Abstract
Reduced caloric intake decreases arterial blood pressure in healthy individuals and improves endothelium-dependent vasodilation in obese and overweight individuals. The SIRT1 protein deacetylase mediates many of the effects of calorie restriction (CR) on organismal lifespan and metabolic pathways. However, the role of SIRT1 in regulating endothelium-dependent vasomotor tone is not known. Here we show that SIRT1 promotes endothelium-dependent vasodilation by targeting endothelial nitric oxide synthase (eNOS) for deacetylation. SIRT1 and eNOS colocalize and coprecipitate in endothelial cells, and SIRT1 deacetylates eNOS, stimulating eNOS activity and increasing endothelial nitric oxide (NO). SIRT1-induced increase in endothelial NO is mediated through lysines 496 and 506 in the calmodulin-binding domain of eNOS. Inhibition of SIRT1 in the endothelium of arteries inhibits endothelium-dependent vasodilation and decreases bioavailable NO. Finally, CR of mice leads to deacetylation of eNOS. Our results demonstrate that SIRT1 plays a fundamental role in regulating endothelial NO and endothelium-dependent vascular tone by deacetylating eNOS. Furthermore, our results provide a possible molecular mechanism connecting the effects of CR on the endothelium and vascular tone to SIRT1-mediated deacetylation of eNOS.
Keywords: calorie restriction, vasorelaxation, silent information regulator 2, resveratrol, deacetylation
Caloric restriction (CR) is a well recognized nonpharmacological approach to reducing arterial blood pressure. CR not only is capable of independently controlling blood pressure of patients with mild hypertension, but also allows a reduction in the number and dosage of medications used to treat hypertension (1, 2). CR and weight loss resulting from CR also improve endothelium-dependent vascular relaxation in obese and overweight individuals with hypertension (3, 4).
In addition, CR prolongs organismal lifespan. In the budding yeast, Saccharomyces cerevisiae, aging of replicating cells is determined by the SIR2 gene (5). Retardation of yeast aging by CR depends on the product of this gene, Sir2 (silent information regulator 2), a class III NAD-dependent histone deacetylase. The mammalian ortholog of Sir2, SIRTUIN 1 (SIRT1), targets histones and many nonhistone proteins (6–10). Resveratrol, a plant polyphenol that stimulates SIRT1 activity (11), activates endothelial nitric oxide synthase (eNOS) (12), improves endothelial function, prevents elevation in blood pressure, and restores vascular eNOS activity in animal models of endothelial dysfunction (13). Hypothesizing that the effects of CR and resveratrol on vascular function are mediated, in part, by SIRT1, we investigated the role of SIRT1 in regulating eNOS activity and endothelium-dependent vascular tone.
Results and Discussion
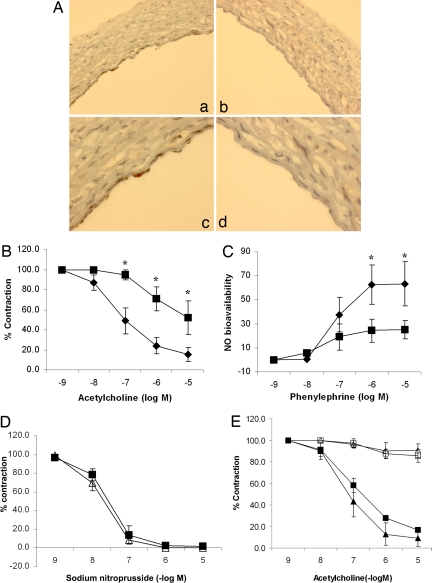
To determine whether SIRT1 plays an important role in regulating endothelium-dependent vascular tone, vasomotor function of rat aortic rings, in which wild-type SIRT1 or a dominant negative SIRT1 mutant (to inhibit endogenous SIRT1) was adenovirally expressed in the endothelium (Fig. 1A), was measured. Control rings were infected with an adenovirus encoding the inert Escherichia coli lacZ gene. Compared with control rings, rings with inhibition of endogenous SIRT1 had impaired acetylcholine-induced, endothelium-dependent vasorelaxation (Fig. 1B) and lower bioavailable NO (Fig. 1C). In contrast, endothelium-independent vasodilation induced by the NO donor sodium nitroprusside was similar in control rings and rings in which SIRT1 was inhibited (Fig. 1D). Moreover, similar to control rings, rings in which wild-type SIRT1 was overexpressed did not show acetylcholine-induced vasodilation in the presence of the NOS inhibitor NG-nitro-l-arginine methyl ester (l-NAME) (Fig. 1E), suggesting that SIRT1 does not activate NOS-independent mechanisms of endothelium-dependent vasorelaxation. Thus, endothelial SIRT1 promotes endothelium-dependent vasodilation and vascular bioavailability of NO through a NOS-dependent mechanism.
Fig. 1.
Endothelial SIRT1 regulates endothelium-dependent vasodilation and bioavailable NO through NOS. (A) A replication-deficient adenovirus encoding the myc-tagged catalytically inactive dominant negative mutant of SIRT1 (AdSIRT1(H363Y)) (a and c) was used to inhibit endogenous SIRT1 in the endothelium of rat aortic rings ex vivo. Control rings were infected with an adenovirus encoding the inert E. coli lacZ gene (AdLacZ) (b and d). Rings were fixed and immunostained with anti-myc antibody 24 h after adenoviral infection. Photomicrographs of representative rings at ×100 (a and b) and ×400 (c and d) are shown. (B) Endothelium-dependent vasodilation was determined by measuring relaxation of rings preconstricted with phenylephrine to the vasodilator acetylcholine. AdSIRT1(H363Y) (■) and AdLacZ (◆) are shown. *, P < 0.001 compared with AdLacZ (n = 7). (C) Bioavailable NO was determined by measuring the difference in phenylephrine-induced vasoconstriction in the presence and absence of the NOS inhibitor l-NAME. AdSIRT1(H363Y) (■) and AdLacZ (◆) are shown. *, P < 0.01 compared with AdSIRT1(H363Y) (n = 7). (D) Endothelium-independent vasodilation was determined by measuring relaxation of rings preconstricted with phenylephrine to the NO donor sodium nitroprusside. AdSIRT1(H363Y) (△) and AdLacZ (■) (n = 4) are shown. (E) Rat aortic rings were infected ex vivo with AdLacZ (■ and □) or AdSIRT1 (△ and ▲). AdSIRT1 encodes wild-type SIRT1. Endothelium-dependent vasodilation, in the presence (□ and △) and absence (■ and ▲) of the NOS inhibitor l-NAME, was determined by measuring relaxation of rings preconstricted with phenylephrine, to the vasodilator acetylcholine. n = 4 rings in each group.
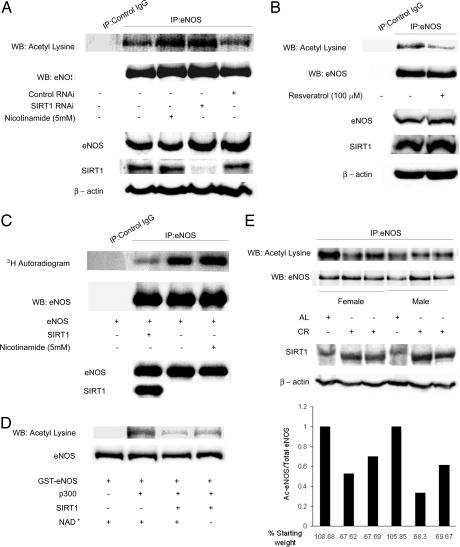
SIRT1 deacetylates ε-acetyllysine residues in proteins. To investigate whether SIRT1 deacetylates eNOS in cells, we determined the acetylation of eNOS in endothelial cells in which endogenous SIRT1 expression was knocked down with RNAi. Decrease in SIRT1 expression increased acetylation of endogenous eNOS on lysine residues (Fig. 2A). Similarly, inhibition of endogenous SIRT1 activity with nicotinamide increased acetyllysine-eNOS (Fig. 2A). Conversely, stimulation of endogenous SIRT1 activity with resveratrol decreased acetylation of eNOS on lysine residues (Fig. 2B). Furthermore, SIRT1 overexpression decreased [3H]acetate incorporation into eNOS expressed in COS7 cells (Fig. 2C), whereas inhibition of endogenous SIRT1 with nicotinamide increased [3H]acetate incorporation into eNOS (Fig. 2C). Finally, to verify that eNOS is a bona fide substrate of SIRT1, we examined whether SIRT1 can deacetylate acetylated eNOS in vitro. Recombinant eNOS was acetylated in vitro with the p300 acetyltransferase and the acetyl group donor acetyl-CoA, and then treated with active SIRT1, in the presence or absence of the SIRT1 cofactor NAD. SIRT1 deacetylated acetylated eNOS in a NAD-dependent fashion (Fig. 2D). Thus, SIRT1 deacetylates eNOS in vitro, and acetylation of eNOS in cells inversely correlates with the activity and/or expression of SIRT1.
Fig. 2.
SIRT1 deacetylates eNOS in cells and in vitro, and calorie restriction is associated with deacetylation of eNOS in mice livers. (A) Acetylation of endogenous eNOS on lysine residues, with and without RNAi-mediated knockdown of endogenous SIRT1, and with and without treatment of cells with the SIRT1 inhibitor nicotinamide. Rat aortic endothelial cells were transfected with SIRT1 RNAi or control RNAi, or treated with nicotinamide. Immunoprecipitates of eNOS from whole-cell lysates were immunoblotted with anti-eNOS and anti-acetyllysine antibodies (top two panels). Immunoprecipitates by using nonimmune IgG were used as a control. Whole-cell lysates were immunoblotted with anti-eNOS, anti-SIRT1, and anti-β-actin antibodies (bottom three panels). (B) Acetylation of endogenous eNOS on lysine residues, with and without treatment with the SIRT1 activator resveratrol. Immunoprecipitated eNOS from whole-cell lysates of rat aortic endothelial cells, with and without treatment with resveratrol, was immunoblotted with anti-acetyllysine and anti-eNOS antibodies. The whole-cell lysates were also immunoblotted with anti-eNOS, anti-SIRT1, and anti-β-actin antibodies. (C) Incorporation of [3H]acetate into eNOS expressed in COS7 cells. Cells transfected with eNOS, with and without SIRT1 overexpression, or treated with nicotinamide, were incubated in medium containing sodium [3H]acetate. Immunoprecipitated eNOS from cell lysates was autoradiographed, and immunoblotted with anti-eNOS antibody. Whole-cell lysates were immunoblotted with anti-eNOS and anti-SIRT1 antibodies. (D) Deacetylation of acetylated eNOS by SIRT1 in vitro. Purified full-length GST-eNOS and GST-eNOS acetylated with the p300 acetyltransferase in the presence of the acetyl group donor acetyl-CoA (Ac-CoA) were incubated with active SIRT1 enzyme, in the presence or absence of the SIRT1 cofactor NAD. Reaction mixes were immunoblotted with anti-acetyllysine and anti-eNOS antibodies. (E) Deacetylation of eNOS in mice livers by CR. Male and female mice were fed ad libitum or a calorie-restricted diet (57.4 kcal/week) for 3 weeks. Immunoprecipitates of eNOS from liver homogenates were immunoblotted with anti-acetyllysine and anti-eNOS antibodies (top two panels), and homogenates immunoblotted with anti-SIRT1 and anti-β-actin antibodies (bottom two panels). The bar graph shows the calculated acetylated/total eNOS ratio, expressed relative to the mouse fed ad libitum in each gender group. The weight of the mice at the end of 3 weeks (as a percentage of starting weight) is shown. AL, ad libitum.
Next, we probed whether acetylation of eNOS is altered during conditions that change SIRT1 expression or activity in vivo. CR stimulates SIRT1 expression in mammals (6). Therefore, we investigated the possibility that the acetylation status of eNOS is modulated by the change in SIRT1 expression in response to caloric intake. We compared acetylation of eNOS on lysine residues in livers of mice that were placed on a calorie-restricted diet with those that were fed ad libitum. CR for a period of 3 weeks led to a decrease in the acetylation of eNOS on lysine residues (Fig. 2E), such that the fraction of eNOS that was acetylated was significantly lower than in the mice that were not on a calorie-restricted diet.
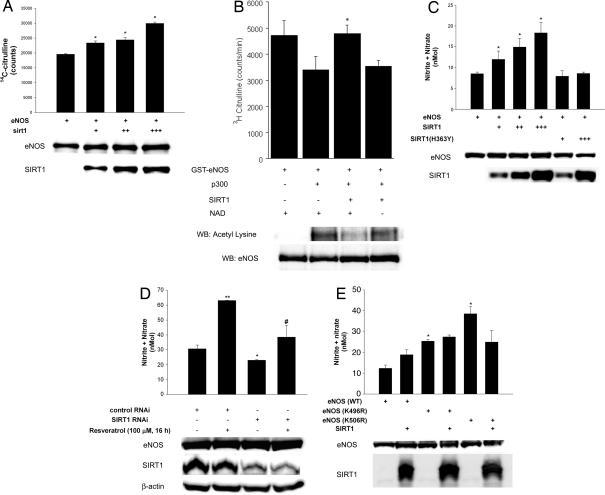
To determine whether SIRT1 regulates eNOS activity, we performed NOS activity assays in lysates of eNOS-expressing COS7 cells in which we overexpressed SIRT1. Increasing SIRT1 expression increased eNOS catalytic activity (Fig. 3A). To show that eNOS activity is subject to direct regulation by acetylation–deacetylation, we also examined NOS catalytic activity in vitro (Fig. 3B). The catalytic activity of recombinant eNOS in vitro was decreased after acetylation with the p300 acetyltransferase. Deacetylation of acetylated eNOS with SIRT1 rescued the decline in NOS catalytic activity in a NAD-dependent fashion. In parallel with an increase in NOS activity, SIRT1 overexpression increased eNOS-derived NO produced by COS7 cells (Fig. 3C). A similar degree of expression of the catalytically inactive dominant negative SIRT1 mutant did not increase NO levels. Moreover, in endothelial cells, resveratrol increased NO levels, an effect that was largely mitigated by knockdown of endogenous SIRT1 (Fig. 3D). To determine the acetyllysine residues in eNOS that are targeted by SIRT1, we performed a structure–function analysis of eNOS. We targeted two lysine residues: lysine 496 and 506 in bovine eNOS (corresponding to lysines 494 and 504 in human eNOS). These residues are in the calmodulin-binding domain (CBD) of eNOS. We chose the CBD because threonine 497 in this domain is phosphorylated by protein kinase C, and dephosphorylation of this residue stimulates eNOS activity (14). We reasoned that because of similar steric and/or charge effects, addition of acetyl moieties to lysine residues in the CBD would have an impact on eNOS activity in a manner similar to addition of a phosphate group to phosphoacceptor residues in the CBD. Therefore, lysine 496 and lysine 506 were changed to nonacetylatable arginine (K496R and K506R). Compared with wild-type eNOS, expression of eNOS(K496R) or eNOS(K506R) in COS7 cells resulted in significantly greater NO levels (Fig. 3E). Moreover, in contrast to wild-type eNOS, overexpression of SIRT1 did not stimulate NO derived from eNOS(K496R) or eNOS(K506R). These findings suggest that both K496 and K506 in wild-type eNOS play important roles in modulating NO production, and the stimulatory effect of SIRT1 on eNOS requires the integrity of these residues.
Fig. 3.
SIRT1 stimulates eNOS activity and increases endothelial NO. (A) NOS catalytic activity (conversion of arginine to citrulline) in lysates of COS7 cells expressing eNOS, with and without SIRT1 overexpression, was measured. Lysates were immunoblotted with anti-SIRT1 and anti-eNOS antibodies. *, P < 0.05 compared with eNOS without SIRT1. (B) NOS catalytic activity in vitro, in the absence and presence of p300, SIRT1, and NAD. Total and acetyllysine eNOS is shown at bottom. *, P < 0.05 compared with eNOS + p300. (C) Metabolites of NO (nitrite and nitrate) were measured in media of COS7 cells expressing eNOS, with and without overexpression of SIRT1, or inactive SIRT1 (H363Y). Whole-cell lysates were immunoblotted with anti-eNOS and anti-SIRT1 antibodies. Endogenous SIRT1 is not seen in the lysates because of underexposure of the blots. *, P < 0.05 compared with eNOS without SIRT1. (D) Nitrite and nitrate were measured in media of rat aortic endothelial cells that were transfected with SIRT1 RNAi or control RNAi, or treated with the SIRT1 activator resveratrol. *, P < 0.05 compared with control RNAi. **, P < 0.01 compared with control RNAi. #, P > 0.05 compared with SIRT1 RNAi. (E) Nitrite and nitrate were measured in COS7 cells expressing eNOS (WT), eNOS (K496R), or eNOS(K506R), with and without SIRT1 overexpression. *, P < 0.05 compared with eNOS (WT).
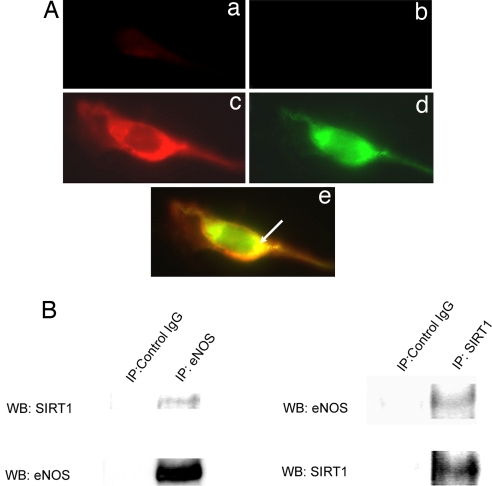
SIRT1 has been described to shuttle between the cytoplasm and nucleus (15). To determine whether SIRT1 associates with eNOS, which is present mostly in the cytoplasm, the localization of eNOS and SIRT1 in endothelial cells was assessed. Double immunofluorescence showed that endogenous SIRT1 and eNOS colocalize in the perinuclear cytoplasm (Fig. 4A). In addition, coimmunoprecipitation showed that SIRT1 and eNOS associate with each other in endothelial cells (Fig. 4B). Thus, a fraction of SIRT1 colocalizes and binds to eNOS in the cytoplasm of endothelial cells.
Fig. 4.
SIRT1 and eNOS localize and associate with each other in endothelial cells. (A) Double immunofluorescence for endogenous eNOS (red) and SIRT1 (green) in human umbilical vein endothelial cells. Fixed, permeabilized cells were stained with anti-eNOS antibody and Texas Red-labeled (red) secondary antibody, followed by staining with anti-SIRT1 antibody and FITC-labeled (green) secondary antibody (c–e). Control cells were stained with the Texas Red-labeled (a) and FITC-labeled (b) secondary antibodies without the anti-eNOS and anti-SIRT1 primary antibodies. Representative red, green, and merged images (×40) captured on a fluorescence microscope are shown. Arrow points to colocalization (yellow) of eNOS and SIRT1 in merged image. (B) Coimmunoprecipitation of endogenous eNOS and SIRT1 from rat aortic endothelial cells. Whole-cell lysates were immunoprecipitated with anti-eNOS or anti-SIRT1 antibodies. Immunoprecipitates were immunoblotted with anti-eNOS and anti-SIRT1 antibodies. Immunoprecipitates with nonimmune IgG were used as controls.
Four lysine residues are within the CBD of eNOS. The identification of two of these residues as putative sites of acetylation suggests that SIRT1 may target more than a single residue within this domain, stimulating enzymatic activity by coordinating deacetylation of these residues. Furthermore, taken in the context of the known role of phosphorylation in eNOS regulation, it suggests that, as for other proteins, acetylation and phosphorylation may occur in an interdependent fashion to modulate eNOS activity (16, 17).
SIRT1 deacetylates members of the FOXO family of transcription factors (18, 19). The LXXLL motif (where L is leucine and X is any amino acid) is conserved in FOXO proteins of different species (20). This motif mediates the interaction of murine FOXO1 with SIRT1 and is essential for SIRT1-induced deacetylation of FOXO1 (20). The LXXLL motif is also found in other established targets of SIRT1, including p53, PGC-1α, and p300/CBP. eNOS of different species contains at least one LXXLL motif, supporting our claim that SIRT1 binds to and targets eNOS for deacetylation.
Amelioration of endothelial dysfunction in obese and overweight subjects by short-term CR (3, 4) may be mediated by corrections in metabolic abnormalities that adversely affect endothelial function. However, in healthy nonobese individuals as well, long-term CR reduces both systolic and diastolic blood pressure (21), suggesting that CR may activate physiological mechanisms of vasorelaxation that are independent of correction of metabolic disorders. Intriguing as these studies may be, it is difficult to attribute the beneficial effect of CR on vascular tone solely to SIRT1. CR also stimulates eNOS expression (22); therefore, its salutary effects on endothelial function may be independent of SIRT1-induced deacetylation of eNOS. Nevertheless, the identification of eNOS as a substrate of SIRT1, combined with the findings that inhibition of endogenous SIRT1 promotes endothelial dysfunction and that CR results in deacetylation of eNOS, suggests an important role for SIRT1 in endothelium-dependent eNOS-mediated vascular homeostasis: a role that may become more prominent during times of low calorie intake.
Materials and Methods
Cells and Cell Transfections.
Human umbilical vein and rat aortic endothelial cells were purchased from Clonetics (San Diego, CA) and Cell Applications (San Diego, CA). COS7 cells were purchased from American Type Culture Collection (Manassas, VA). Cells were transfected with cDNA or RNAi by using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to manufacturer's recommendations.
Antibodies, Immunoprecipitations, and Immunoblotting.
Immunoprecipitations of eNOS and SIRT1 were typically carried out by incubating 2 μg of antibody (rabbit polyclonal anti-eNOS and anti-SIRT1 from Santa Cruz Biotechnology, Santa Cruz, CA) with 1 mg of cell lysate overnight, followed by 40 μl of protein A-Sepharose slurry (Amersham, Piscataway, NJ) for 4 h. After washing, immunoprecipitates were boiled in SDS/PAGE gel loading buffer, subjected to SDS/PAGE, transferred to nitrocellulose filter, and probed with the specified primary antibody and the appropriate peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology). Western blotting of 50 μg of whole-cell lysates was similarly performed by using appropriate primary and secondary antibodies. Chemiluminescent signal was developed by using SuperSignal West Pico or Femto substrate (Pierce, Rockford, IL), and blots were imaged with a Gel Doc 2000 Chemi Doc system (Bio-Rad, Hercules, CA).
RNA Interference.
Validated Stealth siRNA for SIRT1 (5′-GCAACAGCAUCUUGCCUGAUUUGUA-3′, nucleotides 1152–1175 of human SIRT1 mRNA) and the appropriate control RNAi were purchased from Invitrogen. This siRNA were transfected into endothelial cells according to the protocols provided by the manufacturer.
Nitrite + Nitrate Measurements.
COS7 or endothelial cells transfected with the indicated plasmid or RNAi were incubated in phenol red-free complete growth medium for 48 h. Medium was deproteinized by using a 10-kDa cutoff filter (Microcon YM10; Millipore, Billerica, MA). The medium was then processed for the measurement of the NO metabolites nitrite (NO2−) and nitrate (NO3−), the stable breakdown product of NO, using a commercially available kit (Nitrate/Nitrite Fluorometric Assay kit; Cayman Chemicals, Ann Arbor, MI) according to the manufacturer's recommendations. After subtraction of background fluorescence, values were normalized for total cell number (protein amounts).
NOS Catalytic Activity.
NOS activity was measured with a commercially available kit. In brief, the conversion of l-[14C]arginine to l-[14C]citrulline was used to determine NOS activity in whole-cell lysates or recombinant purified GST-eNOS, according to the manufacturer's recommendations (NOS Detect Assay kit; Stratagene, La Jolla, CA). Values were normalized for cell number (total protein).
Recombinant Proteins.
The cDNA of full-length bovine eNOS was cloned into pGEX4T-2 (Amersham) bacterial expression plasmid. Protein was expressed and induced with isopropyl β-d-thiogalactoside (IPTG) (0.1 mM) in BL21-Gold (DE3) pLysS (Stratagene) bacterial host strain. Expressed eNOS was purified by using glutathione Sepharose beads (Amersham Biosciences) following the batch purification protocol recommended by the manufacturer. Purity of the eluted fractions was determined by SDS/PAGE and Coomassie staining.
Dual Immunofluorescence Staining.
Endothelial cells seeded on coverslips were fixed in 4% paraformaldehyde in 1× PBS (pH 7.2–7.4) for 30 min at room temperature. After permeabilization with 10% goat serum, 0.4% Triton X-100, and 10 mM glycine, they were incubated in blocking solution (10% goat serum at room temperature for 1 h), followed by application of the first primary antibody (rabbit polyclonal anti-eNOS at 1:200 dilution) for 1 h at 37°C. Texas red-tagged fluorescent secondary antibody (at 1:50 dilution; Jackson ImmunoResearch Laboratories, West Grove, PA) was then applied for 1 h at 37°C in the dark. The second primary antibody (mouse monoclonal anti-SIRT1 at 1:200 dilution) was then applied for 1 h at 37°C in the dark. FITC-tagged green fluorescent secondary antibody (at 1:50 dilution; Jackson Immuno Research Laboratories) was then added for 1 h at 37°C in the dark. After each step, coverslips were washed three times for 5 min in PBS containing 10 mM glycine. The coverslips were then mounted by using ProLong Gold Antifade Reagent (Invitrogen) and analyzed under a dual fluorescence Axiovert microscope (Zeiss, Jena, Germany). Control coverslips were processed the same way, except for omission of the primary antibodies.
Site-Directed Mutagenesis.
Mutagenesis was performed with commercially available QuikChange kits (Stratagene). All mutations were verified by sequencing.
In Vitro Acetylation and Deacetylation.
Purified GST-tagged recombinant eNOS was desalted and buffer exchanged with acetylation buffer (50 mM Tris·HCl, pH 8.0/137 mM NaCl/0.1 mM EDTA, pH 8.0/10% glycerol/1 mM DTT/0.1 mM PMSF/2 μM trichostatin A) by using Zeba spin columns (Pierce). Five micrograms of GST-eNOS was used per acetylation reaction. It was incubated with acetyl-CoA (20 μM) and p300 acetyltransferase (60 units, Active Motif) at 30°C for 40 min with shaking. The acetylation reaction was immediately followed by the deacetylation reaction by adding deacetylation buffer (25 mM Tris·HCl, pH 8.0/137 mM NaCl/2.7 mM KCl/1 mM MgCl2/1 mg/ml BSA), 1 mM NAD+, and active recombinant SIRT1 (10 units; Biomol, Plymouth Meeting, PA) and incubated at 37°C for 1 h with shaking. The reaction mixture was subjected to SDS/PAGE and immunoblotting.
In Vivo Radiolabeling with [3H]Acetate.
COS7 cells were cotransfected with eNOS and SIRT1 plasmids. Cells were grown overnight in acetate-free medium containing 5 mM nicotinamide and 300 μCi/ml sodium [3H]acetate for 5 h before harvesting. Cells were harvested and lysed in a buffer containing 10 mM nicotinamide and 5 μM trichostatin A. One milligram of protein was immunoprecipitated by using eNOS antibody with an IgG control, solubilized in SDS/PAGE sample buffer, and resolved on 8% SDS/PAGE. The gel was fixed in a solution containing 10% glacial acetic acid and 40% methanol for 1 h and then incubated in a commercial autoradiography enhancing solution (Amplify; Amersham) for another 30 min. The gel was dried and exposed to film at −80°C for 5 days.
CR in Mice.
Six 8-week-old male and female C57BL6 mice were either maintained on a 57.4 kcal/week diet or allowed to feed ad libitum for 3 weeks. Prolab Isopro RMH 3000 rodent chow was used. Mice were weighed weekly. Mice were killed after 3 weeks, and their livers were homogenized. SIRT1 expression was assessed in the homogenates by immunoblotting. Lysine acetylation of eNOS in homogenates was determined by immunoblotting immunoprecipitated eNOS with acetyllysine antibody.
Vascular Reactivity.
Young (3–4 months old) Wistar–Kyoto rats were killed with an overdose of sodium pentobarbital. A midsternal split was quickly performed, and the descending thoracic aorta was carefully excised and placed in ice-cold Krebs buffer (118.3 mM NaCl/4.7 mM KCl/2.5 mM CaCl2/1.2 mM KH2PO4/25 mM NaHCO3/1.2 mM MgSO4/11 mM glucose/0.0026 mM CaNa2EDTA). The aorta was cleaned of excess fat, cut transversely into 5–10 rings (2.0–3.0 mm), each of which was infected with 6 × 1011 viral particles per ml of the AdLacZ, AdSIRT1(H363Y), or AdSIRT1 adenoviral stocks, and incubated at 37°C for 24 h. The next day the vessels were placed in oxygenated chambers (95% O2/5% CO2) superfused with Krebs buffer solution and maintained at 37°C and pH 7.4. Each ring was suspended between two wire stirrups in a 5-ml organ chamber of a four-chamber myograph system (DMT). One stirrup was connected to a three-dimensional micromanipulator and the other to a force transducer. The contractile force was recorded electronically. All rings were stretched to 2,000 mg in 500-mg increments over a 1-h period to optimize the contractile response to KCl. One dose of KCl (60 mM) was administered to verify vascular smooth muscle viability. Cumulative dose–response curve for phenylephrine (10−9 to 10−5 M) was obtained by administering the drug in one-half log doses. Endothelium-dependent vasodilation was determined by generating dose–response curves to acetylcholine. Vasorelaxation evoked by acetylcholine was expressed as percent contraction determined by the percentage of inhibition to the preconstricted tension. Endothelium-dependent NOS-independent vasorelaxation was assessed by generating dose–response curves to acetylcholine in rings pretreated with the NOS inhibitor l-NAME (10−4 M). NO bioavailability was measured physiologically by determining the increase in contractile response to inhibitor l-NAME in rings preconstricted with phenylephrine (10−6 M). Endothelium-independent vasodilation was measured by the vasorelaxation evoked by cumulative sodium nitroprusside in rings preconstricted with phenylephrine (10−6 M).
Acknowledgments
We thank T. Michel (Harvard Medical School, Boston, MA) for the eNOS cDNA and A. Cook (University of Cambridge, Cambridge, U.K.) for the SIRT1 cDNA. This work was supported by the American Heart Association (Pennsylvania–Delaware affiliate).
Abbreviations
- l-NAME
NG-nitro-l-arginine methyl ester
- CR
caloric restriction
- CBD
calmodulin-binding domain.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Joint National Committee on Detection, Evaluation, Treatment of High Blood Pressure. Hypertension. 1986;8:444–467. [PubMed] [Google Scholar]
- 2.Hypertension Prevention Trial Research Group. Arch Intern Med. 1990;150:153–162. [PubMed] [Google Scholar]
- 3.Raitakari M, Ilvonen T, Ahotupa M, Lehtimaki T, Harmoinen A, Suominen P, Elo J, Hartiala J, Raitakari OT. Arterioscler Thromb Vasc Biol. 2004;24:124–128. doi: 10.1161/01.ATV.0000109749.11042.7c. [DOI] [PubMed] [Google Scholar]
- 4.Sasaki S, Higashi Y, Nakagawa K, Kimura M, Noma K, Sasaki S, Hara K, Matsuura H, Goto C, Oshima T, et al. Am J Hypertens. 2002;15:302–309. doi: 10.1016/s0895-7061(01)02322-6. [DOI] [PubMed] [Google Scholar]
- 5.Kaeberlein M, McVey M, Guarente L. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 7.Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, Bultsma Y, McBurney M, Guarente L. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- 8.Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 10.Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 11.Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, et al. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 12.Wallerath T, Deckert G, Ternes T, Anderson H, Li H, Witte K, Forstermann U. Circulation. 2002;106:1652–1658. doi: 10.1161/01.cir.0000029925.18593.5c. [DOI] [PubMed] [Google Scholar]
- 13.Miatello R, Vazquez M, Renna N, Cruzado M, Zumino AP, Risler N. Am J Hypertens. 2005;18:864–870. doi: 10.1016/j.amjhyper.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 14.Fleming I, Fisslthaler B, Dimmeler S, Kemp BE, Busse R. Circ Res. 2001;88:E68–E75. doi: 10.1161/hh1101.092677. [DOI] [PubMed] [Google Scholar]
- 15.Tanno M, Sakamoto J, Miura T, Shimamoto K, Horio Y. J Biol Chem. 2007;282:6823–6832. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- 16.Chen LF, Williams SA, Mu Y, Nakano H, Duerr JM, Buckbinder L, Greene WC. Mol Cell Biol. 2005;25:7966–7975. doi: 10.1128/MCB.25.18.7966-7975.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuzaki H, Daitoku H, Hatta M, Aoyama H, Yoshimochi K, Fukamizu A. Proc Natl Acad Sci USA. 2005;102:11278–11283. doi: 10.1073/pnas.0502738102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, et al. Science. 2004;303:2011–2215. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 19.Daitoku H, Hatta M, Matsuzaki H, Aratani S, Ohshima T, Miyagishi M, Nakajima T, Fukamizu A. Proc Natl Acad Sci USA. 2004;101:10042–10047. doi: 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakae J, Cao Y, Daitoku H, Fukamizu A, Ogawa W, Yano Y, Hayashi Y. J Clin Invest. 2006;116:2473–2483. doi: 10.1172/JCI25518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fontana L, Meyer TE, Klein S, Holloszy JO. Proc Natl Acad Sci USA. 2004;101:6659–6663. doi: 10.1073/pnas.0308291101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nisoli E, Tonello C, Cardile A, Cozzi V, Bracale R, Tedesco L, Falcone S, Valerio A, Cantoni O, Clementi E, et al. Science. 2005;310:314–317. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]