Abstract
Mutations in proprotein convertase subtilisin/kexin type 9 (PCSK9) are strongly associated with levels of low-density lipoprotein cholesterol in the blood plasma and, thereby, occurrence or resistance to atherosclerosis and coronary heart disease. Despite this importance, relatively little is known about the biology of PCSK9. Here, the crystal structure of a full-length construct of PCSK9 solved to 1.9-Å resolution is presented. The structure contains a fully folded C-terminal cysteine-rich domain (CRD), showing a distinct structural similarity to the resistin homotrimer, a small cytokine associated with obesity and diabetes. This structural relationship between the CRD of PCSK9 and the resistin family is not observed in primary sequence comparisons and strongly suggests a distant evolutionary link between the two molecules. This three-dimensional homology provides insight into the function of PCSK9 at the molecular level and will help to dissect the link between PCSK9 and CHD.
Keywords: hypercholesterolemia, low-density lipoprotein receptor, proprotein convertase, x-ray crystallography, adipocytokine
PCSK9 (also known as neural apoptosis-regulated convertase, NARC-1) is a 692-residue extracellular protein expressed primarily in the kidneys, liver and intestines (1) representing the 9th member of the secretory subtilase family. Various genetic observations subsequently mapped PCSK9 as the third gene (along with LDLR and APOB) to cause autosomal dominant hypercholesterolemia (ADH). These studies suggested that gain-of-function mutations increase plasma levels of LDL-c (2–6), whereas nonsense or missense (loss-of-function) mutations, which interfere with folding or secretion of PCSK9, lead to a reduction of plasma levels of LDL-c and an 88% decrease in the risk of coronary heart disease (CHD) (5). In mice, adenoviral overexpression of PCSK9 results in increased plasma LDL-c level in normal mice but not in LDLR-deficient mice (7). Deletion of PCSK9 causes an increase in level of LDLR protein but not mRNA (8). These findings lead to a hypothesis that PCSK9 exerts its role in cholesterol metabolism through posttranslational down-regulation of LDLR, the receptor responsible for clearing LDL-c from plasma. In support of this hypothesis, a recent mouse parabiosis study revealed a nearly complete loss of liver LDLR proteins in wild-type recipients of human PCSK9 parabiosed from a transgenic littermate (9).
Like other secretory subtilisin-like serine proteases, PCSK9 is synthesized in the endoplasmic reticulum (ER) as a precursor that undergoes autocatalytic processing, intracellular trafficking, and, finally, release into the extracellular medium with the capability of decreasing the cellular LDLR level and LDL-c uptake (10). Despite the rapid progress in understanding the genetics and biology of PCSK9, little is known of its structure, substrate specificity, or mode of action of PCSK9 in the regulation of LDLR. The full-length, mature sequence consists of three domains, [the first 30 residues of the N terminus contain signal sequence (Fig. 1A)]; the first two correspond to an inhibitory prodomain (amino acids 31–152) and a serine protease domain (amino acids 153–452) of the proteinase K subfamily member of the subtilisin-like serine proteases (1, 11), respectively. The overall fold of the catalytic domain has been well characterized because of the early x-ray crystallography work done on bacterial subtilisins (11), followed by structures of the catalytic domain in complex with its prodomain (12). Recently, representative structures from eukaryotes have also been solved. These structures, Furin and Kex2 (13, 14) reveal the structure of the ≈150-residue, C-terminal, P (processing)-domain believed to be essential for folding and regulation of the activated protease (15). The equivalent domain in PCSK9 is longer, 239 residues in length (amino acids 453–692), rich in cysteine and histidine residues, and structurally and functionally uncharacterized but thought to play an analogous role to the P-domains.
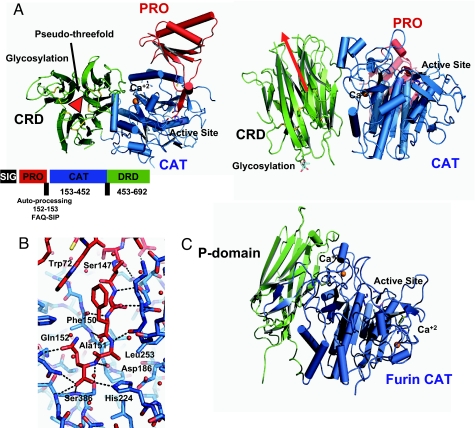
Fig. 1.
Structure of PCSK9 and comparison with Furin. (A) Two views of the full-length PCSK9, rotated relative to one another by 180° and colored according to domain, prodomain in red, protease domain in blue, and resistin-like CRD in green. Cysteine residues and active-site residues are shown as a ball and stick representation. (B) Diagram of autoinhibitory loop of the PCSK9 prodomain within the catalytic domain active site following the color scheme of A. Atoms within hydrogen bonding distance are shown by dotted lines. (C) The structure of Furin (13) in an identical orientation to PCSK9, showing the differences in orientation of the C-terminal domain of the two molecules. Images were produced by PYMOL (43).
Here, the high-resolution full-length structure of PCSK9, at 1.9-Å resolution is presented. The structure not only allows mapping of the gain- and loss-of-function mutations to three-dimensional coordinates but also structurally characterizes the C-terminal domain a unique domain of the proprotein convertases showing structural homology to another plasma protein resistin.
Results
Overall Structure.
The core of the structure conforms to a standard subtilisin-like serine protease domain, an α/β protein consisting of a seven-stranded parallel β-strand sandwiches between sets of helices, inhibited by its prodomain (12). A C-terminal, accessory cysteine-rich domain (CRD) is situated next to the catalytic domain forming a cloverleaf of subdomains displaying a clear pseudothreefold axis (Fig. 1A). The CRD is loosely attached to the catalytic domain (Fig. 1A) occluding a surface area of 1,450 Å2 at the interface between the protease domain. Despite the relatively weak binding between the domains and the presence of a flexible linker between them, it seems likely that this is the physiological conformation of the molecule as indicated by the rigid body refinement of a structure at lower resolution (≈4.0 Å) crystallized at a different pH 6.5 and space group, as opposed to pH 10.5 for the high-resolution structure, which exhibited the same conformation (data not shown). Both chains were well defined in the electron density and were clearly traced between T61-R682. The exceptions are residues of loops 168–175 and 213–219 in the protease domain, the linker residues between the protease and the CRD (449–452), and loops of this domain (572–583, 617–618, 640–641, 660–672) that were undefined in the electron density. A calcium atom is located in one of the known subtilisin calcium-binding sites coordinated by the side chains of D60 and T335 and the carbonyl oxygens of A328, A330, V333, and C358 (Fig. 1).
Comparison with Eukaryotic and Bacterial Homologues.
The structure is closely aligned in both sequence and structure to the subtilisin-like family of serine proteases (11). The structures of both murine Furin and yeast Kexin (Kex2) have been solved (13, 14). As would be expected, the structural homology between Kex2 and Furin is much closer (48% sequence identical) than that of PCSK9, (24% and 25% sequence identity, respectively) with the rmsd between Furin and Kex2 being 0.95 Å (on 301 aligned Cαs) as opposed to 1.57 and 1.51 Å with PCSK9 (on 192 and 172 aligned Cαs).
Seven of the characterized mammalian proprotein convertases, including Furin and Kex2, show a preference for mono- and dibasic substrate residues at the S1 pocket of the enzyme, whereas one other family member, PCSK8 (also known as SKI-1 and Site-1 protease), shows preference for nonbasic substrate residues at the S1 pocket [Seidah (ref. 16 and references therein)], subsite nomenclature according to Schechter and Berger (17). The catalytic triad of PCSK9 (H224, S386, and D186) is conserved and completely superimposable with serine protease active sites, suggesting that PCSK9 could be an active protease. Relative to the other members of the proprotein convertase family, PCSK9 is an outlier, the only known physiologic substrate; the autoprocessing sequence LVFAQ SIPWN (18) and structure (Fig. 1B) suggest a preference in the P1 site for a glutamine or glutamic acid preceded by hydrophobic residues in P2, P3, and P4. The active site of Furin is more occluded at the P1 position largely because of the insertion of two loops relative to PCSK9. In contrast to the Kex2 proteases, the aspartate residue at the bottom of the P1 pocket is substituted by a tyrosine residue (325), excluding the possibility of a basic P1 specificity for PCSK9 (16). The inhibitory P1 residue, Q152 from the prodomain, does not extend far into the S1 pocket, but instead the side chain is directed to the side of the pocket by a side-chain oxygen and main-chain nitrogen (N317) hydrogen bond (2.7 Å) (Fig. 1B). An alanine, positioned at the P2 position of the prodomain peptide, fits tightly within the hydrophobic lined S2 pocket, suggesting a preference for a small residue, although slightly larger residues can be tolerated as detailed by Benjannet (19), where substitution of a leucine resulted in an ≈20% drop in relative processing. The site of the naturally occurring human loss-of-function mutation L253F (5, 20) is present in the catalytic S2 pocket (Figs. 1B and 2), suggesting that this highly conserved residue is essential for correct functioning of the molecule and is sensitive to mutation.
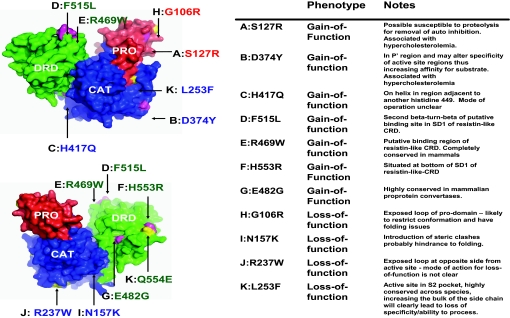
Fig. 2.
Mapping of common natural mutation of PCSK9 to the surface of the molecule. PCSK9 is shown in two orientations related to each other by a 180° rotation around the y axis. Domains are colored as in Fig. 1. Gain-of-function mutations are colored magenta, and loss-of-function mutations are depicted in yellow. Images were produced by PYMOL (43).
Structures of bacterial subtilisin-like proteases complexed with their prodomain inhibitors have been published (12). The proteins are expressed as a single chain, with the prodomain constituting the first 70 or so residues of the N terminus. This prodomain appears to be essential for proper folding of the intact molecule. Maturation of the protease is a two-step process initiated by an autocleavage event, whereby the immature protease cleaves itself into a two-chain molecule, leaving the C terminus of the prodomain in the active site and the prodomain attached to the head of the protease domain (Fig. 1A). The second maturation step is generally achieved by means of further proteolytic processing or a change in environmental stimulus, such as change in pH or increase in Ca2+ concentration. As expected, the PCSK9 catalytic/prodomain pair is structurally very similar to the bacterial structure (rmsds of 1.59 Å with PDB ID codes 1SCJ and 1SPB on 293 aligned Cα atoms). An ordered motif at the N terminus of the PCSK9 prodomain exists relative to the bacterial structures (amino acids 61–76). This extends the central prodomain β-sheet from four to five strands (Fig. 1A) and forms a hood over the active site. These ancillary residues increase the occluded area between the two domains from 2,180 Å2 in the bacterial proteases to 2,638 Å2 in PCSK9 and contribute a number of additional interactions with the catalytic domain (Fig. 1B). It is as yet unknown what physiological event, if any, causes the dissociation of the inhibitory prodomain from the protease domain in PCSK9.
CRD Is Structurally Homologous to Resistin.
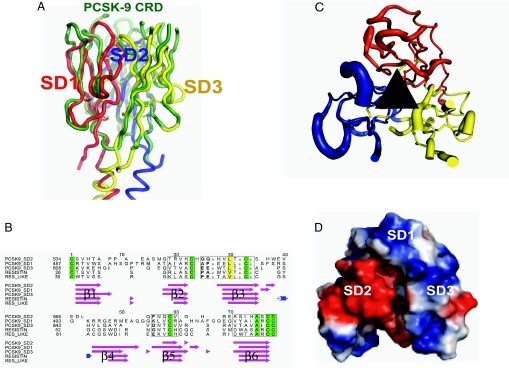
Other members of the subtilisin/kexin family contain a conserved 150-residue P-domain (15), mostly present in eukaryotic proprotein convertases/kexins. Although the function of the P-domain is largely unknown, it appears to be necessary for the correct functioning/folding of the protein (15). Of the two eukaryotic proteases solved, Furin and Kex2 both adopt the same conformation for the 150-residue P-domain, which exhibits a jelly roll-like fold consisting of two four-stranded antiparallel β-sheets with a single helix (Fig. 1C). In contrast to this, PCSK9 has a larger C-terminal domain of ≈240 residues (residues 453–692) containing a large number of cysteine residues paired to form nine disulfide bonds. The C-terminal domains from Furin and PCSK9 both contain jelly-roll structures. However, the C-terminal domain from PCSK9 consists of three, three-stranded β-domains arranged in a pseudothreefold with no helices, whereas the P-domain of Furin contains one helix and one four-stranded antiparallel β-sheet (Figs. 1 B and C and 3). In addition, the cysteine-rich C-terminal domain of PCSK9 is positioned in a different orientation to the P-domain of Furin (Fig. 1C) and does not form the tight binding interface that Furin shares with its P-domain. Each of the subdomains in the CRD of PCSK9 consists of three structurally conserved disulfide bonds, arranged such that the six cysteines are bonded in a 1–6, 2–5, 3–4 pattern between β-sheets β1–β6, β2–β6, and β3–β5, respectively (Fig. 3C). This arrangement produces a consensus pattern of C (amino acids 17–19) C (amino acids 8–9) C (amino acids 17–25) C (amino acids 11–23) CC, when each subdomain is aligned (Fig. 3B). Searching the human proteome for this motif by using a simple flexible regular expression pattern and a five-residue window between each cysteine to allow for insertions/deletions reveals a number of plasma proteins that match [full list and details in supporting information (SI) Text and SI Table 1]. CRDs from fibronectin, the ADAM family of proteins, and the urokinase-type plasminogen activator receptor contain the motif but do not exhibit the same fold. Other proprotein convertase family members also include the motif, proprotein convertase-5 and paired basic amino acid cleaving enzyme 4 (PACE4), contain only one instance. Indeed, only PCSK9 and a hypothetical protein (GenBank accession no. XP_001133083.1) contain this motif three times. Of those proteins that match the cysteine pattern, only the C-terminal 70 residues of resistin, an adipocytokine, (FIZZ3, PDB ID code 1RHF) is structurally homologous. Resistin is the only other six-stranded β-barrel to be known (21–23) and exhibits remarkable homology to the three jelly-roll “subdomains” of the CRD (Fig. 1). Recently, resistin has been linked with type II diabetes and is thought to antagonize insulin secretion (24–27). The resistin protein is characterized by an ≈30-residue three-stranded α-helical coiled-coil topped by a 70-residue six-stranded β-strand jelly-roll structure. The resulting oligomeric state is homotrimeric, each monomer containing five disulfide bonds in the jelly-roll “head” domain (Fig. 3). The remaining disulfides occur in the α-helical coiled-coil “tail” segments the N-terminal disulfide, contributing to an interchain disulfide responsible for the formation and stability of resistin trimers and hexamers (22). In comparison to this, the CRD of PCSK9 contains approximately 210 residues and forms a heteropseudotrimeric structure, completely structurally equivalent to the resistin homotrimer but without the coiled-coil of resistin (Fig. 3 A and B). The sequence identity between the hetero- and homotrimer is extremely low, <15.00%, whereas a structural alignment over the whole hetero-verses homotrimer produces an rmsd of 1.9 Å on aligned Cα atoms (Fig. 3B). This, coupled with the identical positions of the three CRD disulfides within the primary sequence and the structure, insinuates that the two share a common ancestor and indicates a possible receptor-like function for the resistin-like CRD. The resistin-like molecules have an additional two disulfide bonds forming connections between strands β3 and β5 and β4 and β5, respectively (Fig. 3B). These additional covalent bonds are likely to contribute to extra stability within the resistin fold, because equivalent positions in the PCSK9 resistin-like CRD are characterized by regions of high atomic displacement parameters or disorder. Given the shapes of the two molecules, the most obvious candidate region for interaction is the head domain containing the nine loops connecting the β-sheets surrounding the deep cleft at the center of the pseudothreefold axis (Fig. 3D). The topological arrangement of β-strands between the two molecules is identical, but the loops connecting the β-strands to the head of the trimer in resistin of the molecule are conformationally divergent, suggesting an alternative binding partner (Fig. 3 A and C).
Fig. 3.
Structural alignments of the CRD heterotrimer with the resistin homotrimer. (A) Superposition of the homotrimer of resistin and the CRD of PCSK9, oriented so that the putative binding region of resistin/CRD is facing upward. The domains of resistin are superposed individually on the CRD subdomains causing a slight rotation of the nonconserved coiled-coil region depicted at the bottom of the figure. The CRD is colored green, and the three resistin homodimer subdomains (SD1, SD2, SD3) are colored red, blue, and yellow respectively. Disulfides are shown in ball and stick representations. (B) Sequence alignment based on structures of the individual resistin-like subdomains of the PCSK9 resistin-like CRD and monomers of mouse resistin and resistin-like protein (22). Secondary structure is shown below the alignment, β-strands are shaded magenta, helices are shaded in blue. Structurally conserved regions are boxed, regions of sequence identity are shaded green, conserved hydrophobic regions are in yellow, and conserved small residues are in small font. The figure was produced by Stamp (44) and Alscript (45). (C) Representation of the conservation between the PCSK9 heterotrimer and the homotrimer of resistin. The pseudothreefold points toward the reader, depicted as a black triangle. Subdomains are colored according to the resistin colors of A, disulfides are represented as ball and sticks. Thickness of the tube is proportional to the rmsd between like Cαs in a three-residue centered window. Structural diversity is highest within the loops of the “putative” binding region. (D) Electrostatic surface representation of the three subdomains of the CRD oriented identically to B, a deep cleft is apparent in the center of the molecule. Images were produced by PYMOL (43).
Naturally Occurring Gain- and Loss-of-Function Mutations.
A number of genetic studies of naturally occurring variants of PCSK9 have been discovered (2–6, 20, 28). If the general model is that PCSK9 is responsible for plasma LDLR levels and, thus, plasma LDL-c levels, then loss-of-function mutations can be linked with nonsense mutations or those mutation related to processing/folding/active-site abnormalities within PCSK9, which thus increase levels of LDLR (Fig. 2). One example in the resistin-like CRD is the nonsense mutation C679X that occurs in the penultimate disulfide bond, suggesting a disruption in the folding pattern of the CRD and thus incorrect processing or folding of the molecule. Another loss-of-function mutation, R46L (20), has been reported to produce a15% reduction in plasma LDL-c levels for heterozygous individuals. This mutation is located within the N terminus of the prodomain, a region that shows no ordered electron density (amino acids 31–60). The flexibility makes it unclear whether the loss of function is a result of folding or secretion problems. It is possible that the structure becomes ordered in response to a binding partner or a different physiochemical environment. In such circumstances, the loss of function mutation could influence this binding or ordering of the sequence.
Gain-of-function mutations are likely more diverse in phenotypic action, the general hypothesis being that the mutations increase the binding affinity for substrates, thus providing a more active molecule. Gain-of-function mutations are distributed around the surface (Fig. 2): one region occurs at the negatively charged lip of the active site where D374Y presumably influences binding affinity for substrate; one mutation in the prodomain S127R may increase binding or enhance the susceptibility of the domain to cleavage by proteases such as furin; and regions at both the front and back ends of the first subdomain of the resistin-like CRD. Those in the resistin-like CRD are interesting and validate the importance of the resistin-like CRD in function. The two mutations R469W and F515L occur in the putative binding region of the resistin-like CRD at the head of the domain within the first monomer of the heterotrimer (SD1 in Figs. 2 and 3).
Discussion
Despite the obvious biomedical interest, the function of PCSK9 has been enigmatic. The structure of PCSK9 provides valuable insight into its specificity and function and the structural relationship to other cellular proteins involved in similar interactions. The existence of a unique C-terminal domain in PCSK9 relative to other proprotein convertases shows novelty, whereas its homology to resistin, not easily discernable at the primary sequence level, exhibits conservation. This observation indicates that the functions of the proprotein convertases, like their functionally homologous counterparts, the trypsin like serine proteases, arose by exons shuffling and gene duplication [see Patthy (29) for review]. The molecular function of resistin has also been mysterious; the gene has been associated with metabolic diseases [for review, see Steppan (27)], but, as yet, no molecular receptor has been identified. With the relationship between the two molecules, it is tempting to speculate that the two share or compete for a common receptor, LDLR. However, it is more likely, given the low sequence identity, structural divergence of putative binding sites (Fig. 3B), and shape changes due to the nature of the hetero-versus homotrimer structure, that the two molecules have evolved to recognize different binding partners. It is tempting to suggest that the CRD provides a means to localize the protease to the substrate molecule [LDLR (8, 30, 31)] or a receptor that localizes function, allowing the PCSK9 protease domain to facilitate degradation or removal of LDLR. An alternative hypothesis, and one consistent with current knowledge, would be the absence of proteolytic activity altogether, whereby binding of PCSK9 to LDLR (or other receptor) provides a signal to lower LDLR levels. A recent publication by Nassoury et al. (32) has provided evidence that the CRD binds to the cell surface via LDLR. A nonproteolytic mechanism has been shown to exist in proteases of the trypsin family such as hepatocyte growth factor (HGF) (33), whereby the protease has lost catalytic activity but functions as a growth factor receptor. An analogous situation may occur in PCSK9, with the exception that positive selection for the maintenance of the active site is apparent, because the protein must autocleave itself to enable the correct folding and secretion of the protein. This connection between structure and disease may be important to dissecting the complex pathway of PCSK9 in CHD.
While the present manuscript was in review, two structures of PCSK9 were published (34, 35). The orthorhombic crystal form detailed in these two articles is identical to the ones presented in this article, with the exception that growth conditions for the two crystals differed widely, with pHs of 8.4 and 7.6 for the Cunningham (34) and Piper (35) structures, respectively. The Piper article (35) also details a cubic crystal form grown at pH 4.6. This, combined with the fact that our structure crystallized at pH 10.5, strongly suggests that the orientation of the domains relative to one another is independent of pH and suggests that recent studies of differential binding affinities of LDLR at different pHs (36) are due to conformational changes in LDLR or other physiochemical changes. In addition, the structures do not contain a Ca+2 at the conserved calcium-binding site. Because calcium is not a major determinant of PCSK9 activity (19), this suggests that PCSK9 can accommodate a Ca+2 but has little effect on the structure. Both articles mistakenly identify the CRD as a new fold rather than a heterotrimeric arrangement of a resistin-like fold.
Materials and Methods
Expression and Purification.
The full-length DNA sequence encoding PCSK9 was amplified from a full-length construct in the baculovirus expression vector pFastBac1 by PCR. The construct included a fused hexahistadine C-terminal affinity tag, and the honey bee melittin secretion signal substituted for the native signal peptide. The forward primer (5′-CCCAAGCTTGCCGCCACCATGAAATTCTTAGTC AAC-3′) incorporates a 5′ HindIII restriction site and Kozak sequence. The reverse primer (5′-GC TCTAGATCAGTGGTGGTGGTGGTGGTGGTGCTGGAG-3′) includes a 3′ XbaI restriction site and introduces a stop codon. The PCR product was digested with HindIII and XbaI, and cloned into these respective sites of the pRS5a mammalian expression vector. PCSK9 was expressed in the Freestyle 293 expression system (Invitrogen, Carlsbad, CA). One liter of cells was transfected according to the manufacturer's protocol. Forty-eight hours after transfection, cultures were harvested by centrifugation at 400 × g. The supernatant was loaded on to a 4-ml NiNTA column (Qiagen, Valencia, CA) equilibrated with 50 mM Tris·HCl (pH 7.4), 300 mM NaCl, 1 mM CaCl2, and 2 mM 2-mercaptoethanol and washed with 20 ml of equilibration buffer plus 20 mM imidazole. Protein was eluted with 50 mM Tris·HCl (pH 7.4), 250 mM imidazole, 300 mM NaCl, 1 mM CaCl, and 2 mM 2-mercaptoethanol and analyzed by SDS/PAGE. The eluted PCSK9 was dialyzed against 50 mM Tris·HCl (pH 7.4), 150 mM NaCl, 1 mM CaCl2, and 2 mM 2-mercaptoethanol before concentration to 10 mg/ml.
Crystallization, Data Collection, and Structure Solution.
Crystals were grown by sitting-drop vapor diffusion and formed within 3 weeks in crystallization conditions containing 20% PEG 6000, 0.2 M sodium chloride at 20°C in 0.1 M CAPS buffer at pH 10.5. Data were collected at the Advanced Light Source beamline 5.0.3 to a maximum resolution of 1.9 Å. Data were processed with the HKL2000 package (37), and initial phases were derived by using phaser (38) and two ensembles of a prodomain structure (PDB ID codes 1SCJ and 1SPB) and one containing five representatives from the subtilisin-like serine protease family (PDB ID codes 1CNM, 1SH7, 2B6N, 1P8J, and 1R64). Iterative building and refinement was carried out with ARP/WARP, Coot and, Refmac5 (39–42). The R-factor and free R-factor for all data up to 1.9 Å converged at 19.0% and 24%, respectively (SI Table 2).
Supplementary Material
Acknowledgments
We thank A. Kreusch, C. Lee, M. Didonato, and P. Chamberlain for help with data collection; J. Trauger, E. Nigoghossian, C. Tumanut, K.-F. Suen, and J.-A. Gavigan for help with protein expression; D. Yowe, B. Kreider, J. Levell, S. Izumo, C. Hernandez, and G. Liau for useful discussion; and P. Schultz for continued support. Data were collected at the Advanced Light Source (Lawrence Berkeley National Laboratory, Berkeley, CA) on beamline 5.0.3; we thank all of the staff for their help.
Abbreviations
- ADH
autosomal dominant hypercholesterolemia
- CHD
coronary heart disease
- LDL
low-density lipoprotein
- NARC-1
neural apoptosis-regulated convertase 1
- PCSK9
prohormone convertase subtilisin kexin-9
- CRD
cysteine-rich domain.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The structure reported in this paper has been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2QTW).
This article contains supporting information online at www.pnas.org/cgi/content/full/0703402104/DC1.
References
- 1.Seidah NG, Benjannet S, Wickham L, Marcinkiewicz J, Jasmin SB, Stifani S, Basak A, Prat A, Chretien M. Proc Natl Acad Sci USA. 2003;100:928–933. doi: 10.1073/pnas.0335507100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abifadel M, Varret M, Rabes JP, Allard D, Ouguerram K, Devillers M, Cruaud C, Benjannet S, Wickham L, Erlich D, et al. Nat Genet. 2003;34:154–156. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- 3.Leren TP. Clin Genet. 2004;65:419–422. doi: 10.1111/j.0009-9163.2004.0238.x. [DOI] [PubMed] [Google Scholar]
- 4.Allard D, Amsellem S, Abifadel M, Trillard M, Devillers M, Luc G, Krempf M, Reznik Y, Girardet JP, Fredenrich A, et al. Hum Mutat. 2005;26:497. doi: 10.1002/humu.9383. [DOI] [PubMed] [Google Scholar]
- 5.Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. N Engl J Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 6.Berge KE, Ose L, Leren TP. Arterioscler Thromb Vasc Biol. 2006;26:1094–1100. doi: 10.1161/01.ATV.0000204337.81286.1c. [DOI] [PubMed] [Google Scholar]
- 7.Maxwell KN, Breslow JL. Proc Natl Acad Sci USA. 2004;101:7100–7105. doi: 10.1073/pnas.0402133101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rashid S, Curtis DE, Garuti R, Anderson NN, Bashmakov Y, Ho YK, Hammer RE, Moon YA, Horton JD. Proc Natl Acad Sci USA. 2005;102:5374–5379. doi: 10.1073/pnas.0501652102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lagace TA, Curtis DE, Garuti R, McNutt MC, Park SW, Prather HB, Anderson NN, Ho YK, Hammer RE, Horton JD. J Clin Invest. 2006;116:2995–3005. doi: 10.1172/JCI29383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cameron J, Holla OL, Ranheim T, Kulseth MA, Berge KE, Leren TP. Hum Mol Genet. 2006;15:1551–1558. doi: 10.1093/hmg/ddl077. [DOI] [PubMed] [Google Scholar]
- 11.Wright CS, Alden RA, Kraut J. Nature. 1969;221:235–242. doi: 10.1038/221235a0. [DOI] [PubMed] [Google Scholar]
- 12.Jain SC, Shinde U, Li Y, Inouye M, Berman HM. J Mol Biol. 1998;284:137–144. doi: 10.1006/jmbi.1998.2161. [DOI] [PubMed] [Google Scholar]
- 13.Henrich S, Cameron A, Bourenkov GP, Kiefersauer R, Huber R, Lindberg I, Bode W, Than ME. Nat Struct Biol. 2003;10:520–526. doi: 10.1038/nsb941. [DOI] [PubMed] [Google Scholar]
- 14.Holyoak T, Wilson MA, Fenn TD, Kettner CA, Petsko GA, Fuller RS, Ringe D. Biochemistry. 2003;42:6709–6718. doi: 10.1021/bi034434t. [DOI] [PubMed] [Google Scholar]
- 15.Zhou A, Martin S, Lipkind G, LaMendola J, Steiner DF. J Biol Chem. 1998;273:11107–11114. doi: 10.1074/jbc.273.18.11107. [DOI] [PubMed] [Google Scholar]
- 16.Seidah NG, Chretien M. Brain Res. 1999;848:45–62. doi: 10.1016/s0006-8993(99)01909-5. [DOI] [PubMed] [Google Scholar]
- 17.Schechter I, Berger A. Biochem Biophys Res Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 18.Naureckiene S, Maa L, Sreekumarb K, Purandarea U, Loa CF, Huangc Y, Chiangd LW, Grenierd JM, Ozenbergera BA, Jacobsena JS, et al. Arch Biochem Biophys. 2003;420:55–67. doi: 10.1016/j.abb.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 19.Benjannet S, Rhainds D, Essalmani R, Mayne J, Wickham L, Jin W, Asselin MC, Hamelin J, Varret M, Allard D, et al. J Biol Chem. 2004;279:48865–48875. doi: 10.1074/jbc.M409699200. [DOI] [PubMed] [Google Scholar]
- 20.Kotowski IK, Pertsemlidis A, Luke A, Cooper RS, Vega GL, Cohen JC, Hobbs HH. Am J Hum Genet. 2006;78:410–422. doi: 10.1086/500615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banerjee RR, Rangwala SM, Shapiro JS, Rich AS, Rhoades B, Qi Y, Wang J, Rajala MW, Pocai A, Scherer PE, et al. Science. 2004;303:1195–1198. doi: 10.1126/science.1092341. [DOI] [PubMed] [Google Scholar]
- 22.Patel SD, Rajala MW, Rossetti L, Scherer PE, Shapiro L. Science. 2004;304:1154–1158. doi: 10.1126/science.1093466. [DOI] [PubMed] [Google Scholar]
- 23.Rangwala SM, Rich AS, Rhoades B, Shapiro JS, Obici S, Rossetti L, Lazar MA. Diabetes. 2004;53:1937–1941. doi: 10.2337/diabetes.53.8.1937. [DOI] [PubMed] [Google Scholar]
- 24.Holcomb IN, Kabakoff RC, Chan B, Baker TW, Gurney A, Henzel W, Nelson C, Lowman HB, Wright BD, Skelton NJ, et al. EMBO J. 2000;19:4046–4055. doi: 10.1093/emboj/19.15.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohue M, Kato K, Nomura A, Toyosawa K, Furutani Y, Kimura S, Kadowaki T, Kim KH. Diabetes. 2001;50:113–122. doi: 10.2337/diabetes.50.1.113. [DOI] [PubMed] [Google Scholar]
- 26.Steppan CM, Bailey ST, Bhat S, Brown EJ, Banerjee RR, Wright CM, Patel HR, Ahima RS, Lazar MA. Nature. 2001;409:307–312. doi: 10.1038/35053000. [DOI] [PubMed] [Google Scholar]
- 27.Steppan CM, Lazar MA. J Int Med. 2004;255:439–447. doi: 10.1111/j.1365-2796.2004.01306.x. [DOI] [PubMed] [Google Scholar]
- 28.Jirholt P, Adiels M, Borén J. Arterioscler Thromb Vasc Biol. 2004;24:1334–1336. doi: 10.1161/01.ATV.0000133682.97348.ff. [DOI] [PubMed] [Google Scholar]
- 29.Patthy L. Genetica. 2003;118:217–231. [PubMed] [Google Scholar]
- 30.Zhao Z, Tuakli-Wosornu Y, Lagace TA, Kinch L, Grishin NV, Horton JD, Cohen JC, Hobbs HH. Am J Hum Genet. 2006;79:514–523. doi: 10.1086/507488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park SW, Moon YA, Horton JD. J Biol Chem. 2004;279:50630–50638. doi: 10.1074/jbc.M410077200. [DOI] [PubMed] [Google Scholar]
- 32.Nassoury N, Blasiole DA, Olter AT, Benjannet S, Hamelin J, Poupon V, McPherson PS, Attie AD, Prat A, Seidah NG. Traffic. 2007;8:718–732. doi: 10.1111/j.1600-0854.2007.00562.x. [DOI] [PubMed] [Google Scholar]
- 33.Donate LE, Gherardi E, Srinivasan N, Sowdhamini R, Aparicio S, Blundell TL. Protein Sci. 1994;3:2378–2394. doi: 10.1002/pro.5560031222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cunningham D, Danley DE, Geoghegan KF, Griffor MC, Hawkins JL, Subashi TA, Varghese AH, Ammirati MJ, Culp JS, Hoth LR, et al. Nat Struct Mol Biol. 2007;14:413–419. doi: 10.1038/nsmb1235. [DOI] [PubMed] [Google Scholar]
- 35.Piper DE, Jackson S, Liu Q, Romanow WG, Shetterly S, Thibault ST, Shan B, Walker NP. Structure (London) 2007;15:545–552. doi: 10.1016/j.str.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 36.Fisher TS, Lo Surdo P, Pandit S, Mattu M, Santoro JC, Wisniewski D, Cummings RT, Calzetta A, Cubbon RM, Fischer PA, et al. J Biol Chem. 2007;282:20502–20512. doi: 10.1074/jbc.M701634200. [DOI] [PubMed] [Google Scholar]
- 37.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 38.Storoni LC, McCoy AJ, Read RJ. Acta Crystallogr D. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 39.Perrakis A, Harkiolaki M, Wilson KS, Lamzin VS. Acta Crystallogr D. 2001;57:1445–1450. doi: 10.1107/s0907444901014007. [DOI] [PubMed] [Google Scholar]
- 40.Emsley P, Cowtan K. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 42.Collaborative Computing Project Number4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 43.Delano W. PYMOL. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- 44.Russell RB, Barton GJ. Proteins Struct Funct Genet. 1992;14:309–323. doi: 10.1002/prot.340140216. [DOI] [PubMed] [Google Scholar]
- 45.Barton GJ. Protein Eng. 1993;6:37–40. doi: 10.1093/protein/6.1.37. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.