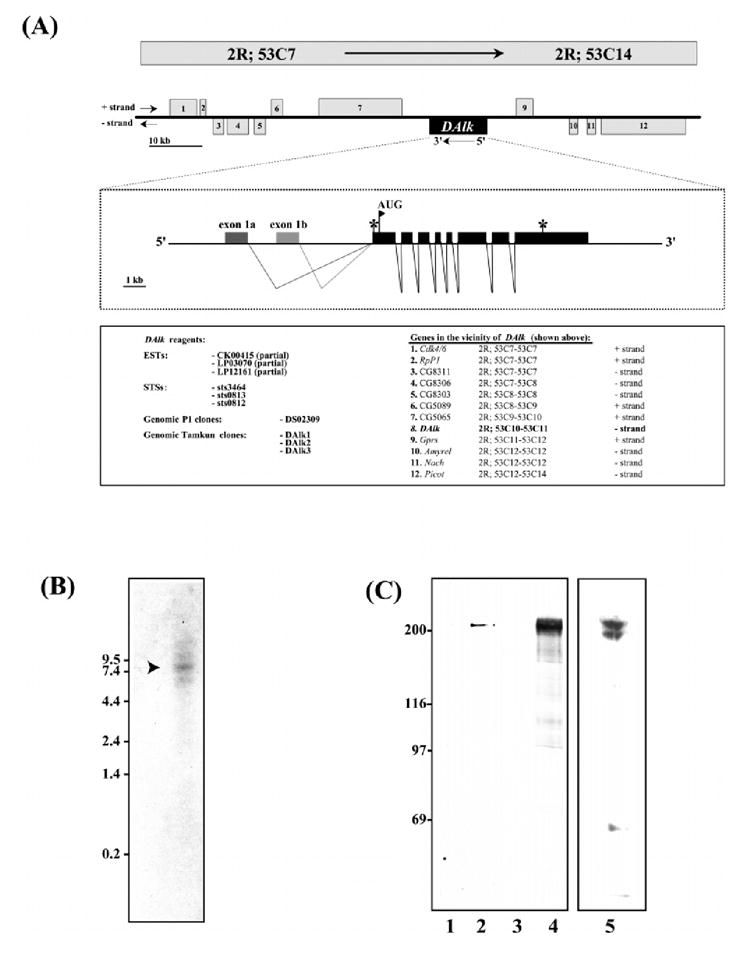
(A) Genomic characterization of DAlk. (Top panel) A map corresponding to the genomic region 53C7–53C14 on the second right chromosome comprising approximately 100 kb of genomic DNA containing the DAlk locus is shown. DAlk (centre) is depicted by the solid black box, and the surrounding genes, numbered 1 through 12, are indicated as grey boxes. (Middle panel) The deduced structure of the ≈15 kb DAlk transcription unit is shown below in expanded form. The 10 exons are represented by solid black boxes, and thin lines represent intron regions. The stop codon preceding the first AUG initiation codon (marked with a flag), is indicated by an asterisk (*), as is the TAA termination codon at the 3′ end of the DAlk transcript. Three EST sequences, CK00415, LP03070 and LP12161, encode partial DAlk sequences. (Bottom panel) ESTs, STSs, Genomic clones and genes in the vicinity of DAlk are indicated. (B) Expression of DAlk mRNA. Northern blot analysis using total RNA from embryos. 20 μg total RNA was loaded. (C) Over-expression of DAlk in 293 cells. 293 cells were cDNA encoding full length DAlk (lanes 2 and 4). Vector DNA alone was used as a control (lanes 1 and 3). DAlk was analysed by SDS-PAGE followed by anti-DAlk immunoblot (lanes 1 and 2) and antiphosphotyrosine immunoblot (lanes 3 and 4). In addition, anti-DAlk antibodies recognize a doublet at 200 kDa in embryo extracts (lane 5).
Figure 1.

Molecular characterization of DAlk. (A) Protein sequence of DAlk. The protein sequence for DAlk is shown. Though not shown here, an in-frame upstream STOP codon precedes the initial methionine residue. The termination STOP codon is indicated by an asterisk (*). The signal sequence, LDLa, MAM, G-rich, transmembrane and PTK domains are shaded, and the consensus GxGxxG and AxK in the ATP binding site is underlined. The originally identified 64 amino acid PCR clone is double underlined. The putative IRS/SHC SH2 binding site (NPNY1170) is boxed. (B) DAlk schematic outline. (C) Schematic comparison of DAlk with the mammalian LTK/ALK family RTKs, ALK. Also shown is the p80-NPM/ALK protein product of the t(2;5) chromosomal translocation found in non-Hodgkins lymphoma. Structural conservation between DAlk, hALK and LTK include the presence of an intracellular PTK domain, an amino-terminal signal sequence, and an extracellular glycine-rich domain. Only DAlk and hALK share extracellular LDLa and MAM domains. (D) Alignment of the deduced amino acid sequences of Drosophila and human ALK. The signal sequence, LDLa, MAM, G-rich, transmembrane and PTK domains are shaded. The putative IRS/SHC SH2 binding site (NPNY1170) is boxed. The amino acid residues which are identical between the two species are marked by asterisks.
DAlk maps to 53C/D on the right arm of the second chromosome
In situ hybridization to polytene chromosomes isolated from third instar larva localized DAlk to region 53 on the second chromosome; therefore we have named this novel Drosophila melanogaster PTK locus DAlk53. Using the in situ mapping information, we have confirmed and further defined the genomic localization of Dalk to region 53C10-C11 in the Drosophila melanogaster genome.
In addition to mapping DAlk53, we have carried out a careful characterization of this region (Fig. 2A). The data here are derived from P1 genomic sequence data from the BDGP (Kimmerly et al. 1996), our own genomic sequence data from four overlapping cosmids covering the DAlk53 locus, multiple DAlk cDNAs, P element data from this laboratory and the BDGP (Spradling et al. 1999; Spradling et al. 1995) as well as STS and EST data from the BDGP (Rubin et al. 2000). These data have recently been confirmed by the published BDGP data (Adams et al. 2000). DAlk maps to an approximately 15 kb genomic fragment between sts3464 and sts0182 within P1 DS02309 and, to the best of our knowledge at this time comprises eight coding exons. Of the multiple DAlk cDNAs obtained, no alternative splicing events within the ORF were observed. However, an analysis of the 5′ UTR of six independent cDNA clones revealed alternate splicing events 5′ of the ORF. Both of these 5′ UTR exons were further characterized and found to map upstream of the ORF encoding exons in the genome (see Fig. 2A). An approximately 8.5 kb DAlk major RNA species was detected by Northern blot analysis (Fig. 2B), in good agreement with our cDNA data, which predicts a cDNA of 8466 bp, inclusive of both 5′ and 3′ UTR regions. The region surrounding DAlk53 contains few P element insertions, none of which are within the DAlk gene, the closest we have characterized being some 40 kb 3′ [l(2)k06503] and within the CDK4/6 gene (Sauer et al. 1996). Other genes in the vicinity of DAlk are noted (Fig. 2A).
To characterize the DAlk protein, pcDNA3:Dalk was transiently expressed in 293 cells. Anti-Dalk antibodies were used to detect DAlk from cell lysates. Lysates which were resolved on SDS-PAGE and analysed by immunoblotting for DAlk. Figure 2 C shows that DAlk antibodies specifically recognized a 200 kDa protein, which is present when the cells were transfected with pcDNA3:DAlk (lane 2), but not with vector alone (lane 1). Lysates were also analysed by anti-phosphotyrosine immunoblotting (Fig. 2C), where DAlk was detected as a 200 kDa tyrosine phosphorylated protein (lane 4), suggesting that DAlk is indeed a PTK. Furthermore, anti-DAlk antibodies recognize a doublet of endogenous DAlk at approximately 200 kDa from whole embryo extracts (Fig. 2C; lane 5). Currently, the nature of this doublet is unknown; it may reflect the phosphorylation status of DAlk, although alternative splicing may also be responsible.
DAlk is expressed in the developing embryonic nervous system: in situ analysis
We have examined the expression of DAlk throughout embryonic development by in situ analysis. Embryos from 0 to 24 h of age were analysed by in situ hybridization using a 3′ digoxigenin DAlk probe (Fig. 3). The DAlk transcript is found in the mesoderm and the dorsal ectoderm earlier in development (stages 10 and 11) (Fig. 3B, C, D). From around 1 h of development to late embryogenesis, DAlk mRNA is concentrated within the developing nervous system, and is observed in both the developing brain and ventral nerve cord (VNC) (Fig. 3E–G).
Figure 3.
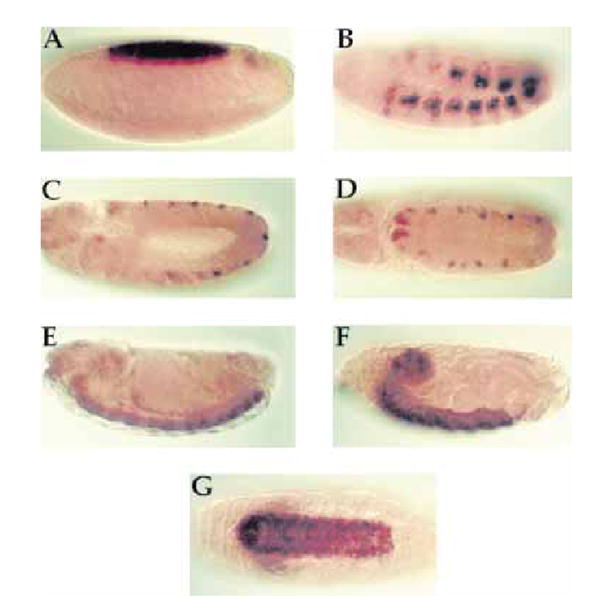
Expression of DAlk during embyrogenesis: in situ hybridization. In situ hybridization of DAlk to whole-mount wild-type embryos showing the expression of DAlk at: (A) stage 6 in the presumptive amnioserosa, (B) stage 10 in a subset of mesodermal cells, (C and D) stage 11 in a subset of ectodermal cells, and (E) stage 14 and (F and G) stage 17 in the developing ventral nerve cord (VNC). Anterior is to the left and dorsal is up, except for (D) which is a dorsal view and (G) which is a ventral view.
DAlk expression in the developing embryo
To further define where DAlk is expressed, we generated monoclonal antibodies to the extracellular portion of the DAlk protein (see Experimental procedures). Immunostaining with anti-DAlk antibodies reveals that DAlk is expressed in a striking pattern throughout development (Fig. 4A). All DAlk expression patterns were abolished by competition with the original DAlk immunogen (data not shown). In particular, prominent DAlk staining is observed in the visceral mesoderm at stage 11. It is first seen as segmental patches, before a fusion of the visceral arches from each segment, and is subsequently observed as a continuous waved line (Fig. 4A, ii.). The uniform pattern of DAlk in the visceral mesoderm suggests that it may not be involved in the directed migration of these cells, but possibly in their differentiation.
Figure 4.
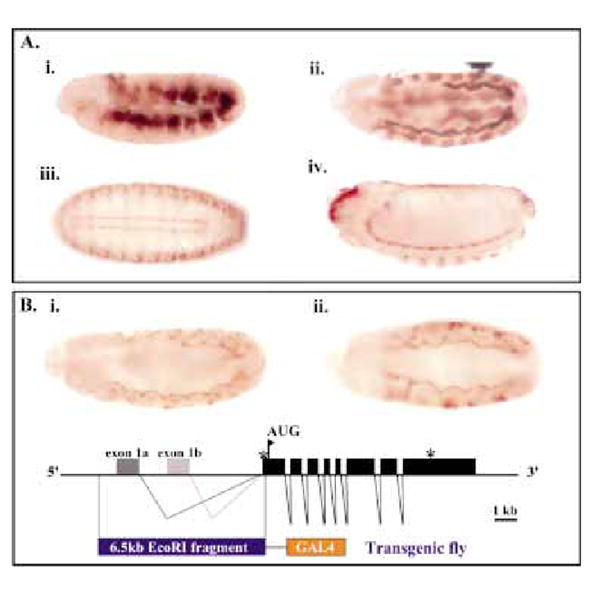
DAlk expression. (A) DAlk expression during embryogenesis. DAlk immunostaining of developing embryos is shown in brown. Embryo orientation is anterior to the left, dorsal up (i, ii, iv), dorsal facing (iii). (i) At stage 10, DAlk is highly expressed within a subset of mesodermal cells. (ii) Stage 11. DAlk protein is found in the visceral mesoderm. Staining can also be observed at the invagination of the tracheal pits. (iii and iv) Strong expression of DAlk is restricted to the brain and the ventral nerve cord. (B) DAlk enhancer promoter region analysis. The DAlk transcription unit is depicted as a schematic diagram. A 6.5 kb EcoRI fragment 5′ of the ORF of DAlk (black box) was placed upstream of Gal4 as shown. Transgenic flies carrying DAlkEI6.5-GAL4 crossed to UAS-nuclearGFP reporter flies displayed a visceral mesoderm expression pattern (i) lateral view, stage 11 embryo (ii) ventral view, stage 11 embryo.
We have further investigated the expression of DAlk through an analysis of the putative enhancer promoter region. The DAlk transcription unit is composed of eight coding exons and two 5′ alternatively spliced noncoding exons spanning approximately 15 kb (Fig. 2). Since both exon1A and exon1B mapped within a 6.5-kb EcoRI fragment 5′ of the ORF of DAlk (Fig. 4B), this fragment was used to generate a transgenic fly in which the putative enhancer region of DAlk was placed upstream of Gal4 in the P-element vector RSBSK (pRSBSK:DAlkEI6.5-GAL4, Fig. 4B, see Experimental procedures). The resulting transgenic flies were then used to drive UAS-GFP reporters. As judged by reporter gene expression, the 6.5 kb EcoRI genomic fragment drives reporter gene expression in the visceral mesoderm (VM) at stages 11/12 in a pattern similar to that seen for the DAlk protein (Fig. 4B). At later stages (from stage 13 onwards) of embryonic development, DAlk was found to be expressed within the developing brain and ventral nerve cord (Fig. 4A, iii and iv). The expression of DAlk within the CNS persists through larval instar stages where DAlk is highly expressed within the brain and ventral nerve cord (data not shown).
DAlk is the ‘unknown’ RTK expressed in the developing visceral mesoderm
A 1997 study conducted by Gabay et al. produced a detailed ‘atlas of MAPK activation’ in vivo (Gabay et al. 1997). These authors used antibodies that were specific for activated phospho-ERK as a tool for dissecting ERK activation throughout Drosophila embryonic development. They noted that most aspects of the phospho-ERK pattern observed could be accounted for by known Drosophila RTK pathways. However, several of the patterns revealed were novel with respect to the receptor they are triggered by. They speculated that these patterns may be induced by unknown RTKs that may activate ERK. In particular, prominent phospho-ERK staining was observed in the visceral mesoderm at stage 11. It was first seen as segmental patches, before fusion of the visceral arches from each segment, and subsequently observed as a continuous waved line. Furthermore, this phospho-ERK pattern in the visceral mesoderm was not dependent upon the Heartless RPTK. Since we had observed DAlk expression in the visceral mesoderm (Fig. 4A, ii), we wished to examine whether or not DAlk expression coincided with the phospho-ERK pattern in the visceral mesoderm in vivo.
In order to confirm that DAlk and phospho-ERK were expressed in the visceral mesoderm during development, wild-type embryos were collected and stained for DAlk and phospho-ERK (Fig. 5A). In both cases, expression was observed in the visceral mesoderm at stages 11/12 in a similar pattern. Subsequently, embryos were collected and double-stained for activated phospho-ERK (Fig. 5B, i) and DAlk (Fig. 5B, iii). Co-localization of both activated phoshpo-ERK and DAlk could clearly be observed in the visceral mesoderm (Fig. 5B, ii).
Figure 5.
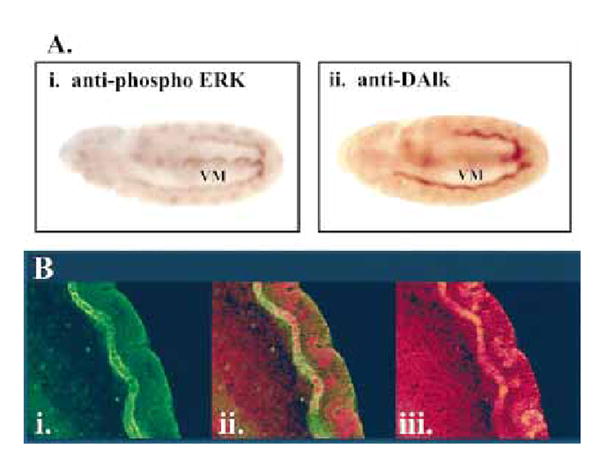
DAlk and phospho-MAPK co-localize in the visceral mesoderm. (A) DAlk and phospho-ERK co-localize in the visceral mesoderm. (i) At stage 11 phospho-ERK is detected in the visceral mesoderm (VM), prior to and after the generation of a continuous visceral stripe. (ii) anti-DAlk staining of a stage 11 embryo, showing DAlk protein in the visceral mesoderm (VM), also prior to and after the generation of a continuous visceral stripe. (B) Stage 11 embryo stained with (i) anti-phosphoERK (red) and (iii) anti-DAlk (green). (iii) Expression of both phospho-ERK and DAlk co-localize and are specifically detected in the visceral mesoderm at this stage (merged image).
So far we have been unable to obtain DAlk mutants (see Discussion) and so we were unable to examine whether DAlk is responsible for ERK activation in the developing visceral mesoderm in vivo. However, we were able to ask if DAlk was capable of driving ERK activation in vivo by utilizing the GAL4-UAS system (Brand & Perrimon 1993). DAlk cDNA was cloned into P element expression vectors under the control of yeast GAL4 upstream activating sequences (UAS) and P element-mediated germ-line transformation was used to generate UAS:DAlk transgenic fly lines. When DAlk was expressed ectopically under the control of the Actin5C promoter driving GAL4 (Actin5C-GAL4) the result was 100% embryonic lethality (data not shown). In order to examine whether the DAlk RTK is capable of driving ERK activation in vivo we employed pGMR-GAL4, which drives expression in all photoreceptor cells (Ellis et al. 1993), to express DAlk in the developing eye disc (Fig. 6 i and ii). We were able to observe a very clear effect of DAlk expression on ERK activation: normally prominent ERK activation is seen within the morphogenetic furrow, with lower levels in the differentiated third instar eye disc ( Fig. 6iii). In contrast, high levels of ERK activation in vivo were observed when DAlk was expressed ( Fig. 6 iv). Further conformation of DAlk driven ERK activation in vivo was achieved using a combination of the FLP-out system and the GAL4-UAS system (Ito et al. 1997). In this system, a fragment of DNA bracketed by FRT sites and containing transcription stop signals is inserted between the Actin5C promoter and GAL4. Heat shock induction of Flippase activity induces recombination in which the transcription stop segment is flipped out, thereby allowing the Actin5C promoter to drive the GAL4 expression. This system allows the creation of clones of cells expressing DAlk, which are marked by GFP expression (Fig. 7). The expression of DAlk, as judged by immunostaining, and GFP were coincident (Fig. 7A), demonstrating that the system works for DAlk as well as establishing the specificity of the anti-DAlk antibodies. While endogenous DAlk protein is expressed in the third instar brain during normal development, levels of DAlk within overexpressing clones are clearly observed over endogenous levels. DAlk over-expressing clones also displayed increased levels of phosphotyrosine, consistent with the over-expressed DAlk being active and either directly or indirectly leading to protein phosphorylation in these clones (Fig. 7B). Furthermore, larger clones were observed to disrupt the normal tissue structure, leading to abnormal disc development (arrow, Fig. 7B). Animals carrying DAlk over-expression clones did not survive to adulthood (data not shown). Further analysis of DAlk clones in discs isolated from third instar larva indicated that DAlk leads to ERK activation in situ (Fig. 7C), consistent with our previous results. Thus, DAlk has the capacity to drive activation of ERK in vivo,and is therefore a prime candidate for the ‘missing’ RTK driving ERK activation within the developing visceral mesoderm in vivo.
Figure 6.
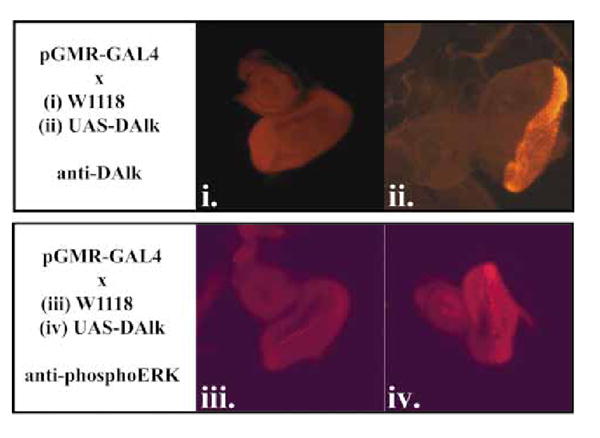
DAlk drives ERK activation in the developing eye disc. Wild-type DAlk is capable of driving ERK activation in vivo. (i) Third instar eye disc from a control animal carrying pGMR-GAL4; anti-DAlk. (ii) Third instar eye disc from an animal carrying pGMR-GAL4 driving UAS-DAlk displaying DAlk overexpression: anti-DAlk. (iii) Third instar eye disc from a control animal carrying pGMR-GAL4; anti-phosphoERK. (iv) Third instar eye disc from an animal carrying pGMR-GAL4 driving UAS-DAlk displaying DAlk over-expression: anti-phosphoERK.
Figure 7.
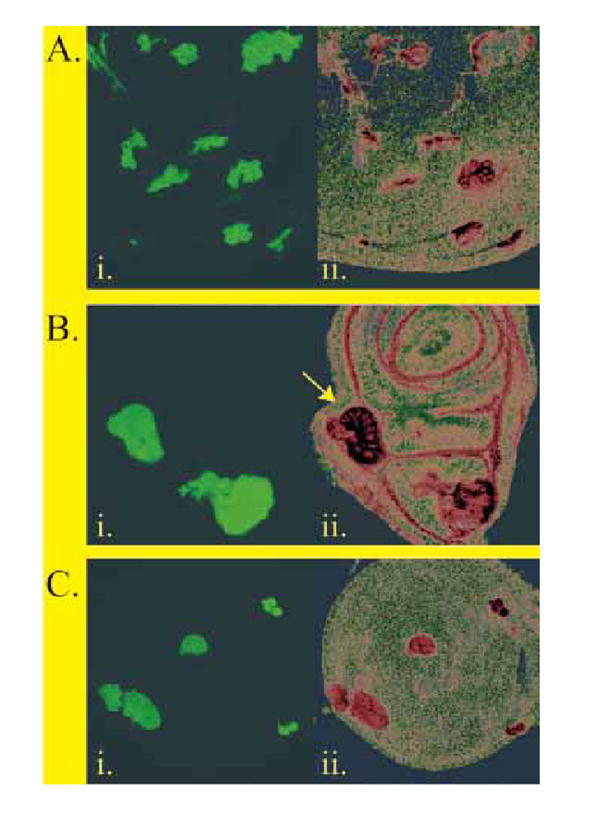
DAlk clonal over-expression in developing imaginal discs. (A) Heat-shock induced clones over-expressing wild-type DAlk under the Actin-5C promoter. (i) Imaginal discs displaying heat-shock induced over-expression clones marked by the overexpression of UAS-GFP. (ii) The DAlk immunostaining of imaginal discs with anti-DAlk antibodies is shown in red. (B) Heat-shock induced clones over-expressing wild-type DAlk under the Actin-5C promoter contain increased levels of phosphotyrosine. (i) Third instar wing disc displaying heat-shock induced over-expression clones marked by the over-expression of UAS-GFP. (ii) Anti-phosphotyrosine immunostaining of the same tissues with 4G10 anti-phosphotyrosine antibodies shown in red. Arrow indicates tissue perturbation observed during analysis of larger DAlk clones. (C) Heat-shock induced clonal overexpression of wild-type DAlk under the Actin-5C promoter results in ERK activation. (i) Third instar wing disc displaying heat-shock induced DAlk over-expression clones marked by the over-expression of UAS-GFP. (ii) Anti-phosphoERK immunostaining of the same tissues antibodies is shown in red.
Discussion
It is estimated that the Drosophila genome encodes 21 RTKs and 11 non-receptor PTKs (G. Plowman, personal communication). Therefore, we now have a finite number of RTK controlled pathways which remain to be elucidated in Drosophila melanogaster. To date, the following RTKs have been described: Breathless (Glazer & Shilo 1991), Heartless (FGFR1/ DFR1) (Beiman et al. 1996; Gisselbrecht et al. 1996; Shishido et al. 1997), DER (EGFR/Torpedo/faint little ball/Ellipse) (Livneh et al. 1985; Wadsworth et al. 1985), Derailed (Linotte) (Callahan et al. 1995; Dura et al. 1995), Dnrk (Oishi et al. 1997), Dror (Dtrk) (Wilson et al. 1993), Sevenless (Hafen et al. 1987), Torso (Casanova & Struhl 1989; Sprenger et al. 1989), dInsR (Fernandez et al. 1995; Ruan et al. 1995) and DEK (Scully et al. 1999). Of these, a substantial proportion are known to be involved in the setting up of a specific system during the developmental process. In this paper we report the identification of a novel RTK in Drosophila–DAlk. Structurally, DAlk falls into the Insulin Receptor superfamily, and more specifically into the LTK/ALK family of mammalian orphan RTKs (Iwahara et al. 1997; Morris et al. 1997). DAlk is the first described Drosophila melanogaster protein to share a strong structural and sequence homology with the mammalian LTK/ALK RTK family members. Indeed, phylogenetic tree analysis suggests that DAlk forms its own group of RTKs in Drosophila melanogaster and that it is more similar to the mammalian ALK and Ltk than to other RTKs thus far described in Drosophila. We have observed evidence of only one ALK family member in Drosophila melanogaster—DAlk; all cDNAs obtained during the screening encoded DAlk, and, while several partial ESTs for DAlk currently exist in the databases, no other related potential DAlk family member exists. DAlk is a Drosophila prototype of the mammalian ALK/LTK family, and this is supported by the fact that it is the only ALK/LTK orthologue in the complete Drosophila melanogaster genome sequence (Adams et al. 2000). Among conserved sequence motifs (see Fig. 1 and Results), we note particularly the previously unrecognized MAM domain in the extracellular domain of the ALK RTKs–both DAlk and hALK (Fig. 1). The presence of this motif, which appears to function in cell/cell interactions, indicates an important function (Beckmann & Bork 1993; Jiang et al. 1993). Currently the ALK RTKs are orphan receptors, and no ligands have been identified to date. The existence of a MAM domain in the extracellular domain suggests that perhaps the ALK RTKs can be activated in a different manner than other RTKs. Furthermore, Iwahara et al. noted that the ALK RTK over-expressed in COS cells may be activated and autophosphorylated without ligand stimulation (Iwahara et al. 1997). We have also noted such a phenomenon when examining the DAlk overexpressed in 293 cells, suggesting that DAlk may also be activated in a ligand-independent manner: furthermore, DAlk appears to be active when it is expressed in the developing eye disc (where it is not normally expressed) (Fig. 5C). In addition to a MAM domain, we have also observed that both DAlk and hALK contain an LDLa motif (Daly et al. 1995; Fass et al. 1997), other functional motifs, such as the PTK domains characterized in vertebrate ALK are conserved in DAlk.
We have mapped DAlk to 53C/D in the Drosophila melanogaster genome and characterized the surrounding genomic region in detail. Since no deficiencies span this region, nor candidate DAlk mutants currently exist, we are now in the process of actively targeting DAlk for disruption. Attempts to target DAlk through local P–element mobilization techniques have so far been unsuccessful.
DAlk mRNA is expressed in a highly regulated pattern in Drosophila embryos; we observe expression in the mesoderm during germ band elongation, while later in embryonic development DAlk mRNA is concentrated within the developing nervous system. We have observed a very similar pattern of DAlk protein expression, where DAlk can be seen as a membrane localized protein in the developing visceral mesoderm and later on is seen to be specifically expressed in the brain and ventral nerve cord. While expression in the embryonic and larval CNS was predicted, the highly dynamic expression in the visceral mesoderm was unexpected. It appears that DAlk may play a previously unrealized important initial role in mesoderm development in vivo, prior to its role in CNS development.
The ‘atlas of MAPK activation’ produced in vivo by Gabay et al. identified patterns of ERK activation throughout Drosophila embryonic development (Gabay et al. 1997). This work noted that most aspects of the phospho-ERK pattern observed can be accounted for by known Drosophila RTK pathways. However, several of the patterns revealed were novel and it is thought that these patterns may be induced by unknown RTKs activating ERK in vivo. They were unable to identify the RTK responsible for the prominent phospho-ERK staining observed in the visceral mesoderm. In this paper we have shown that DAlk and phospho-ERK co-localize in this tissue and thus DAlk appears to be the previously unknown RTK which is responsible for the activation of ERK in the visceral mesoderm in vivo. This implies that DAlk works through an IRS/Shc type pathway (R.H.P. & T.H., unpublished data) to drive the ERK activation in Drosophila. This idea is further supported by the conservation of consensus phosphotyrosine motifs between hALK and DAlk, which, in the case of NPM-ALK, are known to utilize IRS1 and Shc (Fujimoto et al. 1996). It would also be predicted that the activation of a number of specific target genes in the visceral mesoderm will be dependent on DAlk, a subject which we are currently investigating. Since DAlk is also strongly expressed in the Drosophila brain and VNC we would expect that this novel RTK will also play an important role in the development of the CNS in vivo. Currently the role of the ALK RTK family in brain development is not understood, however, we are optimistic that the powerful genetic Drosophila system will help to shed light on such questions.
In conclusion, we have identified a novel Drosophila melanogaster RPTK, which we have named DAlk. DAlk encodes a 200 kDa PTK which is expressed in the developing visceral mesoderm and then in the nervous system of Drosophila melanogaster. At present we do not know anything about the ligands for the ALK RTK family. It may be a soluble factor or a membrane-bound protein; however, the identification of a MAM domain in the extracellular domain of DAlk and hALK raises a serious possibility that ALK RTK may be regulated in a novel ligand-independent manner. Our experiments imply a role for DAlk in the in vivo development of the visceral mesoderm, more specifically, that DAlk may drive a signal transduction pathway leading to the activation of ERK in this tissue. The identification of this novel DAlk RTK in Drosophila will allow exploitation of a genetically tractable system which could be used to improve our understanding of the role of the DAlk RTK in vivo. Indeed, the results presented here should provide the basis for such powerful genetic approaches in the future.
Experimental procedures
Drosophila stocks
Standard Drosophila husbandry procedures were employed. All Drosophila strains were maintained on a standard cornmealmolasses medium. Flies were raised and crossed at room temperature unless otherwise stated. The wild-type strain used was Canton-S. The W1118 strain was used for all microinjections described.
Molecular techniques
Standard molecular biology protocols were used (Sambrook et al. 1989). Degenerate PCR was performed as previously described (Palmer et al. 1999). Sequencing data was collected using the dideoxy chain termination method using 35S-dATP and the Sequenase sequencing kit (US Biochemicals). The sequence was determined on both strands and the resulting sequence data were compiled and assembled using the DNAstar and DNA strider computer packages (Marck 1988). Homology searches were performed using the Internet accessible NCBI GenBank BLAST programs (Altschul et al. 1990) (http://www.ncbi.nlm.nih.gov/). Library screening was performed using Drosophila melanogaster tudor male and female libraries constructed in λZAP and were plated on E. coli strain BB4 (Stratagene). Multiple cDNAs were obtained, all of which were sequenced and further analysed. A 5′ fragment of the longest clone was then employed as a probe and six of the resulting positives were isolated and sequenced. A full length DAlk cDNA, encoding the full DAlk ORF, is described and utilized in these studies. This clone contains a 525-bp 5′ untranslated region with an upstream in-frame stop codon, a 5100 bp ORF and 252 bp of 3′ untranslated sequence. Genomic cosmid clones were isolated by standard techniques from a library containing DNA from the iso-1 line prepared by John Tamkun (Tamkun et al. 1992). P1 genomic clones (Kimmerly et al. 1996) were obtained from the BDGP (Berkeley Drosophila Genome Project). Total RNA was prepared from Drosophila embryos, separated on formaldehyde- containing 1% agarose gels, and blotted on to Hybond membranes. Hybridization was carried out using standard techniques.
Immunostaining
Wild-type embryos were fixed and immunostained as described (Patel 1994). Mouse monoclonal primary antibodies against DAlk were used at 1:100, mouse polyclonal primary antibodies against DAlk were used at 1:1500. Mouse monoclonal antiphosphoERK antibodies (Sigma) were used at 1 : 1000 and rabbit polyclonal anti-phosphoERK antibodies (NEB) were used at 1 : 500. Immunolocalization was visualized with biotinylated anti-mouse IgG and the avidin DH-biotinylated horseradish peroxidase H complex using diaminobenzidine and H2O2 as a substrate (Vector lab) or fluorescent secondary antibodies (Southern Biotechnology Associates). Imaginal discs from wandering third instar larvae were dissected, collected and rinsed in PBS, prior to fixing in 3.5% paraformaldehyde for 60 min. Imaginal discs were incubated overnight at 4 °C with primary antibodies, washed 3 × 10 min in PBS/0.1% TritonX- 100, followed by overnight incubation in secondary antibody at 4°C and washed 3 × 10 min in PBS/0.1% TritonX-100. Imaginal discs were then mounted on polylysine coated slides prior to visualization by confocal microscopy.
In situ analysis
In situ hybridization of whole mount embryos was carried out as previously described (Bourgouin et al. 1992) using the DAlk cDNA to generate digoxigenin-labelled anti-sense RNA probes (Boehringer Mannheim). The sense (control) probe revealed no signal in the embryos.
Generation of transgenic flies
pBluescript:DAlk cDNA was used to generate various P element transformants. The plasmid pUAST:Dalk was constructed by subcloning the DAlk cDNA into the pUAST vector (Brand & Perrimon 1993). The plasmid pRSBSK:DAlkEI6.5-GAL4 was constructed by subcloning the 5′ 6.5 kb EcoRI fragment from DAlk genomic Tamkun DNA into the P-element vector RSBSK. P element transformation was performed by microinjection of pUAST:DAlk or pRSBSK:DAlkEI6.5-GAL4 together with a Δ2.3 transposase containing plasmid into a W1118 Drosophila melanogaster strain (Rubin & Spradling 1982; Spradling & Rubin 1982). Multiple lines were obtained for each construct injected. To drive the expression of UAS constructs, we used pGMR-GAL4 which drives expression in all photoreceptor cells in the developing eye (Ellis et al. 1993). In addition, we have used a clonally inducible GAL4 line that combines the Flippase (FLP)/FRT system and the GAL4/UAS system resulting in the clonal expression of UAS-DAlk under the Actin-5C promoter within wild-type tissue. Female flies homozygous for hsp70-flp (insertion on the X chromosome) were crossed to males homozygous for both AyGal4 (insertion in the second chromosome) and UAS-GFP (also inserted on the second chromosome). Progeny from this cross were then crossed to homozygous UAS-DAlk flies and the resulting larvae heat shocked for 30 min at 37 °C. Thus, the DAlk expressing clones induced in this system are marked by a UAS-GFP reporter (Ito et al. 1997).
Mammalian expression constructs and transient transfection
For transient transfections, we used the cytomegalovirus-based pcDNA3 mammalian expression vector. The coding sequence of DAlk was subcloned into pcDNA3 and human 293 cells were transfected using the calcium phosphate precipitation method. Cells were lysed 36– 48 h post-transfection, and the resulting cell lysates were used for immunoblotting and protein kinase assays.
Anti-DAlk antibodies
DNA encoding DAlk amino acids 30–316 was subcloned into pHIS8-3 to generate a 39 kDa extracellular HIS-DAlk fusion protein. The orientation and reading frame of the HIS-DAlk sequence was subsequently confirmed by DNA sequence analysis. The HIS-DAlk fusion protein was induced and purified from E. coli (BL21(DE3)) bacterial lysates by standard protocols using Ni-NTA agarose (Qiagen). The resulting cleaved HIS-DAlk recombinant protein was used for mouse immunization. Polyclonal antibodies against HIS-DAlk were obtained from the serum of mice numbers 45, 46, 47 and 48 and were used at 1:1500 dilutions for the immunoflouresence analysis. Monoclonal antibodies against HIS-DAlk were obtained from mice 45, 46, 47 and 48 following standard procedures to produce monoclonal cell lines. Three positive monoclonal cell lines (nos 123, 66 and 29) were further characterized and their isotypes analysed. All three were found to be IgM. Following IgM affinity purification from the cell supernatant, monoclonal antibodies from clone no. 123 were used at 1:100 dilution for immunoblotting and immunofluoresence analysis.
Other methods
Autoradiography was carried out at –70 °C using X-OMAT AR (Kodak). SDS/PAGE was carried out according to Laemmli (Laemmli 1970). Protein was determined by the method of Bradford (Bradford 1976). Standard molecular techniques were employed for DNA manipulation (Sambrook et al. 1989).
Accession number
The GenBank nucleotide sequence accession number for the DAlk cDNA is AAF36990.
Acknowledgments
The authors would like to thank the following people: K. Finley, N. Ghbeish, W. Jiang, M. Kanemitsu, J. Meisenhelder and H. Mondala. We are grateful to K. Matthews and T. Laverty for providing the P-element lines and GAL4-driver lines described in this paper. R.H.P. would particularly like to thank B. Hallberg for support and advice. R.H.P. was supported by a Human Frontiers Science Programme Fellowship, a SSMF fellowship and Swedish NFR. T.H. is a Frank and Else Schilling American Cancer Society Research Professor. T.H. and M.M. were supported by NIH grants CA39780 and MH57460, respectively.
References
- Adams MD, Celniker SE, Holt RA, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Beckmann G, Bork P. An adhesive domain detected in functionally diverse receptors. Trends Biochem Sci. 1993;18:40– 41. doi: 10.1016/0968-0004(93)90049-s. [DOI] [PubMed] [Google Scholar]
- Beiman M, Shilo BZ, Volk T. Heartless, a Drosophila FGF receptor homolog, is essential for cell migration and establishment of several mesodermal lineages. Genes Dev. 1996;10:2993–3002. doi: 10.1101/gad.10.23.2993. [DOI] [PubMed] [Google Scholar]
- Bourgouin C, Lundgren SE, Thomas JB. Apterous is a Drosophila LIM domain gene required for the development of a subset of embryonic muscles. Neuron. 1992;9:549–561. doi: 10.1016/0896-6273(92)90192-g. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Callahan CA, Muralidhar MG, Lundgren SE, Scully AL, Thomas JB. Control of neuronal pathway selection by a Drosophila receptor protein-tyrosine kinase family member. Nature. 1995;376:171–174. doi: 10.1038/376171a0. [DOI] [PubMed] [Google Scholar]
- Casanova J, Struhl G. Localized surface activity of torso, a receptor tyrosine kinase, specifies terminal body pattern in Drosophila. Genes Dev. 1989;3:2025–2038. doi: 10.1101/gad.3.12b.2025. [DOI] [PubMed] [Google Scholar]
- Colleoni GW, Bridge JA, Garicochea B, Liu J, Filippa DA, Ladanyi M. ATIC-ALK: a novel variant ALK gene fusion in anaplastic large cell lymphoma resulting from the recurrent cryptic chromosomal inversion, inv(2)(p23q35) Am J Pathol. 2000;156:781–789. doi: 10.1016/S0002-9440(10)64945-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly NL, Scanlon MJ, Djofrdjevic JT, Kroon PA, Smith R. Three-dimensional structure of a cysteine-rich repeat from the low-density lipoprotein receptor. ProcNatl Acad Sci USA. 1995;92:6334–6338. doi: 10.1073/pnas.92.14.6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dura JM, Taillebourg E, Preat T. The Drosophila learning and memory gene linotte encodes a putative receptor tyrosine kinase homologous to the human RYK gene product. FEBS Lett. 1995;370:250–254. doi: 10.1016/0014-5793(95)00847-3. [DOI] [PubMed] [Google Scholar]
- Ellis MC, O’Neill EM, Rubin GM. Expression of Drosophila glass protein and evidence for negative regulation of its activity in non-neuronal cells by another DNA-binding protein. Development. 1993;119:855–865. doi: 10.1242/dev.119.3.855. [DOI] [PubMed] [Google Scholar]
- Fantl WJ, Johnson DE, Williams LT. Signalling by receptor tyrosine kinases. Annu Rev Biochem. 1993;62:453–481. doi: 10.1146/annurev.bi.62.070193.002321. [DOI] [PubMed] [Google Scholar]
- Fass D, Blacklow S, Kim PS, Berger JM. Molecular basis of familial hypercholesterolaemia from structure of LDL receptor module . Nature. 1997;388:691–692. doi: 10.1038/41798. [DOI] [PubMed] [Google Scholar]
- Fernandez R, Tabarini D, Azpiazu N, Frasch M, Schlessinger J. The Drosophila insulin receptor homolog: a gene essential for embryonic development encodes two receptor isoforms with different signaling potential. EMBO J. 1995;14:3373–3384. doi: 10.1002/j.1460-2075.1995.tb07343.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto J, Shiota M, Iwahara T, et al. Characterization of the transforming activity of p80, a hyperpho-sphorylated protein in a Ki-1 lymphoma cell line with chromosomal translocation t(2;5) Proc Natl Acad Sci USA. 1996;93:4181–4186. doi: 10.1073/pnas.93.9.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabay L, Seger R, Shilo BZ. MAP kinase in situ activation atlas during Drosophila embryogenesis. Development. 1997 ;124:3535–3541. doi: 10.1242/dev.124.18.3535. [DOI] [PubMed] [Google Scholar]
- van der Geer P, Hunter T, Lindberg RA. Receptor protein-tyrosine kinases and their signal transduction pathways. Annu Rev Cell Biol. 1994;10:251–337. doi: 10.1146/annurev.cb.10.110194.001343. [DOI] [PubMed] [Google Scholar]
- Gisselbrecht S, Skeath JB, Doe CQ, Michelson AM. heartless encodes a fibroblast growth factor receptor (DFR1/DFGF-R2) involved in the directional migration of early mesodermal cells in the Drosophila embryo. Genes Dev. 1996;10:3003–3017. doi: 10.1101/gad.10.23.3003. [DOI] [PubMed] [Google Scholar]
- Glazer L, Shilo BZ. The Drosophila FGF-R homolog is expressed in the embryonic tracheal system and appears to be required for directed tracheal cell extension. Genes Dev. 1991;5:697–705. doi: 10.1101/gad.5.4.697. [DOI] [PubMed] [Google Scholar]
- Hafen E, Basler K, Edstroem JE, Rubin GM. Sevenless, a cell-specific homeotic gene of Drosophila, encodes a putative transmembrane receptor with a tyrosine kinase domain. Science. 1987;236:55–63. doi: 10.1126/science.2882603. [DOI] [PubMed] [Google Scholar]
- Ito K, Awano W, Suzuki K, Hiromi Y, Hiromi Y, Yamamoto Y. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 1997;124:761– 771. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- Iwahara T, Fujimoto J, Wen D, et al. Molecular characterization of ALK, a receptor tyrosine kinase expressed specifically in the nervous system. Oncogene. 1997;14:439–449. doi: 10.1038/sj.onc.1200849. [DOI] [PubMed] [Google Scholar]
- Jiang YP, Wang H, D’Eustachio P, Musacchio JM, Schlessinger J, Sap J. Cloning and characterization of R-PTP-kappa, a new member of the receptor protein tyrosine phosphatase family with a proteolytically cleaved cellular adhesion molecule-like extracellular region. Mol CellBiol. 1993;13:2942–2951. doi: 10.1128/mcb.13.5.2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmerly W, Stultz K, Lewis S, et al. A P1-based physical map of the Drosophila euchromatic genome. Genome Res. 1996;6:414–430. doi: 10.1101/gr.6.5.414. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680– 685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lai C, Lemke G. An extended family of protein-tyrosine kinase genes differentially expressed in the vertebrate nervous system. Neuron. 1991;6:691–704. doi: 10.1016/0896-6273(91)90167-x. [DOI] [PubMed] [Google Scholar]
- Livneh E, Glazer L, Segal D, Schlessinger J, Shilo BZ. The Drosophila EGF receptor gene homolog: conservation of both hormone binding and kinase domains. Cell. 1985;40:599–607. doi: 10.1016/0092-8674(85)90208-9. [DOI] [PubMed] [Google Scholar]
- Ma Z, Cools J, Marynen P, et al. Inv(2)(p23q35) in anaplastic large-cell lymphoma induces constitutive anaplastic lymphoma kinase (ALK) tyrosine kinase activation by fusion to ATIC, an enzyme involved in purine nucleotide biosynthesis. Blood. 2000;95:2144–2149. [PubMed] [Google Scholar]
- Marck C. ‘DNA Strider’: a ‘C’ program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucl Acids Res. 1988;16:1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SW, Kirstein MN, Valentine MB, et al. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin’s lymphoma. Science. 1994;263:1281–1284. doi: 10.1126/science.8122112. [DOI] [PubMed] [Google Scholar]
- Morris SW, Naeve C, Mathew P, et al. ALK, the chromosome 2 gene locus altered by the t(2;5) in non-Hodgkin’s lymphoma, encodes a novel neural receptor tyrosine kinase that is highly related to leukocyte tyrosine kinase (LTK) Oncogene. 1997;14:2175–2188. doi: 10.1038/sj.onc.1201062. [DOI] [PubMed] [Google Scholar]
- Oishi I, Sugiyama S, Liu ZJ, Yamamura H, Nishida Y, Minami Y. A novel Drosophila receptor tyrosine kinase expressed specifically in the nervous system. Unique structural features and implication in developmental signaling. J Biol Chem. 1997;272 (1):1916–11923. doi: 10.1074/jbc.272.18.11916. [DOI] [PubMed] [Google Scholar]
- Palmer RH, Fessler LI, Edeen PT, Madigan SJ, McKeown M, Hunter T. DFak56 is a novel Drosophila melanogaster focal adhesion kinase. J Biol Chem. 1999;274 (3):5621–35629. doi: 10.1074/jbc.274.50.35621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel NH. Imaging neuronal subsets and other cell types in whole-mount Drosophila embryos and larvae using antibody probes. Meth Cell Biol. 1994;44:445–487. doi: 10.1016/s0091-679x(08)60927-9. [DOI] [PubMed] [Google Scholar]
- Robertson SC, Tynan JA, Donoghue DJ. RTK mutations and human syndromesĐwhen good receptors turn bad. Trends Genet. 2000;16:265–271. doi: 10.1016/s0168-9525(00)02021-7. [DOI] [PubMed] [Google Scholar]
- Ruan Y, Chen C, Cao Y, Garofalo RS. The Drosophila insulin receptor contains a novel carboxyl-terminal extension likely to play an important role in signal transduction. J Biol Chem. 1995;270:4236–4243. doi: 10.1074/jbc.270.9.4236. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Hong L, Brokstein P, et al. A Drosophila complementary DNA resource. Science. 2000;287:2222–2224. doi: 10.1126/science.287.5461.2222. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: a Laboratory Manual Cold Spring Harbour. New York: Cold Spring Harbour Laboratory Press; 1989. [Google Scholar]
- Sauer K, Weigmann K, Sigrist S, Lehner CF. Novel members of the cdc2-related kinase family in Drosophila: cdk4/6, cdk5, PFTAIRE, and PITSLRE kinase. Mol Biol Cell. 1996;7:1759–1769. doi: 10.1091/mbc.7.11.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully AL, McKeown M, Thomas JB. Isolation and characterization of Dek, a Drosophila eph receptor protein tyrosine kinase. Mol Cell Neurosci. 1999;13:337–347. doi: 10.1006/mcne.1999.0752. [DOI] [PubMed] [Google Scholar]
- Shiota M, Fujimoto J, Semba T, Satoh H, Yamamoto T, Mori S. Hyperphosphorylation of a novel 80 kDa protein-tyrosine kinase similar to Ltk in a human Ki-1 lymphoma cell line, AMS3. Oncogene. 1994;9:1567–1574. [PubMed] [Google Scholar]
- Shishido E, Ono N, Kojima T, Saigo K. Requirements of DFR1/Heartless, a mesoderm-specific Drosophila FGF-receptor, for the formation of heart, visceral and somatic muscles, and ensheathing of longitudinal axon tracts in CNS. Development. 1997;124:2119–2128. doi: 10.1242/dev.124.11.2119. [DOI] [PubMed] [Google Scholar]
- Spradling AC, Rubin GM. Transposition of cloned P elements into Drosophila germ line chromosomes. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- Spradling AC, Stern D, Beaton A, et al. The Berkeley Drosophila Genome Project gene disruption project: single P-element insertions mutating 25% of vital Drosophila genes. Genetics. 1999;153:135–177. doi: 10.1093/genetics/153.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling AC, Stern DM, Kiss I, Roote J, Laverty T, Rubin GM. Gene disruptions using P transposable elements: an integral component of the Drosophila genome project. Proc Natl Acad Sci USA. 1995;92 (1):0824–11080. doi: 10.1073/pnas.92.24.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenger F, Stevens LM, Nusslein-Volhard C. The Drosophila gene torso encodes a putative receptor tyrosine kinase. Nature. 1989;338:478–483. doi: 10.1038/338478a0. [DOI] [PubMed] [Google Scholar]
- Tamkun JW, Deuring R, Scott MP, et al. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell. 1992;68:561– 572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- Treisman R. Regulation of transcription by MAP kinase cascades. Curr Opin Cell Biol. 1996;8:205–215. doi: 10.1016/s0955-0674(96)80067-6. [DOI] [PubMed] [Google Scholar]
- Trinei M, Lanfrancone L, Campo E, et al. A new variant anaplastic lymphoma kinase (ALK)-fusion protein (ATIC-ALK) in a case of ALK-positive anaplastic large cell lymphoma. Cancer Res. 2000;60:793–798. [PubMed] [Google Scholar]
- Wadsworth SC, Vincent WS, Bilodeau-Wentworth D. A Drosophila genomic sequence with homology to human epidermal growth factor receptor. Nature. 1985;314:178– 180. doi: 10.1038/314178a0. [DOI] [PubMed] [Google Scholar]
- Whitehead JP, Clark SF, Urso B, James DE. Signalling through the insulin receptor. Curr Opin Cell Biol. 2000;12:222–228. doi: 10.1016/s0955-0674(99)00079-4. [DOI] [PubMed] [Google Scholar]
- Widmann C, Gibson S, Jarpe MB, Johnson GL. Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol Rev. 1999;79:143– 180. doi: 10.1152/physrev.1999.79.1.143. [DOI] [PubMed] [Google Scholar]
- Wilson C, Goberdhan DC, Steller H. Dror, a potential neurotrophic receptor gene, encodes a Drosophila homolog of the vertebrate Ror family of Trk-related receptor tyrosine kinases. Proc Natl Acad Sci USA. 1993;90:7109–7113. doi: 10.1073/pnas.90.15.7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarden Y, Ullrich A. Growth factor receptor tyrosine kinases. Annu Rev Biochem. 1988;57:443–478. doi: 10.1146/annurev.bi.57.070188.002303. [DOI] [PubMed] [Google Scholar]


