Abstract
Organelles are frequently distributed in a nonrandom manner in a cell's cytoplasm. A particular distribution pattern often facilitates a specific function of a cell, whereas its aberrations can lead to cell death. We show that a mutation in the zebrafish mikre oko (mok) locus, which encodes dynactin 1 subunit of the dynactin complex, produces a severe displacement of the photoreceptor cell nucleus toward the synaptic terminus. Interference with the function of other dynein complex constituents, including p50/dynamitin, the Lis1 polypeptide, and the disruption of a nuclear envelope component of the syne gene family in vertebrate photoreceptors also result in the mispositioning of nuclei. Although the overall photoreceptor polarity is not affected, this phenotype is accompanied by a misdistribution of the Bardet–Biedl syndrome 4 polypeptide and a decreased photoreceptor survival. These findings reveal an important mechanism that regulates nuclear position in vertebrate neurons.
Keywords: Bardet–Biedl syndrome, retina, motor complex, neurodegeneration
Cell nuclei are precisely positioned in many tissues. In the vertebrate retina and brain, for example, cell nuclei occupy well defined strata, frequently separated from each other by layers of neuronal processes. A mislocalization of nuclei in the context of the vertebrate CNS is likely to result in profound patterning defects and severely compromised function. Genetic defects of nuclear positioning have been characterized in fungi, Caenorhabditis elegans, and Drosophila. In contrast to invertebrates, nuclear positioning in vertebrate phyla remains poorly characterized. To fill this gap, here we provide genetic analysis of the mechanism that regulates nuclear position in vertebrate photoreceptor cells. To our knowledge, this is the first study of a nuclear localization mechanism in the vertebrate eye and one of very few attempts to understand this process and its significance in the context of the vertebrate nervous system.
Results
mok Mutation Causes a Severe Basal Displacement of Nuclei.
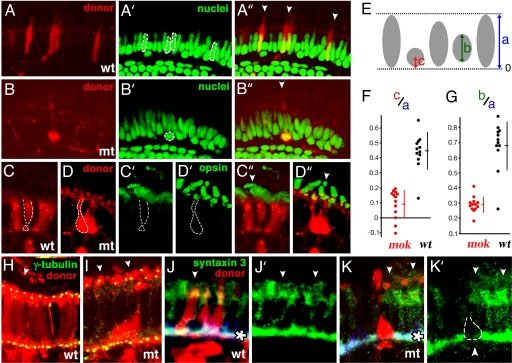
A mutation in the mikre oko (mok) locus results in a rapid loss of nearly all photoreceptor cells by 5 days postfertilization (dpf), one of the most severe photoreceptor defects described in zebrafish so far (1, 2). We showed that in mosaic retinae, mok−/− cells surrounded by wild-type tissue survive markedly longer and develop robust outer segments (2). Photoreceptor morphology is not completely normal, however. Mutant cells retain elongated appearance, but their width is extremely reduced in the central portion of the apico-basal axis, a region normally occupied by the nucleus [Fig. 1B (compare with wild-type in Fig. 1A)]. This shape change suggests that photoreceptor nuclei are mislocalized. As mokm632 photoreceptors die very rapidly during differentiation in homozygous mutant embryos, their phenotype has to be studied in mosaic animals. To determine where nuclei localize in mokm632 mutant cells, we visualized them in mosaic retinae, using a nuclear stain (Fig. 1) and evaluated their positions relative to the basal margin of neighboring nuclei (Fig. 1 A–A″ and B–B″, illustrated schematically in Fig. 1E). Indeed, mokm632 nuclei are severely displaced basally, compared with wild-type ones (Fig. 1F, P < 0.00000007), and, in the most extreme cases, they translocate into the outer plexiform layer, which in the normal retina consists exclusively of neuronal processes and synaptic termini. Nuclei in another zebrafish photoreceptor mutant ovltz288b (3) are not mispositioned, indicating that this phenotype is not a general consequence of photoreceptor degeneration [supporting information (SI) Fig. 5 A–C]. In addition, mokm632 photoreceptor nuclei display a markedly different shape. Although the length of wild-type nuclei along their apico-basal axis equals ≈70% of photoreceptor cell layer thickness, the length of mutant photoreceptor nuclei in the same dimension is only 30% of photoreceptor cell layer thickness [Fig. 1B (compare with Fig. 1A)]; quantitative estimate in Fig. 1G, P < 0.000002). The width to length ratio of nuclei is also severely affected by the mokm632 defect and equals 0.33 in the wild-type (16 measurements) versus 1.19 in the mutant (10 measurements, P < 0.0000001). These observations indicate that mutant nuclei are markedly rounder, compared with wild-type ones.
Fig. 1.
Nuclear positioning defect in mok mutant photoreceptors. All experiments were performed in genetically mosaic retinae. The genotypes of donor-derived cells are indicated in lower right corners. (A and B) Somata of photoreceptors derived from wild-type (A) or mutant (B) donors are visualized in red. (A′ and B′) The same sections as in A and B, stained with YoPro to visualize nuclei. (A″ and B″) Red and green channels shown to the left superimposed. In A′ and B′, the nuclei of donor-derived cells are outlined with white dashes. (C and D) In contrast to wild-type cells, mok photoreceptor nuclei are rounder and displaced basally. Somata of donor-derived wild-type (C) and mutant (D) photoreceptors surrounded by wild-type cells in mosaic retinae. (C′ and D′) The same sections as in C and D stained for rod and blue opsins. (C″ and D″) Green and red channels shown to the left superimposed. In wild-type and mutant photoreceptors alike, opsins are confined to outer segments. (E) To describe the position of nuclei, we used the following parameters: (i) the distance between apical and basal margins of the photoreceptor cell layer; (ii) the length of nuclei along the apico-basal axis; and (iii) the distance between the center of a nucleus and the basal margin of the photoreceptor cell layer. (F) As revealed by their c/a ratios, mok nuclei are displaced basally, compared with those in wild-type cells. (G) As indicated by the b/a ratios, mok nuclei are rounder, compared with those in the wild type. In F and G, each dot represents a single cell nucleus. Vertical bars and short horizontal cross bars indicate standard deviations and averages, respectively. (H and I) γ-tubulin staining of basal bodies (yellow) in mosaic retinae, containing wild-type (H) and mutant (I) donor-derived photoreceptors (in red). In wild-type and mutant donor-derived photoreceptor cells alike, we do not observe basal bodies away from apical cell termini. (J and K) Syntaxin 3 staining (green) in mosaic retinae containing wild-type (J) or mutant (K) donor-derived photoreceptor cells (red). (J′ and K′) Syntaxin staining alone. The presence of mutant but not wild-type photoreceptors correlates with discontinuous syntaxin staining (arrowhead in K′). In all images, apical termini of photoreceptors and retinal pigmented epithelium are up (indicated with arrowheads). White asterisks in J and K indicate the outer plexiform layer.
The correct position of the nucleus on the apico-basal axis of the cell is a major feature of polarity. Does mok gene function in other aspects of apico-basal polarity? One conspicuous component of polarity in photoreceptors is the unidirectional transport of billions of opsin polypeptides from the soma into the outer segment (4). An uninterrupted occurrence of this process is a prerequisite for photoreceptor health (3). To investigate whether mokm632 mutation affects opsin transport, we stained mosaic retinae with a mix of anti-rod opsin and anti-green opsin antibodies, which recognize ≈40% of photoreceptors. In wild-type environment of mosaic retinae, wild-type photoreceptors (n = 21) do not accumulate opsins outside their outer segments (Fig. 1 C–C″). Similarly, in mokm632 homozygous photoreceptors (n = 26), visual pigments localize to outer segments in a manner indistinguishable from wild-type cells, indicating that the mechanism of visual pigment transport is not affected [Fig. 1 D–D″ (compare with wild type in Fig. 1 C–C″)]. To further evaluate whether mokm632 outer segments differentiate correctly, we visualized basal bodies of connecting cilia, using anti-γ-tubulin antibodies. Photoreceptor cilia are positioned apically, connecting the outer segment to the rest of the photoreceptor body. We did not find ectopically localized basal bodies in mokm632 photoreceptor cells [Fig. 1I, n = 13 (compare with wild-type cells in 1H, n = 24)]. These data indicate that in contrast to nuclear positioning, other key aspects of cell polarity, outer segment formation in particular, are normal in mok mutants.
A misplacement of mokm632 photoreceptor nuclei may affect the differentiation of synaptic termini, which normally form basal to nuclei. To investigate whether this is the case, we stained mosaic retinae with antibodies against syntaxin 3, a component of the photoreceptor synaptic apparatus (Fig. 1 J and K). In control retinae, syntaxin 3 staining remains uninterrupted in the vicinity of donor-derived wild-type photoreceptor cells (Fig. 1 J and J′, n = 11). By contrast, syntaxin 3 distribution is frequently disrupted in the vicinity of mutant cells (Fig. 1 K and K′, six of eight cells examined), and we do not observe the characteristic shape of wild-type pedicles in mutant photoreceptors. These results indicate that synaptic differentiation is adversely affected in mutant cells, possibly as a result of nuclear displacement. Synaptic defects may contribute to photoreceptor degeneration in mok mutants, although it has to be noted that even severe synaptic defects in Cacna1f mutant mice do not cause a marked reduction of photoreceptor viability (5).
mok Gene Encodes a Motor Complex Component.
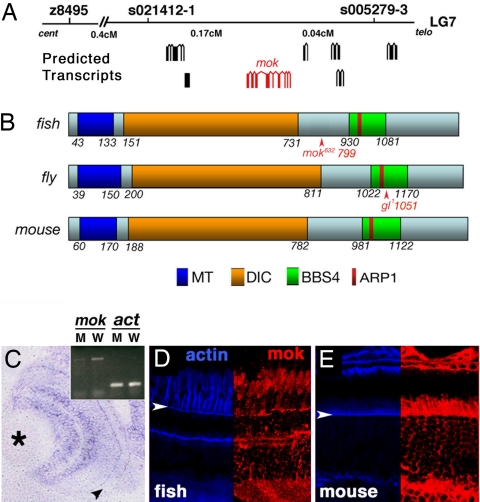
To determine the molecular nature of the mok gene product, we performed positional cloning. Conventional linkage analysis confined mok to an interval of ≈0.21 cM in the telomeric region of LG7 (Fig. 2A). This region contains at least six predicted transcripts, one of which encodes a homolog of the dynactin 1 subunit of the dynein motor complex, also known as p150. To search for mutations in this gene, we cloned its coding region by RT-PCR and RACE. The ORF of this transcript consists of 1,218 aa and is highly homologous both to the mouse dynactin 1 (64% identity, 74% similarity) and to its Drosophila homolog, glued (31% identity, 49% similarity). Both fly and vertebrate proteins contain a microtubule binding domain, a dynein intermediate chain binding domain, and a Bardet–Biedl syndrome (BBS)4 binding domain, which includes a putative ARP-1 binding site (amino acids 955–970; Fig. 2B and SI Text) (6, 7). mok is one of two dynactin 1 paralogs present in the zebrafish genome. The function of the second gene (SI Text), which we term mok2, remains at present unknown. Sequence analysis revealed that the mokm632 allele contains a nucleotide 2395C → T transition in the dynactin 1 gene, which results in the appearance of a premature stop codon at the position 799 of the polypeptide. The mutant transcript is expressed at a much lower level, most likely because of nonsense-mediated decay (Fig. 2C Inset). To confirm that this mutation is responsible for the mokm632 phenotype, we injected anti-dynactin 1 morpholinos into wild-type embryos. Approximately 60% (7 of 11, SI Fig. 6A) of dynactin 1 morpholino-injected fish display photoreceptor loss at 3 dpf, whereas retinae of control morpholino-treated individuals retain the wild-type phenotype (18 of 18, SI Fig. 6B). In parallel, we rescued the mokm632 defect by injecting dynactin 1 mRNA. Mutants treated with GFP mRNA display severe photoreceptor loss, characteristic of mokm632 homozygotes (16 of 16, SI Fig. 6C). In contrast, the injection of full-length mok mRNA rescues photoreceptor loss in 60% (9 of 15, SI Fig. 6D) of mutant animals. The injection of either GFP or dynactin 1 mRNA into wild-type embryos does not result in photoreceptor defects (9 of 9 for each group, data not shown). Based on these results, we conclude that the zebrafish mikre oko locus encodes a dynactin 1 homolog. Although the mokm632 allele is predicted to encode a truncated polypeptide, we did not detect a shortened protein product on Western blots of extracts from homozygous mutant larvae or from adult heterozygotes (data not shown). The Mok polypeptide does not appear to be deposited maternally in a large quantity, because no obvious signal is present on Western blots from embryos at 2 h postfertilization (hpf) (data not shown). Consistent with its mutant phenotype, mok transcript is expressed in the eye, including the photoreceptor cell layer, at 3 dpf (Fig. 2C). Staining of zebrafish and mouse retinae with three different antibodies suggests that although present in the entire photoreceptor cytoplasm, the Mok polypeptide is enriched in the vicinity of cell junctions and in the inner segment (Fig. 2 D and E and data not shown). Staining of outer segments is either weak or absent.
Fig. 2.
Molecular characterization of the mikre oko (mok) gene. (A) Positional cloning of mok. (B) A schematic diagram of the zebrafish Mok polypeptide in comparison with its Drosophila and mouse homologs. In all three species, Mok polypeptide includes domains thought to bind microtubules (MT), dynein intermediate chain (DIC), actin-related protein 1 (ARP1), and BBS protein 4 (BBS4). In both the zebrafish and the fly, mutant alleles produce C-terminal truncations of the polypeptide (arrowheads). (C) A transverse section through the retina showing mok transcript distribution as evaluated by in situ hybridization. Arrowhead indicates the photoreceptor cell layer, and the asterisk indicates the lens. (Inset) RT-PCR reveals that the mok mRNA level is severely reduced in mokm632 mutant homozygotes, compared with their wild-type siblings. β-actin level (act) is unchanged. (D and E) Mok polypeptide distribution in zebrafish (D) and mouse (E) retinae. (Right) Anti-Mok antibody staining (red). (Left) F-actin distribution (blue). Staining is particularly prominent apical to the outer limiting membrane (arrowheads). In D and E, retinal pigmented epithelium is up.
Dynein Complex Regulates Nuclear Position in Vertebrate Photoreceptors.
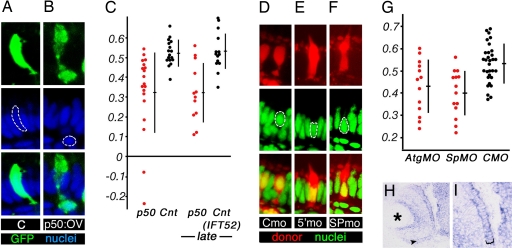
The Mikre oko polypeptide is a component of the dynactin complex, which consists of at least eight proteins and functions as a dynein regulatory subunit (8). The Mok involvement in nuclear positioning could be mediated either via the dynactin complex or by an independent mechanism. In the first case, the disruption of dynactin function by other means would also result in nuclear mispositioning. To test whether or not this is the case, we cloned another dynactin component, the zebrafish p50/dynamitin gene. In several experimental systems, the overexpression of p50 causes a dissociation of the dynactin complex and consequently blocks its function (9, 10). We took advantage of this observation and overexpressed p50/dynamitin in zebrafish photoreceptors by introducing a heat-shock-inducible DNA construct into zebrafish embryos and inducing its expression during photoreceptor differentiation at 2–3 dpf. Similar to mokm632 mutation, p50/dynamitin overexpression, but not the treatment with an empty vector, results in the mispositioning of photoreceptor nuclei, which in the most extreme cases translocate into the outer plexiform layer [Fig. 3B (compare with Fig. 3A, P < 0.0003)]. The zebrafish photoreceptors are well differentiated and functional by 3 dpf. To determine whether the dynein/dynactin complex maintains nuclear position in morphologically mature cells, we applied a series of heat shocks between 4.5 and 5.5 dpf. These tests also produced an obvious basal displacement of nuclei (P < 0.0004, Fig. 3C). These observations, together with the analysis of the mok phenotype, provide strong evidence that the dynactin complex regulates nuclear position in the vertebrate eye.
Fig. 3.
Dynein complex function in nuclear positioning. (A and B) Mosaic overexpression of a control vector (A) or p50 (B). Overexpressing cells are marked with GFP (green). In the most extreme cases (B), photoreceptor nuclei (blue) are displaced into the outer plexiform layer. (C) The distribution of nuclear positions in cells expressing p50 vector (red dots), a control empty vector (black dots, center), or a control ift52 overexpression vector (black dots, rightmost graph). (D–F) YoPro staining of cell nuclei (green) in genetically mosaic retinae, containing cells (red) derived from donor embryos treated with a control morpholino (D), anti-lis1a ATG (5′) morpholino (E), or anti-lis1a splice site (SP) morpholino (F). (G) Both ATG and SP morpholinos produce a statistically significant basal displacement of photoreceptor nuclei. (H and I) In situ hybridization result showing that lis1a is expressed in wild-type photoreceptors. (A, B, and D–F) (Top) Tracer labeling of cell somata in which nuclear positioning was evaluated (red for dextran tracer, green for GFP). (Middle) Nuclear staining (blue for ToPro-3, green for YoPro). (Bottom) Superimposed images from above; apical is up. (H and I) Arrowhead (H) and bracket (I) indicate the photoreceptor cell layer. Graphs in C and G are plotted as described in Fig. 1 legend. Asterisk indicates the lens.
Because of its central role in visual perception and in neurodegenerative disorders of the eye, the vertebrate photoreceptor cell has been the focus of intensive research. Despite these efforts, many aspects of photoreceptor cell biology remain poorly investigated. We decided to investigate further what genes in addition to mok and p50 may be involved in the positioning of photoreceptor nuclei. The lissencephaly-1 (Lis1) protein is a dynein motor cofactor known for its involvement in human cerebral cortex disorders (11–13). We cloned two zebrafish Lis1 homologs, lis1a and lis1b, which display 93% identity and 96% similarity to each other and are highly homologous to the mouse polypeptide (93% identity and 97% similarity for Lis1a). In situ hybridization shows that lis1a is enriched in photoreceptor cells (Fig. 3 H and I). To test its function, we performed antisense morpholino knockdown. A lis1a splice site-directed morpholino nearly completely eliminates the wild-type transcript until at least 48 hpf (SI Fig. 7A and SI Text). Because in both Drosophila and the mouse, loss-of-function mutations in Dlis-1/Lis1 result in embryonic lethality (12, 14), to ameliorate its early embryonic phenotype, we studied lis1a function in mosaic animals. We transplanted blastomeres from anti-lis1a morpholino-treated donor embryos into untreated host blastulae and evaluated nuclear position in donor-derived photoreceptor cells. Treatment with two different anti-lis1a antisense oligonucleotides but not with a control morpholino results in a shift of photoreceptor nuclei to more basal positions (Fig. 3 D–G; P < 0.0002 for SP morpholino; P < 0.008 for ATG). To confirm the validity of these results, we overexpressed the N-terminal region (amino acids 1–87) of Lis1a in photoreceptor cells. We refer to this polypeptide as N-lis1. This protein fragment is known to interfere with Lis1a dimerization, mediated by its N-terminal coiled-coil motif (11, 15). In agreement with morpholino knockdown data, the overexpression of Lis1a N-terminal region produces a basal displacement of photoreceptor nuclei [SI Fig. 5E (compare with SI Fig. 5D and graph in SI Fig. 5F, P < 0.00002)]. These results indicate that one or both lis1 genes regulate nuclear position in photoreceptor cells.
Positioning of Cell Nucleus Requires Nuclear Envelope Components.
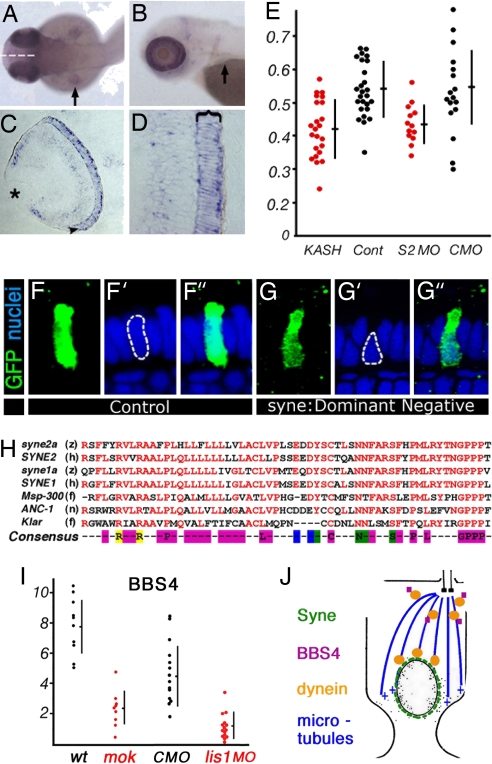
In addition to motor machinery, the regulation of nuclear position must involve factors that anchor the nucleus to the motor apparatus. Several nuclear envelope components are required for the correct localization of nuclei in flies and nematodes. KASH-domain proteins in particular, including ANC-1, MSP-300, and Klarsicht, have been shown to control nuclear position in several tissues (reviewed in ref. 16). The KASH domain is thought to anchor polypeptides to the nuclear envelope, and thus it is likely that KASH family proteins mediate the attachment of nuclei to the cytoskeleton (reviewed in ref. 17). To investigate the role of KASH-domain proteins in vertebrate photoreceptors, we cloned partial sequences of four zebrafish Syne genes: syne1a, syne1b, syne2a, and syne2b. Interestingly, the expression of syne2a is highly enriched in the eye (Fig. 4 A and B), particularly in photoreceptor cells (Fig. 4 C and D). The overexpression of the C-terminal portion of Syne/ANC-1 proteins, a region highly conserved between vertebrate and invertebrate Syne/ANC-1 genes (Fig. 4H), is thought to act in a dominant negative manner (18, 19). To test the function of zebrafish syne2a in nuclear positioning, we overexpressed its KASH domain (69 C-terminal amino acids, we refer to this polypeptide as C-Syne2a) using a heat-shock promoter. This treatment produces a basal displacement of photoreceptor nuclei [Fig. 4 G–G″ (compare with Fig. 4 F–F″); graph in Fig. 4E, P < 0.000008)]. To verify this result, we performed a knockdown of syne2a function, using a splice site-directed morpholino. This reagent eliminates the wild-type syne2a transcript nearly completely until at least 2 dpf (SI Fig. 7B and SI Text). Again in this case, photoreceptor phenotype was analyzed in mosaic animals. syne2a knockdown also causes a basal displacement of nuclei (Fig. 4E, P < 0.0098). These results indicate that syne family proteins function in the positioning of vertebrate photoreceptor nuclei, possibly by providing a mechanical link to motor complexes.
Fig. 4.
The function of syne2a in nuclear positioning. (A and B) Dorsal (A) and lateral (B) views of PTU-treated zebrafish embryos at 50 and 72 hpf, respectively. Arrows indicate the pectoral fin. syne2a expression is strongly elevated in the eye. (C and D) Transverse sections through the zebrafish retina showing syne2a transcript expression detected by in situ hybridization. The photoreceptor cell layer is indicated with an arrowhead in C and with a bracket in D. (E) The distribution of nuclear positions in cells expressing dominant negative syne2a vector (KASH, red dots) or a control empty vector (Cont, black dots) (left side of the graph) and cells treated with an anti-syne2a morpholino (S2, red dots) or a control morpholino (CMO, black dots) (right side of the graph). The positions of nuclei are calculated as defined in Fig. 1. (F–F″ and G–G″) Examples of data used to plot the distribution of nuclear positions in E in cells overexpressing a control vector (F) or a dominant negative syne2a (G). (F and G) GFP staining of cells in which nuclear positioning was evaluated. (F′ and G′) Nuclear staining (ToPro-3). (F″ and G″) Superimposed images; apical is up. (H) The alignment of KASH domains from two zebrafish syne genes to human sequences, and to sequences of invertebrate genes: Msp-300, ANC-1, and Klarsicht. Conserved residues are in red. The consensus sequence is color-coded as follows: magenta, nonpolar amino acids; blue, acidic; green, polar; yellow, basic. Species designations are as follows: z, zebrafish; h, human; f, fly; n, nematode. (I) The ratio of BBS4-positive particles to the number of cell somata in wild-type and mok photoreceptors (right side of the graph). The ratio of BBS4-positive particles to the number of basal bodies in lis1a morphants, and in control morpholino (CMO)-treated animals (left side of the graph). Each dot represents a separate retina. (J) A model of the mechanism that positions nuclei in vertebrate photoreceptors. The interaction of dynein with Syne polypeptide is hypothetical.
Nuclear Mispositioning Is Accompanied by a Decreased Photoreceptor Survival.
Because many abnormalities of the outer segment structure and function lead to a photoreceptor loss (20), we hypothesized that nuclear positioning defects will have a similar outcome. To test whether this is the case, we interfered with the functions of either the dynein motor or nuclear envelope by overexpressing p50, N-lis1, and C-Syne2a polypeptides. Transposon-based Tol2 vector was used to produce expression in a large number of retinal cells, and the use of a heat-shock promoter allowed us to control the timing of expression (SI Fig. 8) (21). To test whether the mispositioning of photoreceptor nuclei is associated with a decreased survival, we induced the expression of p50, N-lis1, and C-Syne2a by heat-shocking larvae either shortly after photoreceptor formation at 48, 60, and 72 hpf, or after photoreceptor cells are fully differentiated and functional at 120, 132, and 144 hpf. In the first case, photoreceptor phenotype was analyzed at 4 and 6 dpf, and in the second case, photoreceptor phenotype was analyzed at 6, 7, and 8 dpf. In all cases, the interference with dynein complex function or syne2a resulted in a decreased percentage of surviving photoreceptor cells. The result was statistically significant for all N-lis1 and C-Syne2a overexpression tests and in two tests involving p50 (SI Fig. 8F). In contrast to photoreceptors, the percentage of ganglion cell layer neurons relative to the number of inner nuclear layer cells did not display consistent changes. The overexpression of each of three constructs resulted in a basal displacement of photoreceptor nuclei at 8 dpf (SI Fig. 8G) but did not affect the localization of synaptic markers, PMCA and syntaxin 3 (data not shown). Cell clone sizes were not obviously affected by overexpression experiments, as compared with control overexpression of GFP (SI Fig. 8F). These results indicate that the mispositioning of photoreceptor cell nuclei is accompanied by a decreased viability of photoreceptor cells.
Dynein Motor Functions in Apical Localization of BBS4.
Mosaic analysis revealed that in contrast to nuclear position, several key aspects of apico-basal polarity remain intact in mok mutant cells (Fig. 1). To test whether nonautonomous aspects of mok function regulate photoreceptor polarity and may be the primary cause of cell degeneration, we stained mok mutant retinae with antibodies that recognize cell junction components, ZO-1, cortactin, and β-catenin, as well as with antibodies directed to apical membrane determinants: Crumbs, Nok, and Has (ref. 22 and references therein). During the initial stages of degeneration, mok mutant photoreceptors retain their normal elongated shape. None of the proteins tested appear to be mislocalized in mutant photoreceptors at this stage [SI Fig. 9D (compare with SI Fig. 9C) and data not shown]. The distribution of mitochondria, which localize apically in wild-type cells, also remains largely normal [SI Fig. 9B (compare with SI Fig. 9A)]. Finally, the localization of two synaptic markers, PMCA and SV2, is unaffected by the mokm632 phenotype (data not shown), suggesting that at later stages synaptic defects in mosaic retinae (Fig. 1 J and K) may be secondary to nuclear displacement.
In contrast to the above findings, the BBS4 polypeptide does not properly localize in mok mutant photoreceptors at stages preceding the loss of elongated morphology. BBS4 is a pericentriolar protein involved in the BBS, a human disorder characterized by photoreceptor loss, and consequently blindness (recently reviewed in ref. 23). In wild-type animals, anti-BBS4 antibodies detect small particles in the vicinity of photoreceptor cilia (SI Fig. 9 E and G). This staining is severely reduced in mok mutant photoreceptors, before their loss of elongated morphology (SI Fig. 9F). Blocking of lis1a function, using a low dose of morpholino oligonucleotides, also results in the loss of BBS4 staining from the apical termini of photoreceptors [SI Fig. 9H (compare with SI Fig. 9G; graph in Fig. 4I, P < 0.000005)]. These data indicate that in addition to its role in nuclear positioning, the mok gene, and most likely the entire dynein motor complex, functions in the apical localization of at least some pericentriolar proteins. Mutations in this mechanism may be responsible for some forms of BBS.
Discussion
The vertebrate photoreceptor is one of the best-studied cells of the nervous system. Yet mechanisms that regulate its morphogenesis are poorly understood. Based on genetic evidence, here we propose a model of a mechanism that determines a key feature of photoreceptor morphology, nuclear position. This model stipulates that nuclear position in vertebrate photoreceptor cells is regulated by the dynein motor complex, which interacts directly or indirectly with nuclear envelope components (Fig. 4J). Importantly, our data suggest that the proper function of this mechanism is necessary for cell viability. Genetic defects in 50–100 loci are known to produce photoreceptor loss and blindness in humans and other vertebrates (for a recent review, see ref. 24). Nearly all of these genetic defects predominantly or exclusively affect the structure or the function of the photoreceptor outer segment. To our knowledge, the mok mutation provides the first example of a sensory neuron loss associated with a nuclear positioning phenotype. It is important to note, however, that mok photoreceptor loss also involves cell nonautonomous components (2). This aspect of the mok phenotype is unlikely to be mediated via nuclear positioning aberrations and may involve defects in cell–cell interactions mediated via cell junctions of the outer limiting membrane or, less likely, synaptic termini.
The nucleus is a bulky organelle that in many cases occupies a substantial fraction of cell volume. In neurons, cell somata are frequently almost entirely occupied by the nucleus, and it is the nuclear position that defines cell body. The regulation of nuclear position is thus a mechanism that contributes to tissue patterning in neuronal formations. The retina alone provides many examples of cells that display nonrandom nuclear positions. For example, although Muller glia span the entire thickness of the retina, their nuclei are positioned in the inner portion of the inner nuclear layer (see, for example, ref. 25). Similarly, cone nuclei are consistently located apical to rod nuclei (26). Experiments presented in this manuscript reveal a mechanism that regulates nuclear position in the photoreceptor cell layer of the retina. Similar mechanisms may be functional in other cell classes: glia and bipolar neurons, for example. Alternatively, forces that do not involve microtubule-based motors may also regulate nuclear position in other cells. Further studies of these mechanisms may be a source of important insights into the patterning of multicellular tissues.
Materials and Methods
Animals.
The maintenance and breeding of zebrafish strains and staging of embryonic development were performed as described previously (27, 28). All animal protocols were approved by the MEEI animal care committee. Embryos were observed by using an Axioscope microscope (Zeiss, Thornwood, NY) or a Leica (Deerfield, IL) MZ12 dissecting scope.
Mosaic Analysis.
Genetically mosaic animals were generated as described in ref. 29. Cells were transplanted into wild-type hosts from the progeny of crosses between mokm632 heterozygotes or from embryos injected with morpholino oligonucleotides. To distinguish mutant donor embryos, we isolated their DNA at 24 hpf and PCR-amplified the genomic fragment containing the mokm632 mutation, using the following two primers: CACACATCGTCAACTATTCCT and TTGTACCTGCTCCACTGCTT. The presence of mutation was determined by sequencing. Additional details can be found in SI Text.
Cryosectioning and Immunohistochemistry.
Sectioning and immunohistochemistry were performed as described in ref. 29. Sources of antibodies are provided in SI Text. We note that antibody detection of Mok polypeptide in zebrafish is complicated because two Dynactin 1 genes are expressed in zebrafish retina. The antibodies that we use may recognize both. To evaluate BBS4 staining, the average thickness of the outer nuclear layer was estimated for each image, and one-fifth of that thickness from the apical margin of the outer nuclear layer was used to determine the area where staining would be examined. BBS-positive particles in this area were counted, and their ratio to the number of cell somata (stained by the Zpr-1 antibody) or to the number of basal bodies (stained by γ-tubulin antibody) was calculated.
Positional Cloning.
A map cross was set up between heterozygous G0 carriers of the mokm632 allele (AB genetic background) and wild-type WIK strain homozygotes. To determine mok position in the genome, we used a panel of 1,200 F2 diploid embryos obtained via incrossing of F1 animals. Genotyping of DNA polymorphisms was performed as described in ref. 3. To clone the full-length zebrafish mok cDNA, human and mouse dynactin 1 coding sequences were obtained from the National Centre for Biotechnology Information public database and used to search the whole-genome zebrafish shotgun sequence database (Sanger Center, Cambridge, U.K.) with a homology search program, BLASTP. Based on partial sequences obtained in this manner, RT-PCR was performed to clone the full-length gene transcript, using conditions described in ref. 3.
Knockdown and Phenotypic Rescue.
Morpholino-modified oligonucleotides (morpholinos; sequences provided in SI Text) were injected into wild-type AB embryos as described in ref. 28. To determine BBS4 distribution in lis1 morphants, a mix of lis1a-ATG and lis1a-SP morpholinos (1.5 mg/ml each) was injected into embryos. In all other experiments, 3.0 mg/ml (1.5–2 nl) of each morpholino was injected separately. To rescue the mutant phenotype, the full-length mikre oko coding sequence was amplified by using the primers CAGTTCAGACGGCGGTGGGCG and TTTACGTCATGAGCAGGTGGT and cloned into the pXT7 vector (figure 1 in ref. 28). Messenger RNA was generated by using standard protocols and injected into embryos at the one cell-stage. Embryos were collected at 3.5 dpf, and their tails were removed, digested in a lysis buffer, and genotyped as above. After genotype determination, the anterior part of each embryo was cryosectioned, and sections were stained with the Zpr1 antibody and the Yo-Pro-1 nuclear stain as above.
Heat-Shock Overexpression.
The following genes or gene fragments were used in overexpression studies: the N terminus of lis1a (first 87 aa), the KASH domain of syne2a (69 C-terminal aa), the entire coding region of p50/dynamitin, and the entire coding region of ift52 (3) (used as a control in p50 overexpression; Fig. 3C). Relevant DNA fragments were cloned into a heat-shock promoter driven zebrafish expression vector pXT7-HS (28). To monitor the distribution of DNA upon injection into embryos, overexpression constructs were mixed with pXT7-HS-GFP expression vector DNA in the 3:1 ratio. Linearized vector DNA was injected into one-cell stage embryos as described in ref. 28. Unless stated otherwise, heat-shock treatments (1 h each, at 38°C) were performed at 48, 60, and 70 hpf. For p50, heat shocks were also applied at 4, 4.5, and 5 dpf. Embryos were collected at 4 or 6 dpf and processed for antibody staining as above. Staining was performed on whole embryos, using an anti-GFP rabbit antibody (1:2,000; Clontech, Palo Alto, CA) in 10% goat serum in PBS Tween-20 overnight at 4°C. Embryos were embedded in JB-4 resin and sectioned, and sections were stained with To-Pro-3 nuclear dye (1:8,000; Molecular Probes, Eugene, OR).
In Situ Hybridization.
Partial sequences of relevant genes were amplified by RT-PCR, cloned into the pCR-II vector (Invitrogen, Carlsbad, CA), and sequenced. RNA probe synthesis, in situ hybridization, washes, and signal detection were carried out by using standard protocols as described in ref. 30. After hybridization, embryos were embedded in JB4 resin (Polysciences, Warrington, PA) and sectioned at 8 μm. Additional details are in SI Text.
Supplementary Material
Acknowledgments
We thank Drs. K. T. Vaughan (University of Notre Dame, Notre Dame, IN) and B. B. Vallee (Worcester Foundation for Biomedical Research, Worcester, MA) for a p150 antibody; Drs. Vihtelic and Hyde (University of Notre Dame) for anti-opsin antibodies; Dr. Nico Katsanis (The Johns Hopkins University, Baltimore, MD) for the anti-BBS4 antibody; and Drs. Pignoni, Dryja, Zon, Katsanis, and Heller for providing comments on earlier versions of this manuscript. This work was supported by a Knights Templar Pediatric Ophthalmology research grant (to M.T.), National Eye Institute Award RO1 EY016859 (to J.M.), and National Eye Institute Core Grant EY14104.
Abbreviations
- BBS
Bardet–Biedl syndrome
- dpf
days postfertilization
- hpf
h postfertilization.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700178104/DC1.
References
- 1.Malicki J, Neuhauss SC, Schier AF, Solnica-Krezel L, Stemple DL, Stainier DY, Abdelilah S, Zwartkruis F, Rangini Z, Driever W. Development (Cambridge, UK) 1996;123:263–273. doi: 10.1242/dev.123.1.263. [DOI] [PubMed] [Google Scholar]
- 2.Doerre G, Malicki J. J Neurosci. 2001;21:6745–6757. doi: 10.1523/JNEUROSCI.21-17-06745.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsujikawa M, Malicki J. Neuron. 2004;42:703–716. doi: 10.1016/s0896-6273(04)00268-5. [DOI] [PubMed] [Google Scholar]
- 4.Pugh E, Lamb T. Handbook of Biological Physics. Vol 3. Amsterdam: Elsevier; 2000. pp. 183–255. [Google Scholar]
- 5.Mansergh F, Orton N, Vessey J, Lalonde M, Stell W, Tremblay F, Barnes S, Rancourt D, Bech-Hansen T. Hum Mol Genet. 2005;14:3035–3046. doi: 10.1093/hmg/ddi336. [DOI] [PubMed] [Google Scholar]
- 6.Swaroop A, Swaroop M, Garen A. Proc Natl Acad Sci USA. 1987;84:6501–6505. doi: 10.1073/pnas.84.18.6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holzbaur EL, Hammarback JA, Paschal BM, Kravit NG, Pfister KK, Vallee RB. Nature. 1991;351:579–583. doi: 10.1038/351579a0. [DOI] [PubMed] [Google Scholar]
- 8.Holleran EA, Karki S, Holzbaur EL. Int Rev Cytol. 1998;182:69–109. doi: 10.1016/s0074-7696(08)62168-3. [DOI] [PubMed] [Google Scholar]
- 9.LaMonte BH, Wallace KE, Holloway BA, Shelly SS, Ascano J, Tokito M, Van Winkle T, Howland DS, Holzbaur EL. Neuron. 2002;34:715–727. doi: 10.1016/s0896-6273(02)00696-7. [DOI] [PubMed] [Google Scholar]
- 10.Echeverri CJ, Paschal BM, Vaughan KT, Vallee RB. J Cell Biol. 1996;132:617–633. doi: 10.1083/jcb.132.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tai CY, Dujardin DL, Faulkner NE, Vallee RB. J Cell Biol. 2002;156:959–968. doi: 10.1083/jcb.200109046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swan A, Nguyen T, Suter B. Nat Cell Biol. 1999;1:444–449. doi: 10.1038/15680. [DOI] [PubMed] [Google Scholar]
- 13.Xiang X, Osmani AH, Osmani SA, Xin M, Morris NR. Mol Biol Cell. 1995;6:297–310. doi: 10.1091/mbc.6.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirotsune S, Fleck MW, Gambello MJ, Bix GJ, Chen A, Clark GD, Ledbetter DH, McBain CJ, Wynshaw-Boris A. Nat Genet. 1998;19:333–339. doi: 10.1038/1221. [DOI] [PubMed] [Google Scholar]
- 15.Ahn C, Morris NR. J Biol Chem. 2001;276:9903–9909. doi: 10.1074/jbc.M010233200. [DOI] [PubMed] [Google Scholar]
- 16.Starr DA, Fischer JA. BioEssays. 2005;27:1136–1146. doi: 10.1002/bies.20312. [DOI] [PubMed] [Google Scholar]
- 17.Starr DA, Han M. J Cell Sci. 2003;116:211–216. doi: 10.1242/jcs.00248. [DOI] [PubMed] [Google Scholar]
- 18.Starr DA, Han M. Science. 2002;298:406–409. doi: 10.1126/science.1075119. [DOI] [PubMed] [Google Scholar]
- 19.Grady RM, Starr DA, Ackerman GL, Sanes JR, Han M. Proc Natl Acad Sci USA. 2005;102:4359–4364. doi: 10.1073/pnas.0500711102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li T. Trends Mol Med. 2001;7:133–135. doi: 10.1016/s1471-4914(00)01902-x. [DOI] [PubMed] [Google Scholar]
- 21.Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. Dev Cell. 2004;7:133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 22.Omori Y, Malicki J. Curr Biol. 2006;16:945–957. doi: 10.1016/j.cub.2006.03.058. [DOI] [PubMed] [Google Scholar]
- 23.Blacque OE, Leroux MR. Cell Mol Life Sci. 2006;63:2145–2161. doi: 10.1007/s00018-006-6180-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hartong DT, Berson EL, Dryja TP. Lancet. 2006;368:1795–1809. doi: 10.1016/S0140-6736(06)69740-7. [DOI] [PubMed] [Google Scholar]
- 25.Peterson RE, Fadool JM, McClintock J, Linser PJ. J Comp Neurol. 2001;429:530–540. doi: 10.1002/1096-9861(20010122)429:4<530::aid-cne2>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 26.Raymond P, Barthel L, Rounsifer M, Sullivan S, Knight J. Neuron. 1993;10:1161–1174. doi: 10.1016/0896-6273(93)90064-x. [DOI] [PubMed] [Google Scholar]
- 27.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 28.Malicki J, Jo H, Wei X, Hsiung M, Pujic Z. Methods. 2002;28:427–438. doi: 10.1016/s1046-2023(02)00262-1. [DOI] [PubMed] [Google Scholar]
- 29.Avanesov A, Malicki J. Methods Cell Biol. 2004;76:333–384. doi: 10.1016/s0091-679x(04)76016-1. [DOI] [PubMed] [Google Scholar]
- 30.Oxtoby E, Jowett T. Nucleic Acids Res. 1993;21:1087–1095. doi: 10.1093/nar/21.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.