Summary
Human UDP-glucuronosyltransferases (UGT) are the dominant phase II conjugative drug metabolism enzymes that also play a central role in the processing of a range of endobiotic compounds. UGTs catalyze the covalent addition of glucuronic acid sugar moieties to a host of therapeutics and environmental toxins, as well as to a variety of endogenous steroids and other signaling molecules. We report the 1.8 Å resolution apo crystal structure of the UDP-glucuronic acid binding domain of human UGT isoform 2B7 (UGT2B7), which catalyzes the conjugative elimination of opioid, antiviral, and anticancer drugs. This is the first crystal structure of any region of a mammalian UGT drug metabolism enzyme. Designed UGT2B7 mutants at residues predicted to interact with the UDP-glucuronic acid cofactor exhibited significantly impaired catalytic activity, with maximum effects observed for amino acids closest to the glucuronic acid sugar transferred to the acceptor molecule. Homology modeling of UGT2B7 with related plant flavonoid glucosyltransferases indicate that human UGTs share a common catalytic mechanism. Point mutations at predicted catalytic residues in UGT2B7 abrogated activity, strongly suggesting that human UGTs also utilize a serine hydrolase-like catalytic mechanism to facilitate glucuronic acid transfer.
Keywords: Drug-metabolism, Catalysis, UGT, Glycosyltransferase, Nucleotide-sugar
Introduction
UDP-glucuronosyltransferases (EC 2.4.1.17; UGTs) are major vertebrate phase II conjugative metabolism enzymes that process and/or detoxify a large number of important endogenous compounds, drugs, and environmental agents (reviewed here 1,2). UGTs are highly promiscuous, catalyzing the covalent addition of a glucuronic acid moiety from the UDP-glucuronic acid (UDPGA) donor ligand to chemically distinct functional groups present on a range of structurally diverse acceptor substrates. This reaction frequently results in a biologically inactive, water soluble glucuronide that is rapidly excreted. Thus, these enzymes play central roles in endobiotic homeostasis and metabolic defense systems.
The UGT superfamily of enzymes in humans consists of 18 members that can be further divided by sequence homology into two main subfamilies: UGT1A and UGT2. The UGT1A family includes nine members (1A1, 1A3, 1A4, 1A5, 1A6, 1A7, 1A8, 1A9, and 1A10) that share a common genetic structure – each is composed of a unique exon 1 spliced to a common set of exons 2-5. While the UGT2 family also includes nine members (2A1, 2A2, 2B4, 2B7, 2B10, 2B11, 2B15, 2B17, and 2B28), each isoform exhibits a unique genetic structure. UGTs are type 1 transmembrane glycoproteins of approximately 530 amino acids localized predominantly to the endoplasmic reticulum (ER) and nuclear membranes3. The majority of the protein is in the ER lumen and is composed of two functional domains, N-terminal and C-terminal, thought to contain the acceptor substrate and UDPGA binding sites, respectively. In addition, the N-terminal domain is thought to contain a cryptic ER retention signal as well as a membrane interacting region 4,5. Human UGTs have also been shown to from both homo- and heterodimers, which may have an effect on function and acceptor ligand specificity 6,7,8,9.
UGTs have highly similar C-terminal domains and highly variable N-terminal domains, thereby imparting to each enzyme a distinct, but often overlapping set of substrate specificities. In particular, all UGT1A proteins, due to common exons 2-5, share an identical C-terminal domain. UGTs are most abundantly expressed in the liver but are also present in variety of other tissues, including the GI tract, kidney, and lung. Thus, UGTs are involved in the metabolism of a wide range of injected, inhaled, and orally administered drugs and other xenobiotics. UGTs are also present in steroid-sensitive tissues like the colon, mammary gland, and prostate, where they regulate the levels of a host of important endobiotics including steroid hormones, retinoids, and bile acids 10,11,12,13, providing protection against the generation and/or progression of certain forms of cancer.
Glycosyltransferases (GTs) have been divided into 87 families (GTx) based on amino acid similarities and the types of donor ligand utilized 14,15 (Carbohydrate-Active Enzymes server at http://afmb.cnrsmrs.fr/CAZY/index.html). Structural examination of GTs has sometimes been hampered by the difficulties associated with their overexpression, purification, and crystallization. In particular, production of soluble, full length human UGT that generates crystals has yet to be reported despite intense efforts by us and other research groups 4,5. Currently, structural information is available for only a limited number of GT families, revealing two distinct structural folds, GT-A and GT-B 16. GT-A proteins consist of a single α/β/α sandwich that resembles a Rossman-like fold and contains a divalent metal that is important for donor ligand binding. GT-B enzymes are composed of two Rossman-like domains that associate to form a catalytic cleft at their interface. While there is cross talk between the domains the majority of the donor ligand contacts are contributed by the C-terminal domain while the majority of the acceptor substrate contacts are contributed by the N-terminal domain. In contrast to GT-A fold-containing GTs, the activity of GT-B enzymes are not dependent on divalent metals; however, these metals enhance the activity of some GT-B enzymes 17,18.
Human UGTs belong to the GT1 family 14,15 and are predicted to adopt a GT-B fold 16. Crystal structures of several GT1 family members have been solved in recent years, including the bacterial Gtf family of enzymes involved with vancomycin synthesis 19,20,21, and two flavonoid GTs from plants 22,23, which are important for the glycosylation of secondary metabolites. We determined the 1.8 Å resolution x-ray crystal structure of the C-terminal domain of human UGT2B7, a major enzyme involved in the metabolism of important endobiotic signaling molecules and therapeutics 24. UGT2B7 regulates the levels of many potent endobiotic signaling molecules like androsterone, estriol, and retinoic acid. UGT2B7 also directly conjugates drugs from all major classes, including cancer therapeutics like epirubicin and the AIDS drug AZT 25. In addition, UGT2B7 is the only human UGT able to metabolize morphine-based opioids, which are central to the management of chronic and acute pain. We present structural and functional data that elucidate numerous aspects of human UGT donor-substrate binding and catalytic activity. Taken together, the results presented here provide a foundation from which the molecular mechanisms behind human UGT donor-substrate binding and catalytic activity can begin to be deciphered.
Results
Overall Structure
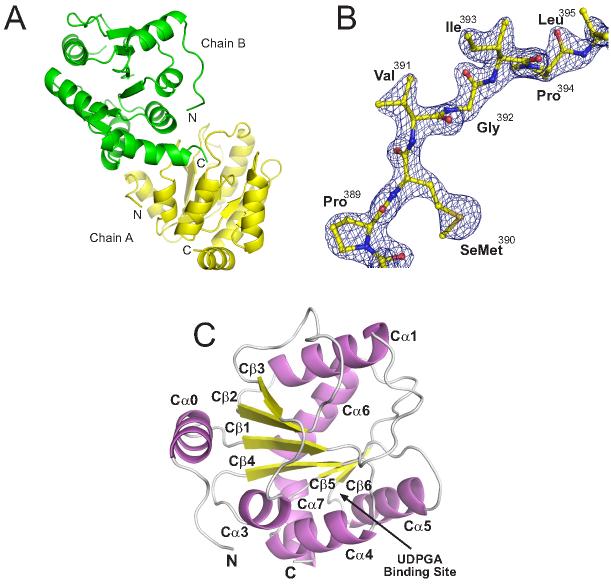
Significant efforts in our laboratory have been directed at producing soluble, human UGTs suitable for crystallographic analyses. Despite employing multiple prokaryotic and eukaryotic expression systems and a wide range of constructs, we have yet to be able to produce soluble forms of a UGT containing both the N-terminal substrate and C-terminal cofactor binding domains. Concurrent with these efforts, we also pursued separate N- and C-terminal domain E. coli expression strategies for UGT2B7. The N-terminal domain proved to be highly unstable. In contrast, the C-terminal domain (2B7CT, residues 285-451) was well behaved and crystallized readily. The three dimensional structure was determined initially to 2.1 Å resolution using multiwavelength anomalous dispersion (MAD) phasing and a single crystal containing selenomethionine-substituted protein (Figure 1A). The resulting experimental electron density map was of high quality for nearly all residues (Figure 1B). MAD methods were utilized because molecular replacement with predicted structural homologs from prokaryotic glycosyltransferases was not successful, and the structures of related glucosyltransferases from plants were not yet available. The 2B7CT structure was ultimately refined to 1.8 Å resolution using the low-remote wavelength data set collected from a different region of the same crystal. Data collection, phasing, and refinement statistics are summarized in Table 1.
Figure 1. Overall Structure of 2B7CT.
(A) Asymmetric dimer present in the crystallographic asymmetric unit. (B) Solvent flattened 2.1 Å experimental electron density map contoured at 2σ. (C) Ribbon cartoon of 2B7CT with labeled secondary structure elements.
Table 1.
Summary of Data Collection, Phasing and Refinement
| Data Collection for UGT2B7 C-terminal Domain | ||||
| Space Group | P6522 | |||
| Unit Cell [abc, Å; αβγ, °] | a,b=75.1 c=218.2 α,γ=90 β=120 |
|||
| Data Set | Low Energy Remote |
Peak | Inflection | High Energy Remote |
| Wavelength [Å] | 0.98089 | 0.97925 | 0.97942 | 0.97174 |
| X-ray Source | APS SER-CAT 22-ID | |||
| Resolution [Å] (Highest shell) | 50−1.80 (1.86−1.80) | 50−2.00 (2.07−2.00) | 50−2.10 (2.18−2.10) | 50−2.00 (2.07−2.00) |
| No. of Reflections [unique] | 33813 | 25127 | 21866 | 25168 |
| Completeness [%] | 97.7 (94.3) | 97.7 (96.9) | 97.8 (97.1) | 97.7 (97.0) |
| Rsym a [%] | 5.6 (39.2) | 9.8 (33.4) | 10.0 (40.6) | 8.7 (42.9) |
| I/σ | 50 (6.5) | 19.2 (6.9) | 18.0 (6.0) | 22.1 (5.9) |
| Redundancy | 14.4 (13.4) | 10.6 (11.0) | 10.7 (11.1) | 10.8 (11.1) |
| MAD Phasing Statistics out to 2.1 Å | ||||
| Se atom sites in AU (observed/total) | 12/14 | |||
| Mean Figure of Merit | ||||
| Solve (Highest shell) | 0.63 (0.41) | |||
| Resolve (Highest shell) | 0.71 (0.60) | |||
| Refinement Summary | ||||
| Resolution [Å] (Highest shell) | 50−1.80 (1.91−1.8) | |||
| Molecules per asymmetric unit (AU) | 2 | |||
| No. of protein atoms/AU | 2673 | |||
| No. of waters/AU | 283 | |||
| Rworking b [%] | 21.9 (29.6) | |||
| Rfree c [%] | 24.5 (34.9) | |||
| Average B factor [Å2] | ||||
| Protein | 28.5 | |||
| Water | 41.3 | |||
| RMS deviations | ||||
| Bonds [Å] | 0.005 | |||
| Angles [°] | 1.30 | |||
| Coordinate error estimates [Å] | ||||
| Luzzati | 0.24 | |||
| SigmaA | 0.21 | |||
| Ramachandran [%] | ||||
| most, allowed, generous, disallowed | 95.3, 3.9 ,0.7, 0.0 | |||
| PDB ID code | 2O6L | |||
Rsym = Σ|I− 〈I〉|/ Σ|I|, where I is the observed intensity and <I> is the average intensity of several symmetry-related observations.
Rworking = Σ||Fo|−|Fc||/ Σ|Fo, where Fo and Fc are the observed and calculated structure factors, respectively.
Rfree = Σ||Fo|−|Fc||/ Σ|Fo for 5% of the data not used at any stage of the structural refinement; 5% data omitted for Rfree
The 2B7CT structure is a globular domain with a Rossman-type fold (Figure 1C). At the core of the protein is a single parallel β-sheet consisting of six individual strands surrounded by seven α-helices. This structure is consistent with the predictions from sequence similarities and fold prediction algorithms like Phyre 26 that human UGTs belong to the GT-B fold family of glycosyltransferases. The asymmetric unit contains two 2B7CT molecules that pack together to form an asymmetric dimer (Figure 1A). These molecules are nearly identical, with a root mean square deviation (RMSD) of 0.38 and 1.38 Å for 157 Cα atoms (residues 285-310, 315-446) and all atoms, respectively. They do contain small regions of differences. Residues 282-284, which are residual expression vector residues, and 447-451 were not modeled in chain A due to poor electron density. Similarly, residues 311-314 from chain B also had poor electron density and were not modeled.
Comparisons to Glycosyltransferase Structures
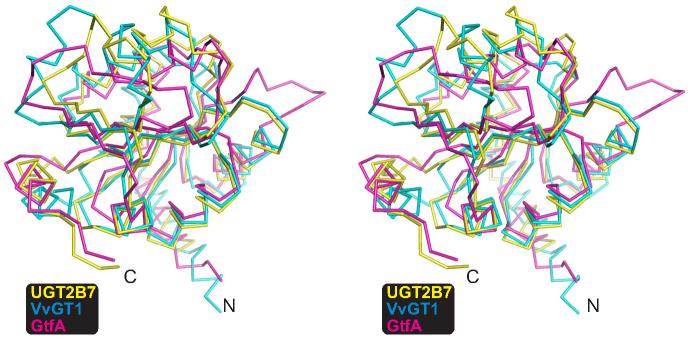
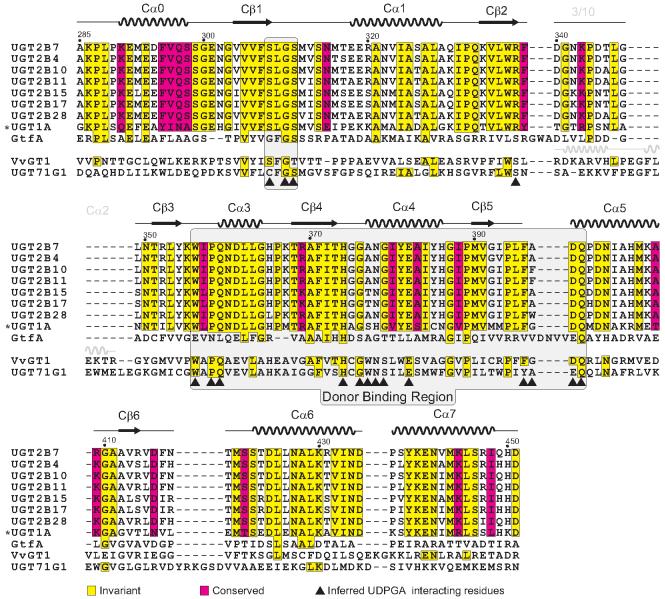
Structures of GT-B fold-containing glycosyltransferases from several GT classes have been determined in recent years 16. These include phage T4 DNA glucosyltransferase (GT63) 27,28, MurG (GT28) 29,30, which is involved in peptidoglycan formation, and the Gtf family of enzymes (GT1) involved with vancomycin synthesis 19,20,21. Two very recent structures of GT1 family plant flavonoid glucosyltransferases, VvGT1 and UGT71G1, have been completed 22,23. Superposition of the C-terminal domains of GtfA and VvGT1 with 2B7CT reveals a high level of structural homology despite their low sequence identity (∼19%), with RMSDs of 1.95 Å over 141 Cα atoms, and 1.77 Å over 147 Cα atoms for each, respectively (Figure 2). A structure based-sequence alignment of representative GT1 family enzymes is shown in Figure 3. Although the majority of secondary structure elements are similar, discrete differences exist. All structures significantly differ in both length and secondary structure for the amino acids connecting Cβ2 and Cβ3. In VvGT1, both a 3/10 helix and α-helix Cα2 are present in this region, while in both 2B7CT and GtfA a shorter loop structure is observed (Figure 3).
Figure 2. Comparison of C-terminal domains of GT1 family enzymes.
Stereo image of the plant glucosyltransferase VvGT1 and bacterial GT GtfA C-terminal domains superimposed to the 2B7CT structure.
Figure 3. Sequence alignment of GT1 family enzymes.
All unique C-terminal domain sequences from human UGTs were aligned to 2B7CT and the conservation highlighted. This conservation mapping was extended to the structure based sequence alignments of representative GT1 family GTs from both bacteria and plants if they shared identity with human sequences. The secondary structure of 2B7CT is depicted above its sequence and in gray above VvGT1 is secondary structure that is present in VvGT1 but missing in 2B7CT. A shaded box is drawn around the regions that are important for donor ligand binding. *-all UGT1A family members share an identical C-terminal domain.
UDPGA Binding Site
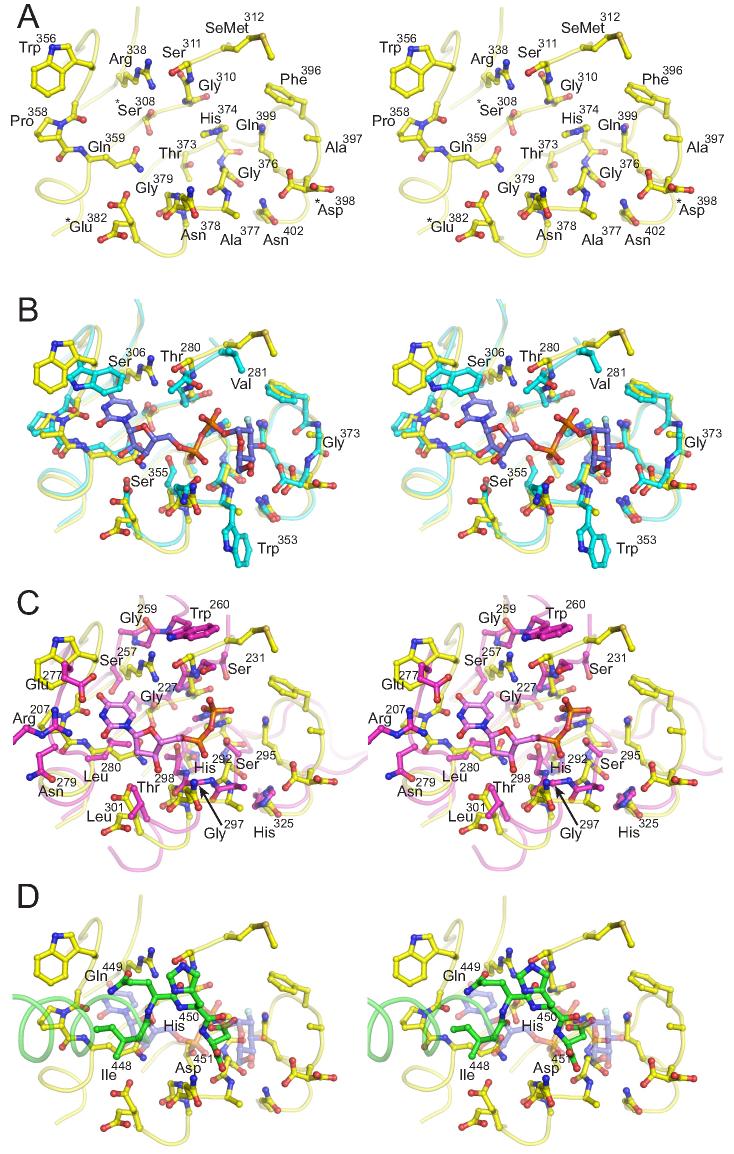
A hallmark feature of GT-B fold-containing enzymes is the di-phosphate nucleotide sugar binding site formed by the C-terminal domain. Structurally characterized GT-B superfamily enzyme nucleotide-sugar binding sites utilize a common structural scaffold; however, the nature of the specific interactions with the donor ligands are varied, even amongst enzymes in the same GT family. Comparison of 2B7CT with other GT1 family enzyme structures from bacteria and plants (GtfA and VvGT1) in complex with donor ligands suggests that human UGT2B7 binds UDPGA with an analogous site (Figure 4A). The presumptive UDPGA binding site of UGT2B7 is remarkably similar to the UDP-glucose binding site of VvGT1 (Figure 4B), with only a handful of amino acid differences. Most notable among them are VvGT1 residues Ser306 and Ser355, which make hydrogen bonds to the uracil base and α-phosphate moieties, respectively. This α-phosphate hydrogen bonding interaction is also seen in other GT-B superfamily enzymes. Equivalent positions in 2B7CT are Arg338 and Gly379, respectively, highlighting key areas of difference that may have functional significance.
Figure 4. UGT2B7 has a nucleotide-sugar binding site most similar to plant flavonoid glucosyltransferases (stereo pairs).
(A) Presumed UDPGA binding site of UGT2B7. Superposition of 2B7CT with the (B) plant glucosyltransferase VvGT1 in complex with UDP-2-deoxy-2-fluro glucose and with (C) the bacterial GT GtfA in complex with dTDP. Only residues that differ with 2B7CT are labeled. (D) 2B7CT asymmetric dimer interface. C-terminus of Chain A (green and residues labeled) packs into the UDPGA binding site of chain A (Yellow). UDP-2-deoxy-2-fluro glucose from the superimposed VvGT1 is shown as a transparent object. *-denotes residues where multiple conformations were observed.
The Gtf family of enzymes in bacteria utilize a dTDP-sugar donor ligand, while plant and human UGTs utilize a UDP-sugar donor ligand. Comparison of this site between GtfA and UGT2B7 revealed several differences (Figure 4C). Ser295 in GtfA hydrogen bonds to the O2G phosphate oxygen; in plants and humans this residue is a glycine, resulting in a different di-phosphate conformation in VvGT1. The thymine moiety is not pinned in place with a Trp residue via a ring-stacking interaction; instead, a salt bridge between Glu277 and Arg207 holds the nucleotide in place. Additionally, the region adjacent to the thymine is more hydrophobic (Leu280/Leu301), which is complemented by the deoxy nature of the ribose. The short loop in 2B7CT/VvGT1 that connects Cα5 and Cβ5 makes direct contacts with the glucose moiety in VvGT1. This loop is both longer and in an extended conformation in the GtfA structure, likely due to only having a dTDP bound. The long length of this loop is thought to help accommodate the large size of Gtf family enzyme acceptor substrates 21. Residues analogous to those in VvGT1 that interact with the glucose moiety are conserved in both human UGTs (Asp398, Gln399) and Gtf family proteins (Figure 3).
All attempts to co-crystallize 2B7CT with UDPGA were unsuccessful. Addition of UDPGA to our crystallization conditions resulted in a concentration dependent inhibition of crystal formation. Similarly, UDPGA soaking experiments with 2B7CT crystals also failed. Once the structure was determined, it was clear that this due in part to the nature of the crystal packing interactions in the asymmetric 2B7CT dimer in the crystallographic asymmetric unit (Figure 1A). The C-terminus of one monomer packs into the predicted UDPGA binding site of the other monomer (Figure 4D). Removal of the C-terminal residues, in an effort to remove this occlusion, produced protein samples that were highly unstable and could not be crystallized. The UDPGA binding site of one 2B7CT monomer in the asymmetric unit is un-obstructed, however. Analysis of difference Fourier electron density maps between 2B7CT data and data from crystals obtained in the presence of 8-fold molar excess UDPGA did not show any significant density at the free nucleotide-sugar binding site (data not shown). This is perhaps not surprising since the UDPGA binding site is only a shallow surface site on 2B7CT in the absence of the N-terminal domain.
Examining the sequence conservation amongst human UGTs at the predicted UDPGA binding site revealed a high level of sequence identity (Figure 3). Mapping this conservation to the molecular surface of 2B7CT reveals that the majority of the binding site residues are invariant (Figure 5), suggesting that all human UGTs interact with UDPGA in an analogous way. The conserved sequence patch beneath the uracil base corresponds to residue 357 (Ile/Val). However, only the mainchain atoms of this residue are predicted to interact with the uracil base; the side chain is directed away from the binding site. Despite the overall low sequence identity between VvGT1 and 2B7CT (19%), many of the residues that contact UDPglucose in VvGT1 are conserved in human UGTs (Figure 3, 4B). In addition, a high degree of structural similarity exists between the C-terminal domain of VvGT1 and 2B7CT at the VvGT1 nucleotide-sugar binding site. Taken together, these data strongly suggest that human UGTs bind nucleotide-sugars in a way that is very similar to plant glucosyltransferases. Utilizing this information we modeled UDPGA into the predicted nucleotide-sugar binding site of 2B7CT (Figure 5).
Figure 5. UDPGA binding site is highly conserved in humans.
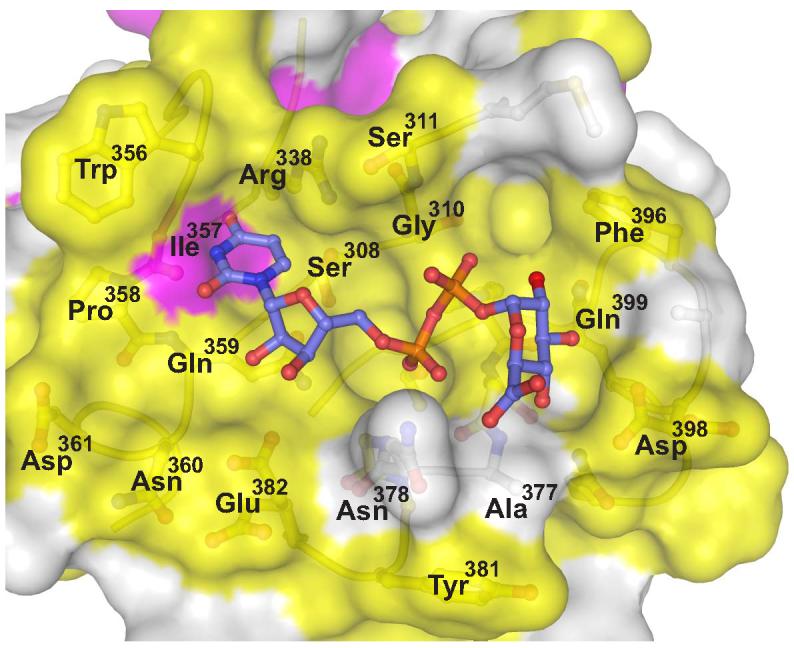
Human UGT sequence conservation (described in Figure 3) is mapped to the molecular surface of 2B7CT. Yellow is invariant, magenta is conserved, and white signifies low conservation. Using the VvGT1 structure as a guide UDPGA was modeled into the 2B7CT structure and is shown in a ball and stick representation.
Mutational Analysis Confirms UDPGA Binding Site
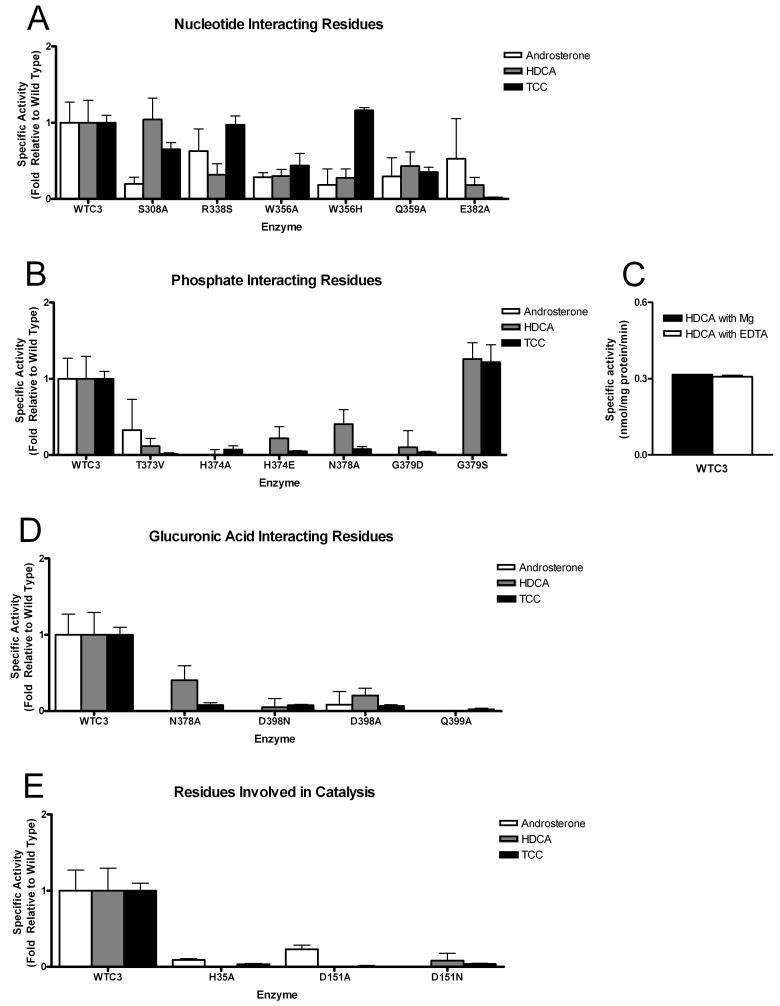
In order to validate our 2B7CT-UDPGA model, a series of enzyme activity assays were conducted with microsomal fractions containing full-length UGT2B7 expressed in HighFive insect cells. Using the 2B7CT-UDPGA model as a guide, the binding site was broken down into three groups: residues predicted to interact with the uracil base, the di-phosphate group, or the glucuronic acid moiety. A panel of point mutants at these positions were generated and their impact on enzyme activity assessed with three distinct substrates (Figure 6).
Figure 6. Mutational analysis of the UGT2B7 UDPGA site.
Activity assays were carried out with three distinct acceptor ligands: androsterone, hyodeoxycholic acid (HDCA) and tetrachlorocatechol (TCC). The activity of each mutant was normalized to wildtype levels and the results graphed according to their predicted interactions with UDPGA (A,B,D,E). (C) Wildtype UGT2B7 activity assay conducted in the presence/absence of EDTA.
Most mutations of residues surrounding the uracil base moderately impaired enzyme activity, while others had only minor effects (Figure 6A). Trp356 is predicted to ring-stack with the uracil base (Figure 4B), which is a type of interaction seen in other GT-B superfamily enzymes 29,31. Mutation of this residue to an alanine or histidine (a smaller aromatic residue) and the resulting attenuated activity demonstrates the importance of this interaction to UGT2B7 function. The foundation provided by Gln359 to the uracil base appears to be of equal importance. Disruption of the predicted hydrogen bond between Glu382 and the ribose 2'-OH via an alanine mutation also impairs function. Residue Ser308 forms part of the uracil binding pocket but is not predicted to make any direct contacts with the uracil base. An alanine mutation at this position has little or no effect on activity, indicating a minor role in enzyme activity. Arg338 was mutated to serine, mimicking the nucleotide binding site found in other GT1 family glycosyltransferases. Restoring this uracil base hydrogen bonding interaction did not result in increased activity, rather it had little or no effect. Interestingly, several mutants (e.g., S308A, R338S, W356H) exhibited stronger effects on glucuronic acid transfer to some substrates (e.g., TCC, HDCA) relative to others (e.g., androsterone). This observation suggests that the positioning of the UDPGA cofactor may have a significant effect on catalytic activity. Subtle alterations in UDPGA binding appear to impact the association of acceptor molecules within the N-terminal region of the enzyme.
Mutations at residues predicted to interact with the di-phosphate moiety of UDPGA had significant effects on enzyme function (Figure 6B). His374, a residue conserved in other GT1 family enzymes, is positioned directly beneath the β-phosphate. Mutation of this residue to alanine or introduction of a negative charge via a glutamic acid mutation virtually eliminated activity. A key contact in the AU dimer of 2B7CT occurs at this site. The negatively charged C-terminal aspartic acid sidechain of one molecule packs where the di-phosphate moiety is predicted to bind (Figure 4D). Taken together, these observations support the idea that His374 is acting to help neutralize the negative charge on the β-phosphate. The N-terminal end of helix Cα4 is predicted to interact with the α-phosphate via its positively charged dipole, a contact common to many GT-B superfamily glycosyltransferases 18. Hydrogen bonding interactions also appear to be important, as mutation of Asn378, predicted to interact with the α-phosphate, to an alanine results in a significant loss of activity.
Due to the presence of a glycine at position 379, a pocket is formed underneath the α-phosphate in 2B7CT. This represents a major point of difference between human and other GT-B superfamily enzymes, in which a highly conserved serine/threonine hydrogen bonds with the α-phosphate. At the base of the pocket in 2B7CT is Thr373, which could interact with the α-phosphate via a water- or ion-mediated contact. Although no divalent metal dependence has been observed for GT-B superfamily enzymes, they have been shown to enhance significantly the activity of some GT-B enzymes. Activity experiments with UGT2B7 were carried out in the presence of EDTA and no appreciable loss or gain in activity was observed (Figure 6C). A network of waters, however, is present in this pocket in the 2B7CT structure. Mutation of Thr373 to a nearly iso-structural valine, which removes the hydrogen bonding potential of the side chain, severely disrupts enzyme activity (Figure 6B) suggesting that water-mediated contacts to the cofactor di-phosphate group are critical to enzyme function. Replacement of Gly379in UGT2B7 with a serine makes another hydrogen bonding partner for UDPGA available and results in an increase of activity compared to wildtype, while an aspartic acid mutation eliminates activity, perhaps by nullifying the positive Cα4 helix dipole and/or direct charge repulsion with the UDPGA di-phosphate moiety.
Several residues that are predicted to hydrogen bond to the glucuronic acid moiety are absolutely critical for UGT2B7 function (Figure 6D). Asp398 and Gln399, which are conserved in other GT1 family glycosyltransferases, are predicted to interact with O3'/O4' and O2'/O3' atoms of glucuronic acid, respectively. Mutation of either residue to an alanine results in loss of enzyme activity. A more conservative mutation of Asp398 to an asparagine also results in a complete loss of activity, suggesting that a negative charge, not just hydrogen bonding potential, is important at this position. Similar mutations at this position in structurally defined plant glucosyltransferases also resulted in a loss of function 22,23. These residues are likely important in defining the sugar specificity of human UGT enzymes. In addition, Asn378, due to its predicted proximity to the glucuronic acid COOH group, may also be involved with donor sugar selectivity.
Taken together, our UGT2B7 structural and functional data demonstrate that mutations predicted to disrupt interactions with either the di-phosphate or glucuronic acid moieties of UGT2B7 tended to abolish activity, while predicted uracil base interacting residues only moderately reduce activity. These observations argue that interactions closest to the phosphate-sugar bond being broken are the most important to enzyme activity.
Insights into the Catalytic Mechanism of Human UGTs
Recently, two groups published crystal structures of plant glucosyltransferases, each proposing similar catalytic mechanisms 22,23. The structure of full length VvGT1 in complex with both a non-hydrolyzable form of UDP-glucose and the acceptor substrate quercetin is shown in Figure 7A. Using this structure as a starting point, the software package loopp was used to generate a homology model of UGT2B7 32. Inspection of this model at the catalytic center revealed that UGT2B7 has residues analogous to plant glucosyltransferases at key catalytic positions (His35, Asp151). This model predicts that human UGTs use a serine hydrolase-like catalytic triad for catalysis. His35 of UGT2B7 deprotonates a suitable atom of the acceptor ligand, facilitating a nucleophilic attack at the C1 atom of glucuronic acid (Figure 7B). The resulting protonated histidine is stabilized by a neighboring aspartic acid at position 151. Mutation of His35 to alanine or Asp151 to either alanine or asparagine resulted in a catalytically inactive enzyme (Figure 6E). Similar mutations in plant glucosyltransferases had equivalent results 22,23, as did analogous mutations in the serine protease members of the serine hydrolase superfamily 33. The histidine residue predicted in the active site of UGT2B7 is invariant in both human and plant enzymes (Figure 7C), while the stabilizing aspartic acid is invariant in humans and the vast majority of plant enzymes. These data suggest that all human UGTs utilize a similar catalytic mechanism of deprotonation by charge relay of an appropriate functional group to carry out their promiscuous conjugative chemistry
Figure 7. UGT2B7 utilizes a serine-hydrolase like catalytic triad for catalysis.
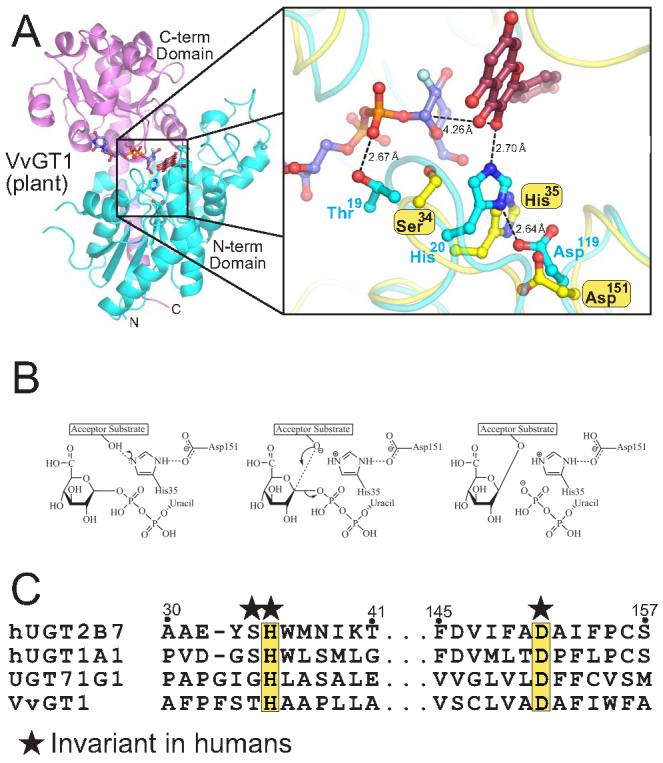
(A) The VvGT1 structure in complex with both donor and acceptor ligands. An enlarged representation of the VvGT1 catalytic region (cyan) is shown superimposed to a homology model of UGT2B7 (yellow). (B) Schematic of a proposed catalytic reaction mechanism for human UGT2B7. (C) Sequence alignment of human and plant GTs in regions important for catalysis.
Discussion
The high resolution crystal structure of the C-terminal domain of human UGT2B7 presented here represents the first structural data on this class of phase II xenobiotic metabolizing enzymes. Structural comparisons with related GT-B superfamily glycosyltransferases and activity data from a panel of UGT2B7 point mutants revealed that human UGTs possess and utilize a nucleotide-sugar binding site similar to those found in plant flavonoid glucosyltransferases. Homology modeling of UGT2B7 further suggested that human UGTs employ a serine hydrolase-like catalytic triad for catalysis, which was experimentally verified with enzyme activity experiments using UGT2B7 mutated at critical catalytic residues (His35, Asp151). In summary, our results provide a wealth of information about human UGT function forming a framework from which the molecular mechanisms behind their donor-substrate binding and catalytic activity can begin to be understood.
Human UGTs are promiscuous enzymes capable of conjugating glucuronic acid to a diverse array of substrates. The predominant functional group used for conjugation is an –OH moiety, although a number of additional functional groups, including sulfurs, amines and acidic carbons, are also conjugated 2,34. This variability is apparently enabled by a serine hydrolase-like catalytic triad, demonstrating the flexibility of this mechanism in human UGTs. Unfortunately, this flexibility comes at the cost of efficiency, a hallmark of promiscuous enzymes, as typical human UGT Kms are in the micromolar range.
The catalytic mechanism of related prokaryotic glycosyltransferases has been studied in some detail. Structural analysis of these enzymes, in contrast to related human and plant enzymes, did not reveal the presence of a His-Asp charge relay 16. Instead these enzymes require only the presence of a general base in equivalent spatial space to deprotonate their acceptor substrates, thus facilitating the sugar transfer. MurG has been shown to have a catalytic histidine 30, while the Gtf family of enzymes was proposed to have two important catalytic residues, a serine/threonine and an aspartic acid 21. Mutation of this aspartic acid residue in GtfD to alanine abolished activity, while a similar alanine mutation of the threonine resulted in attenuated activity 19. Replacement of the catalytic His35 in human UGT2B7 with an aspartic acid, however, resulted in no activity (data not shown).
GT-B superfamily glycosyltransferase activity is divalent metal ion-independent; however, divalent metal ions are required for some GT-B enzymes to achieve optimal activity 17,18. It is clear from the structural data thus far accumulated that a divalent metal is not involved in nucleotide-sugar binding, as it is in GTA superfamily glycosyltransferases. Enhancement of activity by divalent metals could be occurring via interactions with acceptor substrates and their subsequent N-terminal domain binding sites. Both phage T4 DNA β-glucosyltransferase and MurG are activated by divalent metal ions, but structural data describing any kind of acceptor substrate complex is lacking. Alternatively, it has been suggested that these metals could be playing a role in product release 27. Our activity experiments with UGT2B7 in the absence and presence of EDTA did not show any gain or loss of activity, suggesting divalent metals do not play a role with the acceptor substrates tested. Preliminary activity experiments with other insect cell produced human UGTs and liver microsomes, however, suggest that the activity of some UGTs are significantly enhanced by divalent metals (data not shown).
UGT2B7 is one of the predominant drug metabolizing UGTs in human tissues. A variety of important drugs, including the anti-cancer drug epirubicin and the opioid-based pain medicines, are inactivated via UGT2B7-mediated glucuronidation. A UGT inhibitor co-administered with these drugs may allow for smaller doses of such drugs to be administered to patients, resulting in higher efficacy and fewer side effects. The most similar region among all human UGTs is the C-terminal nucleotide-sugar binding site. Some of the most effective UGT inhibitors, which are based on a uridine/UDP moiety scaffold, exploit this characteristic 35. Unfortunately, these inhibitors are likely to have serious side effects, as a number of critical biological processes utilize nucleotide-sugars. Our structural data in conjunction with other GT1 family enzyme structures may facilitate the development of additional scaffolds that are more UGT specific. As additional structural information becomes available, the development of isoform-specific UGT inhibitors may lead to safer, more effective co-therapies for a variety of conditions.
Materials and Methods
Protein Expression and Purification
Residues 285-451, which correspond to the C-terminal ER luminal domain, from of human UGT2B7 cDNA (a generous gift from Dr. Peter Mackenzie, Flinders University, Australia) were cloned into a pET based expression vector using ligation independent cloning techniques 36. This protein was expressed as a six-histidine tagged N-terminal maltose binding protein (MBP) fusion protein via a linker containing a TEV protease site (HIS-MBP-TEV-2B7CT). Protein expression was carried out in the methionine auxotroph E. coli strain B834 (DE3) (Novagen) in SelenoMet media mix (Molecular Dimensions) containing 50 mg/L of selinomethionine according to included protocols. E. coli cells were grown at 37° C to an OD600 of 0.6. The temperature was lowered to 18° C and 0.1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) added to induce protein expression. Ten hours post-induction the cells were collected by centrifugation and resuspended into a lysis buffer containing 50 mM NaH2PO4 pH 8.0, 300 mM NaCl, 0.2% Triton X-100, 25 mM imidazole, and 0.01% Na Azide. The cells were disrupted by sonication and centrifuged at 40,000g for 1hr at 4° C. The resulting supernatant was subjected to affinity chromatography with Ni-6 sepherose media (Amersham Biosciences). Purified fusion protein was eluted with an imidazole gradient and the appropriate fractions pooled. Protein was diluted to 5 mg/ml or less, TEV protease added, and the resulting solution dialyzed against cell lysis buffer minus Triton X-100 for 12 hrs at room temp. Ni-6 sepherose affinity chromatography was used to remove uncleaved fusion and MBP away from free 2B7CT, which was then concentrated and subjected to gel filtration chromatography using a superdex75 column (Amersham Biosciences) equilibrated with 20 mM Hepes pH 7.4, 150 mM NaCl, and 0.01% Na Azide. Appropriate fractions were pooled, dialyzed into a crystallization buffer (100 mM Ammonium Acetate pH 6.88, 75 mM NaCl, 0.01% Na Azide) and concentrated to 7 mg/ml.
To generate protein for UGT activity assays, full length human UGT2B7 (residues 1-529) was cloned into a modified pFastBac DUAL vector (A generous gift from Dr. Daved H. Fremont, Washington University St. Louis, MO) under the control of the polyhedron promoter as a C-terminal enterokinase cleavable sixhistidine tag fusion. The vector modification entails the replacement of the P10 promoter with the pBasic promoter which is used to drive the early (18-24 hours) expression of eGFP, a visual marker for viral titering and infection. UGT2B7 point mutations were generated using site-directed mutagenesis. Recombinant bacmids and virus were generated using the Bac-to-Bac system (Invitrogen) according to manufacturer's protocols. Briefly, recombinant vector is transformed into the E. coli strain DH10BAC and recombination positive colonies are used to prepare recombinant bacmid, which is then transfected into Sf9 insect cells growing in adherent culture using cellfectin. 72 hours post-transfection, the virus containing medium is harvested and subjected to multiple rounds of amplification with a multiplicity of infection (MOI) of 0.1. Full length protein was expressed by infecting HighFive cells at a density of 1.5 × 106 per ml with recombinant virus at an MOI of 5-10. 72 hours post-infection, cells are isolated by centrifugation, flash frozen in liquid nitrogen and stored at −80° C. Microsomal fractions suitable for enzymatic activity assays were prepared as described 37. The microsomes were resuspended in a solution containing 100 mM potassium phosphate pH 7.4, 20% (v/v) glycerol, 0.1 mM EDTA, and 1 mM DTT to a concentration of 10 mg/ml. The resulting samples were normalized with respect to UGT2B7 protein amounts using a combination of BCA assay and an anti-six-histidine western blot.
Crystallization
Initial crystallization conditions were generated from the free screening services offered by the high throughput crystallization lab at the Hauptman-Woodward Institute (Buffalo, NY). Several rounds of iterative refinement were carried out to adapt these initial crystallization hits to hanging drop vapor diffusion methodology. Diffraction quality crystals of 2B7CT were produced by mixing 1 μl of 7 mg/ml protein solution with 1 μl of reservoir solution (12-16% PEG 4000, 100 mM K2CO3, 100 mM Tris pH 7.4) and equilibrating against 500 μl of reservoir solution at 22° C. Crystals appeared within 1 week and grew as large hexagonal rods. Crystals were cryoprotected by addition of 2 μl of well solution plus 20% ethylene glycol to the drop. Crystals were immediately looped and flash-frozen in a 100° K nitrogen stream.
Structure determination
A three-wavelength (peak, inflection point, and high remote of the Se K-edge) SeMet MAD experiment was carried out on beamline ID-22 (SER-CAT) at the Advanced Photon Source at Argonne National Laboratory in Chicago, IL. An additional low-remote wavelength data collection was conducted on a different part of the same crystal. Data were integrated, scaled and reduced using HKL2000 38. SOLVE was used to find the positions of the Se atoms and provide initial phase estimates 39. The resulting experimental electron density map was solvent flattened and traced in an automated fashion using RESOLVE, generating a nearly complete poly-alanine model 40,41. Additional tracing was carried out using COOT and the resulting model refined with CNS using maximum likelihood targets 42,43. Final rounds of model building and refinement were carried out using the high-resolution low-remote data set. A summary of data collection, phasing, and refinement statistics can be found in Table 1. Alternate conformations were observed and built for a number of amino acid side chains. Molecular graphics for figures were generated with Pymol 44.
Enzymatic assays
Glucuronidation of androsterone, hyodeoxycholic Acid (HDCA) and tetrachlorocatechol (TCC) (all from Sigma) were measured using protocols based on previously published procedures 45. Briefly, membrane protein (10 μg) was incubated in 100 μM Tris-HCl (pH 7.4)/5 mM MgCl2/5 mM saccharolactone/2% DMSO with each of the substrates (200 μM), in a total volume of 30 μl. The reactions were started by the addition of [14C]UDPGA (4 mM) and incubated at 37°C for 20 min. The reaction was stopped by addition of 30 μl of ethanol.
Aliquots (40 μl) of each sample were then applied to the preabsorbent layer of channeled silica gel TLC plates (Baker 250Si-PA (19C); VWR Scientific, Sugarland, TX) and glucuronidated products and unreacted substrate were separated by development in chloroform-methanol-glacial acetic acid-water (65:25:2:4, v/v), with the exception of TCC, which was developed in butanol-acetone-glacial acetic acid-30% ammonia-water (70:50:18:1.5:60). Radioactive compounds were then localized on the TLC plates by autoradiography for 3-4 days at −80°C. Silica gel in areas corresponding to the glucuronide bands identified from autoradiograms and corresponding areas from control lanes were scraped from the TLC plate into scintillation vials, and the radioactivity was measured by liquid scintillation counting (Packard TRI-CARB 2100TR, Perkin-Elmer).
Acknowledgements
The authors would like to thank Anna Gallus-Zawada for her excellent technical assistance with this project. The authors would also like to thank Christopher Fleming, Scott Lujan, and Eric Ortlund for helpful advice. This work was supported in part by NIH grants CA98468 to M.R.R., and DK56226 and DK60109 to A.R.-P.
Abbreviations
- UGT
UDP-Glucuronosyltransferase
- UGT2B7
Human UDP-Glucuronosyltransferase isoform 2B7
- UDPGA
UDP-glucuronic acid
- ER
endoplasmic reticulum
- GI
gastrointestinal tract
- GT
glycosyltransferase
- AIDS
acquired immunodeficiency syndrome
- AZT
azidothymidine
- MBP
maltose binding protein
- 2B7CT
C-terminal domain of UGT2B7, residues 285-451
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- eGFP
enhanced green fluorescent protein
- Sf9
Spodoptera frugiperda cell line
- HighFive
Trichoplusia ni cell line
- MOI
multiplicity of infection
- BCA
bicinchoninic acid
- MAD
multiple wavelength anomalous dispersion
- HDCA
hyodeoxycholic acid
- TCC
tetrachlorocatechol
- DMSO
dimethylsulfoxide
- RMSD
root mean square deviation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tukey RH, Strassburg CP. Human UDP-glucuronosyltransferases: metabolism, expression, and disease. Annu Rev Pharmacol Toxicol. 2000;40:581–616. doi: 10.1146/annurev.pharmtox.40.1.581. [DOI] [PubMed] [Google Scholar]
- 2.King CD, Rios GR, Green MD, Tephly TR. UDPglucuronosyltransferases. Curr Drug Metab. 2000;1:143–61. doi: 10.2174/1389200003339171. [DOI] [PubMed] [Google Scholar]
- 3.Radominska-Pandya A, Pokrovskaya ID, Xu J, Little JM, Jude AR, Kurten RC, Czernik PJ. Nuclear UDP-glucuronosyltransferases: identification of UGT2B7 and UGT1A6 in human liver nuclear membranes. Arch Biochem Biophys. 2002;399:37–48. doi: 10.1006/abbi.2001.2743. [DOI] [PubMed] [Google Scholar]
- 4.Meech R, Mackenzie PI. Determinants of UDP glucuronosyltransferase membrane association and residency in the endoplasmic reticulum. Arch Biochem Biophys. 1998;356:77–85. doi: 10.1006/abbi.1998.0750. [DOI] [PubMed] [Google Scholar]
- 5.Ouzzine M, Magdalou J, Burchell B, Fournel-Gigleux S. An internal signal sequence mediates the targeting and retention of the human UDP-glucuronosyltransferase 1A6 to the endoplasmic reticulum. J Biol Chem. 1999;274:31401–9. doi: 10.1074/jbc.274.44.31401. [DOI] [PubMed] [Google Scholar]
- 6.Ghosh SS, Sappal BS, Kalpana GV, Lee SW, Chowdhury JR, Chowdhury NR. Homodimerization of human bilirubin-uridine-diphosphoglucuronate glucuronosyltransferase-1 (UGT1A1) and its functional implications. J Biol Chem. 2001;276:42108–15. doi: 10.1074/jbc.M106742200. [DOI] [PubMed] [Google Scholar]
- 7.Ikushiro S, Emi Y, Iyanagi T. Protein-protein interactions between UDPglucuronosyltransferase isozymes in rat hepatic microsomes. Biochemistry. 1997;36:7154–61. doi: 10.1021/bi9702344. [DOI] [PubMed] [Google Scholar]
- 8.Meech R, Mackenzie PI. UDP-glucuronosyltransferase, the role of the amino terminus in dimerization. J Biol Chem. 1997;272:26913–7. doi: 10.1074/jbc.272.43.26913. [DOI] [PubMed] [Google Scholar]
- 9.Operana TN, Tukey RH. Oligomerization of the UDP-glucuronosyltransferase 1A proteins. Homo- and heterodimerization analysis by fluorescence resonance energy transfer (FRET) and Co-immunoprecipitation. J Biol Chem. 2006 doi: 10.1074/jbc.M609417200. [DOI] [PubMed] [Google Scholar]
- 10.Hum DW, Belanger A, Levesque E, Barbier O, Beaulieu M, Albert C, Vallee M, Guillemette C, Tchernof A, Turgeon D, Dubois S. Characterization of UDPglucuronosyltransferases active on steroid hormones. J Steroid Biochem Mol Biol. 1999;69:413–23. doi: 10.1016/s0960-0760(99)00061-8. [DOI] [PubMed] [Google Scholar]
- 11.Czernik PJ, Little JM, Barone GW, Raufman JP, Radominska-Pandya A. Glucuronidation of estrogens and retinoic acid and expression of UDPglucuronosyltransferase 2B7 in human intestinal mucosa. Drug Metab Dispos. 2000;28:1210–6. [PubMed] [Google Scholar]
- 12.Samokyszyn VM, Gall WE, Zawada G, Freyaldenhoven MA, Chen G, Mackenzie PI, Tephly TR, Radominska-Pandya A. 4-hydroxyretinoic acid, a novel substrate for human liver microsomal UDP-glucuronosyltransferase(s) and recombinant UGT2B7. J Biol Chem. 2000;275:6908–14. doi: 10.1074/jbc.275.10.6908. [DOI] [PubMed] [Google Scholar]
- 13.Kamisako T, Kobayashi Y, Takeuchi K, Ishihara T, Higuchi K, Tanaka Y, Gabazza EC, Adachi Y. Recent advances in bilirubin metabolism research: the molecular mechanism of hepatocyte bilirubin transport and its clinical relevance. J Gastroenterol. 2000;35:659–64. doi: 10.1007/s005350070044. [DOI] [PubMed] [Google Scholar]
- 14.Campbell JA, Davies GJ, Bulone V, Henrissat B. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem J. 1997;326(Pt 3):929–39. doi: 10.1042/bj3260929u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coutinho PM, Deleury E, Davies GJ, Henrissat B. An evolving hierarchical family classification for glycosyltransferases. J Mol Biol. 2003;328:307–17. doi: 10.1016/s0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- 16.Breton C, Snajdrova L, Jeanneau C, Koca J, Imberty A. Structures and mechanisms of glycosyltransferases. Glycobiology. 2006;16:29R–37R. doi: 10.1093/glycob/cwj016. [DOI] [PubMed] [Google Scholar]
- 17.Morera S, Lariviere L, Kurzeck J, Aschke-Sonnenborn U, Freemont PS, Janin J, Ruger W. High resolution crystal structures of T4 phage beta-glucosyltransferase: induced fit and effect of substrate and metal binding. J Mol Biol. 2001;311:569–77. doi: 10.1006/jmbi.2001.4905. [DOI] [PubMed] [Google Scholar]
- 18.Hu Y, Walker S. Remarkable structural similarities between diverse glycosyltransferases. Chem Biol. 2002;9:1287–96. doi: 10.1016/s1074-5521(02)00295-8. [DOI] [PubMed] [Google Scholar]
- 19.Mulichak AM, Lu W, Losey HC, Walsh CT, Garavito RM. Crystal structure of vancosaminyltransferase GtfD from the vancomycin biosynthetic pathway: interactions with acceptor and nucleotide ligands. Biochemistry. 2004;43:5170–80. doi: 10.1021/bi036130c. [DOI] [PubMed] [Google Scholar]
- 20.Mulichak AM, Losey HC, Walsh CT, Garavito RM. Structure of the UDP-glucosyltransferase GtfB that modifies the heptapeptide aglycone in the biosynthesis of vancomycin group antibiotics. Structure. 2001;9:547–57. doi: 10.1016/s0969-2126(01)00616-5. [DOI] [PubMed] [Google Scholar]
- 21.Mulichak AM, Losey HC, Lu W, Wawrzak Z, Walsh CT, Garavito RM. Structure of the TDP-epi-vancosaminyltransferase GtfA from the chloroeremomycin biosynthetic pathway. Proc Natl Acad Sci U S A. 2003;100:9238–43. doi: 10.1073/pnas.1233577100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shao H, He X, Achnine L, Blount JW, Dixon RA, Wang X. Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from Medicago truncatula. Plant Cell. 2005;17:3141–54. doi: 10.1105/tpc.105.035055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Offen W, Martinez-Fleites C, Yang M, Kiat-Lim E, Davis BG, Tarling CA, Ford CM, Bowles DJ, Davies GJ. Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. Embo J. 2006;25:1396–405. doi: 10.1038/sj.emboj.7600970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radominska-Pandya A, Little JM, Czernik PJ. Human UDPglucuronosyltransferase 2B7. Curr Drug Metab. 2001;2:283–98. doi: 10.2174/1389200013338379. [DOI] [PubMed] [Google Scholar]
- 25.Innocenti F, Iyer L, Ramirez J, Green MD, Ratain MJ. Epirubicin glucuronidation is catalyzed by human UDP-glucuronosyltransferase 2B7. Drug Metab Dispos. 2001;29:686–92. [PubMed] [Google Scholar]
- 26.Kelley LA, MacCallum RM, Sternberg MJ. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J Mol Biol. 2000;299:499–520. doi: 10.1006/jmbi.2000.3741. [DOI] [PubMed] [Google Scholar]
- 27.Lariviere L, Gueguen-Chaignon V, Morera S. Crystal structures of the T4 phage beta-glucosyltransferase and the D100A mutant in complex with UDP-glucose: glucose binding and identification of the catalytic base for a direct displacement mechanism. J Mol Biol. 2003;330:1077–86. doi: 10.1016/s0022-2836(03)00635-1. [DOI] [PubMed] [Google Scholar]
- 28.Vrielink A, Ruger W, Driessen HP, Freemont PS. Crystal structure of the DNA modifying enzyme beta-glucosyltransferase in the presence and absence of the substrate uridine diphosphoglucose. Embo J. 1994;13:3413–22. doi: 10.1002/j.1460-2075.1994.tb06646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ha S, Walker D, Shi Y, Walker S. The 1.9 A crystal structure of Escherichia coli MurG, a membrane-associated glycosyltransferase involved in peptidoglycan biosynthesis. Protein Sci. 2000;9:1045–52. doi: 10.1110/ps.9.6.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu Y, Chen L, Ha S, Gross B, Falcone B, Walker D, Mokhtarzadeh M, Walker S. Crystal structure of the MurG:UDP-GlcNAc complex reveals common structural principles of a superfamily of glycosyltransferases. Proc Natl Acad Sci U S A. 2003;100:845–9. doi: 10.1073/pnas.0235749100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buschiazzo A, Ugalde JE, Guerin ME, Shepard W, Ugalde RA, Alzari PM. Crystal structure of glycogen synthase: homologous enzymes catalyze glycogen synthesis and degradation. Embo J. 2004;23:3196–205. doi: 10.1038/sj.emboj.7600324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teodorescu O, Galor T, Pillardy J, Elber R. Enriching the sequence substitution matrix by structural information. Proteins. 2004;54:41–8. doi: 10.1002/prot.10474. [DOI] [PubMed] [Google Scholar]
- 33.Hedstrom L. Serine protease mechanism and specificity. Chem Rev. 2002;102:4501–24. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- 34.Radominska-Pandya A, Czernik PJ, Little JM, Battaglia E, Mackenzie PI. Structural and functional studies of UDP-glucuronosyltransferases. Drug Metab Rev. 1999;31:817–99. doi: 10.1081/dmr-100101944. [DOI] [PubMed] [Google Scholar]
- 35.Grancharov K, Naydenova Z, Lozeva S, Golovinsky E. Natural and synthetic inhibitors of UDP-glucuronosyltransferase. Pharmacol Ther. 2001;89:171–86. doi: 10.1016/s0163-7258(00)00109-1. [DOI] [PubMed] [Google Scholar]
- 36.Stols L, Gu M, Dieckman L, Raffen R, Collart FR, Donnelly MI. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr Purif. 2002;25:8–15. doi: 10.1006/prep.2001.1603. [DOI] [PubMed] [Google Scholar]
- 37.Kurkela M, Garcia-Horsman JA, Luukkanen L, Morsky S, Taskinen J, Baumann M, Kostiainen R, Hirvonen J, Finel M. Expression and characterization of recombinant human UDP-glucuronosyltransferases (UGTs). UGT1A9 is more resistant to detergent inhibition than other UGTs and was purified as an active dimeric enzyme. J Biol Chem. 2003;278:3536–44. doi: 10.1074/jbc.M206136200. [DOI] [PubMed] [Google Scholar]
- 38.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 39.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55(Pt 4):849–61. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr. 2000;56:965–72. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Terwilliger TC. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr D Biol Crystallogr. 2003;59:38–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 43.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 44.DeLano W. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA, USA: 2002. [Google Scholar]
- 45.Pillot T, Ouzzine M, Fournel-Gigleux S, Lafaurie C, Radominska A, Burchell B, Siest G, Magdalou J. Glucuronidation of hyodeoxycholic acid in human liver. Evidence for a selective role of UDP-glucuronosyltransferase 2B4. J Biol Chem. 1993;268:25636–42. [PubMed] [Google Scholar]