Abstract
Chromosome segregation ensures that DNA is equally divided between daughter cells during each round of cell division. The centromere (CEN) is the specific locus on each chromosome that directs formation of the kinetochore, the multiprotein complex that interacts with the spindle microtubules to promote proper chromosomal alignment and segregation during mitosis. CENs are organized into a specialized chromatin structure due to the incorporation of an essential CEN-specific histone H3 variant (CenH3) in the centromeric nucleosomes of all eukaryotes. Consistent with its essential role at the CEN, the loss or up-regulation of CenH3 results in mitotic defects. Despite the requirement for CenH3 in CEN function, it is unclear how CenH3 nucleosomes structurally organize centromeric DNA to promote formation of the kinetochore. To address this issue, we developed a modified chromatin immunoprecipitation approach to analyze the number and position of CenH3 nucleosomes at the budding yeast CEN. Using this technique, we show that yeast CENs have a single CenH3 nucleosome positioned over the CEN-determining elements. Therefore, a single CenH3 nucleosome forms the minimal unit of centromeric chromatin necessary for kinetochore assembly and proper chromosome segregation.
Keywords: CenH3, chromatin, kinetochore
Chromosome segregation is directed by the multiprotein kinetochore (KT) complex that assembles on centromeres (CENs) and interacts with the spindle apparatus to control chromosome movement during cell division (for reviews, see refs. 1 and 2). Although KT function is conserved, phylogenetic analysis of centromeric DNA reveals no common sequence element that would identify a CEN across species. Like all other parts of the genome, the DNA of CENs is organized into chromatin, a higher order structure in which DNA is wrapped around histones to generate nucleosomes. Typical octameric nucleosomes contain two molecules of each of four histones, H2A, H2B, H3, and H4, whereas centromeric nucleosomes are structurally distinct, containing an evolutionarily conserved CEN-specific histone H3 variant (CenH3) in place of canonical H3 (for review, see ref. 3). The conservation of CenH3 across eukaryotes and its strict CEN localization in all species suggest that it is the epigenetic CEN identifier. Consistent with this finding, overexpression of the Drosophila CenH3 leads to its localization in euchromatin and the formation of ectopic KTs, resulting in subsequent defects in genomic stability (4).
Despite the requirement for CenH3 in CEN specification, it is unclear how CenH3 nucleosomes structurally organize centromeric DNA to promote KT formation. In humans, CenH3 associates with centromeric α satellite repeats (5), resulting in megabase expanses of centromeric nucleosomes. These centromeric nucleosomes drive the assembly of the KT structure that interacts with multiple microtubules. However, the number of CenH3 nucleosomes far exceeds the number of microtubules associated with a CEN, making the minimal number and arrangement of CenH3 nucleosomes necessary to assemble a single functional KT unclear. In flies and humans, blocks of CenH3 nucleosomes are interspersed with blocks of H3 nucleosomes (6), suggesting that higher order folding of the centromeric region may be necessary for its function. In contrast to the complex CEN arrangement of higher eukaryotes, Saccharomyces cerevisiae contains a short, defined CEN that associates with a single microtubule (7, 8), making budding yeast an ideal system to molecularly dissect the minimal unit of centromeric chromatin necessary for proper chromosome segregation.
The budding yeast CEN is defined by an ≈200-bp nuclease-resistant region that encompasses the ≈125-bp CEN-determining elements, with regularly spaced nucleosomes positioned on either side (9). However, because of the technical limitations imposed by standard ChIP experiments, in particular the size distribution of chromatin fragments produced by sonication, the precise number and position of CenH3 nucleosomes with respect to the CEN remains controversial. Some models suggest that there is one CenH3 nucleosome residing over the CEN (10), whereas others conclude that there are several CenH3 nucleosomes positioned only in CEN-flanking regions (11), or stretches of CenH3 nucleosomes encompassing a 20-kb region centered on the CEN (12). Although the development of tiling microarrays has provided additional resolution to analyzing ChIP data, centromeric DNA is currently not included on commercially available tiling arrays.
Here, we describe an approach to determine the organization of chromatin at the CEN in budding yeast with single-nucleosome resolution. Our method couples the immunoprecipitation of micrococcal nuclease (MNase)-treated native chromatin with Southern blot analysis, which preserves the information about the size and relative position of centromeric nucleosomes that are lost by popular detection methods such as PCR. This approach demonstrates that a single nucleosome containing the budding yeast CenH3, Cse4, forms the minimal unit of centromeric chromatin necessary for KT assembly and proper chromosome segregation.
Results
ChIP Approach with Single-Nucleosome Resolution.
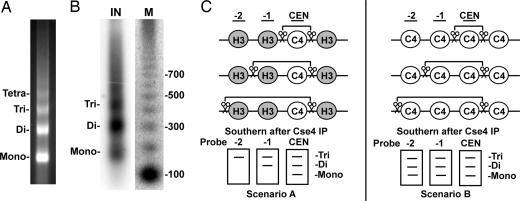
Previous attempts to map the localization of Cse4 with respect to the CEN have relied on the use of PCR or microarray assays after ChIP. The standard ChIP protocol utilizes sonication to shear chromatin before immunoprecipitation, resulting in a distribution of chromatin fragments typically centered at ≈500 bp. With a mean sonicated fragment size much larger than that of the average nucleosomal protected fragment, positive localization signals will be detected provided that the amplicon (PCR) or probe (microarray) exist anywhere in the immunoprecipitated DNA. Because these techniques lose information about the size of the immunoprecipitated DNA and the relative location of the amplicon or probe with respect to the protein of interest, ChIP data reflect the sonicated fragment size distribution around the target protein rather than the protein's true location. To overcome these issues and accurately determine the position of CenH3 nucleosomes in budding yeast, we modified the standard ChIP approach to obtain single-nucleosome resolution. Nuclei were isolated from midlogarithmic-phase cultures and were subjected to partial MNase digestion, which preferentially cuts the linker DNA, generating a nucleosomal ladder (Fig. 1A). Because we were interested in the nucleosome composition of CENs, we verified the presence of centromeric DNA in the solubilized chromatin sample by Southern blot analysis with a probe specific to the CEN of chromosome IV (CEN4) (Fig. 1B). In addition, we confirmed the presence of Cse4 in the chromatin sample by immunoblotting with anti-Cse4 antibody (data not shown).
Fig. 1.
ChIP with single-nucleosome resolution. (A) Ethidium bromide staining of the input material for ChIP. Chromatin was treated with MNase, and the DNA was isolated and subjected to electrophoresis on a 1.5% agarose gel in the presence of ethidium bromide. Partial MNase digestion generates a nucleosomal DNA ladder with visible mono-, di-, tri-, and tetranucleosomal fragments. (B) CEN DNA is present in extracted MNase-treated chromatin used for ChIP. Southern blot analysis was performed on the isolated input DNA (IN) in A by using a probe specific for the CEN4 nucleosome. M, radiolabeled 100-bp ladder marker. (C) Southern blot analysis on Cse4-associated chromatin preserves size and positional information. Chromatin is digested with MNase (scissors), and Cse4-associated chromatin is immunoprecipitated with antibody. If Cse4 localizes only to the CEN, a mononucleosome-sized fragment is detected only with a probe corresponding to the CEN, whereas probes distal to the CEN detect only increasingly larger fragments (scenario A). In contrast, if Cse4 localizes to both the CEN and the flanking nucleosomes, mononucleosome-sized fragments would be detected with the distal probes (scenario B). C4, Cse4; H3, canonical histone H3.
To localize Cse4, solubilized chromatin was immunoprecipitated with anti-Cse4 antibody and analyzed by Southern blot analysis. To obtain single-nucleosome resolution, the Southern probes were designed to hybridize to DNA sequences within a single nucleosome and not to linker DNA between nucleosomes on the basis of published nucleosome positioning data (9, 13) [supporting information (SI) Table 1]. Because Southern blot analysis preserves information about the size of the immunoprecipitated fragment, the use of probes that specifically hybridize to individual nucleosomes can precisely determine the localization of Cse4 with respect to the CEN (Fig. 1C). For example, if Cse4 localizes exclusively to the ≈125-bp centromeric DNA sequence, a mononucleosome-sized fragment will be detected only with a probe corresponding to the CEN, whereas probes distal to the centromeric core will detect only increasingly larger nucleosomal fragments (Fig. 1C, scenario A). In contrast, if Cse4 localizes to both the CEN and the flanking nucleosomes, then mononucleosome-sized fragments will be detected with both the core and distal nucleosomal probes (Fig. 1C, scenario B). Therefore, by using probes that hybridize to DNA within a single nucleosome, the presence of mononucleosome-sized DNA after immunoprecipitation decisively demonstrates that Cse4 associates with DNA at that nucleosomal position.
Budding Yeast CENs Are Composed of a Single Cse4 Nucleosome.
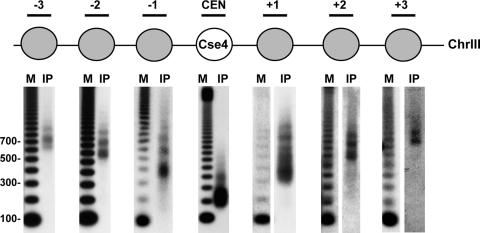
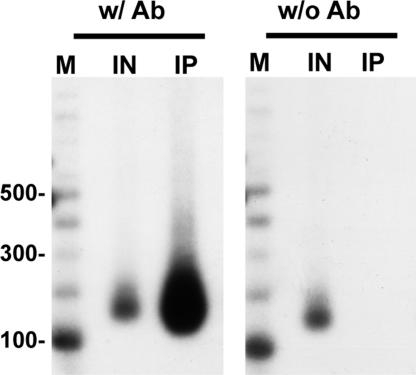
To determine the localization of Cse4 nucleosomes with respect to the CEN, we generated a yeast strain in which the sole source of Cse4 was a functional N-terminal 3xFLAG-epitope-tagged version. Cse4 was immunoprecipitated from solubilized MNase-treated chromatin with anti-FLAG agarose beads, and Southern blot analysis was performed on the associated DNA by using probes specific to nucleosomes at and around the CEN of chromosome III (CEN3), one of the smallest budding yeast chromosomes. As shown in Fig. 2, a mononucleosome-sized DNA fragment was detected only with a probe specific to the centromeric core sequence. We confirmed that the detection of the mononucleosome-sized fragment at the CEN depends on the presence of antibody against Cse4 (Fig. 3). Despite the presence of MNase-protected fragments ranging from monomers to larger oligomers in the input material (SI Fig. 5), probes corresponding to the −1 and +1 nucleosomal positions recognized dinucleosome and larger fragments with no detectable signal for mononucleosome-sized fragments (Fig. 2). Similarly, probes to the −2 and +2 nucleosomal positions recognized trinucleosome and larger fragments, and probes to the −3 and +3 nucleosomal positions recognized tetranucleosome and larger fragments. Because mononucleosomes are not detected at nucleosomal positions distal to the core, these data indicate that Cse4 is stably positioned and precipitated only in larger nucleosomal arrays because of its presence at the CEN. Of special note is that the absence of trinucleosomes after hybridization with probes specific to the −3 and +3 nucleosomal positions relative to the core indicates that Cse4 is not incorporated into the five nucleosomes to the immediate left or right of the centromeric core sequence, distances >1 kb from the CEN. Importantly, quantification of our data has revealed that we are able to readily detect 35-fold differences in intensity in Cse4 localization to various nucleosomal fragments. This difference is striking because changes in protein localization reported as significant by standard ChIP are often 2-fold. Taking these results together, the single-nucleosome resolution provided by our modified ChIP approach indicates that the budding yeast CenH3, Cse4, is present only in the nucleosome formed over the CEN, and these data are consistent with ref. 10.
Fig. 2.
A single Cse4 nucleosome forms over the CEN. MNase-treated chromatin prepared from strain SBY5146 (FLAG-CSE4) was immunoprecipitated with anti-FLAG agarose beads. Cse4-associated DNA was isolated and separated on a 1.5% agarose gel. Southern blotting was performed with probes specific to either the CEN3 or a CEN3-flanking nucleosome. M, radiolabeled 100-bp ladder marker; IP, the DNA bound to Cse4.
Fig. 3.
Detection of a Cse4 mononucleosome at the CEN depends on the inclusion of anti-Cse4 antibody. MNase-treated chromatin prepared from strain SBY3 was immunoprecipitated with protein G Dynabeads in the presence (w/ Ab) and absence (w/o Ab) of anti-Cse4 antibody. Cse4-associated DNA was isolated and separated on a 1.5% agarose gel. Southern blotting was performed with a probe specific to the CEN3. M, radiolabeled 100-bp ladder marker; IN, input DNA; IP, the DNA bound to Cse4.
One Cse4 Nucleosome per Budding Yeast KT-Forming Unit.
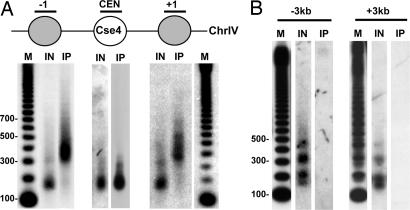
To determine whether one Cse4 nucleosome per CEN is a general feature of budding yeast CENs, we examined the localization of Cse4 to the CEN of chromosome IV (CEN4), the largest budding yeast chromosome. Examining Cse4 localization to CEN4 allowed us to simultaneously test whether the size of centromeric chromatin established by the deposition of Cse4 varies with the size of the chromosome, perhaps to withstand the increase in torsional stress necessary to pull larger chromosomes to the poles (14, 15). Consistent with this hypothesis, chromosome size appears to contribute to the level of CenH3 incorporation at human CENs (16). However, with our single-nucleosome resolution ChIP technique, a mononucleosome-sized DNA fragment was detected only with a probe specific to the CEN4 core sequence, whereas probes to the −1 and +1 nucleosome positions recognized dinucleosome-sized fragments (Fig. 4A). This same Cse4 localization pattern was also observed at CEN6 (data not shown). Therefore, one Cse4 nucleosome per KT-forming unit appears to be a general feature of budding yeast CENs.
Fig. 4.
Budding yeast CENs are composed of a single Cse4 nucleosome, regardless of chromosome size. (A) MNase-treated chromatin prepared from strain SBY5146 (FLAG-CSE4) was immunoprecipitated with anti-FLAG agarose beads. Cse4-associated DNA was isolated and separated on 1.5% agarose gel. Southern blotting was performed with probes specific to either the CEN4 or a CEN4-flanking nucleosome. (B) MNase-treated chromatin prepared from strain SBY5146 (FLAG-CSE4) was immunoprecipitated with anti-FLAG agarose beads. Cse4-associated DNA was isolated and separated on 1.5% agarose gel. Southern blotting was performed with 600-bp probes specific to sequences ≈3 kb to the left (−3 kb) or right (+3 kb) of CEN6. M, radiolabeled 100-bp ladder marker; IN, input; IP, the DNA bound to Cse4.
Throughout the centromeric chromatin of flies and humans, blocks of CenH3 nucleosomes alternate with blocks of H3 nucleosomes (6). Therefore, it remained possible that Cse4 nucleosomes are present at greater distances from the CEN than we have analyzed, a hypothesis consistent with microarray data suggesting that Cse4 localizes to a 20-kb region around the core CEN (12). To address this possibility, we tested for Cse4 localization by using ≈600-bp probes ≈3 kb to the left or right of CEN6, chromosomal positions previously shown to be positive for Cse4 localization (12). Although the use of a larger probe compromises positional information, detection of a mononucleosome-sized fragment is sufficient to indicate the presence of a Cse4 nucleosome within the 600-bp fragment. However, we were unable to detect any Cse4 localization to these genomic loci (Fig. 4B), even though hybridization of these same Southern blots with probes specific to either the −2 or the +1 nucleosome, respectively, showed Cse4 localization to CEN3 (data not shown). Taken together, these data clearly show the generality of one Cse4 nucleosome per KT in budding yeast.
Discussion
Here, we describe a ChIP approach with resolution capable of determining the histone content of individual nucleosomes. With this assay, we were able to unambiguously show that the budding yeast CenH3, Cse4, is present in a single, stably positioned nucleosome over the CEN, a pattern that is independent of chromosome size.
Utilization of a Native ChIP–Southern Blot Assay for Histone Localization.
Conventional ChIP does not provide the single-nucleosome resolution needed to address the precise position and/or composition of nucleosomes, because this technique utilizes sonication to shear DNA, which typically results in average size DNA fragments that can easily accommodate more than one canonical nucleosome. Consequently, the use of sonicated DNA as a template for PCR results in the amplification of genomic regions that may not directly associate with a particular histone. In contrast, our approach relies on the use of partial MNase digestion to generate a distribution of nucleosomal-sized fragments that allows for the determination of the exact size of the DNA associated with a histone when analyzed by Southern blot analysis. We have demonstrated the advantages of this technique in determining the genomic localization of CenH3. This approach will be generally applicable in identifying the position and post-translational modification state of other histones and possibly other DNA binding proteins on a nucleosome-by-nucleosome basis at any genomic locus. In addition, this technique provides an alternative to microarrays when the need for single-nucleosome resolution is limited to a few genomic loci rather than the entire genome. Aside from the localization of Cse4 described here, the extent of localization of other histones or nucleosomal binding proteins could be mistaken due to the inherent mobility of a nucleosome, because our approach did not include a cross-linking reagent. However, ≈70% of yeast nucleosomes are well positioned, reducing such nucleosome delocalization concerns (13). Modifications of our experimental protocol to include a cross-linking reagent should further eliminate reservations over nucleosome mobility.
One Centromeric Nucleosome: Implications for CEN Identity.
The organization of centromeric chromatin is essential not only for KT function during each round of mitosis but also for CEN propagation after each round of genome duplication. After DNA replication, existing histone octamers randomly distribute to each of the daughter strands, while newly synthesized histones fill in the gaps (17, 18). Although such a passive mechanism may suffice for the propagation of CENs composed of large arrays of CenH3s, we have shown that yeast CENs contain only one Cse4 nucleosome. Assuming a conservative segregation of octamers, as it occurs throughout the genome (17, 18), only one sister chromatid would inherit the old Cse4 nucleosome. In this case, CEN identity must be established by the de novo deposition of CenH3 every cell cycle on the other sister chromatid. In fact, there is evidence to suggest that such de novo deposition may not be limited to one sister chromatid, but that there is complete exchange of Cse4 at CENs after DNA replication in budding yeast (19).
Mechanisms to Ensure a Single CEN Nucleosome.
With a single centromeric nucleosome, one way budding yeast could establish CEN identity each cell division would be to prevent canonical H3 from being deposited at CENs. Aside from one discrepancy (20), recent data have shown that canonical H3 is capable of assembling into a nucleosome over the CEN-determining elements (21, 22), an observation that we have also made (S.F. and S.B., unpublished data). Therefore, in addition to the direct deposition of CenH3 at the CEN, CenH3 assembly may also occur by replacing canonical H3. Histone replacement would not be surprising because other histone variants, such as H2A.Z and H3.3, use such a mechanism (23–26). If a replacement mechanism exists, it may explain the observation that budding yeast CENs are one of the earliest replicating regions of the genome (27). Early CEN replication could allow time for canonical H3 to be replaced with CenH3, thus ensuring that KTs are assembled before mitosis.
Budding yeast are unique in that the centromeric DNA is a defined sequence of ≈125 bp that is capable of forming a functional KT (for reviews, see refs. 28 and 29). Yet, DNA sequence alone seems insufficient to ensure proper Cse4 localization because distinct KT proteins are essential for Cse4 localization (22, 30, 31). Consistent with this, KT proteins are also required for CenH3 localization in multicellular eukaryotes (32–36). Although these proteins may play a direct role in CenH3 localization, it is just as likely that they assemble a structure that promotes CenH3 binding or stabilizes CenH3 after deposition.
The deposition of histones requires a protein chaperone to prevent histone aggregation and promote proper nucleosome assembly (for reviews, see refs. 37 and 38); therefore, we anticipate that there is specific Cse4 deposition machinery. Presently, the only protein known to act as a CenH3 chaperone is the RbAp48 protein, which directs the assembly of Drosophila CenH3 and histone H4 in vitro (39) and is required for CenH3 localization in fission yeast and humans (33). To date, the role of RbAp48 in Cse4 localization has not been explored. The recently described Cse4-interacting protein Scm3 (20, 22, 40) has also been proposed to be a potential chaperone (40).
CenH3 may also direct its centromeric localization. Recently, it was shown that loop 1 of CenH3 is sufficient for targeting to CENs (41) and the CEN-targeting domain (CATD) of yeast CenH3, which contains loop 1, is sufficient to target a chimeric canonical H3–CATD protein to the CEN, where it can rescue depletion of CenH3 (42). This domain confers a structural rigidity to a CenH3/H4 tetramer in vitro (43), which has been proposed to generate a specialized CEN chromatin structure that results in the recruitment of new CenH3 molecules to previously assembled CenH3. However, in light of our data showing only one CenH3 nucleosome at the CEN in budding yeast, de novo CEN formation is likely required during each round of cell division, making it unclear how a self-recruitment model would be used.
Structure of Centromeric Nucleosomes: More Questions Than Answers.
In budding yeast, the bulk of DNA is wrapped around a canonical histone octamer composed of two molecules of each of four histones, H2A, H2B, H3, and H4. Cse4 has been shown to copurify with H2A, H2B, and H4, as well as exhibit genetic interactions with H2A and H4 (10, 44, 45), suggesting that centromeric nucleosomes are structurally similar to canonical nucleosomes. However, a recent study suggests that Cse4 and canonical H4 associate with the nonhistone protein Scm3 instead of H2A and H2B to form a novel hexameric nucleosomal core structure at the CEN (20). It will therefore be critical to determine the histone/nonhistone components associated with budding yeast CenH3 by examining their localization to the CEN at precise cell cycle intervals. Such an experiment is also particularly noteworthy in light of several groups reporting the deposition of CenH3 outside of S phase (42, 46–48), a detail that has not been observed in budding yeast (19, 49). Because of the ability to achieve single-nucleosome resolution, utilization of our modified ChIP protocol should clarify the protein content of the centromeric nucleosome.
Conclusions and Perspectives.
Our data show that a single centromeric nucleosome is sufficient for the formation of a functional microtubule binding site in budding yeast, results that have been independently confirmed through similar modifications of the standard ChIP protocol (M. Smith, personal communication). Consequently, it is critical that CenH3 is prevented from mislocalizing to euchromatin, where it could form ectopic KTs (4). The cell routinely establishes chromatin barriers to restrict the spreading of one epigenetic chromatin state into that of another (for review, see ref. 37). Hence, identifying whether such mechanisms prevent CenH3 from spreading into euchromatin is the next step in understanding the link between the structural organization of centromeric chromatin and the maintenance of genomic stability.
Materials and Methods
Microbial Techniques.
Media and microbial techniques were essentially as described in refs. 50 and 51.
Strains.
SBY3: Mata bar1 ura3-1 leu2-3,112 his3-11,15 trp1-1 can1-100 ade2-1. SBY5146: Mata bar1 ura3-1:pCSE4-3xFLAG-CSE4 leu2-3,112 his3-11,15:pCUP1-GFP12-LacI12:HIS3 trp1-1:256lacO:TRP1 can1-100 ade2-1 lys2Δ cse4ΔKanMX. SBY5245: Mata ura3-1 leu2-3,112 his3-11,15 trp1-1 can1-100 ade2-1 RAD5+ hht1Δ-hhf1Δ:HygB hht2Δ-hhf2Δ:NAT (pSB977 T7-HHT2, HHF2, TRP1, CEN). All strains are isogenic with the W303 background.
Yeast Nuclei Preparation.
This technique was taken from refs. 9 and 52 and adapted as described below. Cells were grown to OD600 = 0.7–0.8 in yeast extract/peptone/dextrose (YPD) at room temperature. Cells were harvested at 4,000 rpm for 10 min at 4°C in a Beckman JA-10 rotor and washed once with distilled deionized H2O. The equivalent of 500 ml of cell culture was resuspended in 1.2 M sorbitol/100 mM potassium phosphate buffer, pH 7.5/0.5 mM CaCl2/7 mM 2-mercaptoethanol and incubated at 37°C for 15 min. Cells were then spheroplasted to 80–90% less than the starting OD with 450 μl of 1 mg/ml Zymolyase-100T (Seikagaku, Tokyo, Japan) for ≈10 min at 37°C. Spheroplasting was monitored by measuring the OD600 in 1% SDS. Spheroplasts were pelleted at 3,000 rpm for 5 min at 4°C in Heraeus high conic rotor no. 75003046 (Thermo Fisher Scientific, Waltham, MA). Spheroplasts were washed twice in 25 ml of SPC (1 M sorbitol/20 mM Pipes, pH 6.3/0.1 mM CaCl2) with 1 mM PMSF and 1× LPC (l0 μg/ml each of leupeptin, pepstatin A, and chymostatin) and spun at 3,000 rpm for 5 min at 4°C in between washings. Spheroplasts were then resuspended in SPC and gently added to 25 ml of Ficoll buffer (9% Ficoll 400/20 mM Pipes, pH 6.3/0.5 mM CaCl2) while stirring. Nuclei were collected by spinning at 8,000 rpm for 20 min at 4°C in Heraeus high conic rotor no. 75003046. Pellets were washed twice with SPC/1 mM PMSF/1× LPC, and spun at 6,000 rpm for 10 min at 4°C. The final pellet (≈500 ml of cell equivalent) was resuspended in 5 ml of SPC with 1 mM PMSF and 1× LPC, frozen in liquid nitrogen, and stored at −80°C.
MNase Treatment of Yeast Nuclei.
A 500-ml cell equivalent of yeast nuclei that was resuspended in 5 ml of SPC was thawed, and CaCl2 was added to a final concentration of 2 mM. The nuclei were prewarmed at 37°C for 5 min, and then ≈416 units of MNase (Worthington Biochemicals, Lakewood, NJ) was added at 37°C for 15 min. Reactions were immediately stopped with 10 mM EDTA, and the nuclei were pelleted at 3,000 rpm for 10 min at 4°C in Heraeus high conic rotor no. 75003046. The supernatant (S1) was recovered and Triton X-100 was added to 0.1% before immunoprecipitation. The pellet was resuspended in hypotonic buffer, ≈1.67 ml of 10 mM Tris/1 mM EDTA, pH 7.4 (TE), with 0.1% Triton X-100, 1× LPC, and 1 mM PMSF, and rotated at 4°C overnight to further extract chromatin. This sample was pelleted at 8,000 rpm for 10 min at 4°C in Heraeus high conic rotor no. 75003046, and the second supernatant (S2) was recovered. We obtained the same localization results with S1 and S2.
Immunoprecipitation of MNase-Treated Chromatin.
After the isolation of chromatin, PBS350 (PBS supplemented with NaCl to 350 mM/1× LPC/1 mM PMSF/1 mM EDTA, pH 8.0) was added to the S1 and/or S2 to a final salt concentration of 100 mM. A portion of this sample was set aside (input). Beads were then prepared for immunoprecipitation by washing with PBS100 (PBS adjusted to 100 mM NaCl/1× LPC/1 mM PMSF/1 mM EDTA). FLAG-agarose beads (Sigma–Aldrich, St. Louis, MO) or protein G-coupled Dynabeads (Invitrogen, Carlsbad, CA) with anti-Cse4 antibody (235N) were used for CSE4 immunoprecipitation. Protein G Dynabeads with 2.5 μg of T7-TAG antibody (Novagen, Madison, WI) were used for T7-H3 immunoprecipitation. S1 or S2 supernatant was added to the beads and incubated at 4°C for at least 2 h. After immunoprecipitation, beads were washed twice with PBS100. Proteins were eluted by boiling in 2× sample buffer (160 mM Tris·HCl, pH 6.8/4% SDS/20% glycerol/20 mM EDTA/0.00026% bromophenol blue/1× LPC/ 1 mM PMSF). After the addition of EDTA (5 mM final) and RNase A (250 μg/ml final), the DNA was eluted from the beads by proteinase K digestion. DNA samples were extracted with phenol/chloroform/isoamyl alcohol and then precipitated with ethanol.
Southern Blot Analysis.
After immunoprecipitation, DNA associated with the histone of interest was run on 1.5% agarose gel at 150 V until the loading dye was ≈12–13 cm from wells. Digested genomic DNA prepared from SBY3 was also run as a control. For gels ultimately hybridized with probes specific to CEN3, genomic DNA was digested with SspI, whereas DdeI was typically used for CEN4. Three thousand counts per minute of [32P]dCTP Klenow filled-in 100-bp ladder (Amersham Biosciences, Piscataway, NJ) was used as marker. After electrophoresis, gels were prepared for transfer by rinsing with distilled deionized H2O, and then they were incubated with 0.25 M HCl for 10 min, 0.4 M NaOH/0.6 M NaCl for 25 min, and 1.5 M NaCl/0.5 M Tris, pH 8.0, for 25 min. DNA was wick transferred to nylon membrane (GeneScreen; PerkinElmer, Waltham, MA) overnight in 2× SSC buffer. DNA was cross-linked to the membrane by using a Gene Linker (program C3, 150 mJ) from Bio-Rad (Hercules, CA). Cross-linked membranes were then incubated in hybridization solution for a minimum of 1 h before addition of probe, which was generated by random priming in the presence of either [32P]dATP or [32P]dCTP (53). Blots were incubated with probe overnight at 60°C and then washed three times (2× SSC, 2× SSC/1% SDS, 0.1× SSC) to remove nonspecific signal. Blots were exposed to either film or phosphor-screen. Quantification of Southern blots was performed by using ImageQuant TL (Amersham Biosciences, Piscataway, NJ).
Supplementary Material
Acknowledgments
We thank R. Gardner (University of Washington, Seattle, WA) and T. Tsukiyama (Fred Hutchinson Cancer Research Center) for plasmids and strains; T. Furuyama, Y. Dalal, and T. Tsukiyama for advice on the project; T. Furuyama, T. Tsukiyama, and members of the S.B. laboratory for comments on the manuscript; Paul Megee and Dan Gottschling for discussion; and Carl Wu, Mitch Smith, and Dong Lin for sharing unpublished data. This work was supported by National Institutes of Health Fellowship 5F32GM071259 (to S.F.) and a National Institutes of Health grant to S.B. S.B. is a Leukemia and Lymphoma Society Scholar.
Abbreviations
- Mnase
micrococcal nuclease
- CenH3
centromeric histone H3
- CEN
centromere
- KT
kinetochore.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0706985104/DC1.
References
- 1.Cleveland DW, Mao Y, Sullivan KF. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- 2.Kline-Smith SL, Sandall S, Desai A. Curr Opin Cell Biol. 2005;17:35–46. doi: 10.1016/j.ceb.2004.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carroll CW, Straight AF. Trends Cell Biol. 2006;16:70–78. doi: 10.1016/j.tcb.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 4.Heun P, Erhardt S, Blower MD, Weiss S, Skora AD, Karpen GH. Dev Cell. 2006;10:303–315. doi: 10.1016/j.devcel.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vafa O, Sullivan KF. Curr Biol. 1997;7:897–900. doi: 10.1016/s0960-9822(06)00381-2. [DOI] [PubMed] [Google Scholar]
- 6.Blower MD, Sullivan BA, Karpen GH. Dev Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fitzgerald-Hayes M, Clarke L, Carbon J. Cell. 1982;29:235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- 8.Winey M, Mamay CL, O'Toole ET, Mastronarde DN, Giddings TH, Jr, McDonald KL, McIntosh JR. J Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bloom KS, Carbon J. Cell. 1982;29:305–317. doi: 10.1016/0092-8674(82)90147-7. [DOI] [PubMed] [Google Scholar]
- 10.Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- 11.Espelin CW, Simons KT, Harrison SC, Sorger PK. Mol Biol Cell. 2003;14:4557–4568. doi: 10.1091/mbc.E02-08-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riedel CG, Katis VL, Katou Y, Mori S, Itoh T, Helmhart W, Galova M, Petronczki M, Gregan J, Cetin B, et al. Nature. 2006;441:53–61. doi: 10.1038/nature04664. [DOI] [PubMed] [Google Scholar]
- 13.Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 14.Mellone BG, Allshire RC. Curr Opin Genet Dev. 2003;13:191–198. doi: 10.1016/s0959-437x(03)00019-4. [DOI] [PubMed] [Google Scholar]
- 15.Henikoff S, Dalal Y. Curr Opin Genet Dev. 2005;15:177–184. doi: 10.1016/j.gde.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 16.Irvine DV, Amor DJ, Perry J, Sirvent N, Pedeutour F, Choo KH, Saffery R. Chromosome Res. 2004;12:805–815. doi: 10.1007/s10577-005-5377-4. [DOI] [PubMed] [Google Scholar]
- 17.Leffak IM, Grainger R, Weintraub H. Cell. 1977;12:837–845. doi: 10.1016/0092-8674(77)90282-3. [DOI] [PubMed] [Google Scholar]
- 18.Russev G, Hancock R. Proc Natl Acad Sci USA. 1982;79:3143–3147. doi: 10.1073/pnas.79.10.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pearson CG, Yeh E, Gardner M, Odde D, Salmon ED, Bloom K. Curr Biol. 2004;14:1962–1967. doi: 10.1016/j.cub.2004.09.086. [DOI] [PubMed] [Google Scholar]
- 20.Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 21.Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- 22.Camahort R, Li B, Florens L, Swanson SK, Washburn MP, Gerton JL. Mol Cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 23.Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 24.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 25.Kobor MS, Venkatasubrahmanyam S, Meneghini MD, Gin JW, Jennings JL, Link AJ, Madhani HD, Rine J. PLoS Biol. 2004;2:E131. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krogan NJ, Baetz K, Keogh MC, Datta N, Sawa C, Kwok TC, Thompson NJ, Davey MG, Pootoolal J, Hughes TR, et al. Proc Natl Acad Sci USA. 2004;101:13513–13518. doi: 10.1073/pnas.0405753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCarroll RM, Fangman WL. Cell. 1988;54:505–513. doi: 10.1016/0092-8674(88)90072-4. [DOI] [PubMed] [Google Scholar]
- 28.Biggins S, Walczak CE. Curr Biol. 2003;13:R449–R460. doi: 10.1016/s0960-9822(03)00369-5. [DOI] [PubMed] [Google Scholar]
- 29.McAinsh AD, Tytell JD, Sorger PK. Annu Rev Cell Dev Biol. 2003;19:519–539. doi: 10.1146/annurev.cellbio.19.111301.155607. [DOI] [PubMed] [Google Scholar]
- 30.Lechner J, Carbon J. Cell. 1991;64:717–725. doi: 10.1016/0092-8674(91)90501-o. [DOI] [PubMed] [Google Scholar]
- 31.He X, Rines DR, Espelin CW, Sorger PK. Cell. 2001;106:195–206. doi: 10.1016/s0092-8674(01)00438-x. [DOI] [PubMed] [Google Scholar]
- 32.Takahashi K, Chen ES, Yanagida M. Science. 2000;288:2215–2219. doi: 10.1126/science.288.5474.2215. [DOI] [PubMed] [Google Scholar]
- 33.Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, Yanagida M. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 34.Okada M, Cheeseman IM, Hori T, Okawa K, McLeod IX, Yates Jr, III, Desai A, Fukagawa T. Nat Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- 35.Kline SL, Cheeseman IM, Hori T, Fukagawa T, Desai A. J Cell Biol. 2006;173:9–17. doi: 10.1083/jcb.200509158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oegema K, Desai A, Rybina S, Kirkham M, Hyman AA. J Cell Biol. 2001;153:1209–1226. doi: 10.1083/jcb.153.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kamakaka RT, Biggins S. Genes Dev. 2005;19:295–310. doi: 10.1101/gad.1272805. [DOI] [PubMed] [Google Scholar]
- 38.Jin J, Cai Y, Li B, Conaway RC, Workman JL, Conaway JW, Kusch T. Trends Biochem Sci. 2005;30:680–687. doi: 10.1016/j.tibs.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 39.Furuyama T, Dalal Y, Henikoff S. Proc Natl Acad Sci USA. 2006;103:6172–6177. doi: 10.1073/pnas.0601686103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stoler S, Rogers K, Weitze S, Morey L, Fitzgerald-Hayes M, Baker RE. Proc Natl Acad Sci USA. 2007;104:10571–10576. doi: 10.1073/pnas.0703178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vermaak D, Hayden HS, Henikoff S. Mol Cell Biol. 2002;22:7553–7561. doi: 10.1128/MCB.22.21.7553-7561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Black BE, Jansen LE, Maddox PS, Foltz DR, Desai AB, Shah JV, Cleveland DW. Mol Cell. 2007;25:309–322. doi: 10.1016/j.molcel.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 43.Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, Jr, Cleveland DW. Nature. 2004;430:578–582. [Google Scholar]
- 44.Westermann S, Cheeseman IM, Anderson S, Yates Jr, III, Drubin DG, Barnes G. J Cell Biol. 2003;163:215–222. doi: 10.1083/jcb.200305100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pinto I, Winston F. EMBO J. 2000;19:1598–1612. doi: 10.1093/emboj/19.7.1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schuh M, Lehner CF, Heidmann S. Curr Biol. 2007;17:237–243. doi: 10.1016/j.cub.2006.11.051. [DOI] [PubMed] [Google Scholar]
- 47.Lermontova I, Schubert V, Fuchs J, Klatte S, Macas J, Schubert I. Plant Cell. 2006;18:2443–2451. doi: 10.1105/tpc.106.043174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shelby RD, Monier K, Sullivan KF. J Cell Biol. 2000;151:1113–1118. doi: 10.1083/jcb.151.5.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Collins KA, Furuyama S, Biggins S. Curr Biol. 2004;14:1968–1972. doi: 10.1016/j.cub.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 50.Sherman F, Fink G, Lawrence C. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1974. [Google Scholar]
- 51.Rose MD, Winston F, Heiter P. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1990. [Google Scholar]
- 52.Nelson RG, Fangman WL. Proc Natl Acad Sci USA. 1979;76:6515–6519. doi: 10.1073/pnas.76.12.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dalal Y, Fleury TJ, Cioffi A, Stein A. Nucleic Acids Res. 2005;33:934–945. doi: 10.1093/nar/gki224. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.