Abstract
The kinase Cdc2p is a central regulator of entry into and progression through nuclear division during mitosis and meiosis in eukaryotes. Cdc2p is activated at the onset of mitosis by dephosphorylation on tyrosine-15, the phosphorylation status of which is determined mainly by the kinase Wee1p and the phosphatase Cdc25p. In fission yeast, the forkhead-type transcription factor Mei4p is required for expression of many genes during meiosis, with mei4 mutant cells arresting before meiosis I. The mechanism of cell cycle arrest in mei4 cells has remained unknown, however. We now show that cdc25+ is an important target of Mei4p in control of entry into meiosis I. Forced dephosphorylation of Cdc2p on tyrosine-15 thus induced meiosis I in mei4 mutant cells without a delay, although no spores were formed. We propose that Mei4p acts as a rate-limiting regulator of meiosis I by activating cdc25+ transcription in coordination with other meiotic events.
Keywords: cell cycle, transcription
In meiosis, the replication of DNA is followed by two rounds of chromosome segregation, resulting in the production of functional haploid gametes (1, 2). A high level of homologous recombination occurs after DNA replication but before the first meiotic nuclear division [meiosis I (MI)] (1, 3–7). Progression through meiosis is controlled by orchestration of the expression of various meiotic genes (8–10). In fission yeast, >500 genes, known as middle genes, undergo transcriptional up-regulation during meiotic divisions (10). The meiosis-specific transcription factor Mei4p is not required for premeiotic DNA replication but is essential for MI, sporulation, and recombination (11, 12). Mei4p contains a forkhead DNA-binding domain that binds the consensus sequence GTAAAYA, also known as FLEX (11, 13). Although Mei4p is thought to be required for induction of most middle genes, few targets of this transcription factor have been characterized among these genes. Among the potential Mei4p target genes, spo6+ is required for sporulation (11). mde2+ has been identified as a Mei4p target gene, and its product is required for formation of DNA double-strand breaks (13, 14), a process that is essential for meiotic recombination, consistent with the fact that Mei4p is also required for double-strand break formation (15).
In many eukaryotes, the protein kinase Cdc2p (also known as cyclin-dependent kinase 1, or Cdk1) controls the onset of mitosis in a manner dependent on various internal and external conditions that include nutrient availability and cell size (16). The activity of Cdc2p is determined by the phosphorylation status of its Tyr15 residue and the availability of cyclin (16, 17). Inhibitory phosphorylation of Cdc2p on Tyr15 is catalyzed by the tyrosine kinases Wee1p and Mik1p, and the dephosphorylation of this residue is mediated predominantly by the tyrosine phosphatase Cdc25p (16, 17). Similar to the situation in mitosis, the kinase activity of Cdc2p increases around the start of DNA replication that precedes meiosis and is maximal during meiotic nuclear division (18–20). The phosphorylation of Cdc2p on Tyr15 is apparent around the onset of premeiotic DNA replication and decreases during meiotic nuclear division (18–21). The regulation of Cdc2p phosphorylation on Tyr15 is a rate-limiting step for entry into nuclear division in both mitosis and meiosis, given that such entry is promoted by forced dephosphorylation of this residue (16–18).
No mutants that proceed through meiotic nuclear division in the absence of mei4+ have been identified since the isolation of mei4 mutant cells (12). Although Mei4p is thought to be an important regulator of middle gene expression during meiosis, with many Mei4p target genes thought to be indispensable for progression through meiotic nuclear division, the precise contribution of Mei4p to meiotic nuclear division has remained unknown. We showed (20), however, that Cdc2p is phosphorylated on Tyr15 in mei4 mutant cells. We now show that phosphorylation of Cdc2p on Tyr15 is responsible for the arrest of mei4 mutant cells, given that forced dephosphorylation of this residue induced meiotic nuclear division without a delay. We also show that cdc25+ is an essential target of Mei4p for progression through meiotic nuclear division. On the basis of our results, we propose that Mei4p is a rate-limiting factor for entry into MI and coordinates cell cycle progression with other meiotic events, such as sporulation and double-strand break formation.
Results and Discussion
Phosphorylation of Cdc2p on Tyr15 Is Responsible for the Arrest of mei4 Mutant Cells.
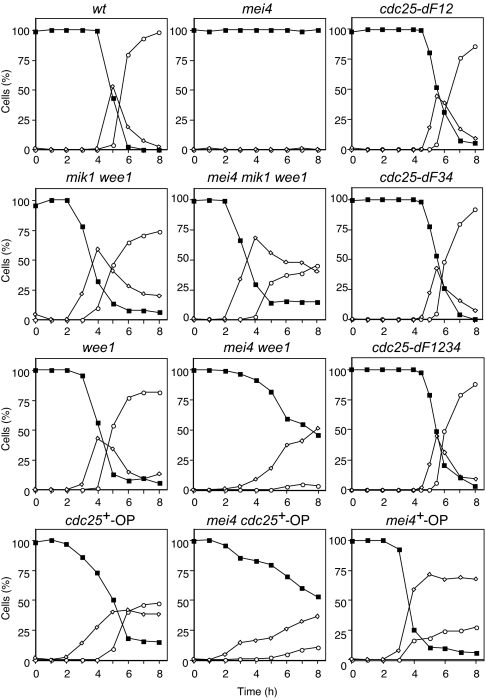
To facilitate the synchronous induction of meiosis, we studied a temperature-sensitive pat1 strain of Schizosaccharomyces pombe. Pat1p is a negative regulator of meiosis, and its inactivation results in the synchronous induction of meiosis with high recombination and viability rates (22). Cells were synchronized in the G1 phase of the cell cycle by nitrogen deprivation and shifted to the restrictive temperature to induce meiosis (Fig. 1). We showed (18) that Tyr15 of Cdc2p is phosphorylated during interphase and dephosphorylated on entry into MI. In addition, this phosphorylation is maintained in mei4 mutant cells (20) [supporting information (SI) Fig. 5]. To investigate whether phosphorylation of Cdc2p on Tyr15 is responsible for the arrest of mei4 mutant cells, we constructed a mei4 mik1 wee1 strain in which this residue of Cdc2p would be expected to be dephosphorylated at the restrictive temperature. We found that mei4 mik1 wee1 cells entered into MI with kinetics almost identical to those of mik1 wee1 cells (Fig. 1). Similar results were obtained with haploid cells (SI Fig. 6). Cdc2p on Tyr15 was dephosphorylated in mei4 mik1 wee1 cells as it was in mik1 wee1 cells (SI Fig. 5) (18). These data suggested that phosphorylation of Cdc2p on Tyr15 is largely responsible for the arrest of mei4 mutant cells. In addition, overexpression of cdc25+ (mei4 cdc25+-OP) or loss of function of wee1+ (mei4 wee1) induced meiotic division in mei4 mutant cells (Fig. 1). Partial suppression in these cells is likely to be attributable to the incomplete dephosphorylation of Cdc2p on Tyr15, which may be caused by an insufficient induction of cdc25+ mRNA for mei4 cdc25+-OP cells or the presence of mik1+ gene for mei4 wee1 cells. Taken together, these data suggest that regulation of cdc25+ and wee1+ is essential for mei4+ function in nuclear division. Cells in which Tyr15 of Cdc2p was replaced with phenylalanine failed or were severely delayed to enter premeiotic DNA replication in pat1-induced meiosis (data not shown).
Fig. 1.
Forced dephosphorylation of Cdc2p on Tyr15 induces meiotic nuclear division in mei4 mutant cells without a delay. Cells of the indicated genotypes (WT, HM1307; mei4, HM2163; mik1 wee1, HM605; mei4 mik1 wee1, HM1959; wee1, HM4739; mei4 wee1, HM3799; cdc25+-OP, HM5220; mei4 cdc25+-OP, HM4835; cdc25-dF12, HM5969; cdc25-dF34, HM5970; cdc25-dF1234, HM5973; and mei4+-OP, HM4582) were grown and then transferred to nitrogen-free medium for 14–16 h at 24°C to induce arrest in the G1 phase. Meiosis was induced by shifting the temperature to 34°C at time 0. Samples were collected at the indicated times thereafter for determination of the proportions of cells containing one nucleus (■), two nuclei (◇), or three or four nuclei (○). Data are from representative experiments.
cdc25+ Is a Target Gene of Mei4p.
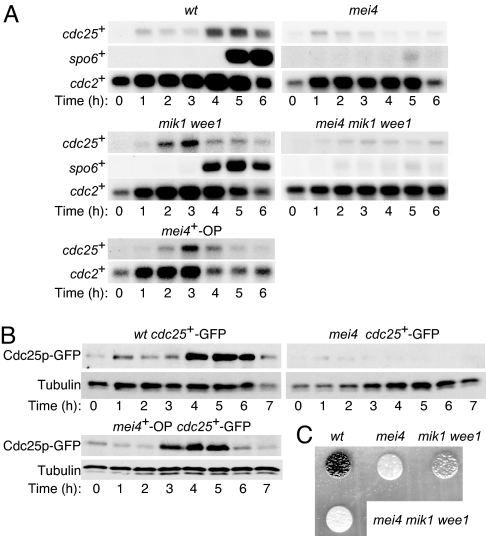
Our data implicated cell cycle-related genes responsible for phosphorylation of Cdc2p on Tyr15 as targets of Mei4p. We therefore next examined the amount of cdc25+ mRNA in both WT and mei4 cells by Northern blot analysis (Fig. 2A). The abundance of cdc25+ mRNA increased around the onset of MI in WT cells, consistent with previous observations (10, 23). In mei4 mutant cells, however, the level of cdc25+ mRNA remained low (23). To exclude the possibility that the lack of induction of cdc25+ mRNA in these latter cells was attributable to arrest of the cell cycle, we measured the amount of cdc25+ transcripts in mei4 mik1 wee1 cells. Although the level of cdc25+ mRNA increased earlier in mik1 wee1 cells than in WT cells, it remained largely unchanged in mei4 mik1 wee1 cells. These results suggested that Mei4p regulates the level of cdc25+ mRNA directly.
Fig. 2.
Expression of cdc25+ and spo6+ during meiosis and effects of mei4+ overexpression or mutation. (A) Cells of the indicated genotypes (WT, HM1307; mei4, HM2163; mik1 wee1, HM605; mei4 mik1 wee1, HM1959; and mei4+-OP, HM4582) were subjected to synchronous induction of meiosis as described in Fig. 1. Samples were collected at the indicated times thereafter and subjected to Northern blot analysis of cdc25+ and spo6+ mRNA. Transcripts of cdc2+ were analyzed as a loading control. (B) Cells expressing GFP-tagged Cdc25p (WT cdc25+-GFP, HM4732; mei4 cdc25+-GFP, HM4833; and mei4+-OP cdc25+-GFP, HM4735) were treated as in A and subjected to immunoblot analysis with antibodies to GFP and to α-tubulin (loading control). (C) Cells of the indicated genotypes were induced to undergo meiosis as in Fig. 1. After incubation for 24 h at 34°C, the cells were examined with staining with iodine.
We also examined the abundance of Cdc25p in both WT and mei4 cells (Fig. 2B). Similar to the results obtained for its mRNA, the amount of Cdc25p increased around the onset of MI in WT cells but remained unchanged in mei4 cells. In addition, the level of Cdc25p increased earlier in mik1 wee1 cells than in WT cells, but it remained largely unchanged in mei4 mik1 wee1 cells. The correlation between the expression of cdc25+ at the mRNA and protein levels suggested that regulation of the abundance of the mRNA is important for physiological gene function. Similar results regarding expression of cdc25+ at both the mRNA and protein levels were obtained with haploid cells (SI Figs. 7 and 8).
Overexpression of Mei4p Induces Early Onset of Meiotic Nuclear Division.
Our data suggested that cdc25+ is an important target of Mei4p in regulation of entry into and progression through meiotic nuclear division. Ectopic expression of Mei4p would therefore be expected to induce an early onset of such division. To test this hypothesis, we engineered expression of mei4+ under the control of the thiamine-sensitive nmt1 promoter (mei4+-OP) in meiotic cells. The cells entered S phase with kinetics similar to those apparent for the control strain (data not shown), but they entered MI ≈1.5 h earlier than did the control strain (Fig. 1). The up-regulation of cdc25+ expression, at both the mRNA and protein levels, was also detected earlier in the manipulated cells (Fig. 2 A and B). These results thus supported the notion that Mei4p is a rate-limiting factor in control of the entry of cells into meiotic nuclear division and that it mediates such control by up-regulating cdc25+ transcription. However, we cannot completely rule out the possibility that there is another mechanism that controls the level of Cdc25p, because the appearance of Cdc25p was slightly delayed compared with its mRNA in the manipulated cells.
Dephosphorylation of Cdc2p on Tyr15 Fails to Suppress the Sporulation Defect of mei4 Cells.
We tested whether dephosphorylation of Cdc2p on Tyr15 suppresses the sporulation defect of mei4 mutant cells. Although no spores were formed by mei4 cells, mik1 wee1 cells formed spores but not as efficiently as did WT cells. No spores were formed by mei4 mik1 wee1 cells (Fig. 2C). Similar results were obtained with haploid cells (SI Fig. 9). Mei4p is required for sporulation at least in part as a result of its induction of spo6+ (11). Consistent with this fact, induction of spo6+ mRNA was largely abolished in mei4 cells (Fig. 2A) (11). Induction of spo6+ mRNA occurred earlier in mik1 wee1 cells than in the WT. However, the up-regulation of spo6+ mRNA was also mostly lost in mei4 mik1 wee1 cells. These observations suggest that dephosphorylation of Cdc2p on Tyr15 suppresses the defect in meiotic nuclear division but not the sporulation defect in mei4 mutant cells.
Mei4p Binds to FLEX Elements Adjacent to cdc25+.
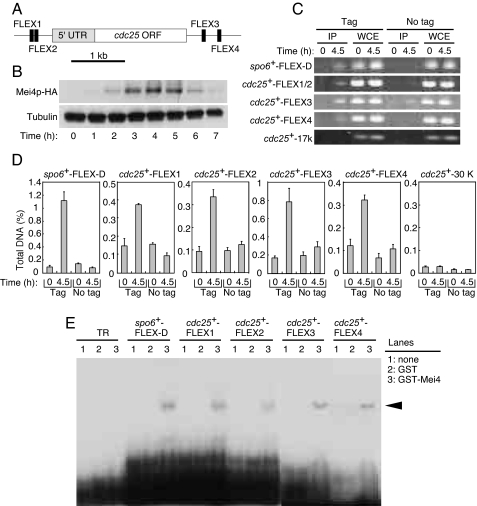
Mei4p binds 27-bp oligonucleotides containing a FLEX heptameric core (11). We searched for FLEX sequences in the genomic regions adjacent to cdc25+. Two FLEX sequences (FLEX1, FLEX2) were detected within a 1.3-kb region upstream of the start codon of cdc25+, and another two FLEX sequences (FLEX3, FLEX4) were found within a 1-kb region downstream of the stop codon of this gene (Fig. 3A).
Fig. 3.
Mei4p binds to the FLEX sequences adjacent to cdc25+ both in vivo and in vitro. (A) Schematic representation of the positions of FLEX sequences associated with cdc25+. UTR, putative untranslated region. (B) Cells expressing HA-tagged Mei4p (HM4832) were subjected to induction of meiosis as described in Fig. 1 and were subjected to immunoblot analysis with antibodies to HA at the indicated times thereafter. (C and D) A ChIP assay for FLEX sites associated with cdc25+ was performed with antibodies to HA at the indicated times after the induction of meiosis in cells expressing (tag, HM4832) or not expressing (no tag, HM1307) Mei4p-HA. The FLEX-D sequence of spo6+ was used as a positive control. cdc25+-17k or cdc25+-30k was ≈17 kb or ≈30 kb downstream of the start codon of cdc25+ and was used as a negative control. (C) After PCR, the samples were subjected to agarose gel electrophoresis. (D) Data represent the percentage of the target DNA precipitated with the antibodies and are means ± SE from at least three independent, quantitative, real-time PCR experiments. (E) EMSA analysis of cdc25+ FLEX sites with a recombinant GST-Mei4p fusion protein. Purified recombinant GST-Mei4p(71–182), GST, or buffer only (none) was incubated with the labeled oligonucleotide probes. A spo6+ FLEX-D probe was used as a positive control. The TR probe is unrelated to the FLEX sequence and was used as a negative control. Arrowheads indicate shifted bands.
We examined whether Mei4p is able to bind to the identified FLEX sequences adjacent to cdc25+ with the use of a chromatin immunoprecipitation (ChIP) assay. We first showed that the abundance of Mei4p peaks around the time of MI in both diploid (Fig. 3B) and haploid (SI Fig. 8) cells. ChIP analysis revealed that Mei4p present in cells around the onset of MI is associated with the FLEX1, FLEX2, FLEX3, and FLEX4 sites of cdc25+ and with the FLEX-D site of spo6+ (Fig. 3 C and D). The extent of such association was greatly reduced for cells in G1 phase, in which the amount of Mei4p is low. Furthermore, the association of Mei4p with a genomic region located ≈17 kb or 30 kb downstream of the start codon of cdc25+ was minimal. These results thus showed that Mei4p binds to the FLEX sites adjacent to cdc25+ in vivo at a time when the abundance of cdc25+ mRNA is high.
To determine whether Mei4p is able to bind directly to the FLEX sites adjacent to cdc25+ in vitro, we performed an EMSA with a fusion protein composed of GST and the forkhead domain of Mei4p (amino acids 71–182) (11) together with radioactive oligonucleotides containing FLEX1, FLEX2, FLEX3, or FLEX4 of cdc25+ as probes. A band shift with each of the four cdc25+ FLEX probes was observed only in the presence of the fusion protein, not in its absence or in the presence of GST alone (Fig. 3E), similar to results obtained with a spo6+ FLEX-D probe (11). A shifted band was not detected with the GST-Mei4 fusion protein and a negative control (TR) oligonucleotide (Fig. 3E). The shifted bands were specific for FLEX1, FLEX2, FLEX3, or FLEX4, given that the corresponding unlabeled oligonucleotides, but not an unrelated oligonucleotide (TR), inhibited binding of the GST-Mei4 fusion protein to the labeled probes (SI Fig. 10). These data thus suggested that Mei4p binds directly in vitro to all four FLEX elements surrounding cdc25+.
Role of cdc25+ FLEX Sites in Determination of mRNA Levels and Meiotic Progression.
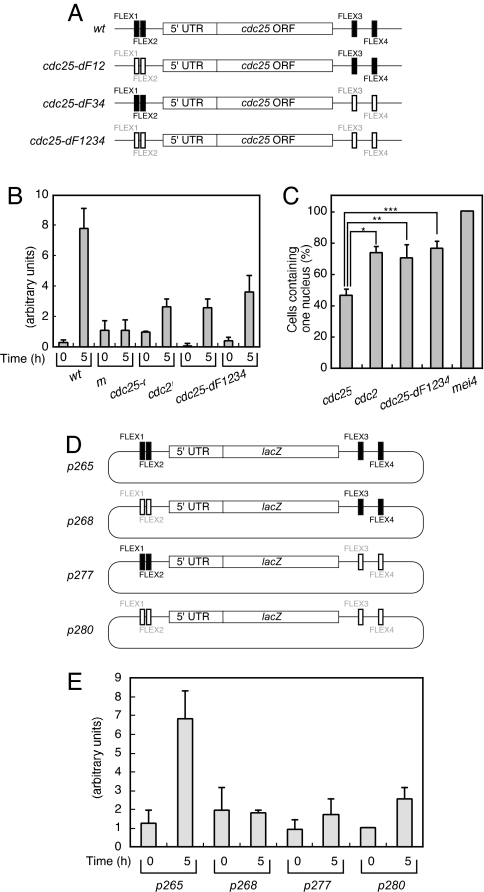
We next examined whether the FLEX sequences associated with cdc25+ function as transcriptional cis elements. We deleted the 7-bp core sequences of both the FLEX1 and FLEX2 sites of cdc25+ (Fig. 4A) and found that the amount of the corresponding mRNA was decreased in the mutant cells (cdc25-dF12) (Fig. 4B). We obtained similar results for cells in which both FLEX3 and FLEX4 (cdc25-dF34) or in which all four FLEX sites (cdc25-dF1234) of cdc25+ had been deleted (Fig. 4B). The absence of an additive effect of deletion of these four FLEX sequences suggests that they function in the same pathway. In budding yeast, transcriptional initiation and termination by RNA polymerase II share some common factors (24), and some genes form a loop structure that juxtaposes promoters and terminators (25). Mei4p might recruit general transcription factors directly when it binds to the upstream region of a gene and do so by looping or bending DNA when it binds to the downstream region.
Fig. 4.
Effects of the FLEX sites of cdc25+ on gene expression and cell cycle progression. (A) Mutation of the FLEX sequences associated with cdc25+. Shaded and open boxes represent intact and deleted FLEX elements, respectively. (B) Cells of the indicated genotypes (WT, HM1307; mei4, HM2163; cdc25-dF12, HM5969; cdc25-dF34, HM5970; and cdc25-dF1234, HM5973) were subjected to induction of meiosis as described in Fig. 1. Samples were collected at the indicated times thereafter for determination of the amount of cdc25+ mRNA by quantitative RT-PCR analysis. Data are means ± SE from at least three independent experiments. (C) Cells treated as in B were analyzed for the proportion containing one nucleus 5 h after the temperature shift. Data are means ± SE from at least three independent experiments. *, P < 0.001; **, P < 0.007; ***, P < 0.001 for the indicated comparisons. (D) Schematic representation of plasmids containing lacZ+ surrounded by genomic DNA fragments corresponding to the upstream and downstream regions of cdc25+. Shaded and open boxes represent intact and deleted FLEX elements, respectively. (E) The various plasmids shown in D were introduced into WT cells (HM5630), which were subsequently induced to undergo meiosis as described in Fig. 1. The amount of lacZ+ mRNA was measured by quantitative RT-PCR at the indicated times thereafter. Data are means ± SE from at least three independent experiments.
Consistent with the low level of induction of cdc25+ mRNA in these various mutant cells, they all showed a marked delay in entry into MI (Figs. 1 and 4C). Together, these results indicated that all four FLEX sites are required for efficient expression of cdc25+ and for timely entry of cells into and progression through meiotic nuclear division. They further suggested that the four FLEX sequences of cdc25+ serve as cis-acting elements for the Mei4p transcription factor. However, the observation that induction of cdc25+ was not completely blocked in all three mutant lines as it was in mei4 mutant cells suggests that, although the four FLEX sites play a major role in such induction by Mei4p, other genomic regions are also required for full induction of gene expression.
It is unusual for the 3′ region of a gene to affect the abundance of the corresponding mRNA in fission yeast (26). It was possible that the ORF of cdc25+ itself or genes located downstream of cdc25+ affected the level of cdc25+ mRNA. It was also possible that the cell cycle delay observed in cells with deletions of the cdc25+ FLEX sites affected the amount of cdc25+ mRNA. To exclude these possibilities, we constructed various plasmids harboring the 5′ region of cdc25+, the lacZ+ gene, and the 3′ region of cdc25+ (Fig. 4D). We introduced these plasmids into WT cells and induced the cells to undergo meiosis. Similar to the pattern observed for cdc25+ mRNA (Fig. 4B), the lacZ+ mRNA produced from a plasmid containing all four cdc25+ FLEX sites (p265) exhibited marked up-regulation 5 h after the induction of meiosis (Fig. 4E). In contrast, the level of lacZ+ mRNA remained low at this time point in cells harboring plasmids in which the FLEX1 and FLEX2 sites (p268), the FLEX3 and FLEX4 sites (p277), or all four FLEX sites (p280) had been deleted (Fig. 4E). These results are similar to those obtained for cdc25+ mRNA after mutation of the FLEX sites at the cdc25+ genomic locus (Fig. 4B), confirming that the regions immediately upstream and downstream of cdc25+ are responsible for induction of its expression. Our finding suggests that these elements function in a manner similar to that of an enhancer in higher eukaryotes (27). The level of lacZ+ mRNA increased in cells having the p280 plasmid when mei4+ was overexpressed (data not shown), suggesting that there are FLEX-like sequences in these regions that Mei4p can bind.
Our study of the role of Mei4p in control of meiotic nuclear division in fission yeast has revealed that: (i) The major target of Mei4p in such control is cdc25+. (ii) cdc25+ mRNA is up-regulated by Mei4p. (iii) FLEX sequences reside near cdc25+ and function as cis-regulatory elements for Mei4p. (iv) Mei4p is a rate-limiting factor for entry into the first meiotic nuclear division and coordinates cell cycle progression with sporulation.
We have shown that forced dephosphorylation of Cdc2p on Tyr15 in mei4 mutant cells induced the first meiotic nuclear division without any delay, indicating that the arrest phenotype of such cells is the result of inactivation of Cdc2p by Tyr15 phosphorylation. Consistent with this notion, we found that cdc25+ is an important target of Mei4p in control of MI. The genes for other proteins that affect the phosphorylation status of Cdc2-Tyr15, including mik1+ and pyp3+, are not likely to be targets for Mei4p. Mik1p is a tyrosine kinase and phosphorylates Cdc2p-Tyr15 (28). The amount of Mik1p peaks during premeiotic DNA replication and is unaffected by mei4 mutation (SI Fig. 8). Pyp3p is a tyrosine phosphatase and dephosphorylates Cdc2p-Tyr15 (29). The amount of pyp3+ mRNA decreases around the time of the first meiotic division (Sanger Center database, www.sanger.ac.uk/). In addition, no Cdc25p homologs appear to be encoded by the fission yeast genome. These observations thus suggest that Mei4p controls entry into MI mainly through regulation of cdc25+ transcription.
Mei4p is thought to be required for the induction of many middle genes, given that the expression of 436 middle genes was found to be affected in mei4 mutant cells (10). In addition, more than half of middle genes harbor FLEX sequences in the 500-bp region upstream of their coding sequence (10). However, not all Mei4p target genes are regulated predominantly at the mRNA level. The B-type cyclin Cdc13p is the product of a middle gene that contains four FLEX sequences in its upstream region (10), and the abundance of at least one species of cdc13+ mRNA during meiosis depends on Mei4p (23). However, the level of Cdc13p is maintained in mei4 mutant cells, suggesting that the abundance of this protein is regulated independently of Mei4p (20).
Why is Mei4p required specifically for meiosis? High levels of recombination and sporulation are required during meiosis but not during the mitotic cycle. Mei4p simultaneously regulates target genes that are required for cell cycle progression (cdc25+), recombination initiation (mde2+), and spore formation (spo6+). Most meiotic events thus occur in a coordinated manner. We propose that Mei4p is specifically required during meiosis because of its role in coordination between meiotic cell cycle progression and other meiotic events. Budding yeast Ndt80 is a meiosis-specific transcription factor required for the first meiotic division, recombination, and spore formation; therefore, this could be a functional counterpart of Mei4p (9). Meiotic expression of several B-type cyclin genes and many genes required for sporulation depends on Ndt80 (9). It seems generally conserved that the meiosis-specific transcription factor is required for the coordination of the meiotic cell cycle and many meiotic events.
At least 50 proteins that belong to the forkhead-box family of transcription factors have been identified in higher eukaryotes (30). FoxM1 is required for both G1–S and G2–M transitions of the cell cycle, and its target genes include those for cyclin B, Cdc25A, and Cdc25B; Aurora B, Plk1, and CENP-F, thereby ensuring proper chromosome stability and segregation during mitosis (31, 32). Mei4p is required for coordination of several meiotic events, including recombination and sporulation. Together, these observations suggest that forkhead transcription factors coordinate various cell cycle events in an evolutionarily conserved manner.
Materials and Methods
Fission Yeast Strains.
Strains used in the present study were constructed according to standard procedures (33) and are listed in SI Table 1. Synchronous meiosis was induced in liquid culture with haploid or diploid temperature-sensitive pat1 mutants as described in ref. 34. Cells were stained with 4′,6-diamidino-2-phenylindole for monitoring of meiotic progression as described in ref. 18. Transformation was performed as described in ref. 35.
Primers and Probes.
Primers and double-stranded oligonucleotides used in the present study are listed in SI Table 2.
Isolation of RNA, Northern Blot Analysis, and Quantitative RT-PCR Analysis.
Total RNA was isolated from synchronized pat1 cells as described in ref. 33, and portions (5–6 μg) of the isolated RNA were subjected to Northern blot analysis. For probes, portions of ORFs were amplified from genomic DNA by PCR with the following sets of primers: 61 and 62 for mei4+, 251 and 252 for cdc25+, 636 and 637 for spo6+, and 1455 and 1456 for cdc2+ (loading control). A portion of the ORF of lacZ+ was amplified from the BamHI–SmaI fragment of the plasmid EcolacZ-pREP3X by PCR with the primers 1334 and 1335. For quantitative real-time RT-PCR analysis, total RNA (1 μg) was incubated with DNase I (Invitrogen, Carlsbad, CA) to eliminate contaminating genomic DNA, and portions of the treated RNA (200 ng) were then subjected to RT with an oligo(dT) primer and a ThermoScript RT-PCR system (Invitrogen). The amounts of cdc25+, lacZ+, and cdc2+ (control) cDNAs were determined by the ΔΔCT method with the use of an ABI prism 7700 instrument and the primers 1470 and 1471, 1477 and 1478, and 1451 and 1452, respectively.
Immunoblot Analysis.
Whole cell extracts (20 μg of protein) were prepared by the “boiling method” (18) and analyzed as described in refs. 18 and 36.
ChIP.
ChIP and quantitative real-time PCR analysis were performed as described in ref. 37. Portions of the precipitated DNA and DNA fragments isolated from 0.4 mg of whole cell extract were subjected to agarose electrophoresis with the following primers: 726 and 727, spo6+; 725 and 737, cdc25+-FLEX1/2; 947 and 948, cdc25+-FLEX3; 949 and 950, cdc25+-FLEX4; 842 and 843, cdc25+-17k or quantitative real-time PCR analysis with the following primers: 1205 and 1206, cdc25+-FLEX1; 1207 and 1208, cdc25+-FLEX2; 1242 and 1243, cdc25+-FLEX3; 1211 and 1212, cdc25+-FLEX4; 1236 and 1237, cdc25+-30k.
EMSA Analysis.
EMSA analysis was performed as described in ref. 38. The purified GST-Mei4 (71–182) fusion protein (400 ng) was prepared according to Horie et al. (11). The double-stranded oligonucleotides used as probes were as follows: TR, 742 and 743; spo6+-FLEX, 740 and 741; cdc25+-FLEX1, 744 and 745; cdc25+-FLEX2, 746 and 747; cdc25+-FLEX3, 943 and 944; cdc25+-FLEX4, 945 and 946.
Deletion of cdc25+ FLEX Elements from the Chromosome.
Two-step sequential gene replacement was performed to obtain a strain of cells congenic with WT cells with the exception of two 7-bp deletions of the core FLEX1 and FLEX2 sequences immediately upstream of cdc25+. First, the chromosomal region containing FLEX1 and FLEX2 of cdc25+ was replaced with a PCR-generated DNA fragment containing the corresponding region interrupted by ura4+ sequences. The resulting strain was subjected to a second gene replacement with a PCR fragment containing two 7-bp deletions of FLEX1 and FLEX2 of cdc25+, and selection was performed with 5-fluoroorotic acid. Genomic PCR analysis was performed at each step to confirm the correct chromosomal gene replacement. The resulting strain was crossed with various other strains to obtain HM5969. Similar methods were used to generate strains HM5970 (cdc25-dF34) and HM5973 (cdc25-dF1234).
Construction of FLEX-lacZ Fusion Plasmids.
Relevant features of the FLEX-lacZ fusion plasmids are described in Fig. 4. In brief, the plasmids contain ars1, LEU2, LacZ, and various FLEX mutants. Exact details are available upon request.
Statistics.
Data are presented as means ± SE and were compared between groups by Student's t test. A P value of <0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank C. Shimoda (Osaka City University, Osaka, Japan) and P. Nurse (The Rockefeller University) for providing yeast strains, J. Kano for the protocols for ChIP and EMSA, and S. Mita for technical assistance. This work was supported in part by grants-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to Y.M.-T. and H.M.).
Abbreviation
- MI
meiosis I.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702906104/DC1.
References
- 1.Roeder GS. Genes Dev. 1997;11:2600–2621. doi: 10.1101/gad.11.20.2600. [DOI] [PubMed] [Google Scholar]
- 2.Davis L, Smith GR. Proc Natl Acad Sci USA. 2001;98:8395–8402. doi: 10.1073/pnas.121005598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Padmore R, Cao L, Kleckner N. Cell. 1991;66:1239–1256. doi: 10.1016/0092-8674(91)90046-2. [DOI] [PubMed] [Google Scholar]
- 4.Smith KN, Penkner A, Ohta K, Nicols A. Curr Biol. 2001;11:88–97. doi: 10.1016/s0960-9822(01)00026-4. [DOI] [PubMed] [Google Scholar]
- 5.Borde V, Goldman AS, Lichten M. Science. 2000;290:806–809. doi: 10.1126/science.290.5492.806. [DOI] [PubMed] [Google Scholar]
- 6.Cervantes MD, Farah JA, Smith GR. Mol Cell. 2000;5:883–888. doi: 10.1016/s1097-2765(00)80328-7. [DOI] [PubMed] [Google Scholar]
- 7.Lee B, Amon A. Curr Opin Cell Biol. 2001;13:770–777. doi: 10.1016/s0955-0674(00)00282-9. [DOI] [PubMed] [Google Scholar]
- 8.Primig M, Williams RM, Winzeler EA, Tevzadze GG, Conway AR, Hwang SY, Davis RW, Esposito RE. Nat Genet. 2000;26:415–423. doi: 10.1038/82539. [DOI] [PubMed] [Google Scholar]
- 9.Chu S, Herskowitz I. Mol Cell. 1998;1:685–696. doi: 10.1016/s1097-2765(00)80068-4. [DOI] [PubMed] [Google Scholar]
- 10.Mata J, Lyne R, Burns G, Bahler J. Nat Genet. 2002;32:143–147. doi: 10.1038/ng951. [DOI] [PubMed] [Google Scholar]
- 11.Horie S, Watanabe Y, Tanaka K, Nishiwaki S, Fujioka H, Abe H, Yamamoto M, Shimoda C. Mol Cell Biol. 1998;18:2118–2129. doi: 10.1128/mcb.18.4.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bresch C, Muller G, Egel R. Mol Gen Genet. 1968;102:301–306. doi: 10.1007/BF00433721. [DOI] [PubMed] [Google Scholar]
- 13.Abe H, Shimoda C. Genetics. 2000;154:1497–1508. doi: 10.1093/genetics/154.4.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gregan J, Rabitsch PK, Sakem B, Csutak O, Latypov V, Lehmann E, Kohli J, Nasmyth K. Curr Biol. 2005;15:1663–1669. doi: 10.1016/j.cub.2005.07.059. [DOI] [PubMed] [Google Scholar]
- 15.Young JA, Hyppa RW, Smith GR. Genetics. 2004;167:593–605. doi: 10.1534/genetics.103.023762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacNeill SA, Nurse P. In: The Molecular Cell Biology of the Yeast Saccharomyces: Life Cycle and Cell Biology. Pringle JR, Broach JR, Jones EW, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab; 1997. pp. 697–763. [Google Scholar]
- 17.Nurse P. Nature. 1990;344:503–508. doi: 10.1038/344503a0. [DOI] [PubMed] [Google Scholar]
- 18.Murakami H, Nurse P. Genes Dev. 1999;13:2581–2593. doi: 10.1101/gad.13.19.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murakami H, Nurse P. Biochem J. 2000;349:1–12. doi: 10.1042/0264-6021:3490001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borgne A, Murakami H, Ayte J, Nurse P. Mol Biol Cell. 2002;13:2080–2090. doi: 10.1091/mbc.01-10-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daya Makin M, Szankasi P, Tang L, MacRae D, Pelech SL. Biochem Cell Biol. 1992;70:1088–1096. doi: 10.1139/o92-154. [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto M, Imai Y, Watanabe Y. In: The Molecular Cell Biology of the Yeast Saccharomyces: Life Cycle and Cell Biology. Pringle JR, Broach JR, Jones EW, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab; 1997. pp. 1037–1106. [Google Scholar]
- 23.Iino Y, Hiramine Y, Yamamoto M. Genetics. 1995;140:1235–1245. doi: 10.1093/genetics/140.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calvo O, Manley JL. Genes Dev. 2003;17:1321–1327. doi: 10.1101/gad.1093603. [DOI] [PubMed] [Google Scholar]
- 25.O'Sullivan JM, Tan-Wong SM, Morillon A, Lee B, Coles J, Mellor J, Proudfoot NJ. Nat Genet. 2004;36:1014–1018. doi: 10.1038/ng1411. [DOI] [PubMed] [Google Scholar]
- 26.Daga RR, Bolanos P, Moreno S. Curr Biol. 2003;23:2015–2024. doi: 10.1016/j.cub.2003.10.061. [DOI] [PubMed] [Google Scholar]
- 27.Kadonaga JT. Cell. 2004;116:247–257. doi: 10.1016/s0092-8674(03)01078-x. [DOI] [PubMed] [Google Scholar]
- 28.Lundgren K, Walworth N, Booher R, Dembski M, Kirschner M, Beach D. Cell. 1991;64:1111–1122. doi: 10.1016/0092-8674(91)90266-2. [DOI] [PubMed] [Google Scholar]
- 29.Millar JB, Lenaers G, Russell P. EMBO J. 1992;11:4933–4941. doi: 10.1002/j.1460-2075.1992.tb05600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Costa RH. Nat Cell Biol. 2005;7:108–110. doi: 10.1038/ncb0205-108. [DOI] [PubMed] [Google Scholar]
- 31.Wang IC, Chen YJ, Hughes D, Petrovic V, Major ML, Park HJ, Tan Y, Ackerson T, Costa RH. Mol Cell Biol. 2005;25:10875–10894. doi: 10.1128/MCB.25.24.10875-10894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laoukili J, Kooistra MR, Bras A, Kauw J, Kerkhoven RM, Morrison A, Clevers H, Medema RH. Nat Cell Biol. 2005;7:126–136. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- 33.Moreno S, Klar A, Nurse P. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 34.Tonami Y, Murakami H, Shirahige K, Nakanishi M. Proc Natl Acad Sci USA. 2005;102:5797–5801. doi: 10.1073/pnas.0407236102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okazaki K, Okazaki N, Kume K, Jinno S, Tanaka K, Okayama H. Nucleic Acids Res. 1990;18:6485–6489. doi: 10.1093/nar/18.22.6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sugimoto I, Murakami H, Tonami Y, Moriyama A, Nakanishi M. J Biol Chem. 2004;279:47372–47378. doi: 10.1074/jbc.M403231200. [DOI] [PubMed] [Google Scholar]
- 37.Kanoh J, Sadaie M, Urano T, Ishikawa F. Curr Biol. 2005;15:1808–1819. doi: 10.1016/j.cub.2005.09.041. [DOI] [PubMed] [Google Scholar]
- 38.Kanoh J, Watanabe Y, Ohsugi M, Iino Y, Yamamoto M. Genes Cells. 1996;1:391–408. doi: 10.1046/j.1365-2443.1996.d01-247.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.