Abstract
Shwachman-Diamond syndrome (SDS) is an autosomal recessive disorder characterized by bone marrow failure, exocrine pancreatic dysfunction, and leukemia predisposition. Mutations in the SBDS gene are identified in most patients with SDS. SBDS encodes a highly conserved protein of unknown function. Data from SBDS orthologs suggest that SBDS may play a role in ribosome biogenesis or RNA processing. Human SBDS is enriched in the nucleolus, the major cellular site of ribosome biogenesis. Here we report that SBDS nucleolar localization is dependent on active rRNA transcription. Cells from patients with SDS or Diamond-Blackfan anemia are hypersensitive to low doses of actinomycin D, an inhibitor of rRNA transcription. The addition of wild-type SBDS complements the actinomycin D hypersensitivity of SDS patient cells. SBDS migrates together with the 60S large ribosomal subunit in sucrose gradients and coprecipitates with 28S ribosomal RNA (rRNA). Loss of SBDS is not associated with a discrete block in rRNA maturation or with decreased levels of the 60S ribosomal subunit. SBDS forms a protein complex with nucleophosmin, a multifunctional protein implicated in ribosome biogenesis and leukemogenesis. Our studies support the addition of SDS to the growing list of human bone marrow failure syndromes involving the ribosome.
Introduction
Shwachman-Diamond syndrome (SDS)1,2 is an autosomal recessive disease characterized by impaired hematopoiesis, exocrine pancreatic insufficiency, and increased leukemia risk. Additional variable clinical features include skeletal, hepatic, immunologic, and cardiac disorders.3,4 Most patients with clinical features of SDS harbor biallelic mutations in the SBDS gene located on chromosome 7.5 SBDS encodes an evolutionarily conserved protein of unknown function. An adjacent conserved pseudogene that shares 97% identity with the SBDS gene is transcribed but fails to encode a full-length protein product. Most SBDS mutations appear to arise from a gene conversion event between the SBDS gene and this adjacent pseudogene.5 The SBDS mRNA and protein is widely expressed throughout different tissues.5–8
Potential functions of the SBDS protein have been inferred from SBDS orthologs.5,9 The archaeal SBDS ortholog lies within a conserved operon that includes genes involved in RNA processing and protein translation.10 In transcriptional profiling studies, the yeast ortholog clusters with other RNA processing genes and ribosomal genes.11 Proteomic analysis of the yeast ortholog protein Ylr022C/Sdo1 suggested an association with other proteins involved in ribosome biogenesis.12 Genetic interactions between the yeast protein Yhr087w, which shares structural homology with the N-terminal domain of SBDS, and other genes involved in RNA and rRNA processing have been described.13 In phylogenetic profiling studies, SBDS clustered with other genes involved in RNA metabolism or translation.9 In support of these findings, we have previously shown that human SBDS is enriched in the nucleolus, the primary cellular site of ribosome biosynthesis.7 Recently, a role for Sdo1 in 60S ribosomal subunit maturation has been described in yeast.14
Studies in primary tissues from SDS patients are essential for our understanding of human disease pathogenesis. Studies in mice indicate that the absence of SBDS expression is lethal.6 Although the early truncating SBDS mutation 183TA>CT is common among SDS patients, no patients homozygous for this mutation have been identified.5,9 Taken together, current data support the hypothesis that SDS patients harbor at least one hypomorphic SBDS allele. To investigate SBDS function in human disease, we embarked on a study of SBDS protein in human cell systems and in primary cells from SDS patients.
Patients, materials, and methods
Cell culture
Dana-Farber Cancer Institute Institutional Review Board (DFCI IRB)–approved informed consent was obtained from participating patients in accordance with the Declaration of Helsinki. Lymphoblast cell lines and primary fibroblasts were maintained in culture as described previously.7 All cells were grown in a humidified 5% CO2 incubator at 37°C. HeLa cells were maintained in culture in Dulbecco modified Eagle medium (DMEM) (Invitrogen, Carlsbad, CA) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Sigma, St Louis, MO), penicillin (100 U/mL), and streptomycin (75 μg/mL) (Sigma). Human embryonic kidney HEK 293T cells were cultured in DMEM/10% FBS. Human skin fibroblasts from American Type Culture Collection (Manassas, VA; GM00038F) were immortalized with SV40 large T antigen and pBABE-hTERT-neo15 and grown in DMEM supplemented with 15% heat-inactivated FBS.
Cell transfection and lentivirus infection
A FLAG-nucleophosmin (NPM) cDNA construct was cloned into the pCMV vector (Stratagene, La Jolla, CA). HEK 293T cells were transfected in 10-cm dishes using TransIT-293 reagent (Mirus, Madison, WI) according to the manufacturer's instructions. Cells were harvested 48 hours after transfection, lysed, and immunoprecipitation performed as described.
Lentivirus vectors bearing a dual-cassette construct expressing GFP under a CMV promoter followed by an internal ribosome entry sequence (IRES) upstream of the SBDS cDNA were used.16 Lentiviral production was performed as described.16 Lentiviral infection efficiency was greater than 90% as confirmed by flow cytometry.
SBDS knockdown was performed using 3 separate small-interfering RNA (siRNA) hairpins in the LentiLox3.7 system.17 The following siRNA sequences were used to target SBDS: AACATGCTGCCATAACTTAGATT, AAGCTTGGATGATGTTCCTGATT, AAGGAAGATCTCATCAGTGCGTT.
Lentiviral infection was achieved using a multiplicity of infection (MOI) of 50, with 5 μg/mL hexadimethrine bromide (Sigma) in 250 μL total volume for lymphoblast cells and in 5 mL total volume for GM00038 fibroblasts.16
Cell survival assays
Equal numbers of lymphoblasts were plated in 96-well dishes in 50 μL RPMI/10% FBS. The indicated concentrations of actinomycin D and cycloheximide were obtained from stock solutions by diluting in RPMI medium and added to the 96-well dish to obtain a final volume of 100 μL. Cells were incubated for 48 to 72 hours at 37°C in triplicate. Cell survival was determined using the CellTiter 96 AQueous One Assay (Promega, Madison, WI) according to manufacturer's instructions. Cell viability was calculated as a percentage of control cell survival (100%), and the mean of triplicate samples was calculated. The standard error is depicted with error bars.
Immunofluorescence
Immunofluorescence analysis was performed as described previously.7 The following antibodies were used for immunofluorescence: anti-SBDS7 (1:400) and anti-NPM (1:800) (Abcam, Cambridge, MA). All secondary antibodies were obtained from Jackson ImmunoResearch Laboratories (West Grove, PA). Slides were mounted with Vectrashield mounting reagent (Vector Laboratories, Burlingame, CA). Slides were visualized with a Zeiss Axioplan 2 microscope (Zeiss, Thornwood, NY) using a 63×/1.4 NA oil-immersion lens. Images were acquired with an Axiocam HRC camera (Zeiss) using OpenLab software (Improvision, Lexington, MA) and Adobe Photoshop, version 10 (San Jose, CA).
Immunoblot analysis
Immunoblot analysis was performed as previously described.7 Cells were lysed in RIPA buffer,7 and lysates were separated on 10% sodium dodecyl sulfate or NuPAGE 4% to 12% Bis-Tris gels (Invitrogen), transferred to nitrocellulose membranes, and probed with the following antibodies: anti-FLAG (Cell Signaling Technologies, Danvers, MA; 1:1000), antitubulin (Sigma; 1:10 000), anti-NPM (Abcam; 1:10 000), and anti-SBDS7 (1:5000).
Immunoprecipitation
For immunoprecipitation from whole-cell extracts, cells were lysed in lysis buffer (20 mM HEPES [pH 7.4], 150 mM KCl, 0.5% NP-40) supplemented with 0.2 mM PMSF, 1 μM DTT, and Complete EDTA-free protease inhibitor cocktail (Roche, Indianapolis, IN). Lysates were precleared with protein G–agarose beads (Roche) for 30 minutes at 4°C. Cleared lysates were then incubated with the indicated antibodies overnight at 4°C, and protein G–agarose beads were added for an additional 3 hours. The beads were washed with lysis buffer 3 times. Immunoprecipitated proteins were eluted from the beads with 100 mM glycine (pH 2.8) and neutralized with 1 M Tris (pH 7.4). Samples were boiled in sample buffer and separated on NuPAGE 4% to 12% Bis-Tris gradient gels (Invitrogen) and analyzed by immunoblot as described previously.
RNA immunoprecipitation
For immunoprecipitation of RNA, nuclear extracts were prepared as described.18 Purified nuclear extracts were incubated with the indicated antibodies precoupled to protein G–agarose beads in buffer (20 mM HEPES [pH 7.4], 100 mM KCl). Immunoprecipitation was performed overnight at 4°C. Beads were washed 3 times in high-salt buffer (20 mM HEPES [pH 7.4], 1.5 mM MgCl2, 1.4 M KCl, 0.2 mM EDTA, 25% glycerol, 0.2 mM PMSF, 0.5 mM DTT). Beads were treated with proteinase K, and RNA was extracted using phenol/chloroform and isopropanol precipitation. The extracted RNA was analyzed on a 1% agarose gel.
Sucrose density gradient fractionation
For analysis of ribosomal subunits and polysomes, cells were grown to confluence, harvested 10 minutes after treatment with 100 μg/mL cycloheximide (Sigma), and resuspended in PBS containing 100 μg/mL cycloheximide. Equal numbers of cells were lysed in 20 mM HEPES [pH 7.4], 100 mM KCl, 5 mM MgCl2, 1% NP-40, 1 mM DTT, 100 μg/mL cycloheximide, 200 μg/mL heparin, 2 μM PMSF, and 100 units of RNAsin per milliliter (Promega, Madison, WI) for 10 minutes at 4°C. The lysates were centrifuged at 12 000g for 2 minutes, and supernatants were layered on 10% to 45% or 10% to 30% (wt/vol) sucrose gradients. The gradients were centrifuged at 178 300g for 3 hours at 4°C in a Beckman SW41Ti rotor and fractionated through an Amersham UV1 (GE Health Care, Chicago, IL) UV monitor connected to a chart recorder for absorbance at 254 nm.
Protein and RNA analysis from sucrose gradients
Proteins were precipitated with 20% trichloroacetic acid (TCA) from equal-volume aliquots of each gradient fraction. Proteins were separated on NuPAGE 4% to 12% Bis-Tris gels and immunoblotted as described in “Immunoblot analysis.” For RNA extraction, gradient fraction aliquots were treated with proteinase K and RNA was extracted with phenol/chloroform and isopropanol precipitation. The RNA was analyzed on a 1% agarose gel.
Metabolic labeling and analysis of ribosomal RNA
Fibroblasts were plated on 6 cm dishes and transfected with the appropriate siRNA (Invitrogen) using Lipofectamine (Invitrogen), according to manufacturer's instructions. After 48 hours, the medium was replaced with phosphate-free DMEM/20% FBS for 2 hours. The cells were then incubated with 40 μCi (1.48 mBq) of 32P-orthophosphate (GE Health Care, Piscataway, NJ) for 75 minutes. The labeled medium was replaced with phosphate-free DMEM/20% FBS containing 25 mM phosphate buffer (pH 7.4). Cells were harvested at the time points indicated, and RNA was extracted using TRIZOL (Invitrogen). Equal amounts (5 μg) of RNA were separated on a 1.2% agarose/0.8 M formaldehyde gel and transferred to HYBOND nylon membrane (GE Health Care, Piscataway, NJ) in 10× sodium saline citrate buffer for 18 hours. The transferred RNA was cross-linked using a UV cross-linker (Stratagene, La Jolla, CA) and analyzed by autoradiography (Bio-Rad, Hercules, CA).
Results
SBDS nucleolar localization is abrogated by actinomycin D
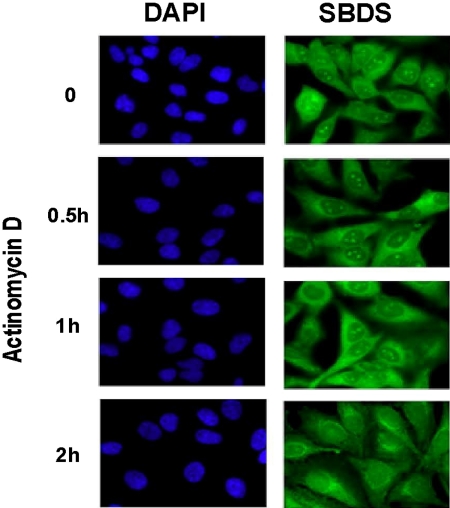
We have previously shown that SBDS, although present throughout the cell, is enriched in the nucleolus, the major cellular site of ribosome biogenesis.7 Numerous studies indicate, however, that the nucleolus is also an important site for additional cellular functions.19 To investigate the possible relationship between SBDS nucleolar localization and ribosome biogenesis, HeLa cells were treated with low levels of actinomycin D. At these low concentrations, actinomycin D primarily exerts its inhibitory effects on RNA polymerase I, which is responsible for transcription of the 18S, 28S, and 5.8S ribosomal RNA precursor.20 SBDS localization was assessed by immunofluorescence at different time points following actinomycin D treatment. SBDS nucleolar localization was markedly diminished by 2 hours following treatment with actinomycin D (Figure 1). These results suggest that SBDS nucleolar localization is dependent on active rRNA transcription.
Figure 1.
Nucleolar localization of SBDS is abrogated by actinomycin D treatment. HeLa cells were treated with 2 nM actinomycin D for the times indicated. Cells were fixed and stained for SBDS (green) and counterstained with DAPI (blue) to visualize nuclei (63× magnification). See “Patients, materials, and methods; Immunofluoresence” for image acquisition details.
SDS patient cells are hypersensitive to actinomycin D
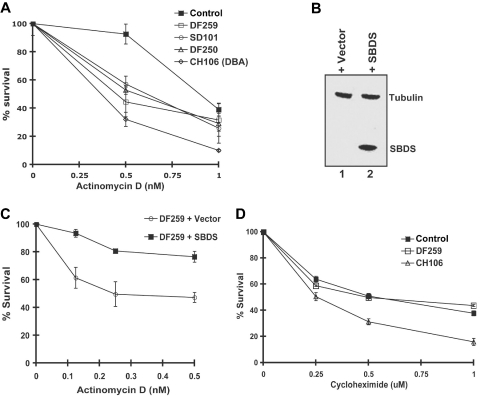
To explore whether ribosome biosynthesis might be functionally impaired in cells from SDS patients, we assayed cell survival in the presence of actinomycin D. We first assessed the effects of actinomycin D on the CH106 lymphoblast cell line derived from a patient with Diamond-Blackfan anemia (DBA) who harbored a heterozygous mutation (IVS1–2, A>G) in the ribosomal protein gene RPS19. Mutations in RPS19 result in impaired processing of 18S rRNA precursors and diminished production of the 40S ribosomal subunit.21–23 As shown in Figure 2A, the CH106 DBA cell line was sensitive to low doses of actinomycin D compared with the normal control. Interestingly, all 3 lymphoblast cell lines derived from SDS patients also exhibited hypersensitivity to actinomycin D relative to the normal control (Figure 2A). The genotypes of the SDS patient mutations are listed in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Figure 2.
Cells from SDS patients are hypersensitive to actinomycin D in an SBDS-dependent manner. (A) Lymphoblasts from a healthy control, SDS patients (DF250, DF259, and SD101), and an RPS19+ DBA patient cell (CH106) were plated in the presence of increasing concentrations of actinomycin D. Cell viability was assayed after 72 hours. Assays were performed in triplicate per experiment and repeated for a minimum of 3 independent experiments. Bars represent the standard error. (B) DF259 cells were infected with lentivirus containing GFP alone or together with wild-type SBDS cDNA downstream of an IRES sequence. Lentiviral infection was greater than 90% as observed by fluorescence microscopy. Cell lysates were analyzed by immunoblotting with antibodies against SBDS and tubulin. (C) DF259 cells infected with lentivirus containing empty vector or full-length SBDS cDNA were plated in the presence of increasing concentrations of actinomycin D. Cell viability was assayed in triplicate per experiment and repeated for 3 independent experiments. Bars represent the standard error. (D) Lymphoblasts (normal control, DF259, and CH106) were plated in the presence of increasing concentrations of cycloheximide. Cell viability was assayed after 72 hours in triplicate for each experiment for a total of 3 independent experiments. Bars represent the standard error.
To investigate whether the actinomycin D sensitivity was the result of SBDS loss, we examined the effect of restoring SBDS expression in SDS patient lymphoblasts. The SDS lymphoblast cell line DF2597 was infected with a lentiviral vector carrying either GFP alone or together with a downstream IRES–wild-type SBDS cDNA sequence. Greater than 90% of the cells were transduced by the lentivirus as determined by quantitating the number of cells exhibiting green fluorescence from the lentiviral GFP cassette. Restoration of SBDS expression following introduction of the SBDS cDNA was confirmed by immunoblotting (Figure 2B, lane 2). Actinomycin D hypersensitivity was corrected following infection with the SBDS lentivirus but not with the empty vector control, consistent with functional complementation (Figure 2C). Thus, the actinomycin D hypersensitivity observed in SDS patient cells is SBDS dependent.
To assess whether SBDS might function in protein translation, we tested the effect of the protein translation inhibitor cycloheximide on SDS patient lymphoblasts. No significant difference in sensitivity between the SDS patient cells and healthy controls following cycloheximide treatment was noted (Figure 2D). However, the DBA cell line CH106 was more sensitive to cycloheximide, as predicted by the presence of a mutation in a ribosomal protein. Taken together, these results suggest that SBDS might function in ribosome biogenesis rather than in general protein translation.
SBDS cosediments with the 60S ribosomal precursor subunit
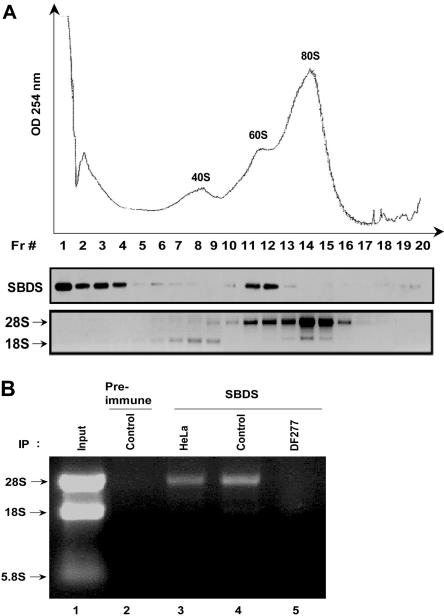
To further investigate a potential role for SBDS in ribosome biogenesis, we asked whether SBDS associates with ribosomal subunits. To begin to address this question, HeLa cell lysates were fractionated through sucrose gradients to separate the 40S small ribosomal subunit, the 60S large ribosomal subunit, and the 80S mature ribosome. Sucrose gradient fractions were blotted for SBDS. A large proportion of SBDS was seen at the top of the gradient (Figure 3A); however, a subset of SBDS cellular protein migrated in a distinct peak in the region of the 60S ribosomal subunit. In contrast, SBDS was largely absent in the regions of the gradient corresponding to the 40S small ribosomal subunit and the 80S mature ribosome (Figure 3A). Immunoblotting of sucrose gradients centrifuged for shorter times to retain the polysomes, which consist of ribosomes assembled onto mRNA, demonstrated that SBDS was absent from the polysome fractions (data not shown). These data were consistent with an association between SBDS and the large ribosomal precursor subunit but not with mature ribosomes.
Figure 3.
SBDS cosediments with the 60S ribosomal precursor subunit and associates with the 28S ribosomal RNA. (A) HeLa cell lysates were fractionated on a 10% to 30% sucrose gradient by ultracentrifugation. Absorbance at 254 nM across the gradient is shown (top panel). Proteins were precipitated from equal aliquots of each fraction and immunoblotted for SBDS (middle panel). RNA was extracted from equal volumes of each fraction and analyzed by agarose gel electrophoresis and ethidium bromide staining (bottom panel). Data shown are representative of results obtained from 3 independent experiments. (B) Endogenous SBDS was immunoprecipitated from nuclear extracts of HeLa cells, healthy control lymphoblasts (control), or DF277 lymphoblasts derived from an SDS patient. Preimmune serum was used as a negative control for the immunoprecipitation (lane 2). RNA was extracted from the immunoprecipitates and analyzed by agarose gel electrophoresis and ethidium bromide staining. Data shown are representative of results obtained from 3 independent experiments.
SBDS associates with 28S rRNA
To determine whether SBDS coprecipitates with components of the 60S ribosomal subunit, SBDS was immunoprecipitated from HeLa cells, control lymphoblasts (Control), or lymphoblasts from a patient with SDS (DF277) (Figure 3B, lanes 3-5). SBDS protein expression was previously shown to be markedly reduced in the DF277 cell line.7 RNA was extracted from the SBDS immunoprecipitate and analyzed by gel electrophoresis. We observed that 28S rRNA coprecipitated with the anti-SBDS antibody but not with preimmune serum (Figure 3B, lane 2) in lysates from HeLa cells or control lymphoblast cell lines, which express SBDS protein (Figure 3B, lanes 3 and 4). 28S rRNA was absent from immunoprecipitates from the SDS patient cell line DF277 (Figure 3B, lane 5), indicating that the coprecipitation of 28S rRNA was dependent on the presence of the SBDS protein. These data further support an association between SBDS and the large 60S ribosomal precursor subunit, which contains the 28S rRNA but not the 18S rRNA.
SBDS forms a protein complex with nucleophosmin
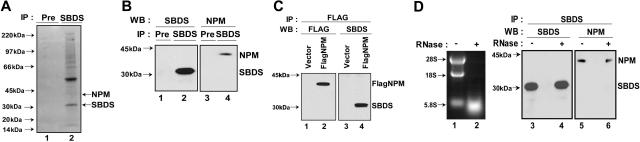
To further understand the function of SBDS, SBDS binding partners were identified using the open-ended approach of immunoprecipitating endogenous human SBDS protein and analyzing coprecipitated proteins by mass spectrometry.24 To minimize nonspecific binding, cell lysates were precleared with preimmune serum (Pre). Background protein immunoprecipitation was low in the absence of SBDS antibody (Figure 4A, lane 1). In contrast, several proteins coprecipitating with SBDS (indicated with an arrow) were visualized with Coomassie staining as shown in Figure 4A, lane 2. Among the SBDS-interacting proteins identified were several ribosomal proteins. An additional protein identified by mass spectrometry was nucleophosmin (NPM). Like SBDS, NPM is found in the nucleolus.25 NPM is a multifunctional protein26 whose functions include a role in ribosome biogenesis.27 Mutations or translocations involving nucleophosmin have been described in acute myelogenous leukemias.28
Figure 4.
SBDS associates with nucleophosmin in an RNA-independent manner. (A) Endogenous SBDS was immunoprecipitated from HeLa nuclear extracts with an anti-SBDS antibody. The immunoprecipitated proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis on a 4% to 12% Bis-Tris gel followed by Coomassie blue staining. Preimmune serum (Pre) was used as a negative control for the immunoprecipitation (lane 1). Protein bands from the SBDS immunoprecipitate (lane 2) were excised and analyzed by tryptic digestion and matrix-assisted laser description/ionisation time of flight (MALDI-TOF) mass spectrometry. (B) HeLa cell lysates were incubated with either anti-SBDS antibody or preimmune serum (Pre). The immunoprecipitates were analyzed by Western blotting for SBDS (lanes 1 and 2) and NPM (lanes 3 and 4). (C) 293T cells were transfected with either FLAG-NPM (lanes 2 and 4) or empty pFLAG-CMV vector (lanes 1 and 3). Cell lysates were immunoprecipitated with an anti-FLAG antibody. The resulting precipitates were immunoblotted for FLAG (lanes 1 and 2) or SBDS (lanes 3 and 4). (D) HeLa cell lysates were incubated at 30°C for 20 minutes in the presence (lane 2) or absence (lane 1) of 4 μg RNase A. RNA was extracted and analyzed by agarose gel electrophoresis and ethidium bromide staining (left panel, lanes 1 and 2). SBDS was immunoprecipitated from the mock-treated and RNase-treated lysates. The resulting pellets were analyzed by immunoblotting (right panel) for SBDS (lanes 3 and 4) and NPM (lanes 5 and 6).
SBDS was immunoprecipitated from cell lysates, and SBDS-associated proteins were analyzed by immunoblotting for nucleophosmin (Figure 4B). SBDS immunoprecipitation by the anti-SBDS antibody but not by preimmune serum (Pre) was confirmed by immunoblotting (Figure 4B, compare lanes 1 and 2). NPM was found to coprecipitate with SBDS (Figure 4B, lane 4) but not with preimmune serum (Figure 4B, lane 3).
To perform the reciprocal immunoprecipitation experiment, plasmids bearing a cDNA for FLAG-tagged NPM (FLAG-NPM) or empty vector plasmid were transfected into 293T cells. The cell lysates were precipitated with an antibody against the FLAG epitope. FLAG-NPM was precipitated by the anti-FLAG antibody from lysates of cells transfected with FLAG-NPM (Figure 4C, lane 2) but not from lysates of cells transfected with empty vector (Figure 4C, lane 1). Immunoblotting revealed the coprecipitation of SBDS only when FLAG-NPM was present (Figure 4C, compare lanes 3 and 4). Thus, SBDS was observed to interact with NPM in 3 separate assays.
Previous studies have reported that NPM associates with 28S rRNA,29 lending support to the notion that, like NPM, SBDS might also play a role in ribosome biogenesis. However, this observation raised the question of whether the coprecipitation of NPM and SBDS might result from independent interactions of each protein with 28S rRNA rather than from an association of SBDS and NPM with each other. To distinguish between these 2 possibilities, HeLa cell lysates were incubated in the presence or absence of RNase prior to SBDS immunoprecipitation. RNA digestion was confirmed by agarose gel electrophoresis demonstrating the loss of rRNA (Figure 4D, lane 2). These lysates were then immunoprecipitated with an antibody against SBDS, and the precipitates were blotted for the presence of SBDS (Figure 4D, lanes 3 and 4) and NPM (Figure 4D, lanes 5 and 6). Nucleophosmin continued to coprecipitate with SBDS even after RNase treatment. Thus, the SBDS-NPM interaction is independent of rRNA.
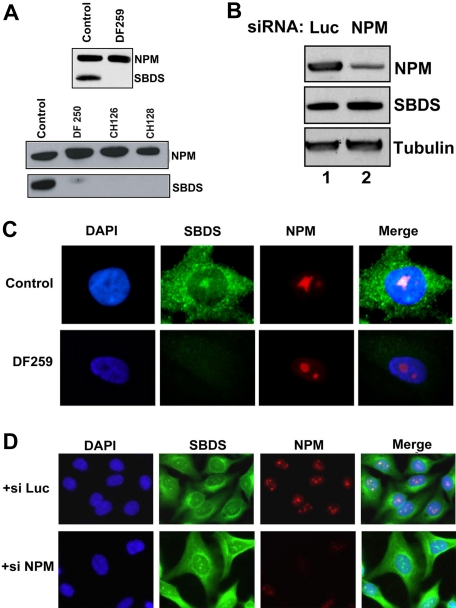
No difference in NPM protein levels was observed between SDS patient cells and normal control cells (Figure 5A). Thus, SBDS does not appear to affect steady state NPM protein stability. Similarly, no difference in SBDS protein levels was noted following transfection of siRNAs directed against NPM to diminish NPM protein levels (Figure 5B). NPM nucleolar localization was intact in SDS patient cells, indicating that nucleolar localization of NPM was not dependent on the present of SBDS (Figure 5C). Inhibition of NPM protein production following transfection with NPM siRNA did not affect SBDS nucleolar localization (Figure 5D). Thus, NPM is not required for SBDS protein stability or nucleolar localization.
Figure 5.
SBDS is not required for NPM protein stability or nucleolar localization. (A) Primary fibroblast lysates from healthy controls or SDS patients (DF259, DF250, CH126, CH128) were immunoblotted for SBDS and NPM. (B) HeLa cells were transfected with siRNA against either luciferase (Luc) or NPM. Cells were lysed 72 hours after transfection and immunoblotted for SBDS, NPM, and tubulin. (C) Normal control and DF259 primary fibroblasts were fixed and stained for SBDS (green) and NPM (red) and counterstained with DAPI (blue) to visualize the nuclei. (D) HeLa cells were transfected with siRNA against either luciferase (++ si Luc) or NPM (+ si NPM) for 48 hours. Cells were fixed and stained for SBDS (green) and NPM (red) and counterstained with DAPI (blue). See “Patients, materials, and methods; Immunofluorescence” for image acquisition details.
Lack of a discrete rRNA processing defect following SBDS loss
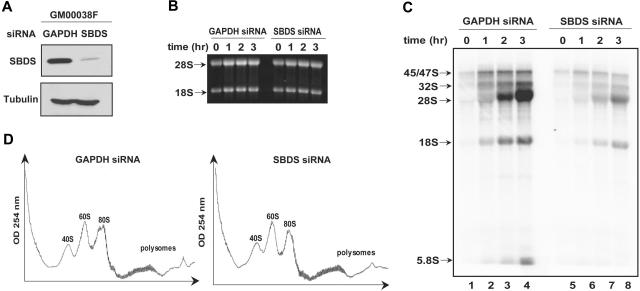
Circumstantial evidence from SBDS orthologs suggested that SBDS might function in RNA processing.5,9 RPS19 gene mutations in the inherited marrow failure syndrome Diamond-Blackfan anemia (DBA) result in inhibition of a specific cleavage step during 18S rRNA processing and diminished 40S ribosomal subunit production.21–23 Because NPM has previously been suggested to play a role in rRNA biogenesis,30,31 the observed interaction of SBDS with NPM and with 28S rRNA raised the possibility that SBDS might play a similar function. To investigate this question, siRNAs directed against SBDS or against GAPDH were introduced into wild-type human skin fibroblasts (GM00038). SBDS protein expression was markedly reduced following the introduction of the SBDS siRNA (Figure 6A). These cells were pulsed with 32P-orthophosphate to label newly synthesized RNA followed by a chase with cold phosphate. RNA was extracted at different time points during the chase and analyzed by gel electrophoresis (Figure 6B) and autoradiography (Figure 6C). Comparable quantities of RNA were loaded on an agarose gel for each time point (Figure 6B). Production of newly synthesized rRNA was markedly reduced following SBDS knockdown (Figure 6C, lanes 5-8) when compared with the control cells (Figure 6C, lanes 1-4). No discrete block in rRNA processing, as evidenced by the lack of accumulation of rRNA precursors relative to the mature 28S and 18S product, was observed. Similar results were obtained when an siRNA against luciferase was used as the control for these experiments (data not shown).
Figure 6.
SBDS loss is not associated with a discrete block in rRNA processing. (A) siRNA against either GAPDH or SBDS was introduced into human skin fibroblasts (GM00038). Cells were lysed 72 hours later and immunoblotted for SBDS and tubulin. GAPDH knock down was confirmed to be greater than 50% by immunoblot densitometry and quantitative PCR (data not shown). (B) GM00038 fibroblasts containing siRNA against GAPDH or SBDS from (A) were metabolically labeled with 32P-orthophosphate for 75 minutes and chased with 25 mM phosphate for the indicated times in hours. RNA was extracted at the indicated time points, resolved on a 1% agarose/formaldehyde gel, and stained with ethidium bromide to confirm equal loading. (C) RNA from the gel in panel B was transferred to a nylon membrane and analyzed by autoradiography. The positions of the 45S/47S and 32S precursor rRNAs and of the mature 28S, 18S, and 5.8S rRNAs are indicated. (D) GM00038 fibroblasts containing siRNA against GAPDH or SBDS were lysed and fractionated on 10% to 45% sucrose gradients by ultracentrifugation. Absorbance at 254 nM was measured across the gradient, and the positions corresponding to the 40S, 60S, and 80S ribosomal particles are indicated. Results shown are representative of 3 independent experiments.
To further investigate the potential role of SBDS in ribosome biogenesis, we compared the sucrose gradient polysome profiles of cells transfected with siRNA directed against GAPDH or SBDS. No consistent differences were observed in the relative amounts of the 40S, 60S, and 80S subunits (Figure 6D, compare left and right panels). Thus, acute loss of SBDS was not associated with an imbalance of ribosomal subunit production by this assay.
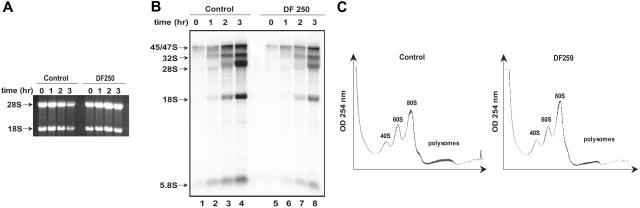
Although acute depletion of SBDS by RNA interference was associated with a global decrease in rRNA synthesis, the possibility that this general reduction of rRNA biogenesis might represent an indirect effect of sudden dramatic SBDS loss rather than a direct function of SBDS cannot be ruled out. To address this possibility, primary fibroblasts from a healthy control and an SDS patient (DF250) were pulsed with 32P-orthophosphate to label newly synthesized RNA followed by a chase with cold phosphate. Comparable quantities of RNA were loaded on an agarose gel for each time point (Figure 7A). Consistent with the results observed following SBDS knockdown, production of newly synthesized rRNA was markedly reduced in the SDS patient cells (Figure 7B, lanes 5-8) when compared with the control cells (Figure 7B, lanes 1-4). In further support of these findings, Northern blot analysis of steady state rRNA levels in control cells versus SDS patient cells also failed to reveal a relative accumulation of rRNA precursor RNAs (data not shown).
Figure 7.
SDS patient cell lines show decreased ribosomal RNA synthesis. (A) Healthy control and SDS patient (DF250) primary fibroblasts were metabolically labeled with 32P-orthophosphate for 75 minutes and chased with 25 mM phosphate for the indicated times in hours. RNA was extracted at the indicated time points, resolved on a 1% agarose/formaldehyde gel, and stained with ethidium bromide. (B) RNA from the gel in panel A was transferred to a nylon membrane and analyzed by autoradiography. The positions of the 45S and 32S rRNA precursor rRNAs and of the mature 28S, 18S, and 5.8S rRNAs are indicated. (C) Lysates from normal control primary fibroblasts or SDS patient primary fibroblasts were sedimented through sucrose gradients as in Figure 6D. This experiment was repeated for a total of 3 independent experiments. No consistent difference in the ratios of the 40S, 60S, and 80S peaks was noted.
We also compared the sucrose gradient polysome profiles of normal control fibroblasts and SDS patient fibroblasts. No consistent differences were observed in the relative amounts of the 40S, 60S, and 80S subunits in SDS patient cells versus normal control cells (Figure 7C). Thus, pathogenic SBDS mutations in SDS patient cells and chronic absence of SBDS were not associated with a consistent imbalance of ribosomal subunit production by this assay. These data indicate that the function of the SBDS protein is distinct from that observed with RPS19 in the inherited marrow failure syndrome DBA.22,23
Discussion
We investigated the potential role of SBDS in ribosome biogenesis using human cell lines and primary cells obtained from patients with Shwachman-Diamond syndrome. We show that cells from SDS patients are hypersensitive to low doses of actinomycin D, consistent with an underlying impairment of ribosome biogenesis. Further data implicating a ribosomal function for SBDS include the observation that SBDS nucleolar localization is abolished by actinomycin D, that SBDS cosediments with the large 60S ribosomal subunit in sucrose gradients, and that SBDS associates with 28S rRNA, a component of the large ribosomal subunit. The lack of rRNA precursor accumulation and the similar ratios of ribosomal precursor subunits observed between normal control cells and SDS patient cells do not support a role for SBDS in rRNA processing. A recent genetic study in yeast suggested a model whereby the yeast SBDS ortholog Sdo1 facilitates Tif6 release from pre-60S ribosomes.14 Tif6 is required for 60S subunit biogenesis and nuclear export.32 The release of the mammalian Tif6 homolog eIF6 from 60S ribosomal subunits is critical to allow the joining of the 60S subunit to the 40S subunit.33 Our findings lend support to this model in human systems, though a full understanding of the role of SBDS in ribosome biogenesis awaits further mechanistic studies.
Using an open-ended approach with immunoprecipitation of endogenous human SBDS followed by mass spectrometry, we observed that human SBDS associates with NPM. The association between SBDS and NPM was confirmed by reciprocal immunoprecipitation-immunoblot assays. NPM is a multifunctional protein implicated in regulation of centrosome duplication,34 chromatin transcription,35 protein chaperoning,36 modulation of apoptosis, stress response,37 and ribosome biogenesis.26 We observed that SBDS shares characteristics previously reported for NPM: both SBDS and NPM bind to 28S rRNA,29 and both are localized to the nucleolus in an actinomycin D–sensitive manner.38 NPM's chaperoning activity may facilitate ribosome assembly36 and transport,39 and the potential role of SBDS in these processes is currently under investigation. Nucleophosmin (NPM) is abundant in highly proliferative or malignant cells, and expression of NPM rises in response to mitogenic stimuli.26,37 NPM expression is reduced during cellular differentiation or apoptosis.37 As seen with SBDS, loss of NPM leads to early embryonic lethality in mice.34 Npm + /- mice exhibit an increased risk of myelodysplasia.34 Translocations and mutations involving the human NPM gene are commonly associated with hematologic malignancies.40 Thus, NPM has been postulated to function as both an oncogene and as a tumor suppressor. Whether NPM might contribute to the increased risk of AML in SDS patients remains an area of active investigation.
The pathologic mechanism(s) whereby defects in ribosome biogenesis might lead to marrow failure and leukemogenesis are currently unclear.41 While an association between ribosome biogenesis and malignant transformation has long been appreciated, the question of causality remains unanswered. Oncogenes such as Myc and tumor suppressors such as Arf have been reported to increase or decrease ribosome production, respectively.42 Ribosome biogenesis is a complex and highly regulated process involving more than 100 different nonribosomal proteins and small RNAs.43 Imbalanced production of ribosomal precursors is poorly tolerated by cells and might contribute to the increased levels of apoptosis seen in the context of marrow failure. An intriguing study recently suggested that lineage-specific regulation of rRNA transcription by the transcription factor Runx2 may contribute to lineage commitment and cell proliferation.44 It is notable that the eukaryotic ribosome contains many more proteins than its prokaryotic counterpart and that the functions of these additional eukaryotic ribosomal proteins remain poorly characterized.45 Extraribosomal functions are being identified for increasing numbers of eukaryotic ribosomal proteins.46–48 These studies continue to broaden our view of the ribosome beyond that of a passive factory for protein translation.
In summary, our studies support the addition of Shwachman-Diamond syndrome to the growing list of inherited bone marrow failure syndromes affecting the ribosome49: Diamond-Blackfan anemia,23,50 dyskeratosis congenital51,52 and cartilage-hair hypoplasia.53 The ribosomal defects targeted in each syndrome appear distinct. In Diamond-Blackfan anemia, mutations in RPS19 result in an impaired rRNA cleavage step during the maturation of 18S ribosomal RNA.22 In dyskeratosis congenita, mutations in DKC1, the gene encoding dyskerin, result in impaired RNA pseudouridylation52 and decreased IRES-mediated protein translation.54 In cartilage-hair hypoplasia, mutations are seen in the RNA component of the RMRP gene, which is involved in rRNA processing.53 However, additional molecular functions have been described for each of these genes. Dyskerin is involved in telomere maintenance,51 RPS19 has been described as a monocyte attractant factor,55 and RMRP is also involved in mRNA degradation.56 Such multiplicity of function is not necessarily mutually exclusive but might additively contribute to disease phenotype. Thus, the elucidation of the molecular contributions of ribosome dysfunction or other functions of SBDS to disease pathogenesis is essential to the development of targeted therapeutics to treat marrow failure and malignancies.
Supplementary Material
Acknowledgments
The authors thank David Nathan for critical advice throughout this project. We are grateful to all the patients and referring physicians for donating samples for this study. We thank the Taplin Biological Mass Spectrometry Facility at Harvard Medical School. We thank Steve Ellis, Rachel Idol, Sara Robledo, Monica Bessler, and Fabrizio Loreni for helpful discussions.
This work was supported by NIH grants (K23 HL068632–03 and R01 HL079582–02) (A.S.), a V Foundation grant (A.S.), and the Gracie Fund.
Footnotes
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: K.A.G. designed and performed research, and wrote the paper; K.M.A., C-S.L., A.D., and M.M.M. performed research; R.R. provided helpful discussion; and A.S. developed and supervised the project, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Akiko Shimamura, University of Washington, Department of Pediatric Hematology/Oncology, 815 Mercer Street, Room 356, Seattle, WA 98109; e-mail: shima2@u.washington.edu.
References
- 1.Shwachman H, Diamond LK, Oski FA, Khaw KT. The syndrome of pancreatic insufficiency and bone marrow dysfunction. J Pediatr. 1964;65:645–663. doi: 10.1016/s0022-3476(64)80150-5. [DOI] [PubMed] [Google Scholar]
- 2.Bodian M, Sheldon W, Lightwood R. Congenital hypoplasia of the exocrine pancreas. Acta Paediatr. 1964;53:282–293. doi: 10.1111/j.1651-2227.1964.tb07237.x. [DOI] [PubMed] [Google Scholar]
- 3.Shimamura A. Shwachman-Diamond syndrome. Semin Hematol. 2006;43:178–188. doi: 10.1053/j.seminhematol.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 4.Dror Y. Shwachman-Diamond syndrome. Pediatr Blood Cancer. 2005;45:892–901. doi: 10.1002/pbc.20478. [DOI] [PubMed] [Google Scholar]
- 5.Boocock GR, Morrison JA, Popovic M, et al. Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat Genet. 2003;33:97–101. doi: 10.1038/ng1062. [DOI] [PubMed] [Google Scholar]
- 6.Zhang S, Shi M, Hui CC, Rommens JM. Loss of the mouse ortholog of the shwachman-diamond syndrome gene (Sbds) results in early embryonic lethality. Mol Cell Biol. 2006;26:6656–6663. doi: 10.1128/MCB.00091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Austin KM, Leary RJ, Shimamura A. The Shwachman-Diamond SBDS protein localizes to the nucleolus. Blood. 2005;106:1253–1258. doi: 10.1182/blood-2005-02-0807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woloszynek JR, Rothbaum RJ, Rawls AS, et al. Mutations of the SBDS gene are present in most patients with Shwachman-Diamond syndrome. Blood. 2004;104:3588–3590. doi: 10.1182/blood-2004-04-1516. [DOI] [PubMed] [Google Scholar]
- 9.Boocock GR, Marit MR, Rommens JM. Phylogeny, sequence conservation, and functional complementation of the SBDS protein family. Genomics. 2006;87:758–771. doi: 10.1016/j.ygeno.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 10.Koonin EV, Wolf YI, Aravind L. Prediction of the archaeal exosome and its connections with the proteasome and the translation and transcription machineries by a comparative-genomic approach. Genome Res. 2001;11:240–252. doi: 10.1101/gr.162001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu LF, Hughes TR, Davierwala AP, Robinson MD, Stoughton R, Altschuler SJ. Large-scale prediction of Saccharomyces cerevisiae gene function using overlapping transcriptional clusters. Nat Genet. 2002;31:255–265. doi: 10.1038/ng906. [DOI] [PubMed] [Google Scholar]
- 12.Savchenko A, Krogan N, Cort JR, et al. The Shwachman-Bodian-Diamond syndrome protein family is involved in RNA metabolism. J Biol Chem. 2005;280:19213–19220. doi: 10.1074/jbc.M414421200. [DOI] [PubMed] [Google Scholar]
- 13.Shammas C, Menne TF, Hilcenko C, et al. Structural and mutational analysis of the SBDS protein family. Insight into the leukemia-associated Shwachman-Diamond syndrome. J Biol Chem. 2005;280:19221–19229. doi: 10.1074/jbc.M414656200. [DOI] [PubMed] [Google Scholar]
- 14.Menne TF, Goyenechea B, Sanchez-Puig N, et al. The Shwachman-Bodian-Diamond syndrome protein mediates translational activation of ribosomes in yeast. Nat Genet. 2007;39:486–495. doi: 10.1038/ng1994. [DOI] [PubMed] [Google Scholar]
- 15.Nakanishi K, Taniguchi T, Ranganathan V, et al. Interaction of FANCD2 and NBS1 in the DNA damage response. Nat Cell Biol. 2002;4:913–920. doi: 10.1038/ncb879. [DOI] [PubMed] [Google Scholar]
- 16.Mostoslavsky G, Fabian AJ, Rooney S, Alt FW, Mulligan RC. Complete correction of murine Artemis immunodeficiency by lentiviral vector-mediated gene transfer. Proc Natl Acad Sci U S A. 2006;103:16406–16411. doi: 10.1073/pnas.0608130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rubinson DA, Dillon CP, Kwiatkowski AV, et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- 18.Krainer AR, Maniatis T, Ruskin B, Green MR. Normal and mutant human beta-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell. 1984;36:993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- 19.Scherl A, Coute Y, Deon C, et al. Functional proteomic analysis of human nucleolus. Mol Biol Cell. 2002;13:4100–4109. doi: 10.1091/mbc.E02-05-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iapalucci-Espinoza S, Franze-Fernandez MT. Effect of protein synthesis inhibitors and low concentrations of actinomycin D on ribosomal RNA synthesis. FEBS Lett. 1979;107:281–284. doi: 10.1016/0014-5793(79)80390-7. [DOI] [PubMed] [Google Scholar]
- 21.Leger-Silvestre I, Caffrey JM, Dawaliby R, et al. Specific role for yeast homologs of the Diamond Blackfan anemia-associated Rps19 protein in ribosome synthesis. J Biol Chem. 2005;280:38177–38185. doi: 10.1074/jbc.M506916200. [DOI] [PubMed] [Google Scholar]
- 22.Flygare J, Aspesi A, Bailey JC, et al. Human RPS19, the gene mutated in Diamond-Blackfan anemia, encodes a ribosomal protein required for the maturation of 40S ribosomal subunits. Blood. 2007;109:980–986. doi: 10.1182/blood-2006-07-038232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choesmel V, Bacqueville D, Rouquette J, et al. Impaired ribosome biogenesis in Diamond-Blackfan anemia. Blood. 2007;109:1275–1283. doi: 10.1182/blood-2006-07-038372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffin TJ, Gygi SP, Rist B, et al. Quantitative proteomic analysis using a MALDI quadrupole time-of-flight mass spectrometer. Anal Chem. 2001;73:978–986. doi: 10.1021/ac001169y. [DOI] [PubMed] [Google Scholar]
- 25.Lischwe MA, Smetana K, Olson MO, Busch H. Proteins C23 and B23 are the major nucleolar silver staining proteins. Life Sci. 1979;25:701–708. doi: 10.1016/0024-3205(79)90512-5. [DOI] [PubMed] [Google Scholar]
- 26.Grisendi S, Mecucci C, Falini B, Pandolfi PP. Nucleophosmin and cancer. Nat Rev Cancer. 2006;6:493–505. doi: 10.1038/nrc1885. [DOI] [PubMed] [Google Scholar]
- 27.Okuwaki M, Tsujimoto M, Nagata K. The RNA binding activity of a ribosome biogenesis factor, nucleophosmin/B23, is modulated by phosphorylation with a cell cycle-dependent kinase and by association with its subtype. Mol Biol Cell. 2002;13:2016–2030. doi: 10.1091/mbc.02-03-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grisendi S, Pandolfi PP. NPM mutations in acute myelogenous leukemia. N Engl J Med. 2005;352:291–292. doi: 10.1056/NEJMe048337. [DOI] [PubMed] [Google Scholar]
- 29.Huang N, Negi S, Szebeni A, Olson MO. Protein NPM3 interacts with the multifunctional nucleolar protein B23/nucleophosmin and inhibits ribosome biogenesis. J Biol Chem. 2005;280:5496–5502. doi: 10.1074/jbc.M407856200. [DOI] [PubMed] [Google Scholar]
- 30.Pinol-Roma S. Association of nonribosomal nucleolar proteins in ribonucleoprotein complexes during interphase and mitosis. Mol Biol Cell. 1999;10:77–90. doi: 10.1091/mbc.10.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Savkur RS, Olson MO. Preferential cleavage in pre-ribosomal RNA byprotein B23 endoribonuclease. Nucleic Acids Res. 1998;26:4508–4515. doi: 10.1093/nar/26.19.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Basu U, Si K, Warner JR, Maitra U. The Saccharomyces cerevisiae TIF6 gene encoding translation initiation factor 6 is required for 60S ribosomal subunit biogenesis. Mol Cell Biol. 2001;21:1453–1462. doi: 10.1128/MCB.21.5.1453-1462.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ceci M, Gaviraghi C, Gorrini C, et al. Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature. 2003;426:579–584. doi: 10.1038/nature02160. [DOI] [PubMed] [Google Scholar]
- 34.Grisendi S, Bernardi R, Rossi M, et al. Role of nucleophosmin in embryonic development and tumorigenesis. Nature. 2005;437:147–153. doi: 10.1038/nature03915. [DOI] [PubMed] [Google Scholar]
- 35.Swaminathan V, Kishore AH, Febitha KK, Kundu TK. Human histone chaperone nucleophosmin enhances acetylation-dependent chromatin transcription. Mol Cell Biol. 2005;25:7534–7545. doi: 10.1128/MCB.25.17.7534-7545.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szebeni A, Olson MO. Nucleolar protein B23 has molecular chaperone activities. Protein Sci. 1999;8:905–912. doi: 10.1110/ps.8.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pang Q, Christianson TA, Koretsky T, et al. Nucleophosmin interacts with and inhibits the catalytic function of eukaryotic initiation factor 2 kinase PKR. J Biol Chem. 2003;278:41709–41717. doi: 10.1074/jbc.M301392200. [DOI] [PubMed] [Google Scholar]
- 38.Yung BY, Busch H, Chan PK. Translocation of nucleolar phosphoprotein B23 (37 kDa/pI 5.1) induced by selective inhibitors of ribosome synthesis. Biochim Biophys Acta. 1985;826:167–173. doi: 10.1016/0167-4781(85)90002-8. [DOI] [PubMed] [Google Scholar]
- 39.Yu Y, Maggi LB, Jr, Brady SN, et al. Nucleophosmin is essential for ribosomal protein L5 nuclear export. Mol Cell Biol. 2006;26:3798–3809. doi: 10.1128/MCB.26.10.3798-3809.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Falini B, Mecucci C, Tiacci E, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352:254–266. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 41.Shimamura A. Inherited bone marrow failure syndromes: molecular features. Hematology Am Soc Hematol Educ Program. 2006:63–71. doi: 10.1182/asheducation-2006.1.63. [DOI] [PubMed] [Google Scholar]
- 42.Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat Rev Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- 43.Brodersen DE, Nissen P. The social life of ribosomal proteins. FEBS J. 2005;272:2098–2108. doi: 10.1111/j.1742-4658.2005.04651.x. [DOI] [PubMed] [Google Scholar]
- 44.Young DW, Hassan MQ, Pratap J, et al. Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2. Nature. 2007;445:442–446. doi: 10.1038/nature05473. [DOI] [PubMed] [Google Scholar]
- 45.Hage AE, Tollervey D. A surfeit of factors: why is ribosome assembly so much more complicated in eukaryotes than bacteria? RNA Biol. 2004;1:10–15. [PubMed] [Google Scholar]
- 46.Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 2003;3:577–587. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- 47.Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 48.Mazumder B, Sampath P, Seshadri V, Maitra RK, DiCorleto PE, Fox PL. Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell. 2003;115:187–198. doi: 10.1016/s0092-8674(03)00773-6. [DOI] [PubMed] [Google Scholar]
- 49.Liu JM, Ellis SR. Ribosomes and marrow failure: coincidental association or molecular paradigm? Blood. 2006;107:4583–4588. doi: 10.1182/blood-2005-12-4831. [DOI] [PubMed] [Google Scholar]
- 50.Cmejlova J, Dolezalova L, Pospisilova D, Petrtylova K, Petrak J, Cmejla R. Translational efficiency in patients with Diamond-Blackfan anemia. Haematologica. 2006;91:1456–1464. [PubMed] [Google Scholar]
- 51.Mochizuki Y, He J, Kulkarni S, Bessler M, Mason PJ. Mouse dyskerin mutations affect accumulation of telomerase RNA and small nucleolar RNA, telomerase activity, and ribosomal RNA processing. Proc Natl Acad Sci U S A. 2004;101:10756–10761. doi: 10.1073/pnas.0402560101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruggero D, Grisendi S, Piazza F, et al. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science. 2003;299:259–262. doi: 10.1126/science.1079447. [DOI] [PubMed] [Google Scholar]
- 53.Ridanpaa M, van Eenennaam H, Pelin K, et al. Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage-hair hypoplasia. Cell. 2001;104:195–203. doi: 10.1016/s0092-8674(01)00205-7. [DOI] [PubMed] [Google Scholar]
- 54.Yoon A, Peng G, Brandenburger Y, et al. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science. 2006;312:902–906. doi: 10.1126/science.1123835. [DOI] [PubMed] [Google Scholar]
- 55.Revollo I, Nishiura H, Shibuya Y, Oda Y, Nishino N, Yamamoto T. Agonist and antagonist dual effect of the cross-linked S19 ribosomal protein dimer in the C5a receptor-mediated respiratory burst reaction of phagocytic leukocytes. Inflamm Res. 2005;54:82–90. doi: 10.1007/s00011-004-1327-4. [DOI] [PubMed] [Google Scholar]
- 56.Thiel CT, Horn D, Zabel B, et al. Severely incapacitating mutations in patients with extreme short stature identify RNA-processing endoribonuclease RMRP as an essential cell growth regulator. Am J Hum Genet. 2005;77:795–806. doi: 10.1086/497708. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.