Abstract
Background
Since prion gene-knockout mice do not contract prion diseases and animals in which production of prion protein (PrP) is reduced by half are resistant to the disease, we hypothesized that bovine animals with reduced PrP would be tolerant to BSE. Hence, attempts were made to produce bovine PRNP (bPRNP) that could be knocked down by RNA interference (RNAi) technology. Before an in vivo study, optimal conditions for knocking down bPRNP were determined in cultured mammalian cell systems. Factors examined included siRNA (short interfering RNA) expression plasmid vectors, target sites of PRNP, and lengths of siRNAs.
Results
Four siRNA expression plasmid vectors were used: three harboring different cloning sites were driven by the human U6 promoter (hU6), and one by the human tRNAVal promoter. Six target sites of bovine PRNP were designed using an algorithm. From 1 (22 mer) to 9 (19, 20, 21, 22, 23, 24, 25, 27, and 29 mer) siRNA expression vectors were constructed for each target site. As targets of siRNA, the entire bPRNP coding sequence was connected to the reporter gene of the fluorescent EGFP, or of firefly luciferase or Renilla luciferase. Target plasmid DNA was co-transfected with siRNA expression vector DNA into HeLaS3 cells, and fluorescence or luminescence was measured. The activities of siRNAs varied widely depending on the target sites, length of the siRNAs, and vectors used. Longer siRNAs were less effective, and 19 mer or 21 mer was generally optimal. Although 21 mer GGGGAGAACTTCACCGAAACT expressed by a hU6-driven plasmid with a Bsp MI cloning site was best under the present experimental conditions, the corresponding tRNA promoter-driven plasmid was almost equally useful. The effectiveness of this siRNA was confirmed by immunostaining and Western blotting.
Conclusion
Four siRNA expression plasmid vectors, six target sites of bPRNP, and various lengths of siRNAs from 19 mer to 29 mer were examined to establish optimal conditions for knocking down of bPRNP in vitro. The most effective siRNA so far tested was 21 mer GGGGAGAACTTCACCGAAACT driven either by a hU6 or tRNA promoter, a finding that provides a basis for further studies in vivo.
Background
Prion diseases are characterized by a prolonged latent period and a distinctive neuropathology that includes spongiform change, gliosis, neuronal loss, and the accumulation of an abnormal prion protein (PrPSc), an isomer of the normal cellular prion protein (PrPC) encoded by the prion gene (PRNP), in affected brains. PrPC, a glycoprotein, is anchored to the outer surface of neurons and to a lesser extent of lymphocytes and other cells. The function of PrPC is not known, but seems to be physiologically important because PRNP has been found in all animals examined (cattle, goats, hamsters, humans, mice, rats, sheep) as well as in the chicken. The conversion of PrPC to PrPSc is believed to occur not as a result of viral or bacterial infection but as a result of interaction with exogenously introduced, self-replicating PrPSc or by a very rare spontaneous event according to the protein-only hypothesis [1].
Although data have been accumulated using BSE-infected mice [2] and Prnp knockout mice as well [3-5], the physiological role of PrPC still remains to be clarified. From the pathological viewpoint, however, it is important that mice devoid of PrPC are resistant to scrapie and fail to propagate prions [6-9] and that the introduction of PrP-encoding transgenes restores susceptibility to the disease [10]. Prnp0/+ mice, which have about half the normal level of PrPC in their brains, show enhanced resistance to scrapie, as revealed by a significant delay in the onset and progression of clinical disease, while in wild-type animals, an increase in prion titer and PrPSc levels was followed within weeks by symptoms of scrapie and death [11]. These findings suggest that the production of BSE-resistant cattle would be possible by knocking down bovine PRNP (bPRNP) using RNA interference (RNAi) technology.
The injection of double-stranded RNA (dsRNA) into the cells of worms led to efficient sequence-specific gene silencing, referred to as RNAi [12]. This phenomenon also occurs in fly and plant cells, but not in mammalian cells. However, Elbashir et al. [13] demonstrated that 21~22-nt dsRNA with 2-nt 3' overhangs (short interfering RNA: siRNA) can induce sequence-specific gene silencing without non-specific inhibition of gene expression in cultured mammalian cells. siRNA expression systems using plasmid vectors are advantageous, because their use makes it possible to make transgenic animals, and the incorporation of one or a few plasmids into the nucleus would provide enough siRNA to induce RNAi. Soon after the discovery by Elbashir et al. [13] of the occurrence of RNAi in mammals, siRNA expression vector systems were developed [14-16]. The current understanding of the mechanisms of RNAi is as follows: dsRNA is digested to siRNA by the actions of Dicer, a family member of RNase III enzymes [17], and one strand of the siRNA unwound with the aid of ATP hydrolysis is incorporated into RISC (RNA-induced silencing complex) which has RNase activity [18]. mRNA with the sequence complementary to the siRNA is cleaved by RISC, knocking-down the gene expression of a specified mRNA (Dykxhoom et al., review [19]). Here we present optimal conditions, including siRNA expression promoters, target sites, and lengths of siRNAs, for knocking down bPRNP.
Results
Target plasmids with the insertion of a stop codon between reporter and target genes
In previous experiments, a full-length mouse Prnp gene with ATG (762 bp) or without ATG (759 bp) and three other fragments (645, 486, and 306 bp from the stop codon) were ligated in-frame into pDsRed2-C1. Cells transfected with the full-length construct and the plasmid with the 486-bp fragment did not show fluorescence, but cells transfected with the plasmids containing the 645-bp and 306-bp fragments emitted fluorescence, indicating that in-frame inserts might stop the production of the fluorescent reporter depending on the sequence. When the full-length bPRNP was inserted into pEGFP-C1 downstream from the EGFP stop codon, cells transfected with the construct emitted fluorescence. Therefore, a stop codon was inserted between the reporter and bPRNP in all the target plasmids, all of which were found to be effective in producing fluorescence in transfected cells (Table 1).
Table 1.
Characteristics of target vectors
| Vector | Promoter 1 | Reporter 1 | Target | Promoter 2 | Reporter 2 | Target |
| pEGFP-bPrP | CMV | EGFP | bPRNP | |||
| pGL3-bPrP | SV40 | Fire-fly luciferase | bPRNP | |||
| pFluc-bPrP-Rluc | SV40 | Fire-fly luciferase | bPRNP | HSV-TK | Renilla luciferase* | |
| pFluc-Rluc-bPrP | SV40 | Fire-fly luciferase* | HSV-TK | Renilla luciferase | bPRNP | |
| pTKFluc-bPrP-Rluc | HSV-TK | Fire fly luciferase | bPRNP | HSV-TK | Renilla luciferase* |
*, used as the internal control.
Dose-response relationship between fluorescence and plasmid DNA
When the dose-response relationship was examined using pEGFP-bPrP, linearity of EGFP fluorescence was obtained below 500 ng/well; a total of 400 ng/well or less was therefore used in subsequent experiments.
Comparison of sensitivity of fluorescence versus luminescence and lengths of siRNAs
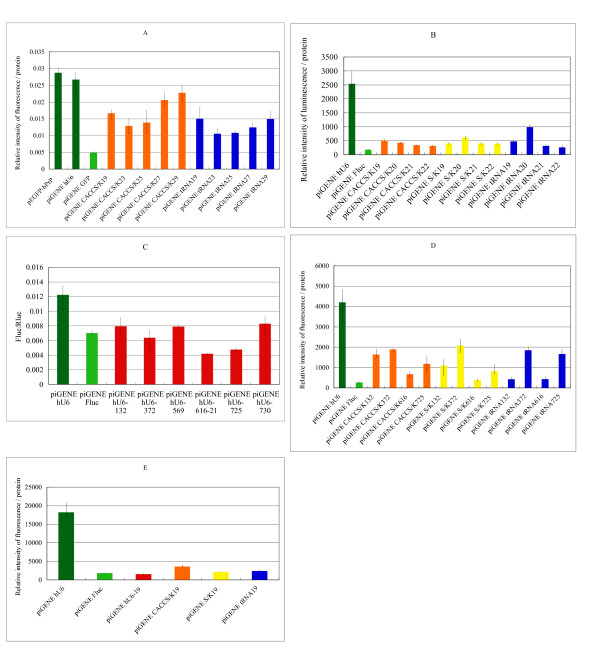
When the sensitivity of the fluorescent reporter of pEGFP-bPrP (Fig. 1A) and the luminescent reporter of pGL3-bPrP (Fig. 1B) was compared, detection of the EGFP fluorescence was found to be less sensitive than detection of the luminescence produced by firefly luciferase. This may be due to the comparatively long life of EGFP protein. Fig. 1A also shows that shorter siRNAs (around 23 mer) were more effective in silensing bPRNP expression than longer ones (27 and 29 mer), and that piGENE tRNA was more effective than piGENE CACC-S/K. Fig. 1B compares a narrow range of lengths of siRNAs expressed by three plasmid vectors. On the whole, 21 or 22 mer was the most effective silencer, and the differences between them were minor.
Figure 1.
Effects of siRNA expression vectors on bPRNP. A and B, effects of different lengths of siRNAs expressed by different vectors. A: piGENE hU6-EGFP was a positive control siRNA vector for EGFP. Numerals after vectors indicate the lengths of siRNA; 19, 23, 25, 27, and 29 correspond to No. 6, 10, 12, 13, and 14 in Table 2, respectively. B: piGENE Fluc is a positive control siRNA vector against firefly luciferase. Numerals after vectors indicate the lengths of siRNA; 19, 20, 21, and 22 correspond to No. 6, 7, 8, and 9 in Table 2, respectively. C and D, effects of different target sites and expression vectors on siRNA activities. C: numerals after vectors indicate the start sites of siRNA; 132, 372, 616, and 725 correspond to No. 2, 4, 9, and 16 in Table 2, respectively. D: as for numerals after vectors, see the legend to C. E: comparison of vector activities. Vectors expressed 19 mer, and the sequence corresponds to No. 6 in Table 2.
Vectors harboring internal control emitters
To determine the effectiveness of siRNAs, measurements of reporter protein levels were made using fluorescence or luminescence reporter assays (Fig. 1A and 1B). Three vectors in which an internal control was integrated into a single target vector, pFluc-bPrP-Rluc, pFluc-Rluc-bPrP, and pTKFluc-bPrP-Rluc, were constructed (Table 1). Since the results obtained with pTKFluc-bPrP-Rluc were almost the same as those for pFluc-Rluc-bPrP and pTKFluc-bPrP-Rluc, the results for pFluc-bPrP-Rluc are shown in Fig. 1C, in which effective target sites are compared (see next section). Renilla luminescence was always much stronger than firefly luminescence driven by either the SV40 or HCV-TK promoter. Although the sensitivity of siRNA effects detected using these double emitter vectors was low compared with that of a single emitter such as pGL3-bPrP, the relative effectiveness of siRNA was reproducible. Therefore, these vectors appeared to be useful for assaying the relative activity.
Effective target sites
Six siRNA target sites were predicted in the bPRNP sequence by using an algorithm [20] (Nos. 1, 3, 5, 6, 15 and 17, Table 2). Six different 22-nt targets (Nos. 2, 4, 5, 8, 16 and 18, Table 2), except for No. 8, where a 21 mer was used, were compared using a single expression vector, piGENE hU6 (Fig. 1C). The target vector was pFluc-bPrP-Rluc and the sensitivity was quite low, as described previously, but relative activities could be determined. No. 8 with the sequence 5'-GGGGAGAACTTCACCGAAACT-3' was the best silencer. Fig. 1D shows differences in siRNA activities examined using combinations of four target sites of 22 nt (Nos. 2, 4, 9, and 16, Table 2) and three expression vectors (piGENE CACC-S/K, piGENE S/K, and piGENE tRNA. Table 3). As for vectors, piGENE S/K was the best, followed by piGENE tRNA; and piGENE CACC-S/K was almost always the worst. As for target sites, sequence No. 9, the 22 mer version of No. 8 (21 mer), was the best and No. 4 was the worst using all three vectors. Differences between 21 mer and 22 mer were minimal in the three vectors (Fig. 1D).
Table 2.
Target sequences examined
| No. | Positiona | sequence | nt | Constructed vectors (○)b | Valuec | |||
| hU6 | CACC-S/K | S/K | tRNA | |||||
| 1 | 132–150 | GGGCAGTCCTGGAGGCAAC | 19 | ○ | ○ | ○ | ○ | 0.779 |
| 2 | 132–153 | GGGCAGTCCTGGAGGCAACCGT | 22 | ○ | ○ | ○ | ○ | |
| 3 | 372–390 | AGGAGCTGCTGCAGCTGGA | 19 | ○ | ○ | ○ | ○ | 0.775 |
| 4 | 372–393 | AGGAGCTGCTGCAGCTGGAGCA | 22 | ○ | ○ | ○ | ○ | |
| 5 | 569–591 | GTGTCAATATCACAGTCAAGGA | 22 | ○ | 0.757 | |||
| 6 | 616–634 | GGGGAGAACTTCACCGAAA | 19 | ○ | ○ | ○ | ○ | 0.852 |
| 7 | 616–635 | GGGGAGAACTTCACCGAAAC | 20 | ○ | ○ | ○ | ○ | |
| 8 | 616–636 | GGGGAGAACTTCACCGAAACT | 21 | ○ | ○ | ○ | ○ | |
| 9 | 616–637 | GGGGAGAACTTCACCGAAACTG | 22 | ○ | ○ | ○ | ○ | |
| 10 | 616–634 | GGGGAGAACTTCACCGAAACTGA | 23 | ○ | ○ | ○ | ○ | |
| 11 | 616–634 | GGGGAGAACTTCACCGAAACTGAT | 24 | ○ | ||||
| 12 | 616–634 | GGGGAGAACTTCACCGAAACTGACA | 25 | ○ | ○ | ○ | ○ | |
| 13 | 616–634 | GGGGAGAACTTCACCGAAACTGACATC | 27 | ○ | ○ | ○ | ○ | |
| 14 | 616–634 | GGGGAGAACTTCACCGAAACTGACATCAA | 29 | ○ | ○ | ○ | ○ | |
| 15 | 725–743 | GTGTGATCCTCTTCTCTTC | 19 | ○ | ○ | ○ | ○ | 0.772 |
| 16 | 725–746 | GTGTGATCCTCTTCTCTTCCCC | 22 | ○ | ○ | ○ | ○ | |
| 17 | 730–748 | ATCCTCTTCTCTTCCCCTC | 19 | ○ | 0.752 | |||
| 18 | 730–751 | ATCCTCTTCTCTTCCCCTCCTG | 22 | ○ | ||||
a, Position 1 starts from A of ATG, the first codon of bPRNP. b, hU6, CC-S/K, S/K, and tRNA indicate piGENE hU6, piGENE CACC-S/K, piGENE S/K, and piGENE tRNA, respectively (Table 3). Open circles designate construction of vectors. c, Values were obtained from the prediction algorithm [20].
Table 3.
Structure of cloning sites of siRNA expression vectors
| Vector | Promoter | Sequence of cloning site |
| piGENE hU6 | hU6 | CACCGTGAGCAGGTGTAAAGCCACCATGGAAGACACCTGCCAAC TTTTTTCAATTGGTCGACCTGCAGGCATGCAAGCTTa |
| piGENE CACC-S/K | hU6 | CACCGAGCTCAGACTCGATATCGGTACCb |
| piGENE S/K | hU6 | GAGCTCAGACTCGATATCGGTACCc |
| piGENE tRNA | tRNA | GAGCTCCAGATCTAATGCGGCCGCTTAGGTACCATAGATATCTTTTTTTCTGCAGGCATGCAAGCTTd |
a: Three Bsp MI sites are underlined. b and c: Sac I, Bgl II, Eco RV, and Kpn I sites are underlined. d: Sac I, Bgl II, Not I, Kpn I, Eco RV, Pst I, and Hind III sites are underlined. T repeats, RNA pol III terminator signal, are italicized.
Comparison of siRNA expression vectors
The structure of the cloning sites of the siRNA expression vectors is shown in Table 3. Fig 1E compares the effectiveness of the siRNA expression vectors. Fig. 1E shows a typical result, and the order of effectiveness was almost always piGENE hU6, piGENE S/K, piGENE tRNA, and piGENE CACC-S/K. The difference between piGENE hU6 and piGENE S/K was minimal and sometimes the order of these two was reversed.
bPRNP silencing as revealed by immunostaining
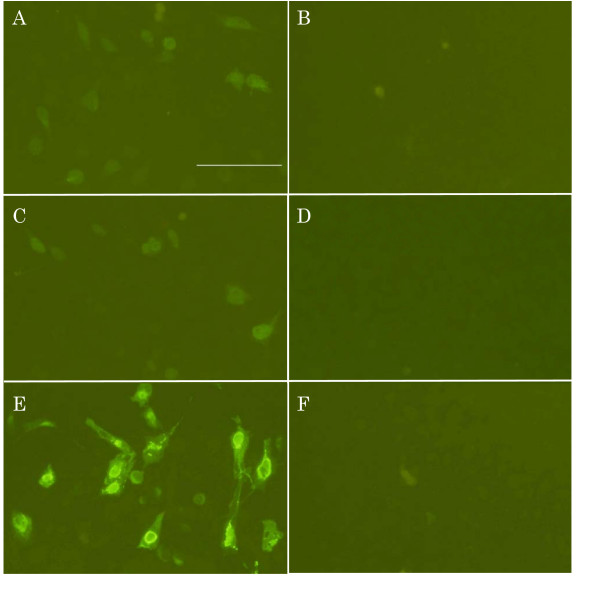
Full-length PrPC and truncated PrPC as targets and three antibodies, SAF32, P6488, and anti-FLAG, recognizing different regions, the upstream, mid-part, and C-terminal tag, respectively, were used for immunostaining experiments. Full-length PrPC could be detected equally using the three antibodies; the fluorescence signal intensities, which were moderate, were almost the same (Fig. 2A and 2C) among the three. The number of fluorescent cells depended on the amount of target DNA applied. When 800 ng/well was applied, approximately 50% of cells were fluorescence-positive, and when 200 ng/well was used, 20–30% were positive. When target DNA was co-transfected with siRNA expression vector DNA, almost no fluorescence was detected (Fig. 2B). This was true for both hU6 and tRNA promoter-driven vectors (Fig. 2B and 2D). Truncated PrPC could be detected using P6488 (Fig. 2E) and anti-FLAG antibody. As expected, SAF32 could not detect truncated PrPC, which lacks the N-terminal region (not shown). The intensity of fluorescence was much stronger than that of full-length PrPC (compare Fig. 2E with A and C). Perinuclear regions were strongly stained (Fig. 2E). The fluorescence almost disappeared when target DNA was co-transfected with siRNA expression plasmid DNA derived from either piGENE hU6 (Fig. 2F) or piGENE tRNA, indicating the effectiveness of siRNA.
Figure 2.
Silencing of b PRNP by siRNA as revealed by immunostaining. HeLaS3 cells were co-transfected with pbPrP- FLAG DNA and the control vector DNA of piGENE tRNA (A) or siRNA expression vector DNA of piGENE tRNA-616-21, the sequence of which is No. 8 in Table 2 (B). Cells were stained with SAF32 antibody. HeLaS3 cells were co-transfected with pbPrP- FLAG DNA and the control vector DNA of piGENE hU6 (C) or siRNA expression vector DNA of piGENE hU6-616-21(D). Cells were stained with P6488 antibody. HeLaS3 cells were co-transfected with pshort-bPrP- FLAG DNA and the control vector DNA of piGENE hU6 (E) or siRNA expression vector DNA of piGENE hU6-616-21 (F). Cells were stained with P6488 antibody.
bPRNP silencing as revealed by Western blotting
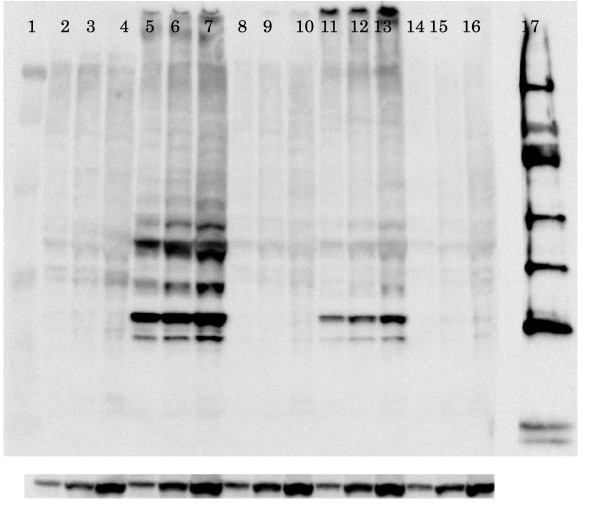
When a full-length PrPC was transiently expressed and detected using the antibody SAF32, 25- to 30-kDa bands were detected, with the 26-kDa band being most intense. When target DNA was co-transfected with siRNA expression vector DNA, almost no bands were detected (Fig 3), indicating the effectiveness of siRNA. This was true for both hU6 (Fig 3, lanes 8–10) and tRNA (Fig 3, lanes14–16) promoter-driven vectors. The bands in lanes 5–7 were darker than those in lanes 11–13. Sonication of the samples in lanes 11–13 seemed to have been insufficient, because intense bands were seen at the sites of sample application. The major band was 26 kDa, non-glycosylated PrPC. Glycosylated PrPC appeared at around 32 kDa, as can be clearly seen in lanes 5–7, and very faintly seen in lanes 11–13. The nature of the strong bands at around 40 kDa in lanes 5–7 is not known. Truncated PrPC could not be detected using SAF32, as expected, but could be detected using P6488 antibody (data not shown).
Figure 3.
Silencing of b PRNP by siRNA as revealed by Western blot analysis. Top, protein size markers (Bio-Rad, lane 1), proteins from HeLaS3 cells (lanes 2–4), proteins from cells co-transfected with pbPrP- FLAG DNA and the control vector DNA of piGENE hU6 (lanes 5–7) or siRNA expression vector DNA of piGENE hU6-616-21, the sequence of which is No. 8 in Table 2 (lanes 8–10), proteins from cells co-transfected with pbPrP- FLAG DNA and the control vector DNA of piGENE tRNA (lanes 11–13) or siRNA expression vector DNA of piGENE tRNA-616-21(lanes 14–16), and protein size markers (150, 100, 75, 35, 25, and 15 kDa from the top, GE Healthcare). Amounts of proteins applied were 1.25 (lanes 2, 5, 8, 11, 14), 2.5 (lanes 3, 6, 9, 12, 15), and 5.0 (lanes 4, 7, 10, 13, 16) μg/lane. The gel was stained with SAF32 antibody. Bottom, actin staining.
Discussion
The hU6 promoter has been widely used to drive siRNA expression in plasmid vectors, but tRNA promoter-driven vectors have rarely been used; therefore, in this study piGENE tRNA was used for comparison with piGENE hU6. These two vectors have different cloning sites, i.e., piGENE hU6 has a Bsp MI cloning site, while the piGENE tRNA has a Sac I, Bgl II, Not I, Kpn I, and Eco RV cloning site, (Table 3). The instructions of the manufacturer are also different: 19 to 22 mer is reccomended for piGENE hU6 and less than or close to 30 mer for piGENE tRNA. siRNA activities are affected by the promoters, siRNA lengths, and cloning site structures. To compare the effectiveness of piGENE hU6 and piGENE tRNA, two other siRNA-expression vectors were constructed; these were piGENE CACC-S/K and piGENE S/K, which had a common cloning site consisting of Sac I, Bgl II, Eco RV, and Kpn I recognition sequences. Using the Sac I and Kpn I sites was convenient because the same constructs could be employed as cassettes for piGENE CACC-S/K, piGENE S/K, and piGENE tRNA (Table 3). CACC was introduced into piGENE CACC-S/K because the natural human U6 promoter habors a G just after CACC, which enhances transcription [20].
siRNA activities can depend on the target genes and their sequences, and therefore definite and universal conclusions cannot be made. However, the present data indicate that a shorter siRNA (around 19 to 22 mer rather than 30 mer) is better for piGENE tRNA. As a whole, piGENE hU6 showed higher levels of siRNA expression activity than piGENE tRNA. piGENE hU6 and piGENE S/K had almost the same levels of activity, but the former seemed to be slightly better. The Sac I recognition site is GAGCTC and the insertion of DNA for siRNA expression occurred just after this site. On the other hand, the inserts come after CACCGAGCTC in piGENE CACC-S/K, the siRNA transcription of which might start from G, A, or G in the Sac I recognition site, and Dicer counts base numbers from these starting sites. An unexpected early start to transcription would leave several bases at the 3' end behind and lead to a shortage of matching bases to target mRNAs, lessening siRNA acrtivity.
As for the length of siRNA, 22 mer was generally the best (Fig. 1B), although 21 mer sometimes showed similar or better activity. However, 20 mer usually showed less activity than 19 mer, 21 mer or 22 mer (Fig. 1B). The reasons for this are not known, but Dicer's mechanisms of action might be involved.
The target sequence No. 6, 5'-GGGGAGAACTTCACCGAAA-3', achieved the highest score among the six targets (Table 2). Principles for the prediction of a favorable siRNA have been proposed [21,22]. One of the key factors in RNAi is the assembly of RISC, which mediates target RNA cleavage. The sense and anti-sense strands of an siRNA duplex are not equally eligible for assembly into RISC. Both the absolute stability and relative stability of the base pairs at the 5' end of siRNA seem to determine which strand takes part in the RNAi pathway. Given that in the conventional way of writing DNA sequences, the sense strand is the upper one and the anti-sense strand is the lower one, the left end should be tight and the right end should be loose for the anti-sense strand to be incorporated into RISC. In such an analysis, No. 6 has a tight left and a loose right end, and thus it is theoretically predicted to be effective, and was in fact found to be effective. This tightness and looseness do not always determine the effectiveness of RNAi, but are important factors.
The purpose of the present study was to knockdown bPRNP using RNAi technology. We used, however, an siRNA expression vector driven by a hU6 promoter and human HeLaS3 cells in an in vitro system. Is the hU6 promoter active in bovine cells? Recently, we [23] cloned chicken U6 promoters and examined their activity in chick cells using the hU6 promoter as a comparative control. Chicken and human promoters showed almost the same level of activity, although the activity levels were not very high. Lambeth et al. [24] cloned a bovine U6 promoter and examined its activity in MDBK (Madin Darby Bovine Kidney) and Vero (African Green monkey kidney) cells together with the mouse U6 promoter. The U6s of both species gave almost identical results, suggesting that the hU6 promoter might also be active in different mammalian cells. Indeed, our experiments showed that siRNA driven by hU6 was active in bovine primary cultured cells (data not shown).
Conclusion
To produce bPRNP-knocked down cattle using RNA interference (RNAi) technology, optimal conditions for knocking down were first investigated in vitro. Four siRNA expression plasmid vectors, six target sites of bPRNP, and various lengths of siRNA from 19 mer to 29 mer were examined. As a target, the bPRNP coding sequence was connected to the reporter of the fluorescent EGFP, firefly luciferase, or Renilla luciferase gene. When target plasmid DNA was co-transfected with siRNA expression vector DNA into HeLaS3 cells, and fluorescence or luminescence was measured, siRNA of 21 mer GGGGAGAACTTCACCGAAACT expressed by a hU6- or tRNA-driven plasmid gave the best knockdown result under the present experimental conditions. The effectiveness of this siRNA was confirmed by immunostaining and Western blotting. These data provide a basis for further studies in vivo.
Methods
Cloning of bovine prion gene PRNP
bPRNP consists of three exons, with the open reading frame (ORF) being located in exon 3 [25,26] (see also GeneBank Accession No. D10612). An ORF consisting of 792 bp encoding 264 amino acid residues was amplified by PCR with a pair of primers, one with a Bgl II site at the terminus and the other with a Hind III site. The PCR product was inserted into a TOPO vector (Invitrogen, Carlsbad, CA).
PRNP target vectors
pEGFP-C1, which harbors a green fluorescent protein gene driven by the CMV promoter, was purchased from Becton, Dickinson and Company, Japan (Tokyo, Japan). The multiple cloning sites of the vector were changed to Bsr GI-Bsp EI-Spe I-Bgl II-Sal I-Afl II-Spl I-Cla I-Apa I-Sma I/Xma I-Bam I-Xba I-Bcl I. This vector has an in-frame stop codon, TAG, in the Spe I recognition site (ACTAGT), which deletes the translation of inserts cloned downstream of the Bgl II site so as not to interfere with the intensity of fluorescence. pGL3 control and pRL-TK, which harbor the firefly luciferase gene (Fluc) driven by the SV40 promoter and the Renilla luciferase gene (Rluc) driven by the HSV-TK promoter, respectively, were purchased from Promega KK (Tokyo, Japan). The multiple cloning sites and Hind III site of the pGL3 control were removed and a new multiple cloning site, Xba I, Aat II, Bgl II, Sal I, Pst I, Apa I, Sma I/Xma I, Spe I, Afl II, and Spl I, was introduced just after the stop codon at the end of Fluc. The bPRNP gene was cloned between the Bgl II and Xma I sites. To use either Fluc or Rluc as an internal control, a combination vector, pRL-TK/SV-FL, was constructed by inserting the Bgl II and Bam HI fragment of the pGL3 control, which encodes the SV40 promoter and Fluc, into the Bam HI site of pRL-TK. Three bPRNP target plasmids were constructed from pRL-TK/SV-FL (Table 1).
PrPC-FLAG and truncated PrPC-FLAG vectors
For immunostaining and Western blot analyses, a FLAG-tagged PrPC expression vector was constructed by introducing the full-length bPRNP into the vector pFLAG-CMV-5 (Sigma-Aldrich, St. Louis, MO) between the Bgl II and Sal I sites. This vector, pbPrP- FLAG, was expected to produce a protein consisting of PrPC (264 amino acid residues) connected with FLAG (8 amino acid residues). Since antibodies to detect PrPC might hinder the identification of PrPC because of similarity in the molecular size, a truncated pPrP-FLAG was also constructed by removing the N-terminal region between Bgl II and Pvu II from PrPC-FLAG. This pshort-bPrP- FLAG vector was expected to produce a protein consisting of the C-terminal PrPC (145 amino acid residues) connected with FLAG (8 amino acid residues).
Selection of favorable target sites and construction of hairpin-type siRNA
Six target sites for siRNA were predicted using an algorithm developed by Taira and Miyagishi [20]. Some derivatives with lengths different from the original six sites were constructed (Table 2). The hairpin loop sequence for these was GTGTGCTGTCC. A few mutations in the sense strand were introduced in all constructs to avoid plasmid instability and difficulty in sequencing.
siRNA expression vectors
piGENE hU6 [14] has the human U6 promoter and a Bsp MI cloning site. piGENE tRNA Pur [27] has the human tRNAVal promoter and a multiple cloning site, Sac I/Bgl II/Not I/Kpn I/Eco RV. Both were obtained from iGENE Therapeutics, Inc. (Tsukuba, Japan). Two vectors modified at the cloning site were made from piGENE hU6. One had a Sac I/Kpn I cloning site and the other a naturally occurring CACC sequence just before the Sac I/Kpn cloning site (Table 3).
Cell culture and transfection
HeLaS3 cells were cultured in Dulbecco's modified Eagle's medium (Sigma, St. Louis, MO) supplemented with 10% fetal calf serum. Cells (0.75 × 105/well) were plated in a 24-well plate and, a day after plating, transfected with plasmid DNA. A typical treatment consisted of 200 μg of target vector DNA and 200 μg of siRNA expression vector DNA. Transfection was performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions.
RNAi assays
Cells transfected with fluorescent-protein-expressing plasmid DNA were examined under a fluorescence microscope (Olympus IX71, Tokyo). To determine the intensity of fluorescence quantitatively, cells were lysed with 100 μL of lysis buffer (Promega, Madison, WI) and aliquots (80 μL) were analyzed using Fluoroskan Ascent FL (Thermo Labosystems, Helsinki, Finland). To determine the amount of protein, aliquots (5 μL) were mixed with 200 μL of 5-fold diluted reagent of the Bio-Rad Protein Assay (Bio-Rad Laboratories-Inc, Hercules, CA), and the absorption was measured using Multiskan Ascent (Thermo Labosystems). Comparisons were made on the basis of fluorescence per μg protein. To measure luminescence, a Dual-luciferase Reporter Assay System (Promega) was used. The detector was a Sirius Luminometer (Berthold Detection Systems, Pforzheim, Germany). In this case, the intensity of the target (Fluc) was divided by that of the internal control (Rluc) or vice versa and the ratios were used for comparison.
Immunostaining of PrPC and truncated PrPC
HeLaS3 cells (4 × 104/well, 24-well plate) were plated and, after 24 h of culturing, pbPrP- FLAG DNA or pshort-bPrP- FLAG DNA (200 ng) and siRNA expression vector DNA or control vector DNA (200 ng) were co-transfected for 4 h. After 24 h, cells were fixed with acetone-methanol (1:1) and washed with TBS (Tris buffered saline) for 10 min 3 times. After being treated with TBST (TBS with 0.05% Triton X) for 10 min and then washed with TBS once, cells were blocked with 10% BSA/TBS for 1 h with shaking. After a brief wash with TBS, cells were stained with a primary antibody for 1 h. The antibody was anti-PrPC or anti-FLAG antibody diluted 2000 to 5000 fold. After another brief wash with TBS, cells were blocked with 3% BSA/TBS for 1 h, and then stained with a secondary antibody (rabbit anti-mouse-IgG-FITC (SIGMA) diluted 10000 fold) for 1 h. After three washes with TBST, cells covered with TBS were examined under a fluorescence microscope (Olympus IX71, Tokyo). One anti-PrPC antibody was SIGMA P6488 (SIGMA; host, mouse; isotype, IgG1), which had been raised against a synthetic peptide corresponding to amino acid residues 146–159 of bovine PrPC. Bovine PrPC was identified as a protein of 33–35 kDa that was immunoreactive with P6488, which also can detect human, sheep, deer, and elk PrPC. Another anti-PrPC antibody was SAF32 (SPI Bio; host, mouse; isotype, IgG2b), which recognizes the octo-repeat region located in the N-terminal part of PrP and cross-reacts with bovine, human, hamster, mouse, and ovine PrPC. Bovine PrPC and the truncated PrPC were also detected using the anti-FLAG antibody M2 (isotype, IgG1) which was one of the components of SIGMA's kit (Carboxy-terminal FLAG mammalian transient expression kit).
Western blot analysis
HeLaS3 cells (7 × 105) were plated in a plastic dish 60 mm in diameter. After 24 h, cells were co-transfected with pbPrP- FLAG DNA or pshort-bPrP- FLAG (4 μg) for 4 h. Cells were washed and cultured in fresh medium. After 24 h, they were washed with PBS once and collected in 100 μL of TBS with a rubber policeman. The cells were lysed by sonication for 3 min on ice. An equal volume of Laemmli sample buffer (Bio-Rad) containing 5% mercaptoethanol was added to the cell lysate, and the mixture was boiled for 5 min. Samples were subjected to SDS-PAGE using the following conditions: electrophoresis buffer, 1 × Tris-glycine-SDS (10 fold dilution of 10 × buffer, Bio-Rad) containing 5% methanol; gel, SuperSep™ 10–20% (Wako Pure Chemical Industries, Inc., Osaka); markers, ECL DualVeu Western blotting markers (GE Health Care, Piscataway, NJ); and electrophoresis conditions, 20 mA for 80 min. Proteins were transferred to a Fluorotrans® sheet (Nippon Genetics, Tokyo) in a Mini Transblot Cell (Bio-Rad) at 100 V for 60 min. The membrane was blocked with 5% skim milk/TBST for 1 h at room temperature. After being washed with TBST for 10 min 3 times, the membrane was stained with the antibody SAF32 (diluted 1/5000 with TBST) for 1 h with shaking. After 3 washes with TBST for 10 min each time, the membrane was stained with a secondary antibody, Anti-mouse IgG-HRP conjugate (idiluted 1/10000 with TBST, ECL Plus Western blotting detection kit, GE Healthcare), for 1 h with shaking. After 3 more washes with TBST for 10 min each time, the membrane was treated with a 40:1 mix of ELC Plus A:B(iWestern blotting detection kit) for 5 min. Photos were taken using a LAS-1000 (Fuji) with 30 sec to 2 min of exposure.
Authors' contributions
SS, MK, and PW carried out all experiments and SS drafted the manuscript. MM predicted target sites. TK and TO participated in the planning, design, and coordination of the research. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
This study was supported in part by a Grant (No. 16310140) from the Ministry of Education and Science of Japan and a research fund to S.S. from Shujitsu University.
Contributor Information
Shizuyo Sutou, Email: sutou@shujitsu.ac.jp.
Miho Kunishi, Email: kunishi@shujitsu.ac.jp.
Toshiyuki Kudo, Email: kudo@shujitsu.ac.jp.
Pimprapar Wongsrikeao, Email: Pimprapar@yahoo.com.
Makoto Miyagishi, Email: makoto-m@sannet.ne.jp.
Takeshige Otoi, Email: otoi@yamaguchi-u.ac.jp.
References
- Prusiner SB. Molecular biology of prion diseases. Science. 1991;252:1515–22. doi: 10.1126/science.1675487. [DOI] [PubMed] [Google Scholar]
- Kempster S, Collins ME, Deacon R, Edington N. Impaired motor coordination on static rods in BSE-infected mice. Behav Brain Res. 2004;154:291–295. doi: 10.1016/j.bbr.2004.01.021. [DOI] [PubMed] [Google Scholar]
- Sakaguchi S, Katamine S, Nishida N, Moriuchi R, Shigematsu K, Sugimoto T, Nakatani A, Kataoka Y, Houtani T, Shirabe S, Okada H, Hasegawa S, Miyamoto T, Noda T. Loss of cerebellar Purkinje cells in aged mice homozygous for a disrupted PrP gene. Nature. 1996;380:528–31. doi: 10.1038/380528a0. [DOI] [PubMed] [Google Scholar]
- Nico PB, de-Paris F, Vinade ER, Amaral OB, Rockenbach I, Soares BL, Guarnieri R, Wichert-Ana L, Calvo F, Walz R, Izquierdo I, Sakamoto AC, Brentani R, Martins VR, Bianchin MM. Altered behavioural response to acute stress in mice lacking cellular prion protein. Behav Brain Res. 2005;162:173–181. doi: 10.1016/j.bbr.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Bueler H, Fischer M, Lang Y, Bluethmann H, Lipp HP, DeArmond SJ, Prusiner SB, Aguet M, Weissman C. Normal development and behavior of mice lacking the neuronal cell-surface PrP protein. Nature. 1992;356:577–82. doi: 10.1038/356577a0. [DOI] [PubMed] [Google Scholar]
- Büeler H, Aguzzi A, Sailer A, Greiner RA, Autenried P, Aguet M, Weissmann C. Mice devoid of PrP are resistant to scrapie. Cell. 1993;73:1339–47. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- Manson JC, Clarke AR, Hooper ML, Aitchison L, McConnell I, Hope J. 129/Ola mice carrying a null mutation in PrP that abolishes mRNA production are developmentally normal. Mol Neurobiol. 1994;8:121–7. doi: 10.1007/BF02780662. [DOI] [PubMed] [Google Scholar]
- Sailer A, Büeler H, Fischer M, Aguzzi A, Weissmann C. No propagation of prions in mice devoid of PrP. Cell. 1994;77:967–8. doi: 10.1016/0092-8674(94)90436-7. [DOI] [PubMed] [Google Scholar]
- Sakaguchi S, Katamine S, Shigematsu K, Nakatani A, Moriuchi R, Nishida N, Kurokawa K, Nakaoke R, Sato H, Jishage K, Kuno J, Noda T, Miyamoto T. Accumulation of proteinase K-resistant prion protein (PrP) is restricted by the expression level of normal PrP in mice inoculated with a mouse-adapted strain of the Creutzfeldt-Jakob disease agent. J Virol. 1995;69:7586–92. doi: 10.1128/jvi.69.12.7586-7592.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M, Rulicke T, Raeber A, Sailer A, Moser M, Oesch B, Brandner S, Aguzzi A, Weissmann C. Prion protein (PrP) with amino-proximal deletions restoring susceptibility of PrP knockout mice to scrapie. EMBO J. 1996;15:1255–64. [PMC free article] [PubMed] [Google Scholar]
- Bueler H, Raeber A, Sailer A, Fischer M, Aguzzi A, Weissmann C. High prion and PrPSc levels but delayed onset of disease in scrapie-inoculated mice heterozygous for a disrupted PrP gene. Mol Med. 1994;1:19–30. [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalein A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalican cells. Nature. 2001;411:494–8. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Miyagishi M, Taira K. U6 promoter-driven siRNA with four uridine 3' overhangs efficiently suppress targeted gene expression in mammalian cells. Nature Biotech. 2002;19:497–500. doi: 10.1038/nbt0502-497. [DOI] [PubMed] [Google Scholar]
- Lee NS, Dohjima T, Bauer G, Li H, Li M-J, Ehsani E, Salvaterra P, Rossi J. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nature Biotech. 2002;20:500–505. doi: 10.1038/nbt0502-500. [DOI] [PubMed] [Google Scholar]
- Paul CP, Good PD, Winer I, Engelke DR. Effective expression of small interfering RNA in human cells. Nature Biotech. 2002;19:505–8. doi: 10.1038/nbt0502-505. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–6. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Nernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediatespost-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–6. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Dykxhoom DM, Novina CD, Sharp PA. Killing the messenger: short RNAs that silence gene expression. Nat Rev Mol Cell Bio. 2003;4:457–67. doi: 10.1038/nrm1129. [DOI] [PubMed] [Google Scholar]
- Taira K, Miyagishi M. Method and apparatus for predicting RNAi effect of siRNA. Japanese Patent Publication number 2005-115662. 2003.
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/S0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–16. doi: 10.1016/S0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Kudo T, Sutou S. Usage of putative chicken U6 promoters for vector-based RNA interference. J Reprod Develop. 2005;51:411–7. doi: 10.1262/jrd.16094. [DOI] [PubMed] [Google Scholar]
- Lambeth LS, Moore RJ, Muralitharan M, Dalrymple BP, McWilliam S, Doran T. Characterisation and application of a bovine U6 promoter for expression of short hairpin RNAs. BMC Biotechnol. 2005;11:5–13. doi: 10.1186/1472-6750-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto J, Iinuma T, Ishiguro N, Horiuchi M, Imamura M, Shinagawa M. Comparative sequence analysis and expression of bovine PrP gene in mouse L-929 cells. Virus Genes. 1992;6:343–56. doi: 10.1007/BF01703083. [DOI] [PubMed] [Google Scholar]
- Mastrangelo P, Westaway D. The prion gene complex encoding PrP(C) and Doppel: insights from mutational analysis. Gene. 2001;275:1–18. doi: 10.1016/S0378-1119(01)00627-8. [DOI] [PubMed] [Google Scholar]
- Kawasaki H, Taira K. Short hairpin type of dsRNAs that are controlled by tRNA(Val) promoter significantly induce RNAi-mediated gene silencing in the cytoplasm of human cells. Nucleic Acids Res. 2003;31:700–7. doi: 10.1093/nar/gkg158. [DOI] [PMC free article] [PubMed] [Google Scholar]