Abstract
The Mycobacterium tuberculosis fatty acid synthase type II (FAS-II) system has the unique property of producing unusually long-chain fatty acids involved in the biosynthesis of mycolic acids, key molecules of the tubercle bacillus. The enzyme(s) responsible for dehydration of (3R)-hydroxyacyl-ACP during the elongation cycles of the mycobacterial FAS-II remained unknown. This step is classically catalyzed by FabZ- and FabA-type enzymes in bacteria, but no such proteins are present in mycobacteria. Bioinformatic analyses and an essentiality study allowed the identification of a candidate protein cluster, Rv0635-Rv0636-Rv0637. Its expression in recombinant Escherichia coli strains leads to the formation of two heterodimers, Rv0635-Rv0636 (HadAB) and Rv0636-Rv0637 (HadBC), which also occurs in Mycobacterium smegmatis, as shown by split-Trp assays. Both heterodimers exhibit the enzymatic properties expected for mycobacterial FAS-II dehydratases: a marked specificity for both long-chain (≥C12) and ACP-linked substrates. Furthermore, they function as 3-hydroxyacyl dehydratases when coupled with MabA and InhA enzymes from the M. tuberculosis FAS-II system. HadAB and HadBC are the long-sought (3R)-hydroxyacyl-ACP dehydratases. The correlation between the substrate specificities of these enzymes, the organization of the orthologous gene cluster in different Corynebacterineae, and the structure of their mycolic acids suggests distinct roles for both heterodimers during the elongation process. This work describes bacterial monofunctional (3R)-hydroxyacyl-ACP dehydratases belonging to the hydratase 2 family. Their original structure and the fact that they are essential for M. tuberculosis survival make these enzymes very good candidates for the development of antimycobacterial drugs.
Keywords: (3R)-hydroxyacyl-ACP dehydratase, hydratase 2, mycolic acid biosynthesis, fatty acid elongation, hot dog fold
Tuberculosis kills nearly two million people every year (i.e., one person every 20 seconds). Emergence of drug-resistant Mycobacterium tuberculosis (Mtb) is one of the main concerns in the treatment of this disease; the prevalence of cases resistant to four main drugs (isoniazid, rifampicin, streptomycin, and ethambutol) is very high in some regions of the world (up to 30% for previously treated cases). Thus, the development of new drugs effective against the resistant strains has become a priority (1).
Mycobacteria possess a very thick lipid-rich envelope, which typically contains very long-chain (C60–C90) α-alkylated β-hydroxylated fatty acids (FAs) called mycolic acids (MAs) (2). These original lipids are found universally in the suborder of Corynebacterineae (including Mycobacterium, Corynebacterium, Rhodoccocus, Nocardia, etc.) where each genus holds molecules of specific chain lengths. MAs are crucial for the architecture and the permeability of the mycobacterial envelope. They also play a role in the virulence and the persistence of the tubercle bacillus within infected organisms (3, 4). Their biosynthetic pathway, which is essential for the survival of mycobacteria, is the target of a frontline antituberculous drug, isoniazid. Therefore, this metabolic pathway represents a valuable source for potential new pharmacological targets (2).
Isoniazid inhibits the elongation process leading to the formation of the main (meromycolic) chain of MAs. The four steps of the elongation cycles are monitored by an acyl carrier protein (ACP)-dependent FA synthase type II (FAS-II) system (5). FAS-II systems are found in plants, bacteria, parasites, and mitochondria, where they usually perform de novo biosynthesis (6). However, the system from Corynebacterineae is unique because it elongates long-chain FAs (C12–C18) into very-long-chain FAs (5). In mycobacteria, enzymes catalyzing three of the four elongation steps have been characterized [supporting information (SI) Fig. 7]: the β-ketoacyl-ACP synthetases KasA and KasB (7, 8), the β-ketoacyl-ACP reductase MabA (9), and the trans-2-enoyl-ACP reductase InhA (10). However, the enzyme(s) involved in the third step of the cycle, corresponding to (3R)-hydroxyacyl-ACP dehydratase(s) (HAD), was still unknown. The peptidic domains or proteins carrying this function within both multifunctional synthases and FAS-II systems have generally been the most difficult to identify (11, 12).
In FAS-II systems, the HAD activity is classically catalyzed by FabZ (dehydratase) or FabA (dehydratase–isomerase) enzymes (6). Yet we have previously shown that no FabZ/FabA-type protein was present in the tubercle bacillus (13). This indicated that the HADs in mycobacteria must present a different or distantly related catalytic sequence motif. After investigation of Mtb genome using bioinformatic tools, a pool of 11 putative (R)-specific enoyl hydratases/3-hydroxyacyl dehydratases has been identified (13, 14). Structural determination or modeling of three of them (13–15) suggested that they all belong or are related to the hydratase 2 protein family, where the underlying 3D structure of FabA/Z-type enzymes is maintained in the so-called hot dog fold but the catalytic site is distinct (16). One of these family members, Rv0636, has been proposed as a candidate for the FAS-II HAD on the basis of essentiality prediction (14). The purpose of the present work was to use complementary experimental strategies to determine which, if any, of these putative proteins could be the missing piece of the FAS-II puzzle.
Results
Selection of HAD Candidates Through in Silico Analyses.
To select in silico candidates for the dehydration step of the FAS-II cycle among the putative (R)-specific hydratases/dehydratases previously identified in Mtb (13, 14), we have defined different criteria (Table 1): (i) the ubiquity of the protein among mycobacteria and related genera (Nocardia and Rhodococcus), (ii) its absence in Corynebacterium that is devoid of FAS-II system (see Search for HadA, HadB, and HadC Orthologs in Other Corynebacterineae), (iii) the occurrence of catalytic motifs, and (iv) the proximity on the chromosome of genes potentially involved in FA/MA biosynthesis or transfer.
Table 1.
In silico analysis of putative (R)-specific hydratases/dehydratases from Mtb
| Protein | Size, kDa | Hydratase 2 motif* | Proximity† | Ubiquity among mycobacteria‡ | Corynebacterium§ | Rhodococcus sp.§ | Nocardia farcinica§ | Known or predicted structure¶ |
|---|---|---|---|---|---|---|---|---|
| Rv0130 | 16.0 | + | fbpC | − | NCgl0284,‖ 44% | Ro03023, 60% | Nfa34100, 58% | SHD |
| Rv0216 | 35.8 | − | + | Ro02873, 68% | DHD | |||
| Rv0241c | 30.2 | ++ | fabG4 | + | JK0821,** 42% | Ro05198, 51% | Nfa54620, 55% | DHD |
| Rv0504c | 18.4 | + | cmaA2 | + | SHD | |||
| Rv0635 HadA | 17.4 | − | mmaA1-mmaA4 | + | Ro01984, 53% | Nfa51180, 53% | SHD | |
| Rv0636 HadB | 14.9 | ++ | mmaA1-mmaA4 | + | Ro01983, 63% | Nfa51180, 63% | SHD | |
| Rv0637 HadC | 18.9 | − | mmaA1-mmaA4 | + | SHD | |||
| Rv2499c | 20.2 | − | − | JK1548,** 75% | Ro06098, 76% | Nfa50400, 78% | SHD | |
| Rv3389c | 30.3 | ++ | cmaA1 | − | DHD | |||
| Rv3538 | 30.2 | ++ | − | Ro04531, 62% | Nfa4620, 59% | DHD | ||
| Rv3542c | 33.9 | + | − | ‖Ro04486, 63% | Nfa4510, 65% | DHD |
*++, strictly conserved motif [YF]-x(1,2)-[LIVG]-[STGC]-G-D-x-N-P-[LIV]-H-x(5)-[AS]; +, motif including at least a conserved basic catalytic motif [D]-x(4)-H; −, absent.
†Presence of ORF(s) demonstrated as or putatively involved in FA/MA biosynthesis or transfer.
‡−, absent or present as a pseudogene; +, conserved in all considered sequenced mycobacterial genomes (see SI Materials and Methods).
§Name of orthologous putative protein with the percentage identity as given by BlastP alignment [on the (quasi-)totality of the sequence]. The following genomes were analyzed: Rhodococcus sp. RHA1, Nocardia farcinicaIFM10152, Corynebacterium diphtheriae NCTC13129, Corynebacterium glutamicum ATCC 13032, Corynebacterium jeikeium K411, and Corynebacterium efficiens YS-314.
¶SHD, single hot dog fold [known (underlined) SHD, see ref. 15]; DHD, double hot dog fold [known (underlined) OHO, see ref. 14].
‖Detected only in C. glutamicum.
**Detected only in C. jeikeium.
Among the six proteins whose genes are present in all mycobacterial genomes sequenced so far, Rv0216 has no obvious catalytic motif (14), and Rv0504c has no ortholog in Rhodococcus or Nocardia (Table 1). In contrast, Rv0241c and Rv0636 are both represented in these genera. They bear a characteristic and well conserved catalytic sequence, called the “hydratase 2 motif” (Table 1 and SI Fig. 8), that includes the catalytic residues D-x(4)-H (13, 14) and is observed in hydratase 2 family (16). Moreover, Rv0241c is adjacent to an ORF encoding a putative β-ketoacyl-ACP reductase (FabG4) distinct from that of FAS-II, MabA (FabG1), and Rv0636 is located near the mmaA1-4 gene cluster involved in the biosynthesis of oxygenated MAs (3). However, an Rv0241c ortholog was found in one Corynebacterium species (Table 1). Thus, Rv0636 alone satisfies all selection criteria and represents the best candidate. Moreover, on Mtb chromosomes, Rv0635 overlaps Rv0636 and Rv0637 is 4 bp away from Rv0636, implying that these three genes form an operon. The structural prediction data for Rv0636 (14), Rv0635, and Rv0637 (Table 1) suggested that each of these putative proteins of 15–19 kDa would be composed of a single hot dog fold. Proteins with such topology always form dimers (at least as a basic structural motif) (16, 17). Because Rv0635 and Rv0637 lack the hydratase 2 catalytic motif (Table 1 and SI Fig. 8), we hypothesized that Rv0636 protein needs to associate in heterodimers with either Rv0635 or Rv0637 to be functional. It is noteworthy that Rv0635 and Rv0637 share 45% sequence identity, whereas they are both poorly related to Rv0636 (13–15% identity) (SI Fig. 8), suggesting that Rv0635 and Rv0637 might have a similar role, distinct from that of Rv0636.
Essentiality of the Rv0635-Rv0637 Cluster.
We used a two-step homologous recombination strategy to demonstrate that the Rv0635-Rv0637 gene cluster is essential in vitro. A deletion delivery vector in which the entire operon was deleted was constructed. In addition, a merodiploid strain was constructed in which the entire operon was integrated by using an L5-based vector. Attempts to generate an unmarked deletion of the three genes were made in both the merodiploid and WT backgrounds (SI Fig. 9). In the latter we were unable to isolate any deletion mutants. Forty double crossover (DCO) strains were analyzed by PCR, and all had the WT gene. In contrast, in the merodiploid background we were able to delete the normal chromosomal copy; one of eight DCOs had both the WT (integrated) and the deletion alleles (SI Fig. 9), as confirmed by Southern blot analysis (data not shown). This provided direct evidence that one or several genes of Rv0635-Rv0637 cluster are essential in vitro. In contrast, a Rv0241c deletion was easily obtained in the WT background by using the same methodology (T.P., unpublished results). Because MAs are components essential for mycobacterial growth, the gene(s) encoding the HAD(s) of the FAS-II system are likely to be essential, like kasA, mabA, and inhA (18–20). Thus, these data are consistent with the Rv0635-Rv0636-Rv0637 cluster being the best candidate for the long-sought dehydratase. For greater convenience, these proteins will be called HadA, HadB, and HadC, respectively.
Expression and Purification of HabA-C Proteins: Analysis of Their Quaternary Structure in Vitro.
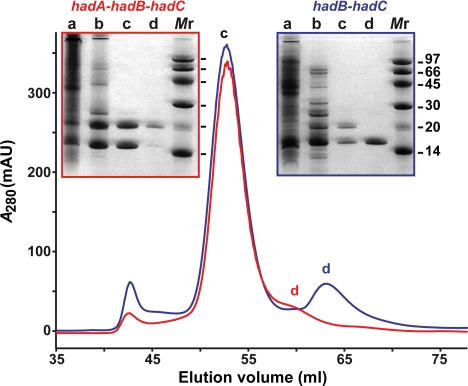
HadB gene or hadA-hadB-hadC and hadB-hadC gene clusters were cloned into a TOPO expression vector downstream of a His tag coding sequence. Proteins were produced in Escherichia coli and purified by using a two-step chromatography procedure on Ni Sepharose and Superdex 75 columns. It is noteworthy that, when hadB was expressed alone, most of the His-tagged product [His-tagged HadB (H-HadB)] was insoluble, and the purified protein was relatively unstable in solution. In contrast, with hadA-hadB-hadC clone, a significant amount of protein could be isolated. Strikingly, His-tagged HadA (H-HadA) copurified with untagged HadB, whereas the presence of HadC was undetectable (Fig. 1). Moreover, when H-HadB and HadC were coproduced, H-HadB coeluted with untagged HadC (Fig. 1). These data strongly suggested that there were interactions between HadA and HadB and between HadB and HadC.
Fig. 1.
Protein purification. SDS/PAGE and gel filtration chromatograms. The cloned genes are mentioned. Lane a, total soluble proteins; lane b, Ni Sepharose fraction; lanes c and d, Superdex 75 fractions. Monomeric sizes of proteins: H-HadA, 18.3 kDa; HadB, 14.8 kDa; H-HadB, 15.9 kDa; HadC, 18.9 kDa.
During gel filtration chromatography, performed at 4°C, the pool of Ni-column fractions containing both H-HadB and HadC displayed a profile with a major peak estimated at 65 kDa and a minor peak at ≈33 kDa (Fig. 1). Analyses by SDS/PAGE (Fig. 1) and peptide mapping demonstrated that the first elution peak corresponded to equimolar amounts of H-HadB and HadC, and the second one corresponded to H-HadB alone. A similar chromatogram was obtained with the fractions containing H-HadA and HadB, the major peak (at 65 kDa) holding both proteins and the minor peak holding only H-HadA (Fig. 1).
Data suggested that the heterocomplexes Rv0635-Rv0636 (HadAB) and Rv0636-Rv0637 (HadBC) formed tetramers. The quaternary structure of HadAB was further examined by dynamic light scattering, which confirmed that it was a tetramer at a low temperature (8°C) (SI Table 2). However, at a higher temperature (20°C) it behaved as a dimer.
Study of Protein–Protein Interactions in Vivo by Using Split-Trp Method.
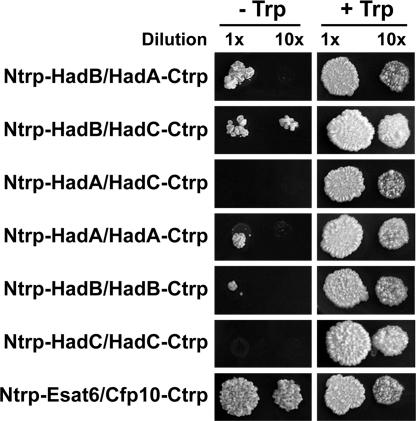
To investigate the occurrence of interactions between proteins encoded by the hadA-C cluster in mycobacteria in vivo, the split-Trp method was used in Mycobacterium smegmatis. Briefly, a tryptophan auxotrophic ΔhisA strain of M. smegmatis was cotransformed with a pair of plasmids, each one encoding a target protein in fusion with either the N-terminal fragment (Ntrp) or the C-terminal fragment (Ctrp) of Saccharomyces cerevisiae protein Trp1p involved in Trp biosynthesis. If an in vivo interaction occurs between the target proteins encoded by the pair of plasmids, Trp1p function will be restored and the recombinant strain will grow without exogenous Trp supply. In our experiments, heterotypic interactions (at 25°C) were observed in M. smegmatis between HadA and HadB and between HadB and HadC, but not between HadA and HadC (Fig. 2). Weaker homotypic interactions were detected for HadA and HadB, although not consistently for the latter.
Fig. 2.
Split-Trp growth assay in M. smegmatis. Recombinant M. smegmatis ΔhisA-expressing pairs of proteins fused to Ntrp and Ctrp were diluted 1- or 10-fold in water and spotted in parallel onto minimal medium and medium plus Trp. The pair of Ntrp-Esat6 and Cfp10-Ctrp was used as a positive control (the early secreted T cell antigens Esat6 and Cfp10 from Mtb form a tight, 1:1 complex). Negative controls and extra positive controls were performed (see SI Materials and Methods). Images were taken after a 3-week incubation.
These results were in full agreement with the in vitro data. They altogether led to the conclusion that HadB interacts in vitro and in vivo with either HadA or HadC to form heterodimers (at a temperature ≥20°C).
Enzymatic Activity of HadAB and HadBC Heterodimers.
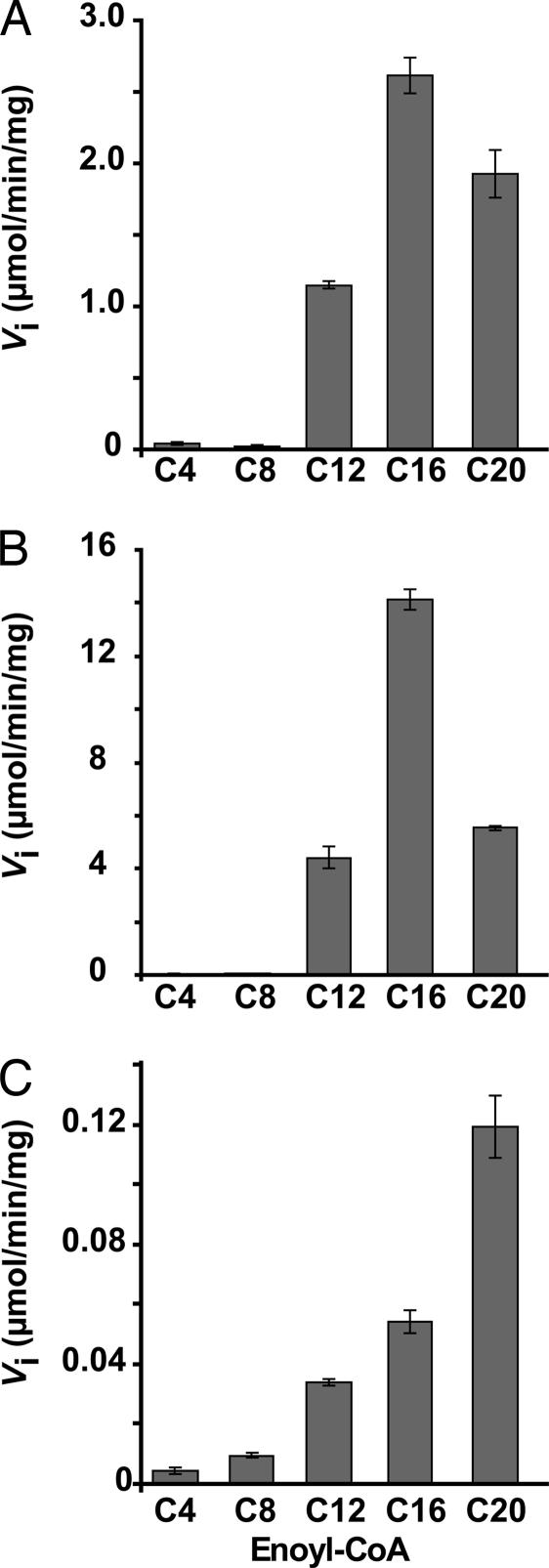
Enzymes belonging to the (R)-specific enoyl hydratase/hydroxyacyl dehydratase family preferentially catalyze the hydration reaction when they are isolated from their enzymatic complex (6). Thus, their activities are most often studied in the presence of enoyl derivatives in vitro. The activities of HadAB and HadBC heterodimers were first measured in the presence of short-chain CoA derivatives, 3-hydroxybutyryl-CoA (C4), crotonoyl-CoA (C4:1), and trans-2-octenoyl-CoA (C8:1). HadBC did not display any significant activity with either substrate in the experimental conditions used, even by varying the enzyme concentration over a large range (see SI Materials and Methods). A slight activity of HadAB could be detected only in the presence of the C8 substrate: 0.506 ± 0.001 μmol/min per milligram of protein at the highest enzyme concentration tested (280 nM).
Kinetic experiments were then realized in the presence of longer-chain derivatives, C12–C20 trans-2-enoyl-CoAs. To minimize the solubility problems encountered with such amphipathic molecules, the experiments were first performed at a fixed low substrate concentration (2.5 μM). HadAB heterodimer proved to be active in the presence of these medium- to long-chain molecules (Fig. 3A). In contrast, no signal was detected with HadBC in these conditions; it exhibited activity only at a higher substrate concentration (25 μM). The activity of HadBC increased with increasing chain length (Fig. 3C), whereas HadAB behaved differently with a specificity centered on the hexadecenoyl-CoA (C16) (Fig. 3 A and B).
Fig. 3.
Chain length specificity profiles of HadAB and HadBC heterodimers. Assays were performed at fixed concentrations of substrate and enzyme. (A and B) HadAB with 2.5 μM enoyl-CoA (A) or 25 μM enoyl-CoA (B). (C) HadBC with 25 μM enoyl-CoA. Data are means ± SD. The activities measured for both heterodimers in the presence of short chain substrates (C4 and C8) were not significantly above the control values (without enzyme). The large SDs obtained sometimes for long-chain substrates are due to solubility problems.
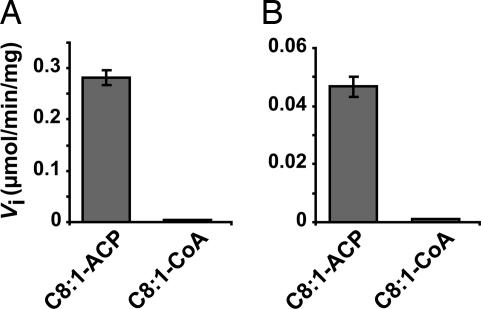
The behaviors of the enzymes in the presence of ACP or CoA derivatives were then compared by using either trans-2-octenoyl-ACP or trans-2-octenoyl-CoA as substrates. Both heterodimers displayed a strict specificity toward the enoyl-ACP (Fig. 4). Interestingly, HadB homodimer was not able to metabolize C8:1-ACP in conditions identical to those used for the heterodimers (data not shown).
Fig. 4.
Comparison of the specific activities in the presence of octenoyl-CoA and octenoyl-ACP. Assays were performed at 2 μM substrate and fixed concentrations of enzymes HadAB (A) and HadBC (B). Data are means ± SD. The activities measured for both heterodimers in the presence of C8:1-CoA were not significantly above the control values (without enzyme).
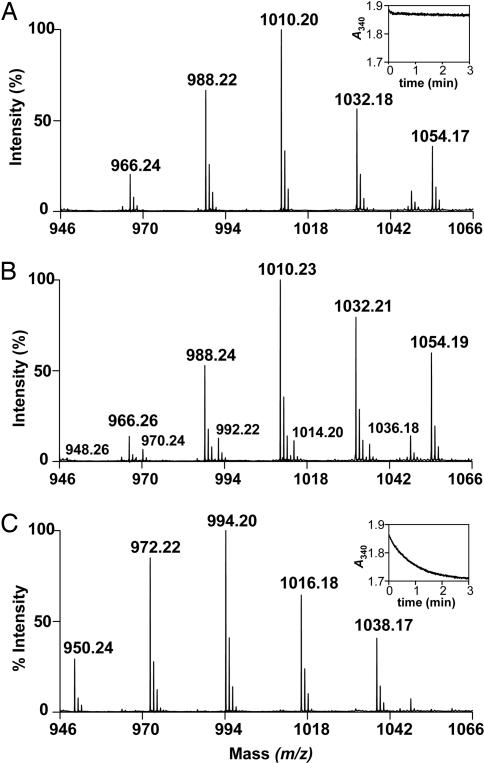
Finally, both heterodimers were tested for their ability to function in coupled reactions in the presence of MabA and InhA, the FAS-II reductases that catalyze the reactions upstream and downstream of the dehydration step in the cycle (SI Fig. 7). The MabA reaction was performed first in the presence of 3-ketododecanoyl-CoA (C12), and, after completion of the reaction, one of the heterodimers plus InhA were added. Aliquots of the reaction mixtures were taken at different time points (see SI Materials and Methods). The MS analyses demonstrate that dodecanoyl-CoA, the saturated product of InhA, was very quickly formed (within 3 min) in the presence of HadAB (Fig. 5C). This biosynthesis did not occur in the absence of the heterodimer (Fig. 5A). In the absence of InhA, a small proportion of the intermediate dodecanoyl-CoA, a product of the dehydration reaction catalyzed by HadAB, appeared (Fig. 5B). This type of profile is reminiscent of what has been described for other HADs (6): in the absence of enoyl reductase, the reaction equilibrium is in favor of the hydration reaction. By comparison, the complete reaction in the presence of HadBC heterodimer went very slowly: the product of InhA began to appear after 30 min only, and the reaction arrived to completion between the 3-h and 24-h time points (SI Fig. 10). This weak activity correlates with the relatively poor specific activity of HadBC in the presence of medium-chain CoA derivatives (see above, earlier in this paragraph).
Fig. 5.
Coupled assay of HadAB in the presence of MabA and InhA. Shown are the results of MALDI-TOF MS analyses of the reaction media containing 3-ketododecanoyl-CoA, NADPH, NADH, and MabA plus InhA (A), or plus 280 nM HadAB (B), or plus both HadAB and InhA (C). In A and B, the peaks at m/z 966, 988, 1,010, 1,032, and 1,054 stand for [M+H]+, [M+Na]+, [M-H+2Na]+, [M-2H+3Na]+, and [M-3H+4Na]+ ions of 3-hydroxydodecanoyl-CoA (product of MabA), respectively. In B, the minor peaks at m/z 948, 970, 992, 1,014, and 1,036 stand for [M+H]+ ion and the monosodium to tetrasodium adducts of the unsaturated species, dodecenoyl-CoA. In C, the peaks at m/z 950, 972, 994, 1,016, and 1,038 stand for [M+H]+ ion and the monosodium to tetrasodium adducts of the saturated species, dodecanoyl-CoA. Insets in A and C display the kinetics of the respective reactions followed at 340 nm (oxidation of InhA coenzyme, NADH). The three spectra correspond to the time points of 3 min of reaction.
In conclusion, the properties of HadAB and HadBC heterodimers are reminiscent of those described for the mycobacterial FAS-II system and for the individual enzymes that are part of this complex, i.e., a marked specificity for medium- to long-chain substrates and for ACP derivatives (7–10). Furthermore, they exhibit a (3R)-hydroacyl dehydratase activity in the presence of both reductases of the FAS-II complex.
Search for HadA, HadB, and HadC Orthologs in Other Corynebacterineae.
To gather other putative information on the respective roles of HadAB and HadBC within the FAS-II cycle, and consequently during the formation of the meromycolic chain, the presence and organization of the hadA-C cluster were examined in three other Corynebacterineae genera, whose genomes were available (see the Table 1 legend). The chromosomal region surrounding the hadA-C gene cluster is very well conserved within mycobacteria as well as in related genera, namely Corynebacterium, Rhodococcus, and Nocardia (SI Fig. 11). However, in Corynebacterium the ORFs corresponding to hadA, hadB, and hadC are missing, and no orthologs were detected in another region by BLAST searches (Table 1), as for the known FAS-II enzymes. Indeed, Corynebacterium produces short MAs (C22–C36), which result from the simple condensation of classical FAs (C8–C18), without any FAS-II-linked FA elongation steps. Rhodococcus and Nocardia possess “intermediate-size” MAs (C34–C48 and C44–C60, respectively) made of a medium-length meromycolic chain (C22–C30 and C32–C42, respectively), shorter than that found in mycobacteria (C52–C64) (2). Consistently, genes orthologous to hadA and hadB, as well as inhA, mabA, and kasA, are present in Rhodococcus and Nocardia (Table 1 and SI Fig. 11). Interestingly, the orthologs of hadA and hadB are fused in a long unique gene in Nocardia. In contrast, hadC ORF is missing in both Nocardia and Rhodococcus (SI Fig. 11), and no ortholog could be found elsewhere in these genomes (Table 1). Likewise, BLAST searches revealed that these genera do not hold any protein orthologous to KasB (data not shown).
Discussion
No typical FabZ or FabA enzymes universally found so far in the bacterial FAS-II systems are present in Mtb or other sequenced mycobacterial genomes. Our previous investigations to seek potential (R)-specific hydratases/dehydratases in the tubercle bacillus have led us (13, 14) to another protein family that include bacterial and eukaryotic enoyl-CoA hydratases 2 (16, 17). If it shares the same global topology and probably the same reaction mechanism as the FabA/FabZ family, it exhibits very poor sequence similarity with the latter and its catalytic motif is distinct (17). Nonetheless, the HAD domain of the multifunctional FAS-I polypeptides from Corynebacterineae, Fungi, and Archaeoglobus do have a hydratase 2 catalytic motif (13, 14, 21). Similarly, we could imagine that the (3R)-hydroxyacyl-ACP dehydratation step of the elongation cycles monitored by the mycobacterial FAS-II system could be catalyzed by an enzyme of the hydratase 2 family.
Among the putative hydratases 2 and related proteins identified in Mtb, the protein cluster HadA-HadB-HadC, probably encoded by an operon, appeared as a good candidate for the HAD of FAS-II. The complete set of experimental data reported in the present study points clearly to HadAB and HadBC heterodimers as the missing pieces of the elongation system. Unlike Rv3389c, a previously studied Mtb hydratase 2 protein (13), HadB-containing enzymes display both ACP dependence and specificity for long-chain substrates that are the characteristic properties of FAS-II components (5).
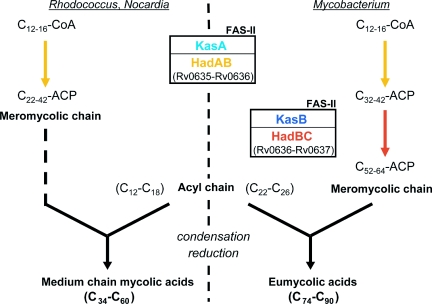
When coproduced in E. coli or in M. smegmatis, HadB protein associates in heterodimers with either HadA or HadC. These partnerships substantially change the behavior, because HadAB and HadBC heterodimers exhibit distinct chain length specificity profiles. Strikingly, we observed through genome analyses that both hadA and hadB genes were conserved among other mycolate-producing genera, Rhodococcus and Nocardia. Consistently, both genes either overlap or fuse in a long unique gene, suggesting that their cotranscription and the association of their products are crucial for the physiology of the Corynebacterineae. Furthermore, we demonstrated that at least one gene of the hadA-C cluster was essential for Mtb survival, as is the case for most genes encoding FAS-II enzymes (18–20). This is consistent with transposon mutagenesis-based predictions of essentiality for hadA and hadB genes (22). Thus, like KasA (4, 23), HadAB is most likely involved in the early FA elongation cycles catalyzed by FAS-II in mycobacteria, but also in Nocardia and Rhodococcus, leading to the formation of intermediate-size meromycolic chains (Fig. 6). The preference of HadBC for longer substrates, as compared with HadAB, and the absence of HadC ortholog in genera bearing medium-chain MAs, and where a KasB ortholog is also absent, strongly suggest that HadBC would be implicated, as described for KasB (4, 23), in the late steps of the meromycolic chain biosynthesis occurring in mycobacteria (Fig. 6). Additionally, the hadC gene does not seem to be essential for Mtb growth in vitro (22). Consistently, kasB was shown to be nonessential in Mtb and Mycobacterium marinum (4, 23). Furthermore, HadC seems ubiquitous among mycobacteria. From these data, we propose that HadBC is necessary for the formation of the complete meromycolic chains of MA in mycobacteria (Fig. 6).
Fig. 6.
Model of the roles of HadAB and HadBC heterodimers in the MA biosynthesis pathway. HadAB would take part, like KasA, in the early FA elongation cycles catalyzed by the FAS-II system, leading to the formation of the intermediate-size (C22–C42) meromycolic chains and consequently to medium-length MAs found in Rhodococcus and Nocardia. In mycobacteria, HadBC, like KasB, would elongate further the intermediate-size meromycolic chains to full-size molecules (C52–C64) during the late elongation cycles performed by FAS-II, resulting in the synthesis of eumycolic acids (C74–C90). For clarity, only some proteins of the FAS-II system are mentioned.
From our sequence analysis, only HadB bears a hydratase 2 motif (13, 14) and therefore would be the catalytic subunit of HadAB and HadBC heterodimers. Furthermore, the structural predictions strongly suggest that each of the three subunits has a single hot dog fold. Thus, they would organize as asymmetrical heterodimers that are reminiscent of the asymmetrical double hot dog fold of the R-hydratase domain of MFE-2 enzymes and probably of Rv3389c R-hydratase/dehydratase protein (13, 17). In such double hot dog folds, one domain contains the catalytic apparatus whereas the other would be necessary to stabilize the long acyl chain of the substrates. A role analogous to the latter can be proposed for HadA and HadC subunits. Consistently, structural predictions suggest that HadAB and HadBC would hold open active site tunnels (required for the long-chain meromycolate precursors), although a detailed description must await crystallographic data. In contrast, a closed substrate-binding pocket has been predicted for HadB homodimer (15). Furthermore, the absence of activity of HadB heterodimer in the presence of octenoyl-ACP suggests that the heterotypic partnerships between HadB and HadA or HadC may also play a role in the interaction with the ACP moiety of the substrate. To our knowledge, no asymmetrical hot dog folded heterodimer has been reported so far. From their organization into a single fused gene, it can be deduced that the orthologs of HadA and HadB in Nocardia are covalently bound. They would therefore adopt a topology equivalent to the predicted structure of Rv3389c (13, 14), with an intervening bridge between the two domains.
The present work is the first description of bacterial monofunctional HADs belonging to the hydratase 2 enzyme family. Thanks to an elegant experimental strategy, the first eukaryotic HAD has been discovered recently, that of the yeast mitochondrial FAS-II system (12). Strikingly, it corresponds to a hydratase 2-related double hot dog enzyme. The high level of sequence divergence between the mycobacterial and eukaryotic enzymes, together with the importance of this essential step in a metabolic pathway specific to Mycobacterium and related genera, makes HadAB and HadBC exciting new targets for drug discovery.
Materials and Methods
Sequence analyses were done as described (13). The Mtb Rv0635-Rv0637-KO strain was constructed based on a previously reported strategy (20). DCOs were isolated from the single crossover (SCO) and merodiploid strains as previously described (24). Protein production and purification of HadB, HadAB, and HadBC were performed as described (14), except that expressions were induced at 37°C, no Triton was added in lysis buffers (see SI Table 3), and a Ni Sepharose FF (GE Healthcare) column was used. The split-Trp experiments were adapted from the procedure described in yeast (25). Enzyme assays and MS analyses of the reaction media were performed as described (13). Short-chain CoA derivatives were used at 25 μM (and also 75 μM for hydroxybutyryl-CoA). The octenoyl-ACP was prepared by using the commercially available E. coli holo-ACP (Sigma, St. Louis, MO), trans-2-octenoic acid, and E. coli acyl-ACP synthase as reported (26). The mycobacterial FAS-II system can use the E. coli ACP instead of its natural ACP, AcpM (SI Fig. 7) (5). See additional details in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank F. Laval, H. Montrozier, F. Viala (Institut de Pharmacologie et de Biologie Structurale), and L. Henry (Ecole Polytechnique Federale de Lausanne) for their precious help during MS analyses, substrate synthesis, figure preparation, and split-Trp experiments, respectively; A. Hartridge (Institute of Cell and Molecular Science) for technical assistance and the Expression Proteomics Facility (Uppsala University) for MS analyses; J. Shanklin (Brookhaven National Laboratory, Upton, NY) and J. Walker (Medical Research Council, Cambridge, U.K.) for their kind gifts of plasmid pAasH and E. coli C41(DE3), respectively; and S. Cole (Institut Pasteur, Paris, France) for total Mtb DNA and cosmid MTCY20H10.This work was supported in part by a doctoral fellowship (to E.S.) from the Fondation Pour la Recherche Médicale, by a Marie Curie fellowship, and by grants from the European Community (QLK2-CT-2001-02018 and LSHP-CT-2005-018923), the Foundation for Strategic Research, and the Swedish Research Council.
Abbreviations
- ACP
acyl carrier protein
- FA
fatty acid
- FAS
FA synthase
- HAD
(3R)-hydroxyacyl-ACP dehydratase
- H-HadA
His-tagged HadA
- H-HadB
His-tagged HadB
- MA
mycolic acid
- Mtb
M. tuberculosis.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704132104/DC1.
References
- 1.World Health Organization. Anti-Tuberculosis Drug Resistance in the World, Report No. 3. Geneva: WHO; 2004. [Google Scholar]
- 2.Barry CE, III, Lee RE, Mdluli K, Sampson AE, Schroeder BG, Slayden RA, Yuan Y. Prog Lipid Res. 1998;37:143–179. doi: 10.1016/s0163-7827(98)00008-3. [DOI] [PubMed] [Google Scholar]
- 3.Dubnau E, Chan J, Raynaud C, Mohan VP, Lanéelle MA, Yu K, Quémard A, Smith I, Daffé M. Mol Microbiol. 2000;36:630–637. doi: 10.1046/j.1365-2958.2000.01882.x. [DOI] [PubMed] [Google Scholar]
- 4.Bhatt A, Fujiwara N, Bhatt K, Gurcha SS, Kremer L, Chen B, Chan J, Porcelli SA, Kobayashi K, Besra GS, Jacobs WR., Jr Proc Natl Acad Sci USA. 2007;104:5157–5162. doi: 10.1073/pnas.0608654104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bloch K. Adv Enzymol Relat Areas Mol Biol. 1977;45:1–84. doi: 10.1002/9780470122907.ch1. [DOI] [PubMed] [Google Scholar]
- 6.Rock CO, Cronan JE. Biochim Biophys Acta. 1996;1302:1–16. doi: 10.1016/0005-2760(96)00056-2. [DOI] [PubMed] [Google Scholar]
- 7.Schaeffer ML, Agnihotri G, Volker C, Kallender H, Brennan PJ, Lonsdale JT. J Biol Chem. 2001;276:47029–47037. doi: 10.1074/jbc.M108903200. [DOI] [PubMed] [Google Scholar]
- 8.Kremer L, Dover LG, Carrere S, Nampoothiri KM, Lesjean S, Brown AK, Brennan PJ, Minnikin DE, Locht C, Besra GS. Biochem J. 2002;364:423–430. doi: 10.1042/BJ20011628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marrakchi H, Ducasse S, Labesse G, Montrozier H, Margeat E, Emorine L, Charpentier X, Daffé M, Quémard A. Microbiology. 2002;148:951–960. doi: 10.1099/00221287-148-4-951. [DOI] [PubMed] [Google Scholar]
- 10.Quémard A, Sacchettini JC, Dessen A, Vilchèze C, Bittman R, Jacobs WR, Jr, Blanchard JS. Biochemistry. 1995;34:8235–8241. doi: 10.1021/bi00026a004. [DOI] [PubMed] [Google Scholar]
- 11.Joshi AK, Smith S. J Biol Chem. 1993;268:22508–22513. [PubMed] [Google Scholar]
- 12.Kastaniotis AJ, Autio KJ, Sormunen RT, Hiltunen JK. Mol Microbiol. 2004;53:1407–1421. doi: 10.1111/j.1365-2958.2004.04191.x. [DOI] [PubMed] [Google Scholar]
- 13.Sacco E, Legendre V, Laval F, Zerbib D, Montrozier H, Eynard N, Guilhot C, Daffé M, Quémard A. Biochim Biophys Acta. 2007;1774:303–311. doi: 10.1016/j.bbapap.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 14.Castell A, Johansson P, Unge T, Jones TA, Bäckbro K. Protein Sci. 2005;14:1850–1862. doi: 10.1110/ps.051442305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johansson P, Castell A, Jones TA, Bäckbro K. Protein Sci. 2006;15:2300–2309. doi: 10.1110/ps.062309306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hisano T, Tsuge T, Fukui T, Iwata T, Miki K, Doi Y. J Biol Chem. 2003;278:617–624. doi: 10.1074/jbc.M205484200. [DOI] [PubMed] [Google Scholar]
- 17.Koski MK, Haapalainen AM, Hiltunen JK, Glumoff T. J Biol Chem. 2004;279:24666–24672. doi: 10.1074/jbc.M400293200. [DOI] [PubMed] [Google Scholar]
- 18.Vilchèze C, Morbidoni HR, Weisbrod TR, Iwamoto H, Kuo M, Sacchettini JC, Jacobs WR., Jr J Bacteriol. 2000;182:4059–4067. doi: 10.1128/jb.182.14.4059-4067.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhatt A, Kremer L, Dai AZ, Sacchettini JC, Jacobs WR., Jr J Bacteriol. 2005;187:7596–7606. doi: 10.1128/JB.187.22.7596-7606.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parish T, Roberts G, Laval F, Schaeffer M, Daffé M, Duncan K. J Bacteriol. 2007;189:3721–3728. doi: 10.1128/JB.01740-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qin YM, Haapalainen AM, Kilpeläinen SH, Marttila MS, Koski MK, Glumoff T, Novikov DK, Hiltunen JK. J Biol Chem. 2000;275:4965–4972. doi: 10.1074/jbc.275.7.4965. [DOI] [PubMed] [Google Scholar]
- 22.Sassetti CM, Boyd DH, Rubin EJ. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 23.Gao LY, Laval F, Lawson EH, Groger RK, Woodruff A, Morisaki JH, Cox JS, Daffé M, Brown EJ. Mol Microbiol. 2003;49:1547–1563. doi: 10.1046/j.1365-2958.2003.03667.x. [DOI] [PubMed] [Google Scholar]
- 24.Parish T, Stoker NG. Microbiology. 2000;146:1969–1975. doi: 10.1099/00221287-146-8-1969. [DOI] [PubMed] [Google Scholar]
- 25.Tafelmeyer P, Johnsson N, Johnsson K. Chem Biol. 2004;11:681–689. doi: 10.1016/j.chembiol.2004.02.026. [DOI] [PubMed] [Google Scholar]
- 26.Rock CO, Garwin JL. J Biol Chem. 1979;254:7123–7128. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.