Abstract
The molecular nature of many plant disease resistance (R) genes is known; the largest class encodes nucleotide-binding site-leucine-rich repeat (NBS-LRR) proteins that are structurally related to proteins involved in innate immunity in animals. Few genes conferring disease susceptibility, on the other hand, have been identified. Recent identification of susceptibility to the fungus Cochliobolus victoriae in Arabidopsis thaliana has enabled our cloning of LOV1, a disease susceptibility gene that, paradoxically, is a member of the NBS-LRR resistance gene family. We found LOV1 mediates responses associated with defense, but mutations in known defense response pathways do not prevent susceptibility to C. victoriae. These findings demonstrate that NBS-LRR genes can condition disease susceptibility and resistance and may have implications for R gene deployment.
Keywords: Cochliobolus victoriae, disease susceptibility, nucleotide binding site-leucine-rich repeat, victorin
In the 1940s, a disease epidemic occurred in oats because of wide-spread planting of “Victoria-type” oats, which contain the Pc-2 gene for resistance to the rust fungus, Puccinia coronata. Oats containing Pc-2 proved to be universally susceptible to a new disease, Victoria blight, caused by the fungus Cochliobolus victoriae (1, 2). Pathogenicity of C. victoriae depends on the production of a toxin called victorin, and in oats, both toxin sensitivity and Victoria blight disease susceptibility are conferred by the dominant Vb gene. Despite extensive efforts, rust resistance (Pc-2) and Victoria blight susceptibility (Vb) have not been genetically separated and are surmised to share identity (3, 4), thus suggesting an unexpected relationship between plant disease resistance and susceptibility.
In recent years, knowledge of genes regulating plant disease resistance has dramatically increased (5), but characterization of genes conferring disease susceptibility is limited (6–8). The information gap between the nature of resistance and that of susceptibility is likely due to differences in their genetic tractability. Gene-for-gene type resistance (9) is typically triggered by activation of a genetically dominant resistance gene product by a dominant, pathogen-derived, avirulence (Avr) gene product. Avr proteins often act as virulence determinants in the absence of their R protein partners (5), indicating that their primary role is in virulence and that recognition by R genes evolved out of this role. The largest class of R genes encodes nucleotide-binding site-leucine-rich repeat (NBS-LRR) proteins. The only known function of these proteins in plants is in conditioning disease resistance (10). In animals, structurally related proteins mediate the innate immune response (11, 12). R gene-mediated signaling pathways, identified through mutant screens, generally require salicylic acid (SA), jasmonic acid, and/or ethylene (13) and often include activation of hypersensitive cell death (HR).
For the majority of plant diseases, the genetics of susceptibility are less tangible. Host susceptibility is typically defined in the context of a gain or loss of resistance (5, 14, 15) and pathogens often possess multiple virulence factors (called effectors), each contributing incrementally to the disease phenotype. A notable exception is Os8N3, a genetically dominant rice gene that is up-regulated by a bacterial type-III effector protein, and that confers gene-for-gene-specified disease susceptibility (7). Likewise, for Victoria blight of oats and a handful of other diseases caused by necrotrophic pathogens (pathogens that incite cell death during pathogenesis), susceptibility is conditioned in a gene-for-gene manner by a single dominant locus in the host and a single pathogen-derived host-selective toxin (HST) (4).
C. victoriae produces the HST, victorin, and the oat gene Vb conditions both victorin sensitivity and disease susceptibility. Interestingly, although victorin is causal to disease susceptibility, it rapidly induces resistance-like physiology in oats, including callose deposition, a respiratory burst, lipid peroxidation, ethylene evolution, extracellular alkalinization, phytoalexin synthesis, K+ efflux, and apoptotic-like cell death (4). This reinforces the idea that Victoria blight susceptibility and rust resistance are regulated by the same gene and that the physiology of disease susceptibility can resemble resistance. Victoria-type oats are allohexaploid and not readily amenable to molecular genetic studies. However, identification of victorin sensitivity and Victoria blight susceptibility in Arabidopsis thaliana (16) has enabled our investigation of this disease susceptibility pathway. We report similarities between this susceptibility response and disease resistance, including identification of the susceptibility locus, LOV1, as a coiled-coil NBS-LRR, R gene family member. These findings provide a unique platform for discussion of disease susceptibility and may have implications for deployment of R genes.
Results
LOV1 Encodes a CC-NBS-LRR Protein with Extensive Similarity to the RPP8 Resistance Gene Family.
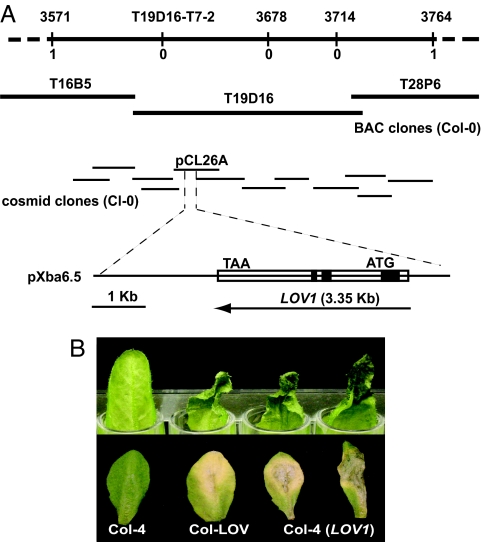
A locus conferring disease susceptibility to C. victoriae in Arabidopsis, called LOV1, was mapped to the interval between Nga63 and NCCI on Chromosome 1 (16). We fine-mapped LOV1, using an additional 200 victorin-insensitive F2 progeny of a cross between a victorin-sensitive line, LOV1 (ecotype Cl-0), and victorin-insensitive Col-4. PCR markers were created by identifying small insertions/deletions that are polymorphic between Ler and Col-0 (17), designing primers flanking each insertion/deletion, and testing for polymorphisms between LOV1 and Col-4. Polymorphic markers were subsequently used to map LOV1 to a 193-kb interval between markers 3571 and 3764 (Fig. 1A).
Fig. 1.
Isolation of LOV1, a gene that confers sensitivity to victorin and susceptibility to C. victoriae in A. thaliana. (A) Map-based cloning of the LOV1 locus from A. thaliana ecotype Cl-0. SSLP markers created for mapping LOV1, designated by their kilobase location on chromosome 1, and CAPS marker T19D16-T7-2 are shown above the line. The number of recombinants found between each marker and LOV1 is shown below the line. BAC clones spanning the mapped interval were obtained from The Arabidopsis Information Resource, and cosmid clones covering the interval were isolated from a Cl-0 genomic library. Clones containing LOV1 are designated pCL26A and pXba6.5, respectively. The LOV1 ORF is shown as an open rectangle. Introns are indicated in black. (B) Arabidopsis leaves 36 h after treatment with 10 μg/ml victorin (Upper) and 5 days after inoculation with 10 μl of 1 × 105/ml C. victoriae spores (Lower). Genotypes include Col-4 (victorin insensitive), Col-LOV (Col-4 near-isogenic for LOV1), and Col-4 (LOV1) (Col-4 transgenic for LOV1).
A genomic library was prepared from DNA of LOV1 (Cl-0) in the binary vector pCLD04541 (18), and clones comprising a contig of the 193-kb interval were isolated and introduced into victorin-insensitive Arabidopsis (Col-4) (Fig. 1B). One cosmid clone, pCL26A, and the subclone of this cosmid, pXba6.5, conferred victorin sensitivity and C. victoriae susceptibility to Col-4 (Fig. 1 A and B). DNA sequencing of clone pXba6.5 revealed a single ORF corresponding to a pseudogene (At1g10920) in the annotated genome of Arabidopsis ecotype Col-0. Six polymorphisms occur between At1g10920 (Col-0) and LOV1 (LOV1, Cl-0), including a base pair change eliminating a stop codon and a frameshift insertion in LOV1 (GenBank accession no. EF472599). Together, these changes result in LOV1 encoding an ORF for a complete CC-NBS-LRR protein with extensive similarity (86%) and 70% identity to members of the RPP8 resistance gene family [see supporting information (SI) SI Fig. 5]. Previously identified RPP8 family members with known function, RPP8, HRT, and RCY1, confer resistance to Hyaloperonospora (Peronospora) parasitica Emco5 (19), turnip crinkle virus (20), and cucumber mosaic virus (CMV-Y) (21), respectively. In contrast, LOV1 confers susceptibility to C. victoriae (Fig. 1B).
LOV1 Conditions Victorin-Dependent Induction of Defense-Associated Proteins.
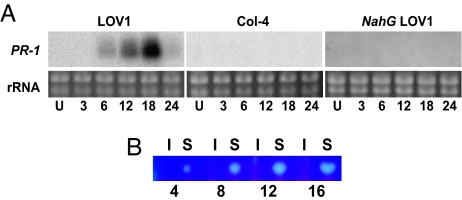
Because victorin induces resistance-like physiology in oats in a genotype-specific manner, and LOV1 not only confers genotype-specific victorin sensitivity in Arabidopsis but also encodes an R-like protein, we evaluated Arabidopsis for LOV1-specific expression of defense responses after treatment with victorin. Expression profiles of the resistance-associated gene PR-1 and production of the phytoalexin, camalexin, are presented in Fig. 2. Victorin rapidly induced PR-1 expression and camalexin production in a genotype-specific manner in Arabidopsis line LOV1 (Cl-0) but not in Col-4. PR-2, PR-5, and PDF1.2, resistance-associated genes known to be induced by other necrotrophic fungi (22), were not induced in either plant genotype (data not shown). PR-1 induction is SA-dependent because PR-1 expression was not induced in NahG plants, in which SA is degraded (23).
Fig. 2.
Victorin elicitation of LOV1-mediated defense response in A. thaliana. Expression of pathogenicity-related protein gene PR-1 (A) and thin-layer chromatography showing camalexin accumulation (B) in Arabidopsis leaves hours (numbers below panels) after infiltration with 30 μg/ml victorin. U, untreated; S, victorin-sensitive LOV1; I, victorin-insensitive Col-4.
Multiple Defense Signaling Pathways Are Dispensable for C. victoriae Susceptibility.
We assessed signaling requirements for LOV1-conditioned disease susceptibility by examining LOV1 genotypes having mutations in SA- (NahG, EDS1, NDR1, NPR1), jasmonic acid- (COI1, JAR1), and ethylene- (EIN2) mediated pathways, in the pathway for biosynthesis of the phytoalexin, camalexin (PAD3), and in the defense-related gene DND1, which is required for cell death during the HR (24). Susceptibility of victorin-sensitive Arabidopsis to C. victoriae was independent of all of these mutations (SI Table 1). Because disease appeared unaffected by these mutations, we pursued a more sensitive test by evaluating contributions of these signaling pathways to victorin sensitivity.
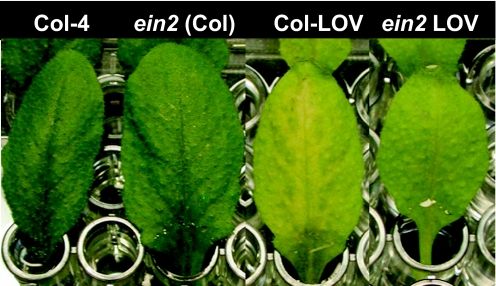
LOV1 conditions incomplete dominance to victorin sensitivity. Plants heterozygous for LOV1 are slightly less sensitive to victorin than are homozygous plants. Subtle effects of signaling pathways on victorin sensitivity, therefore, might be seen by assessing dilute concentrations of toxin on both homozygous and heterozygous LOV1 Arabidopsis genotypes. When plants heterozygous for LOV1 in various mutant backgrounds (SI Table 1) were evaluated with victorin, a slight attenuation of victorin sensitivity was evident in ein2 mutants at a concentration of 5 μg/ml victorin (Fig. 3), but no consistent alteration of victorin sensitivity occurred in other mutant backgrounds (SI Table 1). This finding indicates ethylene may play a subtle role in victorin-induced disease susceptibility. Ethylene also plays a subtle role in RPP8 family member, RCY1-mediated resistance to cucumber mosaic virus. Cucumber mosaic virus resistance was reduced by 8% in ein2 Arabidopsis (20).
Fig. 3.
Effects of ein2-1 mutation on LOV1-mediated victorin sensitivity. A. thaliana leaves 36 h after treatment with 5 μg/ml victorin.
Complete resistance mediated by several R genes requires RAR1, SGT1b, and/or HSP90 to maintain NBS-LRR steady-state levels (25–30). RAR1 and HSP90 are thought to act as cochaperones that positively regulate NBS-LRR levels (26, 28–30). SGT1b may play a role in cellular protein degradation and can function antagonistically to RAR1 in RPP8-conditioned disease resistance (25, 28). HSP90 and SGT1b have been shown to physically interact with several R proteins (26, 31). We evaluated RAR1, SGT1b, and HSP90 contributions to LOV1-mediated victorin sensitivity. Effects of mutations rar1-1 and sgt1b on victorin sensitivity were assessed in a LOV1 background (SI Table 1). Geldanamycin, an HSP90-specific inhibitor, was used to assess the requirement for HSP90 in victorin sensitivity. Slight attenuation of victorin sensitivity was observed in rar1 (Ws) plants at 36 h after victorin treatment but was not consistently reproducible. By 48–72 h this phenotype was not discernible. Neither sgt1b nor inhibition of HSP90 with geldanamycin reduced Arabidopsis sensitivity to victorin (SI Table 1 and data not shown). RPP8-mediated disease resistance also does not require functional SGT1b or HSP90, and it is only slightly affected by mutation in RAR1 (28). In summary, the LOV1-conditioned disease susceptibility response appears similar to the RPP8-conditioned disease resistance response in that known defense response signaling pathways, individually, are not required for either response, and proteins that affect steady-state levels of some R proteins (RAR1, SGT1b, and HSP90) are not required to keep LOV1 at a presumed threshold level (28).
C. victoriae Infection Process of Arabidopsis.
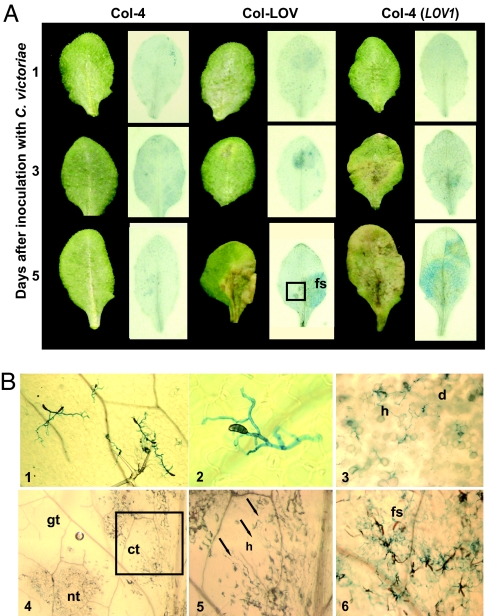
During infection of oat and Arabidopsis, C. victoriae develops appressoria and penetrates tissue of compatible hosts, but in incompatible plant genotypes, fungal penetration stops following appressorium development (16, 32). Analysis of C. victoriae infection of oat suggests that the fungus may penetrate host tissue before host cells are dying, implying the fungus does not merely gain nutrients from dead or dying cells (32). We compared symptom development and fungal growth over time in victorin-sensitive and victorin-insensitive Arabidopsis to correlate host symptom development with C. victoriae growth in host tissue (Fig. 4). Macroscopic observations of disease progress over a 5-day period in victorin-insensitive Col-4, near-isogenic line Col-LOV, and transgenic Col-4 (LOV1) are shown in Fig. 4A. Microscopic observations of select trypan-blue-stained leaves in Fig. 4A are shown in Fig. 4B. At 24 h after inoculation, fungal spores had germinated and formed appressoria on leaves of C. victoriae-susceptible and resistant Arabidopsis, but macroscopic symptoms were not evident (Fig. 4A). Col-4 plants remained symptomless for the duration of the experiment, and fungal hyphae on Col-4 leaves were limited to the leaf surface (Fig. 4 A and B). Interestingly, although C. victoriae did not penetrate Col-4 tissue, hyphae continued to grow over the leaf surface, repeatedly forming appressoria in an apparent attempt to penetrate tissue (Fig. 4B). By 3 days after inoculation, chlorotic, dying and dead cells were visible on Col-LOV and transgenic Col-4 (LOV1) plants [dead and dying cells stain blue-black with trypan blue (19), and fungal hyphae appear bright blue]. Symptom development clearly preceded hyphal invasion of tissue. Comparisons of live and stained tissue indicate that hyphae were absent from chlorotic tissue and were evident around some, but not all, stained, dead, or dying cells. By 5 days after inoculation, hyphae were abundant and sporulating from dead tissue, and hyphal invasion of new tissue appeared more aggressive. Hyphae extended into some chlorotic areas close to tissue that appeared green. However, healthy green tissue did not appear to contain hyphae (Fig. 4 A and B). In summary, C. victoriae acts as a necrotroph early in infection. Symptom development and cell death appear to precede fungal proliferation. Further, based on the interaction on the insensitive genotypes, it is likely that cell death is necessary for penetration. Later, when infection is well established in necrotic tissue, expanding hyphae invade tissue more aggressively and enter chlorotic regions in which cells do not appear to be dead.
Fig. 4.
Genotype-specific symptom development and C. victoriae ingress in A. thaliana. (A) Leaves of C. victoriae-resistant (Col-4) and C. victoriae-susceptible [Col-LOV and Col-4 (LOV1) transgenic] plants days after inoculation with 10 μl of 1 × 105/ml C. victoriae spores are shown before (Left) and after (Right) staining with trypan blue. Dead and dying cells appear blue-black and fungal hyphae appear bright blue. (B) Select leaves from A, depicted with Nomarski (panels 1–3 and 6) or stereoscope (panels 4 and 5) microscopy; Col-4, 5 days after inoculation, panel 1 (magnification: ×40), and 2 (magnification: ×400); Col-LOV, at 3 days, panel 3 (magnification: ×100); and 5 days after inoculation, panel 4 (magnification: ×25), panel 5 (magnification: ×63), and panel 6 (magnification: ×80). The boxed region of the Col-LOV leaf at 5 days after inoculation in A, contains green, chlorotic, and necrotic tissues and is shown at 25× magnification (4). A higher magnification of the boxed region in 4 is shown in 5. Arrows point to hyphae in chlorotic tissue. Hyphae are abundant and sporulation is occurring from the same leaf, to the right of the midvein (6). fs, fungal sporulation; h, hyphae; d, dead or dying cells; gt, green tissue; ct, chlorotic tissue; nt, necrotic tissue.
Discussion
We mapped and cloned a dominant disease susceptibility gene called LOV1 from Arabidopsis (ecotype Cl-0) (Fig. 1). We found that LOV1 encodes a CC-NBS-LRR protein, a class of proteins that, previously, was only known to function in conditioning plant disease resistance. LOV1 is a member of the RPP8 disease resistance gene family (SI Fig. 5). However, whereas other functional family members, RPP8, HRT, and RCY1, confer resistance to H. (Peronospora) parasitica Emco5 (19), turnip crinkle virus (20), and cucumber mosaic virus (21), respectively, LOV1 confers susceptibility to C. victoriae. We demonstrate that like other NBS-LRR proteins, LOV1 mediates responses associated with disease resistance. LOV1 conditions rapid, SA-dependent induction of the pathogenicity-related protein gene, PR-1, and production of the phytoalexin, camalexin (Fig. 2). Additionally, victorin-inducible electrolyte leakage and DNA degradation associated with HR-like cell death in sensitive oat tissue (33) have also been shown to occur in a LOV1-dependent manner in Arabidopsis (T.A.S. and T.J.W., unpublished data). Collectively, these findings strongly suggest that LOV1-conditioned disease susceptibility shares features of disease resistance responses. This also raises a conundrum. How does rapidly eliciting “resistance” result in disease susceptibility? How does victorin perception, directly or indirectly, by a CC-NBS-LRR protein overcome resistance when, in fact, it induces resistance-like physiology?
During incompatible interactions of C. victoriae with oat or Arabidopsis (absence of Vb or LOV1, respectively), appressoria develop, but host tissue is not penetrated (refs. 32 and 16 and Fig. 4). Because C. victoriae produces a “toxin” and has been considered to be a necrotroph, its penetration of tissue may be arrested by basal and/or victorin-induced resistance until victorin-induced cell death occurs, then allowing the fungus to invade the dead and dying tissue. We provide some evidence for this scenario in that early in infection, fungal growth lags behind chlorosis and necrosis. Hyphae align with some but not all dying cells (Fig. 4). However, later in infection, hyphal invasion was evident in chlorotic tissue in which cells did not appear to be dead. In investigations of C. victoriae infection of oat, susceptible mesophyll cells were reported to contain hyphae before host cells were visibly affected (32). Separation of pathogenesis and host cell death has also been reported for HST-producing Alternaria alternata (4) and is implicated for the closely related, HST-producing pathogen, C. carbonum. In this latter case, HC-toxin is not a toxin per se, but rather functions as a cytostatic agent (3). Taken together, these findings indicate that the role of host cell death in susceptibility to HST-producing pathogens is not clearly resolved. The precise temporal relationship between fungal invasion, defense response and cell death for these pathogens, including C. victoriae, requires further investigation.
C. victoriae may not be avoiding resistance (by penetrating after cell death) but, alternatively, might be immune to negative effects of, and actually benefit from, the host resistance response. Several lines of evidence suggest that some fungi are tolerant to aspects of plant disease resistance physiology and actually use it for nutritional gain. For example, susceptibility to B. cinerea is decreased in a dnd1 background but increased in Arabidopsis undergoing a P. syringae-induced HR response (34). Furthermore, reactive oxygen species (ROS) are known to play a role in resistance to biotrophic and hemibiotrophic pathogens, but in susceptibility to the necrotrophic pathogens, B. cinerea, S. sclerotiorum, and Cercospora species (4). Although oat Pc-2 evolved to halt infection of P. coronata, C. victoriae may be exploiting this resistance pathway to gain nutrition during pathogenesis.
As one approach to evaluate the role of defense in the C. victoriae–Arabidopsis interaction, we conducted analysis of defense-associated signaling mutations in a LOV1 background. These analyses revealed that LOV1-mediated susceptibility, similar to the RPP8-conditioned resistance response (35), does not require SA, jasmonic acid, or ethylene-mediated signaling pathways. Also, these signaling pathways did not contribute to resistance to C. victoriae, because susceptibility did not appear to be enhanced in mutant backgrounds (SI Table 1). Nonetheless, SA and ethylene do participate in the LOV1-mediated response. LOV1-specified, victorin-induced PR-1 expression is SA-dependent (Fig. 2), and victorin-induced expression of ATTRX5, a gene required for LOV1-conditioned C. victoriae susceptibility is also SA-dependent (36). Furthermore, ethylene involvement has been demonstrated in Vb-conditioned susceptibility of oats to C. victoriae (33), and a slight attenuation of LOV1-mediated victorin sensitivity occurs in ein2 Arabidopsis (Fig. 3). SA, ethylene, and camalexin synthesis pathways are also activated during RPP8-mediated resistance (37), although they are not required (35). A requirement for (or dispensability of) these commonly activated signaling pathways is clearly pathogen dependent (13). Signal requirements for several R gene-mediated pathways remain unknown. Novel pathways have been proposed for RPP7- and RPP8-mediated resistance (19), and known signaling pathways account for only a portion of the resistance mediated by RCY1 (21). Given the similarity of LOV1 to RPP8 and RCY1 and the finding that an extensive screen for suppressors of LOV1 did not reveal genes previously known to be required for defense (36), LOV1 could also function in an uncharacterized pathway, one that is sufficient for resistance to some pathogens (such as obligate biotrophs, as is the case for RPP8 and RCY1) but readily exploited for susceptibility by other pathogens such as C. victoriae. Such opposing functions in host response have been reported for MLO proteins, which are required for compatible interactions of powdery mildew pathogens with barley and Arabidopsis but contribute to resistance to necrotrophic and hemibiotrophic pathogens of Arabidopsis (8).
The dispensability of defense-associated signaling in victorin sensitivity and disease susceptibility could also indicate that LOV1 does not function through a defense-related pathway. As an alternative, LOV1 could be mediating a condition of disease susceptibility analogous to Os8N3, a rice susceptibility gene that is activated by a bacterial type III effector protein (7). With few exceptions (6, 8), disease susceptibility has been considered a suppression or absence of resistance (5), but for Os8N3, disease susceptibility is dominant, disease resistance is recessive, and susceptibility is conferred by positive action of a virulence effector. However, if the function of LOV1 and Vb are analogous to Os8N3, then in the C. victoriae interaction with oat, the victorin/Vb-mediated response would necessarily be distinct from the P. coronata /Pc-2 resistance response, which contradicts genetic evidence (4). Furthermore, like other NBS-LRR R proteins, activation of LOV1 is necessary for its function. Missense mutations resulting in amino acid changes in the P-loop domain of LOV1 at residue 192 or 199 abolish its function (T.A.S. and T.J.W., unpublished data). Given this and the findings that LOV1 and Vb/Pc-2 both mediate defense-associated physiology and that LOV1 is highly related to RPP8 resistance family members, the possibility that LOV1 signals in a susceptibility pathway that is unrelated to resistance seems unlikely. Furthermore, the necrotrophic nature of C. victoriae pathogenesis early in infection (Fig. 4) suggests that an HR could condition disease susceptibility.
In animal systems, TLR (Toll/interleukin 1 receptor (TIR) domain, LRR motif) proteins are one class of proteins that mediate innate immunity and are structurally related to NBS-LRR proteins (12). Although TLR proteins typically modulate innate immunity, in some instances fungal, bacterial and viral pathogens, by various mechanisms, have exploited their interaction with TLR proteins to increase pathogen virulence/host susceptibility (12). Similarly, LOV1, an NBS-LRR R protein family member, is targeted by a pathogen effector, victorin, and this interaction results in disease susceptibility, regardless of the specific mechanism by which this occurs. Furthermore, this phenomenon is probably not unique. An NBS-LRR gene has also been implicated in susceptibility to Milo disease (38). Pathogen effectors, first identified genetically as avirulence determinants, have since been shown to target defense machinery and play a role in virulence (5). Likewise, R genes, first identified as a result of their genetic dominance, could now prove to be targets of pathogen effectors and have roles in susceptibility. Identifying LOV1 as a gene encoding a NBS-LRR protein strongly suggests that a disease resistance gene and a disease susceptibility gene can share identity and supports the prospect of Pc-2/Vb identity in oat. A role for R genes in disease susceptibility could have implications for engineering of disease resistance in plants and R gene deployment.
Materials and Methods
Plant Material, Growth Conditions, and Victorin Sensitivity Assays.
Seeds of A. thaliana Col-4, Ws, and mutant lines, jar1-1 (39), npr1-1 (40), ein2-1 (41), pad3 (42), and NahG (23), all in Col-0, were obtained from the Ohio State University Arabidopsis Biological Resource Center (Columbus, OH). LOV1 (16) and Col-LOV (36) were generated in our laboratory. Other seeds were obtained from the following individuals: Sainsbury Laboratory [eds1-1 Ws (43)], B. Staskawicz (University of California, Berkeley, CA) [ndr1-1 (44)], B. Kunkel (Washington University, St. Louis, MO) [coi1-35 (45)], A. Bent (University of Wisconsin, Madison, WI) [dnd1 (24)], and J. Chang [rar1-1 (25)]. Seeds were placed in 0.1% agar for 5 days at 4°C, pipetted onto soil, and grown at 22°C under a long-day photoperiod (16 h light, 8 h dark). Victorin sensitivity assays were conducted as described in ref. 16, by using 1–20 μg/ml victorin C, which was purified as described in ref. 46.
Fine-Mapping and Cloning of LOV1.
Initial mapping of LOV1 to the Nga63 and NCCI interval of Chromosome 1 was described in ref. 16. For fine-mapping, an additional 200 victorin-insensitive, F2 progeny of a LOV1 × Col-4 cross were subjected to analysis with PCR. DNA for PCR was purified from leaf tissue according to Edwards et al. (47).
A genomic library was prepared in the binary vector pCLD04541 (18). DNA was isolated from LOV1 with a CTAB method (48) and further purified by ultracentrifugation in cesium chloride. DNA was partially digested with Sau3A and ligated into the BamHI site of pCLD04541. Ligations were packaged with Gigapack III XL extracts and transformed into XL1-Blue MR cells (Stratagene, La Jolla, CA). The resultant genomic library was screened (48) with 32P-labeled DNA of BAC clone T19D16 and selected regions of BAC clones T16B5 and T28P6, which were obtained from ABRC. Positive clones from this screen were end-sequenced, arranged into a contig, and transformed into Agrobacterium tumefaciens strain GV3101 by triparental mating (49). Electroporation does not work well with pCLD04541.
Col-4 plants were transformed by using the floral dip method (50). Seed from dipped plants were plated on nutrient agar (16) supplemented with 100 μg/ml kanamycin and 100 μg/ml cefatoxamine. Transgenic seedlings were transplanted to soil after 1 week. Transgenic plants were assayed for victorin sensitivity as described. A 6.5-kb XbaI fragment containing LOV1 was subcloned from pCL26A into pCB302 (51) and electroporated into A. tumefaciens strain GV3101. Transgenic plants were made as described above, except putative transgenic seed were planted directly in soil wet with 0.02% glufosinate-ammonium.
Isolation of LOV1 Signal Transduction Mutants.
F2 and F3 progeny from crosses of LOV1 and signal transduction mutant lines (NahG, npr1-1, ndr1-1, ein2, coi1-35, jar1, pad3, dnd1, respectively) were screened for both victorin sensitivity (as described above) and for PCR markers linked to respective loci of interest. PCR markers for following the segregation of above loci were marker 3571 for LOV1 (see fine-mapping above), gene-specific primers for NDR1-1 (forward, AATCTACTACGACGATGTCCAC; reverse, GTAACCGATGGCAACTTTCAC) and NahG (forward, CAGAAGGTATCGCCCAATTC; reverse, ACCTTCCAGCACATGGCTAC) or markers linked to each locus selected from sequence information available at The Arabidopsis Information Resource (www.arabidopsis.org). The presence of several mutations could also be confirmed by visible phenotypes, such as lack of PR-1 expression in npr1-1 and NahG, dwarfism of dnd1, leaky male-sterility of coi1-35, and triple-mutant response of ein2-1 grown on MS agar with 0.5 mM ACC. eds1-1 and rar1 are in ecotype Ws, which is toxin sensitive and could be tested directly for victorin sensitivity.
Approximately 100 F2 plants for each cross (above) were screened for victorin sensitivity and by PCR. Individual plants homozygous for or heterozygous for LOV1, and also homozygous for each mutant allele, were allowed to self-fertilize and also tested in the F3 generation for victorin sensitivity and presence of the appropriate PCR markers.
C. victoriae Susceptibility Assays.
C. victoriae spores were prepared as described in ref. 16. Initially, for Col-0, LOV1, ein2-1 (Col-0), ein2LOV1, NahG (Col-0), NahGLOV1, npr1-1 (Col-0), and npr1LOV1, C. victoriae (106 spores per ml) C. victoriae spores were sprayed onto 32 3- to 4-week-old plants with an aspirator until runoff. Plants were then incubated at 100% humidity in a growth chamber at 22°C under a long-day photoperiod (16 h light, 8 h dark) until symptoms appeared. After finding that no plant lacking the LOV1 gene had any disease lesions, and all plants having the LOV1 gene (LOV1, ein2LOV1, NahGLOV1, and npr1LOV1) appeared equally susceptible, we used a simpler detached leaf assay.
Six detached leaves of approximately the same age (third through sixth true leaves) from each plant genotype were placed in a sealed Petri dish lined with moist filter paper. For time-course experiments shown in Fig. 4, leaves were not detached but remained on plants which were covered to maintain humidity. Ten microliters of C. victoriae spores, washed as described and resuspended to a concentration of 105 spores per ml, were pipetted onto the center of each leaf. Leaves were observed daily for up to 10 days for symptom development and signs of fungal growth. Again, leaves from plant genotypes lacking LOV1 showed no symptom development. Most leaves of the LOV1 genotype showed chlorosis, and then necrosis and visible hyphae, which eventually sporulated. The infection varied somewhat from leaf to leaf depending on leaf age and shape, which affected fungal distribution and wetness and consequently, the area of initial infection (e.g., leaves that curled up from the surface were less wet in spots and moisture pooled in other spots). Because of this variability, and because the only obvious differences in disease development occurred between LOV1- and lov1-containing genotypes, we simply scored plants as susceptible or resistant. An exception to this was that Col-4 transgenic for LOV1 typically appeared more sensitive to victorin and slightly more susceptible to C. victoriae, likely due to gene copy number. Also, the Ws and Ws-0 ecotypes were somewhat less sensitive to victorin and less susceptible to C. victoriae (SI Table 1).
The rar1-1 mutation was reported to be in Ws-0 (25). However, because the rar1-1 plants looked like the Ws ecotype, we checked Ws, Ws-0, and rar-1 for the sequence of LOV1 and evaluated polymorphisms for seven SSLP markers dispersed throughout the genome. The rar1-1 plants had the same LOV1 sequence and SSLP profile as Ws, whereas Ws-0 plants had a slightly different LOV1 sequence and were polymorphic for 4 SSLP markers in comparison to rar1-1 plants. For this reason we used both Ws and Ws-0 as controls for our rar1-1 assays and obtained equivalent results with each. Results for Ws are presented.
Visualization of fungi was according to Lorang et al. (16) as modified from Keogh et al. (52). Specimens were visualized with stereoscope and Nomarski optics microscopy.
Geldanamycin Treatment.
Sensitivity to victorin after treatment with geldanamycin was assayed with the detached leaf assay described above, except that test leaves were subjected to a 2-h pretreatment with 10 μM geldanamycin (EMD Biosciences, Inc., La Jolla, CA) in a final concentration of 0.2% DMSO. Control leaves were pretreated with 0.2% DMSO only. To detect subtle effects of geldanamycin on victorin sensitivity, multiple victorin concentrations of 0.5, 5, and 10 μg/ml were evaluated. In LOV1 plants with or without geldanamycin treatment, victorin-elicited leaf symptoms were obvious within 24 h but were less pronounced in the 0.5 μg/ml toxin treatment. Control leaves treated with geldanamycin only showed phytotoxic affects of treatment (leaf yellowing) at 60–72 h after treatment. This was not a problem, because at this time victorin treated plants had already displayed clear symptoms.
Northern Analyses and Camalexin Extraction.
Northern analyses were according to Sweat and Wolpert (36). Plasmids containing cDNA clones of PR-1, PR-2, and PR-5 were obtained from Syngenta Biotechnology (Research Triangle Park, NC). The cDNA inserts were excised by using EcoRI/XhoI for PR-1 and PR-2 and BamHI/KpnI for PR-5 and gel-purified for use as probes. A gene-specific probe for PDF1.2 was PCR-amplified from genomic DNA by using the primers 5′-GCAATGGTGGAAGCACAGAA-3′ and 5′-CTCATAGAGTGACAGAGACT-3′. Camalexin was extracted and analyzed similar to the way described in ref. 42. After incubation for the indicated times, leaf disks (7 mm) were prepared from detached leaves, using a no. 3 cork borer. Three-leaf disks were combined for each repetition at each time point.
Supplementary Material
Acknowledgments
We thank Anita Brent for technical assistance, Dr. Vlado Macko, and the Ohio State University Arabidopsis Biological Resource Center for providing seed stocks and cDNA clones. This work was supported in part the National Research Initiative of the U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service Grant 2005-35319-15361.
Abbreviations
- HR
hypersensitive cell death
- HST
host-selective toxin
- NBS-LRR
nucleotide- binding site-leucine-rich repeat
- SA
salicylic acid.
Footnotes
The authors declare no conflict of interest.
The sequence reported in this paper has been deposited in the GenBank database (accession no. EF472599).
This article contains supporting information online at www.pnas.org/cgi/content/full/0702572104/DC1.
References
- 1.Meehan F, Murphy HC. Science. 1946;104:413–414. doi: 10.1126/science.104.2705.413. [DOI] [PubMed] [Google Scholar]
- 2.Litzenberger SC. Phytopathology. 1949;39:300–318. [Google Scholar]
- 3.Walton JD. Plant Cell. 1996;8:1723–1733. doi: 10.1105/tpc.8.10.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolpert TJ, Dunkle LD, Ciuffetti LM. Annu Rev Phytopathol. 2002;40:251–285. doi: 10.1146/annurev.phyto.40.011402.114210. [DOI] [PubMed] [Google Scholar]
- 5.Chisholm ST, Coaker G, Day B, Staskawicz BJ. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 6.Van Damme M, Andel A, Huibers RP, Panstruga R, Weisbeek PJ, Van den Ackerveken G. Mol Plant-Microbe Interact. 2005;18:583–592. doi: 10.1094/MPMI-18-0583. [DOI] [PubMed] [Google Scholar]
- 7.Yang B, Sugio A, White FF. Proc Natl Acad Sci USA. 2006;103:10503–10508. doi: 10.1073/pnas.0604088103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Consonni C, Humphry ME, Hartmann HA, Livaja M, Durner J, Westphal L, Vogel J, Lipka V, Kemmerling B, Schulze-Lefert P, et al. Nat Genet. 2006;38:716–720. doi: 10.1038/ng1806. [DOI] [PubMed] [Google Scholar]
- 9.Flor HH. J Agric Res. 1946;73:335–357. [Google Scholar]
- 10.Nimchuk Z, Eulgem T, Holt BF, III, Dangl JL. Annu Rev Genet. 2003;37:579–609. doi: 10.1146/annurev.genet.37.110801.142628. [DOI] [PubMed] [Google Scholar]
- 11.Belkhadir Y, Subramaniam R, Dangl JL. Curr Opin Plant Biol. 2004;7:391–399. doi: 10.1016/j.pbi.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 12.Akira S, Uematsu S, Takeuchi O. Cell. 2006;123:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 13.Glazebrook J. Annu Rev Phytopathol. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- 14.Parker JE, Holub EB, Frost LN, Falk A, Gunn ND, Daniels MJ. Plant Cell. 1996;8:2033–2046. doi: 10.1105/tpc.8.11.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vogel J, Sommerville S. Proc Natl Acad Sci USA. 2000;97:1897–1902. doi: 10.1073/pnas.030531997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lorang JM, Carkaci-Salli N, Wolpert TJ. Mol Plant Microbe Interact. 2004;17:577–582. doi: 10.1094/MPMI.2004.17.6.577. [DOI] [PubMed] [Google Scholar]
- 17.Jander G, Norris SR, Rounsley SD, Bush DF, Levin IM, Last RL. Plant Physiol. 2002;129:440–450. doi: 10.1104/pp.003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bent AF, Kunkel BN, Dahlbeck D, Brown KL, Schmidt R, Giraudat J, Leung J, Staskawicz BJ. Science. 1994;265:1856–1860. doi: 10.1126/science.8091210. [DOI] [PubMed] [Google Scholar]
- 19.McDowell JM, Dhandaydham M, Long TA, Aarts MG, Goff S, Holub EB, Dangl JL. Plant Cell. 1998;10:1861–1874. doi: 10.1105/tpc.10.11.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dempsey DA, Pathirana SM, Wobbe KK, Klessig DF. Plant J. 1997;11:301–311. doi: 10.1046/j.1365-313x.1997.11020301.x. [DOI] [PubMed] [Google Scholar]
- 21.Takahashi H, Miller J, Nozaki Y, Takeda M, Shah J, Hase S, Ikegami M, Ehara Y, Dinesh-Kumar SP, Sukamto Plant J. 2002;32:655–667. doi: 10.1046/j.1365-313x.2002.01453.x. [DOI] [PubMed] [Google Scholar]
- 22.Penninckx IA, Eggermont K, Terras FR, Thomma BP, De Samblanx GW, Buchala A, Metraux JP, Manners JM, Broekaert WF. Plant Cell. 1996;8:2309–2323. doi: 10.1105/tpc.8.12.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E, Ryals J. Science. 1994;266:1247–1250. doi: 10.1126/science.266.5188.1247. [DOI] [PubMed] [Google Scholar]
- 24.Yu I-C, Parker J, Bent AF. Proc Natl Acad Sci USA. 1998;95:7819–7824. doi: 10.1073/pnas.95.13.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Azevedo C, Betsuyaku S, Peart J, Takahashi A, Noel L, Sadanandom A, Casais C, Parker J, Shirasu K. EMBO J. 2006;25:2007–2016. doi: 10.1038/sj.emboj.7601084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bieri S, Mauch S, Shen QH, Peart J, Devoto A, Casais C, Ceron F, Schulze S, Steinbiss HH. Plant Cell. 2004;16:3480–3495. doi: 10.1105/tpc.104.026682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muskett PR, Kahn K, Austin MJ, Moisan LJ, Sandandom A, Shirasu K, Jones JDG, Parker JE. Plant Cell. 2002;14:979–992. doi: 10.1105/tpc.001040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holt BF, III, Belkhadir Y, Dangl JL. Science. 2005;309:929–932. doi: 10.1126/science.1109977. [DOI] [PubMed] [Google Scholar]
- 29.Lu R, Malcuit I, Moffett P, Ruiz MT, Peart J, Wu AJ, Rathjen JP, Bendahmane A, Day L, Baulcombe DC. EMBO J. 2003;22:5690–5699. doi: 10.1093/emboj/cdg546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hubert DA, Tornero P, Belkhadir Y, Krishna P, Takahashi A, Shirasu K, Dangl JL. EMBO J. 2003;22:5679–5689. doi: 10.1093/emboj/cdg547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lui Y, Burch-Smith T, Schiff M, Feng S, Dinesh-Kumar SP. J Biol Chem. 2004;279:2101–2108. doi: 10.1074/jbc.M310029200. [DOI] [PubMed] [Google Scholar]
- 32.Yoder OC, Scheffer RP. Phytopathology. 1969;59:1954–1959. [Google Scholar]
- 33.Navarre R, Wolpert TJ. Plant Cell. 1999;11:237–249. doi: 10.1105/tpc.11.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Govrin EM, Levine A. Curr Biol. 2000;10:751–757. doi: 10.1016/s0960-9822(00)00560-1. [DOI] [PubMed] [Google Scholar]
- 35.McDowell JM, Cuzick A, Can C, Beynon J, Dangl JL, Holub EB. Plant J. 2000;22:523–529. doi: 10.1046/j.1365-313x.2000.00771.x. [DOI] [PubMed] [Google Scholar]
- 36.Sweat TA, Wolpert TJ. Plant Cell. 2007;19:673–687. doi: 10.1105/tpc.106.047563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eulgem T, Weigman VJ, Chang HS, McDowell JM, Holub EB, Glazebrook J, Zhu T, Dangl JL. Plant Physiol. 2004;135:1129–1144. doi: 10.1104/pp.104.040444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagy ED, Lee TC, Ramakrishna W, Xu Z, Klein PE, SanMiguel P, Cheng CP, Li J, Devos KM, Schertz K, et al. Theor Appl Genet. 2007;114:961–970. doi: 10.1007/s00122-006-0481-1. [DOI] [PubMed] [Google Scholar]
- 39.Staswick PE, Yuen GY, Lehman CC. Plant J. 1998;15:747–754. doi: 10.1046/j.1365-313x.1998.00265.x. [DOI] [PubMed] [Google Scholar]
- 40.Cao H, Bowling SA, Gordon S, Dong X. Plant Cell. 1994;6:1583–1592. doi: 10.1105/tpc.6.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guzmán P, Ecker JR. Plant Cell. 1990;2:513–523. doi: 10.1105/tpc.2.6.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Glazebrook J, Ausubel FM. Proc Natl Acad Sci USA. 1994;91:8955–8959. doi: 10.1073/pnas.91.19.8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parker J. Trends Plants Sci. 2003;8:245–247. doi: 10.1016/S1360-1385(03)00105-5. [DOI] [PubMed] [Google Scholar]
- 44.Century KS, Holub EB, Staskawicz BJ. Proc Natl Acad Sci USA. 1995;92:6597–6601. doi: 10.1073/pnas.92.14.6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Staswick PE, Tiryaki I, Rowe M. Plant Cell. 2002;14:1405–1415. doi: 10.1105/tpc.000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolpert TJ, Macko V, Acklin W, Jaun B, Seibl J, Meili J, Arigoni D. Experientia. 1985;41:1524–1529. [Google Scholar]
- 47.Edwards K, Johnstone C, Thompson C. Nucleic Acids Res. 1991;19:1349. doi: 10.1093/nar/19.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1997. [Google Scholar]
- 49.Keen NT, Shen H, Cooksey DA. In: Molecular Plant Pathology: A Practical Approach. Gurr SJ, McPherson MJ, Bowles DJ, editors. Oxford: IRL; 1992. pp. 45–50. [Google Scholar]
- 50.Clough SJ, Bent AF. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 51.Xiang C, Han P, Lutziger I, Wang K, Oliver DJ. Plant Mol Biol. 1999;40:711–717. doi: 10.1023/a:1006201910593. [DOI] [PubMed] [Google Scholar]
- 52.Keogh RC, Deverall FJ, Mcleod S. Trans Br Mycol Soc. 1980;74:329–333. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.