Abstract
The objective of this study was to determine if there are innate differences in gene expression in selected CNS regions between inbred alcohol-preferring (iP) and —non-preferring (iNP) rats. Gene expression was determined in the nucleus accumbens (ACB), amygdala (AMYG), frontal cortex (FC), caudate-putamen (CPU), and hippocampus (HIPP) of alcohol-naïve adult male iP and iNP rats, using Affymetrix Rat Genome U34A microarrays (n = 6/strain). Using Linear Modeling for Microarray Analysis with a false discovery rate threshold of 0.1, there were 16 genes with differential expression in the ACB, 54 in the AMYG, 8 in the FC, 24 in the CPU, and 21 in the HIPP. When examining the main effect of strain across regions, 296 genes were differentially expressed. Although the relatively small number of genes found significant within individual regions precluded a powerful analysis for over-represented Gene Ontology categories, the much larger list resulting from the main effect of strain analysis produced 17 over-represented categories (P <.05), including axon guidance, gliogenesis, negative regulation of programmed cell death, regulation of programmed cell death, regulation of synapse structure function, and transmission of nerve impulse. Co-citation analysis and graphing of significant genes revealed a network involved in the neuropeptide Y (NPY) transmitter system. Correlation of all significant genes with those located within previously established rat alcohol QTLs revealed that of the total of 313 significant genes, 71 are located within such QTLs. The many regional and overall gene expression differences between the iP and iNP rat lines may contribute to the divergent alcohol drinking phenotypes of these rats.
Keywords: Alcohol-preferring P rat, Alcohol—non-preferring NP rat, Microarrays, Gene expression, Nucleus accumbens, Amygdala, Frontal cortex, Hippocampus, Caudate-putamen
1. Introduction
Alcoholism and alcohol abuse are complex disorders that result from a combination of genetic and environmental factors. Selective breeding strategies for ethanol preference have yielded divergent rat lines that possess different frequencies of genes that impact ethanol preference, whereas the frequency of trait-irrelevant genes remains randomly distributed (Lumeng et al., 1977). The alcohol-preferring (P) and alcohol—non-preferring (NP) rat lines were established from a randomly bred, closed colony of Wistar rats using free-choice access to 10% (vol/vol) ethanol and water (Lumeng et al., 1977). P rats meet the proposed criteria (Cicero, 1979) for an animal model of alcoholism (reviewed in McBride & Li, 1998; Murphy et al., 2002). In brief, the P line of rats (1) consumes in excess of 5 g ethanol/kg body weight/day, attaining blood alcohol concentrations in the range of 50-200 mg%; (2) works to obtain ethanol when food and water are freely available; (3) consumes ethanol for its pharmacological effects, and not solely for caloric value nor taste or odor properties; (4) develops functional and metabolic tolerance; (5) develops physical dependence; and (6) demonstrates robust relapse ethanol drinking after a period of abstinence. On the other hand, NP rats consume less than 1 g ethanol/kg/day and do not attain measurable blood alcohol concentrations under free-choice conditions. Compared to NP rats, P rats are more sensitive to the low-dose stimulating effects of ethanol (Rodd et al., 2004; Waller et al., 1986), less sensitive to the high-dose motor impairing effects of ethanol (Lumeng et al., 1982), and develop acute tolerance more rapidly (Waller et al., 1983).
Innate differences in neurotransmitter and receptor systems in several brain regions have been reported between the selectively bred P and NP rat lines (reviewed in McBride & Li, 1998; Murphy et al., 2002). P rats have reduced serotonin (5-HT) and dopamine (DA) innervations (Zhou et al., 1991, 1994a, 1994b, 1995), as well as differences in 5-HT (McBride et al., 1993a, 1994, 1997; Wong et al., 1993), DA (McBride et al., 1993b), and opioid (McBride et al., 1998; Strother et al., 2001) receptors. Furthermore, neuropeptide Y (NPY) (Ehlers et al., 1998), corticotropin-releasing factor (Ehlers et al., 1992), neurotensin (Ehlers et al., 1999), substance P, and neurokinin levels (Slawecki et al., 2001) are all significantly lower in CNS regions of P compared to NP rats. Additionally, higher functional neuronal activity has been found in numerous brain regions of the P rat compared to the NP rat (Smith et al, 2001; Strother et al., 2005).
Witzmann et al. (2003) examined differences in protein levels in the hippocampus (HIPP) and nucleus accumbens (ACB) of alcohol-naïve inbred-P (iP) and inbred-NP (iNP) rats, and found that almost all of the proteins that differed were lower in the iP rats compared to the iNP rats. Those proteins that could be identified were involved in many key aspects of neuronal function such as metabolism, cell signaling, and protein transport, which may suggest that there are basic differences in synaptic transmission mechanisms between the two rat strains (Witzmann et al., 2003). Edenberg et al. (2005) compared gene expression differences in the HIPP of two different strains of iP and iNP rats, when microarray analyses were conducted several months apart. The results indicated excellent repeatability of the assay. Genes involved in cell growth and adhesion, cellular stress reduction and antioxidation, protein trafficking, cellular signaling pathways, and synaptic function were differentially expressed in the HIPP (Edenberg et al., 2005). Worst et al. (2005) reported on the transcriptome analysis in the anterior cerebral cortex of alcohol-naïve Alko, alcohol (AA) and Alko, nonalcohol (ANA) rats, and found differences in mRNA levels between the AA and ANA rats that could alter transmitter release (e.g., vesicle-associated membrane protein 2, syntaxin 1, syntaxin binding protein). Kerns et al. (2005) examined gene expression differences in response to acute ethanol in the ACB, prefrontal cortex, and ventral tegmental area of C57BL/6J and DBA/2J mice, which have high and low alcohol drinking characteristics, respectively. Ethanol-regulated genes were region specific and involved in glucocorticoid signaling, neurogenesis, myelination, neuropeptide signaling, and retinoic acid signaling. Gene expression profiles were also reported for whole brain of inbred long-sleep and inbred short-sleep mice (Xu et al., 2001). A total of 41 genes or expressed sequence tags (ESTs) displayed significant differences between these inbred strains of mice. Expression of genes encoding tyrosine protein kinase and ubiquitin carboxyl terminal hydrolase was higher in the brain of inbred long-sleep compared to short-sleep mice. In a comprehensive transcriptome meta-analysis of different mice strains, Mulligan et al. (2006) identified several cis-regulated candidate genes for an alcohol preference quantitative trait loci (QTL) on chromosome 9.
Portions of the present data have been used for comparison analyses in two recently published studies, although none of the present data have been presented in duplicate form. In one study, the present HIPP data were used with other HIPP data to evaluate the reliability of the microarray analysis, when assays were conducted months apart (Edenberg et al., 2005). In the second study (Rodd et al., 2006), the data were used as part of a convergent functional genomics approach to identify common genes across different experimental approaches and between human and animal findings. In that study, however, the iP—iNP data were not analyzed using rigorous statistical criteria (for a gene to be considered significant, an false discovery rate (FDR)-uncorrected P <.05 was considered sufficient), and the results were presented only in a summarized format, which were then integrated with information from other studies. As the P and NP lines are well established animal models in the alcohol field, we believe it is important that the present findings, derived using rigorous region-by-region analyses, are presented because they yield a much more complete and statistically reliable picture of the genetic factors involved in the high and low alcohol drinking behavior in these rat lines.
The objective of the present study was to determine if there are innate differences between inbred P and NP rats in the expression of functionally relevant genes in selected brain regions. The current study focuses on five distinct brain regions: the ACB, caudate-putamen (CPU), amygdala (AMYG), HIPP, and frontal cortex (FC). These regions were selected based on their inclusion in the mesolimbic and mesocortical systems, both of which are critically important in the initiation and maintenance of goal directed and reward mediated behaviors (reviewed in Bonci et al., 2003; and Maldonado, 2003).
2. Materials and methods
2.1. Animals and RNA preparation
Inbred adult male rats, 90-100 days old, from the iP-5C and iNP-1 strains were used in these experiments. Inbreeding by brother—sister mating was initiated after the S30 generation of mass selection and was in the F37 generation at the start of these experiments. It should be noted that the iP and iNP rats have not been characterized to the extent to which the parent selected lines have been studied. However, preliminary studies indicate that alcohol intake (Bell et al., 2004), and differences in sweet preference, anxiety, spontaneous motor activity, and the development of rapid tolerance (Stewart et al., 2004) are similar to the parent lines.
Animals were received in our facilities 3 weeks prior to the experiment. Rats were double housed on a 12:12-h light dark cycle with lights on at 0700 h. Rats had water and rat chow ad libitum. Animals were habituated to handling and to the guillotine daily between 0900 and 1000 h for 10 days prior to sacrifice. The animals used in these experiments were maintained in facilities fully accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care. All research protocols were approved by the Institutional Animal Care and Use Committee and are in accordance with the guidelines of the Institutional Animal Care and Use Committee of the National Institute on Drug Abuse, NIH, and the Guide for the Care and Use of Laboratory Animals (Institute of Laboratory Animal Resources, Commission on Life Sciences, National Research Council 1996).
Animals were sacrificed by decapitation between 0900 and 1000 h over consecutive days, with equal numbers of animals from each strain sacrificed each day. This minimized differences in time of sacrifice and dissection, and maintained the experimental balance across the two strains. The head was immediately placed in a cold box maintained at -15°C, where the brain was rapidly removed and placed on a glass plate for dissection. All equipment used to obtain tissue was treated with RNAse Zap (Ambion, Inc. Austin, TX) to prevent RNA degradation. The ACB, CPU, FC, AMYG, and HIPP were dissected according to the coordinates of Paxinos and Watson (1998). Briefly, the ACB, CPU, and FC were dissected from a 2 mm section generated by a coronal cut at 2 mm anterior to the optic chiasm (Bregma 1.70 mm) and a coronal cut at the optic chiasm (Bregma -0.26 mm). The AMYG was dissected by a cut at the lateral borders of the lateral hypothalamus (Bregma -2.12 mm) and ventral of the rhinal fissure, with cortical tissue then trimmed at the lateral edges of the dissected slice. The entire HIPP was dissected from the remaining brain by a midline incision between the hemispheres and gently rolling the HIPP out of the cerebral cortex. We have previously demonstrated the consistency of dissection of discrete brain regions in our laboratory (Liang et al., 2003; Witzmann et al., 2003). Dissected tissues were immediately homogenized in Trizol reagent (Invitrogen, Carlsbad, CA) and processed according to the manufacturer’s protocol, but with twice the suggested ratio of Trizol to tissue. Ethanol precipitated RNA was further purified through RNeasy® columns (Qiagen, Valencia, CA) according to the manufacturer’s protocol. The yield, concentration, and purity of the RNA were excellent, and were determined by running a spectrum from 210 to 350 nm and analyzing the ratio of large and small ribosomal bands using an Agilent Bioanalyzer.
2.2. Labeling and hybridization
RNA from each individual rat was labeled and analyzed separately on an Affymetrix Rat Genome U34A microarray. Starting with 10 μg of total RNA from each animal, first-and second-strand cDNA synthesis was carried out according to the standard protocol (Affymetrix: GeneChip® Expression Analysis Technical Manual. Santa Clara, CA: Affymetrix; 2001). Biotinylated cRNA was synthesized in vitro from the double-stranded cDNA using the ENZO Bio-Array High Yield RNA Transcript Labeling Kit (ENZO Diagnostics, Inc., Farmingdale, NY) according to the Affymetrix protocol. Fragmented, biotinylated cRNA (15 μg) was mixed into 300 μl of hybridization cocktail, of which 200 μl were used for each hybridization. Hybridization was for 17 h at 42°C. Washing, staining, and scanning were carried out according to the standard protocol.
To minimize potential systematic errors, all stages of the experiment were balanced across phenotypes. That is, equal numbers of iP and iNP animals were sacrificed each day, and equal numbers of RNA preparations from iP and iNP animals were processed through the labeling, hybridization, washing, and scanning protocols on each day, in different alternating orders. Whenever possible, common premixes of reagents were used to minimize effects due to differences in reagent preparation.
2.3. Data and neuroinformatic analyses
Each GeneChip® was scanned using an Affymetrix Model 3000 scanner and underwent image analysis using Affymetrix GCOS software. Microarray data are available from the National Center for Biotechnology Information’s Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/geo/, (Barrett et al., 2005; Edgar et al., 2002) under series accession no. GSE4494 (GSM100999... GSM101057). Raw cell files were then imported into the statistical programming environment R (R Development Core Team, 2006) for further analysis with tools available from the Bioconductor Project (Gentleman et al., 2004), themselves further expanded by the authors using the R language. Expression data from 59 arrays were normalized and converted to log2 as a set using the Robust Multi-chip Average (RMA) method (Irizarry et al., 2003) implemented in the Bioconductor package RMA. As a standardization step to facilitate later comparisons with other experiments, expression levels were scaled such that the mean expression of all arrays was log2(1000). To increase power and decrease the FDR (see below), probe sets were subjected to further filtering steps. As we were primarily concerned with identifying genes that could be subjected to further bioinformatic analysis, all probe sets currently annotated by Affymetrix as “expressed sequence tags” or whose gene names contain the words “riken,” “predicted,” or “similar to” were filtered out. We next filtered out probe sets with a very low likelihood of actual expression in our samples, accomplished with the Bioconductor package “genefilter.” Probe sets that did not have at least 25% of samples with normalized, scaled expression greater than 64 were filtered out. Finally, we wished to filter out, in an unbiased manner, probe sets with very low variability across samples. This was accomplished by constructing a distribution of the interquartile range of all probe sets from all samples of both groups and then filtering out those probe sets that fell in the lowest 25% of this distribution using the Bioconductor package genefilter according to the method described by Scholtens and von Heydebreck (2005). Linear modeling to calculate gene-wise P values was performed using the package Linear Modeling for Microarray Analysis (LIMMA) (Smyth, 2004) P values were then corrected for FDR by the method of Benjamini & Hochberg (1995). Probe sets were considered to be differentially expressed if the FDR adjusted P value was P <.10 (FDR 10%). Using this method, the data were analyzed two ways. In the first analysis, each individual brain region was analyzed separately, using only strain as a factor in the linear model. In the second analysis, the five brain regions tested were combined in the linear model, using factors of region and strain, and the main effect of strain was examined. This analysis will be referred to as the “overall analysis.”
Testing for over-representation of Gene Ontology (GO) (Ashburner et al., 2000; Harris et al., 2004) biologic process categories was performed using the Bioconductor package GOstats (Gentleman, 2004). Briefly, for each gene set tested, a list of unique Entrez-Gene identifiers was constructed. This list was then compared to the list of all known Entrez-Gene identifiers that are represented on the Affymetrix chipset RGU34A. Identification of over-represented GO categories was then accomplished within GOstats using the hypergeometric distribution. To filter out uninteresting categories, those categories with less than 9 or greater than 300 genes represented on the chipset were excluded from the analysis, as were categories with less than five significant genes. Categories were called significant for P <.05.
Co-citation analysis was accomplished within R using a modification of the package “MedlineR” (Lin et al., 2004). Gene aliases were obtained from the Rat Genome Database, http://rgd.mcw.edu/. A list of all PubMed, http://www.pubmedcentral.nih.gov/, occurrences of genes and their aliases in abstracts and keywords was constructed for each gene, from which a co-citation matrix was constructed. The Bioconductor packages “graph” (Gentleman et al., 2004) and “Rgraphviz” (Gentry, 2006) were used to graphically display networks of co-citated genes.
2.4. Quantitative real-time PCR
Real-time (RT) polymerase chain reaction (PCR) was carried out using SybrGreen chemistry and the ABI Prism 7700 Sequence Detection System (Applied Biosystems). The amplification primers were designed using Vector NTI software. Total RNA, isolated for the microarray analyses, was used for these analyses. Following reverse transcription of the RNA (TaqMan Reverse Transcription Reagents, Applied Biosystems), an aliquot of each reverse transcription reaction was amplified in triplicate. This reaction was repeated to generate six values for each test group. Two control reactions were run for each RNA preparation: (1) a reverse transcription and PCR reaction with no added RNA to control for contamination of the reagents and (2) a PCR reaction without the reverse transcription reaction in the presence of RNA to detect DNA contamination of the RNA preparation. To correct for sample-to-sample variation, an endogenous control, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was amplified with the target and served as an internal reference to normalize the data. The average GAPDH Ct values for iP and iNP were the same in each brain region tested, making this an appropriate control gene to normalize the expression of the candidate genes of interest. Relative quantification of data from the ABI Prism 7700 Sequence Detection System was performed using the standard curve method (Applied Biosystems, User Bulletin #2; http://www.appliedbiosystems.com). Quantitative real-time polymerase-chain-reaction (qRT-PCR) measurements were conducted on genes to verify differences observed with microarray hybridization. Genes were selected on the basis of significant differential expression in at least one brain region and reasonable fold changes. When this list was compiled, initial analysis had been completed using MAS5 background correction; after the RMA algorithm had been substituted for MAS5, mRNA samples were no longer of sufficient quality to select a new list.
3. Results
Principal component analysis, using all probe sets that passed through the filters described in Methods, indicated that regional differences in gene expression were greater than strain differences, as illustrated by a biplot of the first and second principal components (Fig. 1). The clusters representing arrays from the ACB, CPU, and FC are tightly grouped, with the exception of one outlier in each region. Those from the HIPP give a fairly good grouping, but not as tight as the other three regions; the AMYG gives the least grouping of data, with one outlier that groups with the HIPP cluster. As can be seen from the figure, there is no clear pattern of separation of the iP and iNP lines within brain region. Visualization of the third and fourth principal components (not shown) also did not resolve line differences within regions.
Fig. 1.
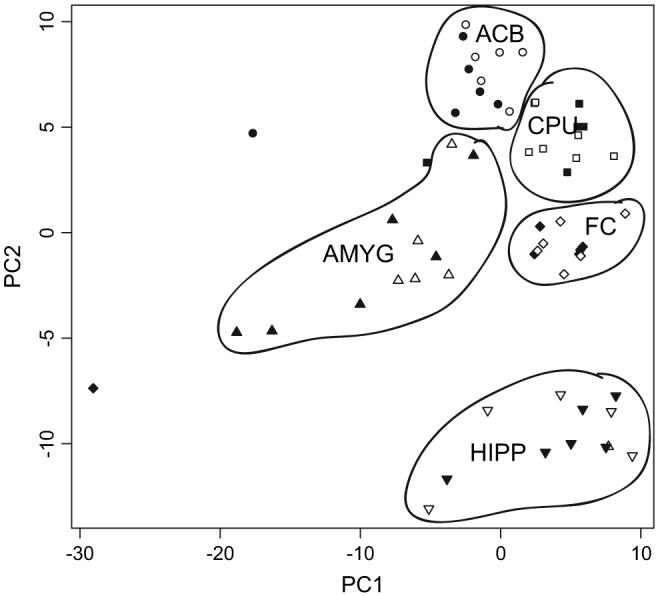
Principal components plot of all 59 arrays. Only processed, filtered data were used (see Methods for details) in the computation. Principal component 1 is plotted on the horizontal axis and principal component 2 is plotted on the vertical axis. Clusters of arrays corresponding to specific brain regions are as outlined. Open symbols represent arrays for inbred alcohol-preferring rats; closed symbols are for arrays from inbred alcohol —non-preferring rats.
We used LIMMA (Smyth, 2004) on RMA preprocessed (Irizarry et al., 2003), log2 transformed, filtered data (see Methods for details), to determine differences between the inbred strains. The FDR, as calculated by the method of Benjamini and Hochberg (1995), was set at ≤0.10. In an analysis of individual brain regions (n = 6 iNP, 6 iP in all regions except the FC, in which data were missing from one iP animal), significant differential gene expression was found in the five brain regions (Tables 1 and 2). The number of differences between the two inbred lines in each of the five individual regions ranged from 8 to 63, with the order of number of differences being AMYG > CPU > HIPP > ACB > FC (Table 1). Across regions, the total number of genes that demonstrated differential expression ranged from 8 to 54; the number of genes that were located within established alcohol QTLs (Bice et al., 1998; Carr et al., 1998, 2003; Foroud et al., 2002, 2003; Radcliffe et al., 2004; Terenina-Rigaldie et al., 2003) ranged from 1 to 16. Table 2 lists the significant genes that were different within each region along with individual fold changes and gene symbols. Table 3 contains detailed expression levels and standard deviations for all the regionally significant probe sets.
Table 1.
Summary of biostatistical and bioinformatic analyses for individual regions and for the combined regions examining the main effect of straina
| Accumbens | Amygdala | Frontal cortex | Hippocampus | Caduate-putamen | Combined regions | |
|---|---|---|---|---|---|---|
| Total significant genes (FDR < 0.10) | 16 | 54 | 8 | 21 | 24 | 351 |
| iP>iNP | 7 | 32 | 3 | 7 | 10 | 198 |
| iNP>iP | 9 | 22 | 5 | 14 | 14 | 153 |
| Minimum significant fold difference (absb) | 1.37 | 1.18 | 1.39 | 1.21 | 1.19 | |
| Minimum significant fold difference (absb) | 2.11 | 2.08 | 2.08 | 2.28 | 2.24 | |
| Significant genes in alcohol QTLs | 2 | 16 | 2 | 1 | 8 | 70 |
| Significant GO categories (P <.05) | 0 | 0 | 0 | 0 | 0 | 12 |
| Significant gene—gene co-citations | 0 | 0 | 0 | 0 | 0 | 121 |
FDR = false discovery rate; iP = inbred alcohol—preferring rat; iNP = inbred alcoholenon-preferring rat; GO = Gene Ontology.
See Methods for details of each analysis.
Absolute value.
Table 2.
Regionally significant genes (FDR < 0.10)
| Symbol | Gene description | iP vs. iNP fold change |
|---|---|---|
| Accumbens | ||
| Ahi1 | Abelson helper integration site 1 | -1.54 |
| Gsta4 | glutathione S-transferase, alpha 4 | -1.52 |
| Lin7c | lin-7 homolog C (C. elegans) | -1.51 |
| Rps2 | ribosomal protein S2 | -1.48 |
| RT1-Aw2a | RT1 class Ib, locus Aw2 | -1.37 |
| Sephs2 | selenophosphate synthetase 2 | -1.61 |
| Stk38 | serine/threonine kinase 38 | -1.37 |
| Scn3a | sodium channel, voltage-gated, type III, alpha polypeptide | -1.38 |
| Slc5a3 | solute carrier family 5 (inositol transporters), member 3 | -1.40 |
| Akap11 | A kinase (PRKA) anchor protein 11 | 1.50 |
| Ampd3 | adenosine monophosphate deaminase 3 | 1.37 |
| Cat | Catalase | 1.42 |
| Lyz | Lysozyme | 2.11 |
| Pgk1 | Phosphoglycerate kinase 1 | 2.05 |
| Psma5 | proteasome (prosome, macropain) subunit, alpha type 5 | 1.44 |
| RT1-Bb | RT1 class II, locus Bb | 1.46 |
| Amygdala | ||
| Hmgcr | 3-hydroxy-3-methylglutaryl-Coenzyme A reductase | -1.28 |
| Phgdha | 3-phosphoglycerate dehydrogenase | -1.36 |
| Ahi1 | Abelson helper integration site 1 | -1.35 |
| Csnk1d | casein kinase 1, delta | -1.18 |
| Casp3 | caspase 3, apoptosis related cysteine protease | -1.21 |
| Ddr2 | discoidin domain receptor family, member 2 | -1.18 |
| Fgfr1 | Fibroblast growth factor receptor 1 | -1.21 |
| Fn1a | fibronectin 1 | -1.32 |
| Gsta4a | glutathione S-transferase, alpha 4 | -1.78 |
| Nyw1 | ischemia related factor NYW-1 | -1.38 |
| Kdr | kinase insert domain protein receptor | -1.50 |
| Lin7c | lin-7 homolog C (C. elegans) | -1.27 |
| Lox | lysyl oxidase | -1.22 |
| Pak2 | p21 (CDKN1A)-activated kinase 2 | -1.20 |
| Rps2a | ribosomal protein S2 | -1.63 |
| RT1-Aw2a | RT1 class Ib, locus Aw2 | -1.41 |
| Sephs2 | selenophosphate synthetase 2 | -1.39 |
| Scn3aa | sodium channel, voltage-gated, type III, alpha polypeptide | -1.44 |
| Slc28a2 | solute carrier family 28 (sodium-coupled nucleoside transporter), member 2 | -1.25 |
| Slc5a3 | solute carrier family 5 (inositol transporters), member 3 | -1.77 |
| Shc3 | src homology 2 domain-containing transforming protein C3 | -1.29 |
| Tagln | Transgelin | -1.22 |
| Atic | 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase | 1.32 |
| Acsl1 | acyl-CoA synthetase long-chain family member 1 | 1.27 |
| Amigo | amphoretin-induced gene and ORF | 1.22 |
| Argbp2a | Arg/Abl-interacting protein ArgBP2 | 1.22 |
| Asns | asparagine synthetase | 1.25 |
| Atrn | Attractin | 1.33 |
| Btg2 | B-cell translocation gene 2, antiproliferative | 1.19 |
| Celsr2 | cadherin EGF LAG seven-pass G-type receptor 2 | 1.25 |
| Cd24a | CD24 antigen | 1.32 |
| Chgb | chromogranin B | 1.25 |
| Eif2b1a | eukaryotic translation initiation factor 2B, subunit 1 alpha | 1.29 |
| Fth1 | ferritin, heavy polypeptide 1 | 1.29 |
| Gfm | G elongation factor | 1.30 |
| Irak2 | interleukin-1 receptor-associated kinase 2 | 1.22 |
| Ldhb | lactate dehydrogenase B | 1.23 |
| Lcp2 | lymphocyte cytosolic protein 2 | 1.18 |
| Lyz | Lysozyme | 1.81 |
| Nrn1 | Neuritin | 1.19 |
| Nptx1 | neuronal pentraxin 1 | 1.32 |
| Pdlim3 | PDZ and LIM domain 3 | 1.28 |
| Pmpcb | peptidase (mitochondrial processing) beta | 1.26 |
| Pcm1 | pericentriolar material 1 | 1.52 |
| Pgk1 | phosphoglycerate kinase 1 | 2.08 |
| Prps1 | phosphoribosyl pyrophosphate synthetase 1 | 1.19 |
| Pbef1 | pre-B-cell colony enhancing factor 1 | 1.25 |
| Psma5 | proteasome (prosome, macropain) subunit, alpha type 5 | 1.57 |
| RT1-N3 | RT1 class Ib gene, H2-TL-like, grc region (N3) | 1.26 |
| RT1-Bb | RT1 class II, locus Bb | 1.39 |
| Sh3gl3 | SH3 domain protein 2 C1 | 1.18 |
| Spats1 | spermatogenesis associated, serine-rich 1 | 1.29 |
| Scp2 | sterol carrier protein 2 | 1.25 |
| Thrsp | thyroid hormone responsive protein | 1.30 |
| Frontal cortex | ||
| Phgdh | 3-phosphoglycerate dehydrogenase | -1.42 |
| Gsta4 | glutathione S-transferase, alpha 4 | -1.75 |
| Rps2 | ribosomal protein S2 | -1.76 |
| Scn3a | sodium channel, voltage-gated, type III, alpha polypeptide | -1.39 |
| Slc5a3 | solute carrier family 5 (inositol transporters), member 3 | -1.44 |
| Gfap | glial fibrillary acidic protein | 1.44 |
| Pgk1 | phosphoglycerate kinase 1 | 2.08 |
| Psma5 | proteasome (prosome, macropain) subunit, alpha type 5 | 1.78 |
| Hippocampus | ||
| Phgdha,b | 3-phosphoglycerate dehydrogenase | -1.39 |
| Azi2 | 5-azacytidine induced gene 2 | -1.28 |
| Ahi1 | Abelson helper integration site 1 | -1.48 |
| B2m | beta-2 microglobulin | -1.31 |
| Fn1 | fibronectin 1 | -1.55 |
| Gsta4a | glutathione S-transferase, alpha 4 | -2.07 |
| Lxn | Latexin | -1.34 |
| Rab5ab | RAB5A, member RAS oncogene family | -1.29 |
| Rps2 | ribosomal protein S2 | -1.53 |
| RT1-Aw2b | RT1 class Ib, locus Aw2 | -1.41 |
| Sephs2b | selenophosphate synthetase 2 | -1.47 |
| Stk38 | serine/threonine kinase 38 | -1.21 |
| Slc5a3 | solute carrier family 5 (inositol transporters), member 3 | -1.47 |
| Tagln | Transgelin | -1.23 |
| Akap11b | A kinase (PRKA) anchor protein 11 | 1.32 |
| Dcnb | Decorin | 1.29 |
| Grb1b4 | growth factor receptor bound protein 14 | 1.32 |
| Iag2b | implantation-associated protein | 1.31 |
| Pgk1b | phosphoglycerate kinase 1 | 2.28 |
| RT1-N3 | RT1 class Ib gene, H2-TL-like, grc region (N3) | 1.35 |
| Slit1 | slit homolog 1 (Drosophila) | 1.32 |
| Caudate-putamen | ||
| Phgdha | 3-phosphoglycerate dehydrogenase | -1.33 |
| Ahi1 | Abelson helper integration site 1 | -1.46 |
| Apobec1 | apolipoprotein B editing complex 1 | -1.23 |
| Fn1 | fibronectin 1 | -1.35 |
| Gsta4a | glutathione S-transferase, alpha 4 | -2.00 |
| Lin7c | lin-7 homolog C (C. elegans) | -1.31 |
| Prkag1 | protein kinase, AMP-activated, gamma 1 noncatalytic subunit | -1.19 |
| Rpl6 | ribosomal protein L6 | -1.21 |
| Rps2 | ribosomal protein S2 | -1.64 |
| RT1-Aw2a | RT1 class Ib, locus Aw2 | -1.39 |
| Sephs2 | selenophosphate synthetase 2 | -1.45 |
| Scn3a | sodium channel, voltage-gated, type III, alpha polypeptide | -1.46 |
| Slc5a3 | solute carrier family 5 (inositol transporters), member 3 | -1.41 |
| Shc3 | src homology 2 domain-containing transforming protein C3 | -1.39 |
| Akap5 | A kinase (PRKA) anchor protein 5 | 1.39 |
| Akr1b4 | aldo-keto reductase family 1, member B4 (aldose reductase) | 1.21 |
| Agt | Angiotensinogen | 1.30 |
| Asns | asparagine synthetase | 1.20 |
| Cryab | crystallin, alpha B | 1.35 |
| Hspa2 | heat shock protein 2 | 1.24 |
| Nefh | neurofilament, heavy polypeptide | 1.44 |
| Pgk1 | phosphoglycerate kinase 1 | 2.24 |
| Psma5 | proteasome (prosome, macropain) subunit, alpha type 5 | 1.63 |
| Znf386 | zinc finger protein 386 (Kruppel-like) | 1.22 |
FDR = false discovery rate; iP = inbred alcohol-preferring rat; iNP = inbred alcoholenon-preferring rat.
Genes represented by significant differential expression (FDR < 0.10) of more than one probe set. The largest absolute fold change for the gene among significant probe sets is listed. In all cases, the significant probe sets expressed differential expression in the same direction.
Gene also significant in Edenberg et al. (2005)
Table 3.
Detailed expression levels and S.D.s for all regionally significant probe sets
| Probe set | Unigene | Entrez gene | Symbol | Gene description | Fold difference | FDR | Mean iP | S.D. iP | Mean iNP | S.D. iNP |
|---|---|---|---|---|---|---|---|---|---|---|
| Accumbens | ||||||||||
| rc_AI639111_at | Rn.93081 | 308923 | Ahi1 | Abelson helper integration site 1 | -1.54 | 0.00 | 810 | 107 | 527 | 60 |
| X62660mRNA_g_at | Rn.57528 | 300850 | Gsta4 | glutathione S-transferase, alpha 4 | -1.52 | 0.00 | 1092 | 184 | 719 | 125 |
| AF090136_at | Rn.44269 | 60442 | Lin7c | lin-7 homolog C (C. elegans) | -1.51 | 0.00 | 160 | 14 | 106 | 11 |
| L81138exon_i_at | Rn.2115 | 83789 | Rps2 | ribosomal protein S2 | -1.48 | 0.00 | 438 | 30 | 299 | 53 |
| M10094_g_at | Rn.40130 | 24737 | RT1-Aw2 | RT1 class Ib, locus Aw2 | -1.36 | 0.00 | 392 | 19 | 288 | 25 |
| M11071_f_at | Rn.40130 | 24737 | RT1-Aw2 | RT1 class Ib, locus Aw2 | -1.37 | 0.00 | 2623 | 290 | 1915 | 148 |
| Y00766_at | Rn.87394 | 25657 | Scn3a | sodium channel, voltage-gated, type III, alpha polypeptide | -1.38 | 0.00 | 643 | 82 | 464 | 50 |
| rc_AA799700_at | Rn.100471 | 308993 | Sephs2 | selenophosphate synthetase 2 | -1.61 | 0.00 | 354 | 62 | 221 | 43 |
| AJ001290cds_at | Rn.79242 | 114507 | Slc5a3 | solute carrier family 5 (inositol transporters), member 3 | -1.40 | 0.00 | 784 | 43 | 566 | 85 |
| rc_AA800712_g_at | Rn.13142 | 361813 | Stk38 | serine/threonine kinase 38 | -1.37 | 0.00 | 855 | 44 | 632 | 105 |
| U48288_at | Rn.10557 | 25228 | Akap11 | A kinase (PRKA) anchor protein 11 | 1.50 | 0.00 | 312 | 54 | 464 | 46 |
| U90888_at | Rn.11106 | 25095 | Ampd3 | adenosine monophosphate deaminase 3 | 1.37 | 0.00 | 102 | 12 | 139 | 19 |
| rc_AA926149_g_at | Rn.3001 | 24248 | Cat | catalase | 1.42 | 0.00 | 197 | 33 | 278 | 34 |
| rc_AA892775_at | Rn.2283 | 25211 | Lyz | lysozyme | 2.11 | 0.00 | 1185 | 159 | 2524 | 501 |
| rc_AA892797_at | Rn.108127 | 24644 | Pgk1 | phosphoglycerate kinase 1 | 2.05 | 0.00 | 2189 | 729 | 4392 | 1061 |
| rc_AA891383_i_at | Rn.1276 | 29672 | Psma5 | proteasome (prosome, macropain) subunit, alpha type 5 | 1.44 | 0.00 | 123 | 17 | 176 | 16 |
| U65217_i_at | Rn.33311 | 309622 | RT1-Bb | RT1 class II, locus Bb | 1.46 | 0.00 | 48 | 5 | 69 | 7 |
| Amygdala | ||||||||||
| rc_AI639111_at | Rn.93081 | 308923 | Ahi1 | Abelson helper integration site 1 | -1.35 | 0.00 | 818 | 118 | 602 | 60 |
| U84410_s_at | Rn.10562 | 25402 | Casp3 | caspase 3, apoptosis related cysteine protease | -1.21 | 0.00 | 200 | 12 | 166 | 15 |
| rc_AI013513_at | Rn.8046 | 64462 | Csnk1d | casein kinase 1, delta | -1.18 | 0.00 | 730 | 23 | 621 | 24 |
| AF016247_at | Rn.92730 | 83573 | Ddr2 | discoidin domain receptor family, member 2 | -1.18 | 0.00 | 142 | 7 | 120 | 5 |
| D12498_s_at | Rn.9797 | 79114 | Fgfr1 | Fibroblast growth factor receptor 1 | -1.21 | 0.00 | 671 | 45 | 556 | 27 |
| U82612cds_g_at | Rn.1604 | 25661 | Fn1 | fibronectin 1 | -1.25 | 0.00 | 445 | 57 | 354 | 22 |
| X05834_at | Rn.1604 | 25661 | Fn1 | fibronectin 1 | -1.32 | 0.00 | 1070 | 95 | 812 | 109 |
| X62660mRNA_at | Rn.57528 | 300850 | Gsta4 | glutathione S-transferase, alpha 4 | -1.32 | 0.00 | 714 | 35 | 545 | 72 |
| X62660mRNA_g_at | Rn.57528 | 300850 | Gsta4 | glutathione S-transferase, alpha 4 | -1.78 | 0.00 | 1182 | 55 | 679 | 163 |
| X55286_g_at | Rn.9437 | 25675 | Hmgcr | 3-hydroxy-3-methylglutaryl-Coenzyme A reductase | -1.28 | 0.00 | 363 | 32 | 283 | 16 |
| U93306_at | Rn.88869 | 25589 | Kdr | kinase insert domain protein receptor | -1.50 | 0.00 | 121 | 18 | 82 | 15 |
| AF090136_at | Rn.44269 | 60442 | Lin7c | lin-7 homolog C (C. elegans) | -1.27 | 0.00 | 139 | 11 | 109 | 6 |
| S66184_s_at | Rn.11372 | 24914 | Lox | lysyl oxidase | -1.22 | 0.00 | 204 | 16 | 167 | 9 |
| rc_AI176253_at | Rn.133954 | 59319 | Nyw1 | ischemia related factor NYW-1 | -1.38 | 0.00 | 122 | 15 | 89 | 13 |
| U35345_s_at | Rn.3840 | 29432 | Pak2 | p21 (CDKN1A)-activated kinase 2 | -1.20 | 0.00 | 145 | 9 | 121 | 7 |
| X97772_at | Rn.6872 | 58835 | Phgdh | 3-phosphoglycerate dehydrogenase | -1.36 | 0.00 | 1710 | 150 | 1261 | 142 |
| X97772_g_at | Rn.6872 | 58835 | Phgdh | 3-phosphoglycerate dehydrogenase | -1.28 | 0.00 | 1466 | 111 | 1150 | 119 |
| L81136cds_f_at | Rn.2115 | 83789 | Rps2 | ribosomal protein S2 | -1.20 | 0.00 | 1150 | 36 | 959 | 44 |
| L81138exon_i_at | Rn.2115 | 83789 | Rps2 | ribosomal protein S2 | -1.63 | 0.00 | 484 | 36 | 298 | 29 |
| M11071_f_at | Rn.40130 | 24737 | RT1-Aw2 | RT1 class Ib, locus Aw2 | -1.41 | 0.00 | 2488 | 163 | 1767 | 141 |
| M31018_f_at | Rn.40130 | 24737 | RT1-Aw2 | RT1 class Ib, locus Aw2 | -1.23 | 0.00 | 1191 | 70 | 971 | 86 |
| Y00766_at | Rn.87394 | 25657 | Scn3a | sodium channel, voltage-gated, type III, alpha polypeptide | -1.44 | 0.00 | 527 | 99 | 363 | 33 |
| Y00766_g_at | Rn.87394 | 25657 | Scn3a | sodium channel, voltage-gated, type III, alpha polypeptide | -1.41 | 0.00 | 1620 | 407 | 1130 | 112 |
| rc_AA799700_at | Rn.100471 | 308993 | Sephs2 | selenophosphate synthetase 2 | -1.39 | 0.00 | 332 | 31 | 239 | 23 |
| AB001453_at | Rn.59227 | 114858 | Shc3 | src homology 2 domain-containing transforming protein C3 | -1.29 | 0.00 | 1166 | 154 | 903 | 87 |
| U66723_s_at | Rn.10140 | 60423 | Slc28a2 | solute carrier family 28 (sodium-coupled nucleoside transporter), member 2 | -1.25 | 0.00 | 350 | 35 | 280 | 19 |
| AJ001290cds_at | Rn.79242 | 114507 | Slc5a3 | solute carrier family 5 (inositol transporters), member 3 | -1.77 | 0.00 | 839 | 48 | 479 | 85 |
| M83107_at | Rn.34397 | 25123 | Tagln | transgelin | -1.22 | 0.00 | 507 | 42 | 416 | 27 |
| D90109_at | Rn.6215 | 25288 | Acsl1 | acyl-CoA synthetase long-chain family member 1 | 1.27 | 0.00 | 444 | 30 | 563 | 49 |
| rc_AA891631_at | Rn.116338 | 295365 | Amigo | amphoretin-induced gene and ORF | 1.22 | 0.00 | 559 | 47 | 680 | 50 |
| AF026505_at | Rn.24612 | 114901 | Argbp2 | Arg/Abl-interacting protein ArgBP2 | 1.21 | 0.00 | 441 | 16 | 535 | 53 |
| rc_AA891194_s_at | Rn.24612 | 114901 | Argbp2 | Arg/Abl-interacting protein ArgBP2 | 1.22 | 0.00 | 637 | 19 | 779 | 83 |
| U07201_at | Rn.11172 | 25612 | Asns | asparagine synthetase | 1.25 | 0.00 | 348 | 27 | 435 | 39 |
| D89514_at | Rn.15114 | 81643 | Atic | 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase | 1.32 | 0.00 | 318 | 27 | 419 | 34 |
| rc_AA859645_at | Rn.53846 | 83526 | Atrn | attractin | 1.33 | 0.00 | 477 | 44 | 635 | 59 |
| M60921_g_at | Rn.27923 | 29619 | Btg2 | B-cell translocation gene 2, antiproliferative | 1.19 | 0.00 | 254 | 12 | 302 | 20 |
| U49062_at | Rn.6007 | 25145 | Cd24 | CD24 antigen | 1.32 | 0.00 | 895 | 99 | 1185 | 122 |
| U49062_g_at | Rn.6007 | 25145 | Cd24 | CD24 antigen | 1.24 | 0.00 | 476 | 43 | 589 | 53 |
| rc_AA875414_at | Rn.2912 | 83465 | Celsr2 | cadherin EGF LAG seven-pass G-type receptor 2 | 1.25 | 0.00 | 957 | 103 | 1199 | 104 |
| AF019974_at | Rn.11090 | 24259 | Chgb | chromogranin B | 1.25 | 0.00 | 3207 | 381 | 4004 | 293 |
| rc_AI031019_at | Rn.9181 | 64514 | Eif2b1 | eukaryotic translation initiation factor 2B, subunit 1 alpha | 1.17 | 0.00 | 346 | 21 | 406 | 12 |
| rc_AI031019_g_at | Rn.9181 | 64514 | Eif2b1 | eukaryotic translation initiation factor 2B, subunit 1 alpha | 1.29 | 0.00 | 254 | 13 | 328 | 26 |
| rc_AI169802_at | Rn.54447 | 25319 | Fth1 | ferritin, heavy polypeptide 1 | 1.29 | 0.00 | 4188 | 348 | 5445 | 782 |
| L14684_at | Rn.10913 | 114017 | Gfm | G elongation factor | 1.30 | 0.00 | 212 | 21 | 275 | 24 |
| rc_AA799581_at | Rn.17123 | 362418 | Irak2 | interleukin-1 receptor-associated kinase 2 | 1.22 | 0.00 | 583 | 58 | 710 | 37 |
| rc_AA892316_at | Rn.1057 | 155918 | Lcp2 | lymphocyte cytosolic protein 2 | 1.18 | 0.00 | 131 | 7 | 154 | 9 |
| U07181_g_at | Rn.1785 | 24534 | Ldhb | lactate dehydrogenase B | 1.23 | 0.00 | 4975 | 374 | 6116 | 291 |
| rc_AA892775_at | Rn.2283 | 25211 | Lyz | lysozyme | 1.81 | 0.00 | 1393 | 121 | 2562 | 518 |
| rc_AI072943_at | Rn.54707 | 266777 | Nptx1 | neuronal pentraxin 1 | 1.32 | 0.00 | 429 | 40 | 566 | 66 |
| U88958_at | Rn.3546 | 83834 | Nrn1 | neuritin | 1.19 | 0.00 | 4572 | 160 | 5426 | 249 |
| rc_AA891220_at | Rn.7264 | 297508 | Pbef1 | pre-B-cell colony enhancing factor 1 | 1.25 | 0.00 | 301 | 22 | 377 | 36 |
| U95920_at | Rn.98622 | 81740 | Pcm1 | pericentriolar material 1 | 1.52 | 0.00 | 488 | 47 | 737 | 49 |
| AF002281_at | Rn.13361 | 114108 | Pdlim3 | PDZ and LIM domain 3 | 1.28 | 0.00 | 125 | 10 | 160 | 16 |
| rc_AA892797_at | Rn.108127 | 24644 | Pgk1 | phosphoglycerate kinase 1 | 2.08 | 0.00 | 2158 | 476 | 4493 | 1110 |
| D13907_g_at | Rn.841 | 64198 | Pmpcb | peptidase (mitochondrial processing) beta | 1.26 | 0.00 | 321 | 36 | 405 | 36 |
| X16554_at | Rn.9761 | 29562 | Prps1 | phosphoribosyl pyrophosphate synthetase 1 | 1.19 | 0.00 | 535 | 25 | 639 | 49 |
| rc_AA891383_i_at | Rn.1276 | 29672 | Psma5 | proteasome (prosome, macropain) subunit, alpha type 5 | 1.57 | 0.00 | 123 | 14 | 196 | 38 |
| U65217_i_at | Rn.33311 | 309622 | RT1-Bb | RT1 class II, locus Bb | 1.39 | 0.00 | 51 | 4 | 72 | 8 |
| L23128_g_at | Rn.107070 | 24750 | RT1-N3 | RT1 class Ib gene, H2-TL-like, grc region (N3) | 1.26 | 0.00 | 583 | 40 | 732 | 22 |
| M62763complete_seq_at | Rn.31887 | 25541 | Scp2 | sterol carrier protein 2 | 1.25 | 0.00 | 352 | 32 | 442 | 46 |
| AF009604_at | Rn.5909 | 81921 | Sh3gl3 | SH3 domain protein 2 C1 | 1.18 | 0.00 | 1346 | 80 | 1590 | 66 |
| Rc_AI639256_at | Rn.40672 | 301255 | Spats1 | spermatogenesis associated, serine-rich 1 | 1.29 | 0.00 | 159 | 12 | 205 | 16 |
| K01934mRNA#2_at | Rn.81140 | 25357 | Thrsp | thyroid hormone responsive protein | 1.30 | 0.00 | 384 | 33 | 499 | 53 |
| Frontal cortex | ||||||||||
| X62660mRNA_g_at | Rn.57528 | 300850 | Gsta4 | glutathione S-transferase, alpha 4 | -1.75 | 0.00 | 1075 | 121 | 622 | 151 |
| X97772_at | Rn.6872 | 58835 | Phgdh | 3-phosphoglycerate dehydrogenase | -1.42 | 0.00 | 1485 | 105 | 1052 | 130 |
| L81138exon_i_at | Rn.2115 | 83789 | Rps2 | ribosomal protein S2 | -1.76 | 0.00 | 475 | 31 | 269 | 11 |
| Y00766_at | Rn.87394 | 25657 | Scn3a | sodium channel, voltage-gated, type III, alpha polypeptide | -1.39 | 0.00 | 493 | 22 | 355 | 26 |
| AJ001290cds_at | Rn.79242 | 114507 | Slc5a3 | solute carrier family 5 (inositol transporters), member 3 | -1.44 | 0.00 | 779 | 67 | 542 | 71 |
| AF028784mRNA#1_s_at | Rn.91512 | 24387 | Gfap | glial fibrillary acidic protein | 1.44 | 0.00 | 6593 | 636 | 9536 | 1118 |
| rc_AA892797_at | Rn.108127 | 24644 | Pgk1 | phosphoglycerate kinase 1 | 2.08 | 0.00 | 2464 | 483 | 5107 | 862 |
| rc_AA891383_i_at | Rn.1276 | 29672 | Psma5 | proteasome (prosome, macropain) subunit, alpha type 5 | 1.78 | 0.00 | 119 | 19 | 215 | 56 |
| Hippocampus | ||||||||||
| rc_AI639111_at | Rn.93081 | 308923 | Ahi1 | Abelson helper integration site 1 | -1.48 | 0.00 | 705 | 89 | 476 | 45 |
| rc_AA893602_at | Rn.14812 | 316051 | Azi2 | 5-azacytidine induced gene 2 | -1.28 | 0.00 | 642 | 51 | 501 | 21 |
| rc_AI170268_at | Rn.1868 | 24223 | B2m | beta-2 microglobulin | -1.31 | 0.00 | 2242 | 272 | 1704 | 162 |
| X05834_at | Rn.1604 | 25661 | Fn1 | fibronectin 1 | -1.55 | 0.00 | 694 | 62 | 448 | 31 |
| X62660mRNA_at | Rn.57528 | 300850 | Gsta4 | glutathione S-transferase, alpha 4 | -1.36 | 0.00 | 718 | 76 | 526 | 38 |
| X62660mRNA_g_at | Rn.57528 | 300850 | Gsta4 | glutathione S-transferase, alpha 4 | -2.07 | 0.00 | 1099 | 219 | 526 | 49 |
| X76985_at | Rn.11404 | 59073 | Lxn | latexin | -1.34 | 0.00 | 904 | 89 | 672 | 45 |
| X97772_at | Rn.6872 | 58835 | Phgdh | 3-phosphoglycerate dehydrogenase | -1.33 | 0.00 | 1704 | 167 | 1279 | 133 |
| X97772_g_at | Rn.6872 | 58835 | Phgdh | 3-phosphoglycerate dehydrogenase | -1.39 | 0.00 | 1522 | 166 | 1094 | 39 |
| rc_AA800305_at | Rn.44477 | 64633 | Rab5a | RAB5A, member RAS oncogene family | -1.29 | 0.00 | 463 | 31 | 362 | 42 |
| L81138exon_i_at | Rn.2115 | 83789 | Rps2 | ribosomal protein S2 | -1.53 | 0.00 | 469 | 27 | 307 | 16 |
| M11071_f_at | Rn.40130 | 24737 | RT1-Aw2 | RT1 class Ib, locus Aw2 | -1.41 | 0.00 | 2351 | 271 | 1660 | 100 |
| rc_AA799700_at | Rn.100471 | 308993 | Sephs2 | selenophosphate synthetase 2 | -1.47 | 0.00 | 382 | 33 | 260 | 17 |
| AJ001290cds_at | Rn.79242 | 114507 | Slc5a3 | solute carrier family 5 (inositol transporters), member 3 | -1.47 | 0.00 | 846 | 81 | 583 | 117 |
| rc_AA800712_g_at | Rn.13142 | 361813 | Stk38 | serine/threonine kinase 38 | -1.21 | 0.00 | 836 | 43 | 691 | 36 |
| M83107_at | Rn.34397 | 25123 | Tagln | transgelin | -1.23 | 0.00 | 435 | 34 | 353 | 17 |
| U48288_at | Rn.10557 | 25228 | Akap11 | A kinase (PRKA) anchor protein 11 | 1.32 | 0.00 | 309 | 30 | 408 | 29 |
| Z12298cds_s_at | Rn.106103 | 29139 | Dcn | decorin | 1.29 | 0.00 | 477 | 49 | 614 | 48 |
| AF076619_at | Rn.30028 | 58844 | Grb14 | growth factor receptor bound protein 14 | 1.32 | 0.00 | 342 | 30 | 451 | 35 |
| AF008554_at | Rn.43578 | 116967 | Iag2 | implantation-associated protein | 1.31 | 0.00 | 173 | 17 | 228 | 27 |
| rc_AA892797_at | Rn.108127 | 24644 | Pgk1 | phosphoglycerate kinase 1 | 2.28 | 0.00 | 2064 | 556 | 4664 | 923 |
| L23128_g_at | Rn.107070 | 24750 | RT1-N3 | RT1 class Ib gene, H2-TL-like, grc region (N3) | 1.35 | 0.00 | 581 | 46 | 781 | 34 |
| AB011530_at | Rn.30002 | 65047 | Slit1 | slit homolog 1 (Drosophila) | 1.32 | 0.00 | 640 | 42 | 846 | 42 |
| Striatum | ||||||||||
| rc_AI639111_at | Rn.93081 | 308923 | Ahi1 | Abelson helper integration site 1 | -1.46 | 0.00 | 745 | 75 | 511 | 76 |
| L07114_at | Rn.10002 | 25383 | Apobec1 | apolipoprotein B editing complex 1 | -1.23 | 0.00 | 146 | 5 | 118 | 4 |
| X05834_at | Rn.1604 | 25661 | Fn1 | fibronectin 1 | -1.35 | 0.00 | 810 | 42 | 605 | 99 |
| X62660mRNA_at | Rn.57528 | 300850 | Gsta4 | glutathione S-transferase, alpha 4 | -1.35 | 0.00 | 651 | 18 | 482 | 34 |
| X62660mRNA_g_at | Rn.57528 | 300850 | Gsta4 | glutathione S-transferase, alpha 4 | -2.00 | 0.00 | 1047 | 109 | 523 | 34 |
| AF090136_at | Rn.44269 | 60442 | Lin7c | lin-7 homolog C (C. elegans) | -1.31 | 0.00 | 148 | 11 | 113 | 7 |
| rc_AI008677_s_at | Rn.6872 | 58835 | Phgdh | 3-phosphoglycerate dehydrogenase | -1.33 | 0.00 | 741 | 45 | 562 | 66 |
| X97772_at | Rn.6872 | 58835 | Phgdh | 3-phosphoglycerate dehydrogenase | -1.28 | 0.00 | 1600 | 95 | 1250 | 73 |
| X97772_g_at | Rn.6872 | 58835 | Phgdh | 3-phosphoglycerate dehydrogenase | -1.29 | 0.00 | 1369 | 90 | 1062 | 73 |
| U42413_at | Rn.11089 | 25520 | Prkag1 | protein kinase, AMP-activated, gamma 1 noncatalytic subunit | -1.19 | 0.00 | 1211 | 33 | 1020 | 52 |
| S71021_s_at | Rn.2660 | 117042 | Rpl6 | ribosomal protein L6 | -1.21 | 0.00 | 5829 | 167 | 4808 | 255 |
| L81138exon_i_at | Rn.2115 | 83789 | Rps2 | ribosomal protein S2 | -1.64 | 0.00 | 427 | 35 | 260 | 16 |
| M11071_f_at | Rn.40130 | 24737 | RT1-Aw2 | RT1 class Ib, locus Aw2 | -1.39 | 0.00 | 2183 | 79 | 1574 | 93 |
| M31018_f_at | Rn.40130 | 24737 | RT1-Aw2 | RT1 class Ib, locus Aw2 | -1.26 | 0.00 | 1039 | 38 | 825 | 91 |
| Y00766_at | Rn.87394 | 25657 | Scn3a | sodium channel, voltage-gated, type III, alpha polypeptide | -1.46 | 0.00 | 632 | 56 | 439 | 84 |
| rc_AA799700_at | Rn.100471 | 308993 | Sephs2 | selenophosphate synthetase 2 | -1.45 | 0.00 | 328 | 44 | 225 | 23 |
| AB001453_at | Rn.59227 | 114858 | Shc3 | src homology 2 domain-containing transforming protein C3 | -1.39 | 0.00 | 1361 | 103 | 985 | 125 |
| AJ001290cds_at | Rn.79242 | 114507 | Slc5a3 | solute carrier family 5 (inositol transporters), member 3 | -1.41 | 0.00 | 791 | 61 | 566 | 89 |
| M12112mRNA#3_s_at | Rn.6319 | 24179 | Agt | angiotensinogen | 1.30 | 0.00 | 477 | 42 | 621 | 44 |
| U67136_at | Rn.122003 | 171026 | Akap5 | A kinase (PRKA) anchor protein 5 | 1.39 | 0.00 | 163 | 12 | 229 | 35 |
| M60322_g_at | Rn.107801 | 24192 | Akr1b4 | aldo-keto reductase family 1, member B4 (aldose reductase) | 1.21 | 0.00 | 537 | 35 | 650 | 37 |
| U07201_at | Rn.11172 | 25612 | Asns | asparagine synthetase | 1.20 | 0.00 | 334 | 15 | 402 | 22 |
| M55534mRNA_s_at | Rn.98208 | 25420 | Cryab | crystallin, alpha B | 1.35 | 0.00 | 869 | 155 | 1161 | 83 |
| X15705cds_at | Rn.112579 | 60460 | Hspa2 | heat shock protein 2 | 1.24 | 0.00 | 130 | 6 | 161 | 11 |
| rc_AA818677_at | Rn.108194 | 24587 | Nefh | neurofilament, heavy polypeptide | 1.44 | 0.00 | 586 | 68 | 841 | 62 |
| rc_AA892797_at | Rn.108127 | 24644 | Pgk1 | phosphoglycerate kinase 1 | 2.24 | 0.00 | 2151 | 294 | 4847 | 896 |
| rc_AA891383_i_at | Rn.1276 | 29672 | Psma5 | proteasome (prosome, macropain) subunit, alpha type 5 | 1.63 | 0.00 | 122 | 24 | 197 | 26 |
| U67082_at | Rn.10663 | 25165 | Znf386 | zinc finger protein 386 (Kruppel-like) | 1.22 | 0.00 | 194 | 11 | 236 | 13 |
S.D. = standard deviation; FDR = false discovery rate; iP = inbred alcohol-preferring rat; iNP = inbred alcohole—non-preferring rat; RMA = Robust Multi-Chip Average.
Positive fold difference is iP/iNP; negative is iNP/iP.
Expression levels are those obtained after RMA background correction and scaling as indicated in Methods.
Over-represented GO biologic process categories (P <.05) were statistically sought using the Bioconductor package GOstats (Gentleman et al., 2004). Briefly, GO categories of significant genes were tested for over-representation using the hypergeometric distribution. An analysis of each of the five individual brain regions did not reveal any over-represented GO biologic process categories, perhaps because there were too few differentially expressed genes in each region.
An overall analysis of all brain regions, which used a linear model with strain and brain region as factors, demonstrated significant differential expression of 296 genes (351 probe sets) (FDR ≤0.10). Of these, 173 genes (198 probe sets) demonstrated higher and 123 genes (153 probe sets) lower expression levels in the iP rats than in the iNP rats (Table 4). Although almost all of the genes identified as being differentially expressed within the individual regions were also identified in the overall analysis, the latter method detected many that fell below the threshold of statistical detection for individual regions, most likely because of the increased power gained by including a much larger number of arrays in the linear model (Table 5). There were 53 genes (71 probe sets) found significant in at least one individual region and in the overall analysis; 241 genes (280 probe sets) were found significant only in the overall analysis. Additionally, there were 15 genes (17 probe sets) found significant in the AMYG that were not significant in the overall analysis, one of which was also significant in the ACB.
Table 4.
Complete list of probe sets that were significantly different between iP and iNP rats in the overall analysis
| Probe set | Unigene | Entrez gene | Symbol | Gene description | Fold difference | FDR | Mean iP | S.D. iP | Mean iNP | S.D. iNP |
|---|---|---|---|---|---|---|---|---|---|---|
| M33648_at | Rn.29594 | 24450 | Hmgcs2 | 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 | -1.07 | 0.00 | 726 | 74 | 686 | 89 |
| M33648_g_at | Rn.29594 | 24450 | Hmgcs2 | 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 | -1.15 | 0.00 | 410 | 105 | 366 | 117 |
| rc_AI008677_s_at | Rn.6872 | 58835 | Phgdh | 3-phosphoglycerate dehydrogenase | -1.27 | 0.00 | 718 | 78 | 570 | 77 |
| X97772_at | Rn.6872 | 58835 | Phgdh | 3-phosphoglycerate dehydrogenase | -1.33 | 0.00 | 1642 | 158 | 1244 | 159 |
| X97772_g_at | Rn.6872 | 58835 | Phgdh | 3-phosphoglycerate dehydrogenase | -1.31 | 0.00 | 1426 | 156 | 1095 | 124 |
| rc_AA893602_at | Rn.14812 | 316051 | Azi2 | 5-azacytidine induced gene 2 | -1.23 | 0.00 | 633 | 49 | 518 | 60 |
| rc_AI639111_at | Rn.93081 | 308923 | Ahi1 | Abelson helper integration site 1 | -1.46 | 0.00 | 736 | 121 | 510 | 89 |
| D00512_at | Rn.4054 | 25014 | Acat1 | acetyl-coenzyme A acetyltransferase 1 | -1.11 | 0.00 | 274 | 19 | 248 | 13 |
| rc_AI170212_s_at | Rn.11007 | 140667 | Ap3m2 | adaptor-related protein complex 3, mu 2 subunit | -1.12 | 0.01 | 2016 | 190 | 1829 | 313 |
| rc_AA891812_at | Rn.5788 | 24170 | Add1 | adducin 1 (alpha) | -1.09 | 0.00 | 3262 | 284 | 2993 | 279 |
| X83715_at | Rn.5788 | 24170 | Add1 | adducin 1 (alpha) | -1.14 | 0.00 | 1377 | 148 | 1232 | 207 |
| S47609_s_at | Rn.11180 | 25369 | Adora2a | adenosine A2a receptor | -1.08 | 0.00 | 918 | 359 | 862 | 361 |
| D17309_at | Rn.25716 | 192242 | Akr1d1 | aldo-keto reductase family 1, member D1 | -1.09 | 0.00 | 214 | 24 | 197 | 28 |
| D86297_at | Rn.32517 | 25748 | Alas2 | aminolevulinic acid synthase 2 | -1.14 | 0.00 | 934 | 146 | 818 | 135 |
| AF029107_at | Rn.62687 | 83610 | Apba2 | amyloid beta (A4) precursor protein-binding, family A, member 2 | -1.13 | 0.01 | 1429 | 187 | 1291 | 266 |
| L07114_at | Rn.10002 | 25383 | Apobec1 | apolipoprotein B editing complex 1 | -1.15 | 0.00 | 146 | 15 | 127 | 12 |
| J04024_at | Rn.2305 | 29693 | Atp2a2 | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 | -1.13 | 0.00 | 501 | 44 | 450 | 81 |
| U34963_s_at | Rn.10323 | 24888 | Bcl2l1 | Bcl2-like 1 | -1.11 | 0.00 | 324 | 36 | 295 | 45 |
| U72350_at | Rn.10323 | 24888 | Bcl2l1 | Bcl2-like 1 | -1.07 | 0.00 | 749 | 76 | 700 | 73 |
| AF065431_s_at | Rn.82709 | 64547 | Bcl2l11 | BCL2-like 11 (apoptosis facilitator) | -1.08 | 0.01 | 95 | 10 | 88 | 9 |
| Rc_AI170268_at | Rn.1868 | 24223 | B2m | beta-2 microglobulin | -1.18 | 0.00 | 2407 | 338 | 2059 | 373 |
| D63860_s_at | Rn.53974 | 25667 | Bmp3 | bone morphogenetic protein 3 | -1.06 | 0.01 | 141 | 13 | 133 | 10 |
| D83349_at | Rn.108785 | 29182 | Cdh22 | cadherin 22 | -1.43 | 0.00 | 1807 | 436 | 1380 | 565 |
| J02844_s_at | Rn.4896 | 83842 | Crot | carnitine O-octanoyltransferase | -1.12 | 0.00 | 187 | 23 | 166 | 15 |
| U26033_at | Rn.4896 | 83842 | Crot | carnitine O-octanoyltransferase | -1.15 | 0.00 | 96 | 11 | 83 | 12 |
| Rc_AI013513_at | Rn.8046 | 64462 | Csnk1d | casein kinase 1, delta | -1.10 | 0.00 | 765 | 72 | 696 | 76 |
| Rc_AI639196_at | Rn.33757 | 66013 | Arhgef9 | Cdc42 guanine nucleotide exchange factor (GEF) 9 | -1.22 | 0.01 | 742 | 78 | 644 | 181 |
| U07619_at | Rn.9980 | 25584 | F3 | coagulation factor III | -1.19 | 0.00 | 150 | 18 | 126 | 21 |
| L41275cds_s_at | Rn.10089 | 114851 | Cdkn1a | cyclin-dependent kinase inhibitor 1A | -1.11 | 0.00 | 357 | 36 | 321 | 34 |
| L33894_at | Rn.10058 | 79127 | Dck | deoxycytidine kinase | -1.12 | 0.00 | 86 | 10 | 77 | 9 |
| Z46882cds_at | Rn.2889 | 25416 | Dpysl2 | dihydropyrimidinase-like 2 | -1.10 | 0.00 | 381 | 30 | 349 | 52 |
| D85035_g_at | Rn.133148 | 81656 | Dpyd | dihydropyrimidine dehydrogenase | -1.11 | 0.00 | 97 | 17 | 87 | 12 |
| Rc_AA894273_at | Rn.7398 | 64157 | Ddah1 | dimethylarginine dimethylaminohydrolase 1 | -1.19 | 0.00 | 212 | 18 | 179 | 22 |
| AJ011607_at | Rn.11846 | 301323 | Prim2 | DNA primase, p58 subunit | -1.13 | 0.00 | 161 | 14 | 142 | 11 |
| AF013144_at | Rn.10877 | 171109 | Dusp5 | dual specificity phosphatase 5 | -1.07 | 0.00 | 423 | 45 | 396 | 49 |
| X94185cds_s_at | Rn.4313 | 116663 | Dusp6 | dual specificity phosphatase 6 | -1.28 | 0.01 | 1483 | 235 | 1238 | 411 |
| Rc_AA893770_g_at | Rn.144669 | 245963 | Egfl7 | EGF-like domain 7 | -1.07 | 0.01 | 546 | 34 | 511 | 52 |
| X02610_at | Rn.4236 | 24333 | Eno1 | enolase 1, alpha | -1.15 | 0.00 | 6777 | 494 | 5917 | 561 |
| X02610_g_at | Rn.4236 | 24333 | Eno1 | enolase 1, alpha | -1.11 | 0.00 | 11825 | 997 | 10717 | 1427 |
| X59290mRNA_at | Rn.62934 | 60589 | Epha8 | Eph receptor A8 | -1.08 | 0.00 | 332 | 37 | 307 | 29 |
| X78689cds_at | Rn.24569 | 79208 | Epha5 | EphA5 | -1.11 | 0.01 | 160 | 24 | 145 | 22 |
| D89730_at | Rn.8037 | 305604 | Efemp1 | epidermal growth factor-containing fibulin-like extracellular matrix protein 1 | -1.15 | 0.00 | 535 | 196 | 456 | 146 |
| D89730_g_at | Rn.8037 | 305604 | Efemp1 | epidermal growth factor-containing fibulin-like extracellular matrix protein 1 | -1.13 | 0.00 | 64 | 17 | 56 | 12 |
| D12498_s_at | Rn.9797 | 79114 | Fgfr1 | Fibroblast growth factor receptor 1 | -1.11 | 0.00 | 813 | 248 | 753 | 278 |
| M91599mRNA_at | Rn.24104 | 25114 | Fgfr4 | fibroblast growth factor receptor 4 | -1.09 | 0.00 | 168 | 21 | 154 | 14 |
| L00191cds#1_s_at | Rn.1604 | 25661 | Fn1 | fibronectin 1 | -1.27 | 0.00 | 410 | 118 | 318 | 72 |
| M28259cds_at | Rn.1604 | 25661 | Fn1 | fibronectin 1 | -1.13 | 0.00 | 116 | 23 | 101 | 11 |
| Rc_AA955600_at | Rn.1604 | 25661 | Fn1 | fibronectin 1 | -1.07 | 0.01 | 69 | 6 | 65 | 6 |
| U82612cds_g_at | Rn.1604 | 25661 | Fn1 | fibronectin 1 | -1.15 | 0.00 | 439 | 52 | 381 | 47 |
| X05834_at | Rn.1604 | 25661 | Fn1 | fibronectin 1 | -1.34 | 0.00 | 952 | 203 | 722 | 220 |
| L27421_at | Rn.62653 | 65153 | Freq | frequenin homolog (Drosophila) | -1.22 | 0.00 | 183 | 35 | 150 | 33 |
| M92076_at | Rn.41715 | 24416 | Grm3 | glutamate receptor, metabotropic 3 | -1.31 | 0.00 | 1399 | 384 | 1117 | 457 |
| X96790_at | Rn.10409 | 81672 | Grm7 | glutamate receptor, metabotropic 7 | -1.08 | 0.00 | 72 | 6 | 67 | 7 |
| X62660mRNA_at | Rn.57528 | 300850 | Gsta4 | glutathione S-transferase, alpha 4 | -1.29 | 0.00 | 680 | 65 | 526 | 57 |
| X62660mRNA_g_at | Rn.57528 | 300850 | Gsta4 | glutathione S-transferase, alpha 4 | -1.81 | 0.00 | 1099 | 146 | 613 | 135 |
| X55246_at | Rn.10109 | 25674 | Glra1 | glycine receptor, alpha 1 subunit | -1.11 | 0.00 | 983 | 132 | 883 | 104 |
| L02896_at | Rn.7044 | 58920 | Gpc1 | glypican 1 | -1.12 | 0.00 | 369 | 51 | 332 | 48 |
| L06986_at | Rn.10935 | 24388 | Gfi1 | growth factor independent 1 | -1.09 | 0.00 | 103 | 9 | 95 | 8 |
| Rc_AA859837_at | Rn.24783 | 83585 | Gda | guanine deaminase | -1.28 | 0.00 | 3722 | 794 | 3130 | 1013 |
| Rc_AA859837_g_at | Rn.24783 | 83585 | Gda | guanine deaminase | -1.21 | 0.00 | 9381 | 1873 | 8042 | 2310 |
| M17526_g_at | Rn.90161 | 50664 | Gnao | guanine nucleotide binding protein, alpha o | -1.26 | 0.01 | 5794 | 1240 | 4845 | 1689 |
| D13417_g_at | Rn.19727 | 29577 | Hes1 | hairy and enhancer of split 1 (Drosophila) | -1.08 | 0.00 | 404 | 32 | 376 | 37 |
| X56325mRNA_s_at | Rn.107334 | 25632 | Hba-a1 | hemoglobin alpha, adult chain 1 | -1.19 | 0.00 | 25130 | 5747 | 21396 | 6048 |
| M94918mRNA_f_at | Rn.36966 | 24440 | Hbb | hemoglobin beta chain complex | -1.21 | 0.00 | 14307 | 4436 | 11865 | 4004 |
| U89695_at | Rn.29981 | 84017 | Hmga2 | high mobility group AT-hook 2 | -1.07 | 0.01 | 107 | 12 | 100 | 7 |
| AB007689_g_at | Rn.30014 | 29547 | Homer2 | homer homolog 2 (Drosophila) | -1.09 | 0.00 | 140 | 18 | 128 | 13 |
| Rc_AA893384_g_at | Rn.1499 | 292892 | Irf3 | interferon regulatory factor 3 | -1.06 | 0.00 | 1298 | 106 | 1223 | 114 |
| Rc_AI639313_at | Rn.87882 | 84009 | Kalrn | kalirin, RhoGEF kinase | -1.09 | 0.01 | 260 | 23 | 240 | 31 |
| U93306_at | Rn.88869 | 25589 | Kdr | kinase insert domain protein receptor | -1.28 | 0.00 | 95 | 19 | 74 | 14 |
| K02814_at | Rn.128333 | 24903 | Kng1 | kininogen 1 | -1.09 | 0.00 | 758 | 66 | 695 | 61 |
| AF071204_at | Rn.44216 | 60427 | Kitl | kit ligand | -1.07 | 0.01 | 79 | 7 | 74 | 7 |
| AF071204_g_at | Rn.44216 | 60427 | Kitl | kit ligand | -1.09 | 0.00 | 93 | 10 | 86 | 12 |
| X76985_at | Rn.11404 | 59073 | Lxn | latexin | -1.19 | 0.00 | 881 | 304 | 750 | 277 |
| AF090134_at | Rn.31766 | 85327 | Lin7a | lin-7 homolog a (C. elegans) | -1.16 | 0.00 | 633 | 96 | 549 | 105 |
| AF090136_at | Rn.44269 | 60442 | Lin7c | lin-7 homolog C (C. elegans) | -1.35 | 0.00 | 152 | 20 | 112 | 8 |
| M64795_f_at d | — | 24735 | RT1@ | Major histocompatibility locus | -1.14 | 0.00 | 918 | 56 | 808 | 76 |
| M15944_at | Rn.33598 | 24590 | Mme | membrane metallo endopeptidase | -1.05 | 0.01 | 70 | 6 | 66 | 5 |
| AF095741_at | Rn.11802 | 171451 | Mg87 | Mg87 protein | -1.11 | 0.00 | 950 | 91 | 856 | 69 |
| Rc_AI227608_s_at | Rn.2455 | 29477 | Mapt | microtubule-associated protein tau | -1.29 | 0.00 | 6120 | 525 | 5022 | 1247 |
| Rc_AA892864_at | Rn.40396 | 29254 | Mgll | monoglyceride lipase | -1.11 | 0.01 | 1353 | 202 | 1228 | 260 |
| X06564_at | Rn.11283 | 24586 | Ncam1 | neural cell adhesion molecule 1 | -1.20 | 0.01 | 1023 | 142 | 892 | 241 |
| L14851_at | Rn.10926 | 116508 | Nrxn3 | neurexin 3 | -1.11 | 0.00 | 1187 | 216 | 1081 | 218 |
| M82826_i_at | Rn.10686 | 24592 | Nf1 | neurofibromatosis 1 | -1.16 | 0.00 | 1786 | 191 | 1556 | 255 |
| V01543mRNA_at | Rn.2865 | 25247 | Nsg1 | neuron specific gene family member 1 | -1.07 | 0.00 | 1376 | 126 | 1292 | 107 |
| M15880_at | Rn.9714 | 24604 | Npy | neuropeptide Y | -1.14 | 0.00 | 2120 | 345 | 1857 | 327 |
| U16845_at | Rn.10075 | 50864 | Hnt | neurotrimin | -1.43 | 0.00 | 855 | 178 | 676 | 289 |
| M86742cds_s_at | Rn.44225 | 25730 | Ntf5 | neurotrophin 5 | -1.06 | 0.01 | 378 | 28 | 358 | 22 |
| D63673_at | Rn.10675 | 117265 | Pex6 | peroxisomal biogenesis factor 6 | -1.08 | 0.01 | 854 | 103 | 786 | 67 |
| M98826mRNA_g_at | Rn.10399 | 29353 | Phkg1 | phosphorylase kinase gamma 1 | -1.08 | 0.00 | 226 | 20 | 209 | 14 |
| U77697_at | Rn.1878 | 29583 | Pecam | platelet/endothelial cell adhesion molecule | -1.14 | 0.00 | 215 | 23 | 188 | 17 |
| J02776_s_at | Rn.9346 | 29240 | Polb | polymerase (DNA directed), beta | -1.08 | 0.00 | 176 | 25 | 162 | 16 |
| L14003UTR#1_f_at | Rn.54456 | 25046 | Pigr | polymeric immunoglobulin receptor | -1.07 | 0.00 | 273 | 25 | 255 | 20 |
| AJ007627_at | Rn.144567 | 27150 | Kcnh3 | potassium voltage-gated channel, subfamily H (eag-related), member 3 | -1.09 | 0.01 | 2628 | 383 | 2392 | 276 |
| rc_AA799421_at | Rn.34966 | 29340 | Prkce | protein kinase C, epsilon | -1.29 | 0.00 | 1503 | 262 | 1264 | 437 |
| U42413_at | Rn.11089 | 25520 | Prkag1 | protein kinase, AMP-activated, gamma 1 noncatalytic subunit | -1.13 | 0.00 | 1181 | 72 | 1042 | 68 |
| U47031_at | Rn.7176 | 29659 | P2rx4 | purinergic receptor P2X, ligand-gated ion channel 4 | -1.08 | 0.00 | 373 | 31 | 343 | 29 |
| rc_AA894207_g_at | Rn.28921 | 246324 | Rab31 | RAB31, member RAS oncogene family | -1.18 | 0.01 | 5493 | 959 | 4781 | 1121 |
| AF072935_at | Rn.44477 | 64633 | Rab5a | RAB5A, member RAS oncogene family | -1.10 | 0.00 | 644 | 65 | 586 | 49 |
| rc_AA800305_at | Rn.44477 | 64633 | Rab5a | RAB5A, member RAS oncogene family | -1.15 | 0.00 | 446 | 73 | 390 | 63 |
| rc_AA900900_s_at | Rn.7107 | 84014 | Ralbp1 | ralA binding protein 1 | -1.11 | 0.01 | 369 | 41 | 335 | 60 |
| rc_AA893471_s_at | Rn.98353 | 24718 | Reln | reelin | -1.14 | 0.00 | 1103 | 160 | 975 | 178 |
| AF035151_s_at | Rn.9796 | 54292 | Rgs12 | regulator of G-protein signaling 12 | -1.07 | 0.01 | 771 | 114 | 723 | 94 |
| AB015191_at | Rn.1943 | 60414 | Rhced | Rhesus blood group CE and D | -1.09 | 0.01 | 458 | 51 | 420 | 46 |
| AB015191_g_at | Rn.1943 | 60414 | Rhced | Rhesus blood group CE and D | -1.08 | 0.00 | 292 | 33 | 268 | 22 |
| rc_AA799672_s_at | Rn.2660 | 117042 | Rpl6 | ribosomal protein L6 | -1.15 | 0.00 | 9605 | 697 | 8390 | 658 |
| S71021_s_at | Rn.2660 | 117042 | Rpl6 | ribosomal protein L6 | -1.14 | 0.00 | 5383 | 511 | 4715 | 479 |
| L81136cds_f_at | Rn.2115 | 83789 | Rps2 | ribosomal protein S2 | -1.15 | 0.00 | 1095 | 77 | 957 | 56 |
| L81138exon_i_at | Rn.2115 | 83789 | Rps2 | ribosomal protein S2 | -1.60 | 0.00 | 459 | 37 | 287 | 34 |
| M24026_f_at | Rn.145396 | 309600 | RT1-CE12 | RT1 class I, CE12 | -1.17 | 0.00 | 2027 | 223 | 1734 | 202 |
| AF074609mRNA_f_at | Rn.40130 | 24737 | RT1-Aw2 | RT1 class Ib, locus Aw2 | -1.14 | 0.00 | 791 | 126 | 691 | 88 |
| M10094_g_at | Rn.40130 | 24737 | RT1-Aw2 | RT1 class Ib, locus Aw2 | -1.20 | 0.00 | 370 | 32 | 307 | 30 |
| M11071_f_at | Rn.40130 | 24737 | RT1-Aw2 | RT1 class Ib, locus Aw2 | -1.38 | 0.00 | 2376 | 260 | 1724 | 160 |
| M24324_f_at | Rn.40130 | 24737 | RT1-Aw2 | RT1 class Ib, locus Aw2 | -1.09 | 0.00 | 2890 | 191 | 2660 | 224 |
| M31018_f_at | Rn.40130 | 24737 | RT1-Aw2 | RT1 class Ib, locus Aw2 | -1.22 | 0.00 | 1141 | 126 | 936 | 121 |
| AF025308_f_at | Rn.160576 | 24977 | RT1-Cl | RT1 class Ib, locus Cl | -1.12 | 0.00 | 823 | 70 | 733 | 57 |
| AF029240_g_at | Rn.143874 | 294228 | RT1-S3 | RT1 class Ib, locus S3 | -1.06 | 0.00 | 406 | 26 | 382 | 36 |
| rc_AI235890_s_at | Rn.143874 | 294228 | RT1-S3 | RT1 class Ib, locus S3 | -1.21 | 0.00 | 236 | 33 | 195 | 24 |
| X56596_at | Rn.33311 | 309622 | RT1-Bb | RT1 class II, locus Bb | -1.16 | 0.00 | 240 | 21 | 206 | 18 |
| rc_AA799700_at | Rn.100471 | 308993 | Sephs2 | selenophosphate synthetase 2 | -1.45 | 0.00 | 341 | 47 | 234 | 28 |
| Y07534cds_s_at | Rn.91257 | 24794 | Spin2b | Serine protease inhibitor | -1.07 | 0.00 | 995 | 84 | 930 | 71 |
| rc_AA800712_g_at | Rn.13142 | 361813 | Stk38 | serine/threonine kinase 38 | -1.17 | 0.00 | 810 | 62 | 695 | 82 |
| U90261UTR#1_g_at | Rn.107226 | 84357 | Sh3kbp1 | SH3-domain kinase binding protein 1 | -1.10 | 0.00 | 2509 | 473 | 2303 | 500 |
| AB011531_at | Rn.12298 | 83467 | Slit3 | slit homolog 3 (Drosophila) | -1.08 | 0.00 | 579 | 59 | 534 | 53 |
| Y00766_at | Rn.87394 | 25657 | Scn3a | sodium channel, voltage-gated, type III, alpha polypeptide | -1.36 | 0.00 | 536 | 116 | 393 | 69 |
| Y00766_g_at | Rn.87394 | 25657 | Scn3a | sodium channel, voltage-gated, type III, alpha polypeptide | -1.38 | 0.00 | 1529 | 337 | 1117 | 235 |
| U66723_s_at | Rn.10140 | 60423 | Slc28a2 | solute carrier family 28 (sodium-coupled nucleoside transporter), member 2 | -1.15 | 0.00 | 341 | 47 | 294 | 19 |
| AF004017_at | Rn.11114 | 84484 | Slc4a4 | solute carrier family 4, member 4 | -1.24 | 0.00 | 1156 | 249 | 950 | 248 |
| AJ001290cds_at | Rn.79242 | 114507 | Slc5a3 | solute carrier family 5 (inositol transporters), member 3 | -1.49 | 0.00 | 808 | 64 | 548 | 93 |
| M96601_at | Rn.9968 | 29464 | Slc6a6 | solute carrier family 6 (neurotransmitter transporter, taurine), member 6 | -1.37 | 0.00 | 339 | 85 | 259 | 97 |
| AB001453_at | Rn.59227 | 114858 | Shc3 | src homology 2 domain-containing transforming protein C3 | -1.28 | 0.00 | 1213 | 183 | 941 | 110 |
| rc_AA875411_s_at | Rn.37542 | 65041 | Skd3 | suppressor of K+ transport defect 3 | -1.06 | 0.00 | 696 | 55 | 654 | 48 |
| rc_AA891445_at | Rn.37542 | 65041 | Skd3 | suppressor of K+ transport defect 3 | -1.11 | 0.00 | 591 | 71 | 532 | 75 |
| AF053938_s_at | Rn.9908 | 192117 | Syngap1 | synaptic Ras GTPase activating protein 1 homolog (rat) | -1.15 | 0.00 | 496 | 36 | 429 | 27 |
| U20105_at | Rn.48074 | 60565 | Syt6 | synaptotagmin 6 | -1.09 | 0.00 | 138 | 21 | 127 | 19 |
| U20106_at | Rn.91884 | 59267 | Syt7 | synaptotagmin 7 | -1.13 | 0.00 | 138 | 13 | 122 | 15 |
| U85513_at | Rn.74259 | 60567 | Syt10 | synaptotagmin X | -1.07 | 0.01 | 100 | 11 | 93 | 7 |
| D12519_s_at | Rn.9943 | 116470 | Stx1a | syntaxin 1A (brain) | -1.10 | 0.01 | 3399 | 943 | 3090 | 954 |
| U88630_at | Rn.2398 | 81813 | Tieg | TGFB inducible early growth response | -1.07 | 0.00 | 146 | 13 | 136 | 11 |
| D30041_at | Rn.87066 | 25233 | Akt2 | thymoma viral proto-oncogene 2 | -1.11 | 0.01 | 304 | 45 | 274 | 46 |
| rc_AA818982_at | Rn.3364 | 25359 | Tmpo | Thymopoietin | -1.16 | 0.01 | 161 | 29 | 141 | 32 |
| rc_AA899854_at | Rn.90996 | 360243 | Top2a | topoisomerase (DNA) 2 alpha | -1.09 | 0.01 | 276 | 32 | 254 | 34 |
| rc_AA893702_s_at | Rn.1829 | 64365 | Tcn2 | transcobalamin 2 | -1.07 | 0.00 | 1309 | 122 | 1224 | 125 |
| rc_AI639517_at | Rn.23354 | 84382 | Tcf4 | transcription factor 4 | -1.20 | 0.00 | 136 | 18 | 114 | 23 |
| U09228_at | Rn.23354 | 84382 | Tcf4 | transcription factor 4 | -1.18 | 0.01 | 421 | 55 | 371 | 102 |
| rc_AA874857_at | Rn.114933 | 498407 | pur-beta | transcription factor Pur-beta | -1.16 | 0.00 | 111 | 20 | 97 | 20 |
| M83107_at | Rn.34397 | 25123 | Tagln | Transgelin | -1.17 | 0.00 | 482 | 101 | 408 | 81 |
| rc_AI639532_at | Rn.43529 | 296369 | Tnnc2 | troponin C2, fast | -1.15 | 0.00 | 412 | 210 | 349 | 183 |
| rc_AI169370_at | Rn.99661 | 64158 | Tuba1 | tubulin, alpha 1 | -1.09 | 0.00 | 16608 | 1110 | 15260 | 1350 |
| rc_AI230748_at | Rn.36610 | 116646 | Tpt1 | tumor protein, translationally controlled 1 | -1.11 | 0.00 | 6404 | 796 | 5832 | 817 |
| rc_AI639378_at | Rn.40168 | 362760 | Galntl1 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 1 | -1.11 | 0.00 | 112 | 11 | 102 | 15 |
| rc_AI071435_at | Rn.51354 | 25218 | Vps52 | vacuolar protein sorting 52 (yeast) | -1.22 | 0.00 | 852 | 79 | 701 | 72 |
| M32167_at | Rn.1923 | 83785 | Vegfa | vascular endothelial growth factor A | -1.09 | 0.00 | 1166 | 100 | 1074 | 89 |
| M32167_g_at | Rn.1923 | 83785 | Vegfa | vascular endothelial growth factor A | -1.16 | 0.00 | 104 | 25 | 88 | 18 |
| D89514_at | Rn.15114 | 81643 | Atic | 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase | 1.25 | 0.00 | 344 | 47 | 431 | 51 |
| U48288_at | Rn.10557 | 25228 | Akap11 | A kinase (PRKA) anchor protein 11 | 1.24 | 0.00 | 345 | 51 | 429 | 59 |
| U67136_at | Rn.122003 | 171026 | Akap5 | A kinase (PRKA) anchor protein 5 | 1.23 | 0.00 | 139 | 21 | 175 | 46 |
| J00692_at | Rn.82732 | 29437 | Acta1 | actin, alpha 1, skeletal muscle | 1.10 | 0.00 | 366 | 35 | 404 | 68 |
| S56508_s_at | Rn.33697 | 117243 | Acsl6 | acyl-CoA synthetase long-chain family member 6 | 1.15 | 0.00 | 617 | 81 | 711 | 121 |
| U90888_at | Rn.11106 | 25095 | Ampd3 | adenosine monophosphate deaminase 3 | 1.27 | 0.00 | 96 | 14 | 123 | 26 |
| rc_AA945321_at | Rn.34353 | 24186 | Alb | Albumin | 1.06 | 0.00 | 427 | 33 | 454 | 49 |
| AF001898_at | Rn.6132 | 24188 | Aldh1a1 | aldehyde dehydrogenase family 1, member A1 | 1.20 | 0.00 | 944 | 179 | 1129 | 178 |
| M60322_g_at | Rn.107801 | 24192 | Akr1b4 | aldo-keto reductase family 1, member B4 (aldose reductase) | 1.19 | 0.00 | 520 | 59 | 620 | 95 |
| rc_AA891631_at | Rn.116338 | 295365 | Amigo | amphoretin-induced gene and ORF | 1.11 | 0.00 | 572 | 53 | 638 | 103 |
| M12112mRNA#3_s_at | Rn.6319 | 24179 | Agt | Angiotensinogen | 1.20 | 0.00 | 745 | 259 | 884 | 260 |
| rc_AA943892_at | Rn.6319 | 24179 | Agt | Angiotensinogen | 1.16 | 0.00 | 1780 | 577 | 2057 | 600 |
| D42137exon_s_at | Rn.3318 | 25673 | Anxa5 | annexin A5 | 1.10 | 0.00 | 1157 | 173 | 1277 | 258 |
| X55572_at | Rn.11339 | 25239 | Apod | apolipoprotein D | 1.25 | 0.00 | 2018 | 598 | 2492 | 665 |
| U07201_at | Rn.11172 | 25612 | Asns | asparagine synthetase | 1.19 | 0.00 | 356 | 41 | 425 | 61 |
| rc_AA859645_at | Rn.53846 | 83526 | Atrn | Attractin | 1.26 | 0.00 | 493 | 50 | 622 | 96 |
| M60921_g_at | Rn.27923 | 29619 | Btg2 | B-cell translocation gene 2, antiproliferative | 1.13 | 0.00 | 247 | 21 | 279 | 30 |
| rc_AA925846_s_at | Rn.89639 | 117271 | Bid3 | BH3 interacting (with BCL2 family) domain, apoptosis agonist | 1.09 | 0.00 | 673 | 79 | 747 | 151 |
| AJ000347_g_at | Rn.8453 | 64473 | Bpnt1 | bisphosphate 30-nucleotidase 1 | 1.13 | 0.00 | 680 | 65 | 772 | 115 |
| U30831_at | Rn.31973 | 192189 | Bk | brain and kidney protein | 1.10 | 0.00 | 2315 | 350 | 2559 | 519 |
| rc_AA800190_at | Rn.1518 | 25739 | Pygb | brain glycogen phosphorylase | 1.10 | 0.01 | 1522 | 220 | 1672 | 209 |
| U16802_at | Rn.88197 | 26989 | Cadps | Ca2+-dependent secretion activator | 1.08 | 0.01 | 1803 | 221 | 1935 | 261 |
| rc_AA875414_at | Rn.2912 | 83465 | Celsr2 | cadherin EGF LAG seven-pass G-type receptor 2 | 1.14 | 0.00 | 963 | 149 | 1095 | 204 |
| M31178_g_at | Rn.3908 | 83839 | Calb1 | calbindin 1 | 1.16 | 0.00 | 1252 | 317 | 1475 | 502 |
| S83194_s_at | Rn.4851 | 60341 | Camkk1 | calcium/calmodulin-dependent protein kinase kinase 1, alpha | 1.12 | 0.00 | 2788 | 905 | 3100 | 1056 |
| X13933_s_at | Rn.4166 | 24242 | Calm1 | calmodulin 1 | 1.07 | 0.01 | 6373 | 726 | 6809 | 851 |
| rc_AA944422_at | Rn.57635 | 54321 | Cnn3 | calponin 3, acidic | 1.16 | 0.00 | 804 | 140 | 932 | 155 |
| S66024_at | Rn.10251 | 25620 | Crem | cAMP responsive element modulator | 1.07 | 0.00 | 146 | 14 | 157 | 17 |
| rc_AI638943_at | Rn.43383 | 298657 | Car6 | carbonic anhydrase 6 | 1.09 | 0.00 | 162 | 13 | 178 | 22 |
| M11670_at | Rn.3001 | 24248 | Cat | Catalase | 1.18 | 0.00 | 111 | 14 | 132 | 20 |
| rc_AA926149_g_at | Rn.3001 | 24248 | Cat | Catalase | 1.32 | 0.00 | 185 | 45 | 242 | 60 |
| U49062_at | Rn.6007 | 25145 | Cd24 | CD24 antigen | 1.18 | 0.00 | 1045 | 236 | 1227 | 274 |
| U49062_g_at | Rn.6007 | 25145 | Cd24 | CD24 antigen | 1.09 | 0.00 | 591 | 145 | 634 | 116 |
| X53517_at | Rn.2357 | 29185 | Cd37 | CD37 antigen | 1.08 | 0.00 | 351 | 20 | 381 | 29 |
| D29646_at | Rn.11414 | 25668 | Cd38 | CD38 antigen | 1.13 | 0.00 | 94 | 11 | 106 | 14 |
| X13016_at | Rn.3705 | 245962 | Cd48 | CD48 antigen | 1.18 | 0.00 | 88 | 16 | 105 | 29 |
| rc_AA818025_g_at | Rn.1231 | 25407 | Cd59 | CD59 antigen | 1.10 | 0.01 | 1730 | 220 | 1909 | 327 |
| X13044_at | Rn.33804 | 25599 | Cd74 | CD74 antigen (invariant polpypeptide of major histocompatibility class II antigen-associated) | 1.21 | 0.00 | 349 | 126 | 432 | 178 |
| X13044_g_at | Rn.33804 | 25599 | Cd74 | CD74 antigen (invariant polpypeptide of major histocompatibility class II antigen-associated) | 1.18 | 0.00 | 453 | 105 | 547 | 177 |
| X14254cds_g_at | Rn.33804 | 25599 | Cd74 | CD74 antigen (invariant polpypeptide of major histocompatibility class II antigen-associated) | 1.12 | 0.00 | 451 | 77 | 509 | 106 |
| AB009999_g_at | Rn.18983 | 81925 | Cds1 | CDP-diacylglycerol synthase 1 | 1.09 | 0.01 | 1216 | 182 | 1333 | 305 |
| rc_AI176308_at | Rn.60067 | 64465 | Cdc42 | cell division cycle 42 homolog (S. cerevisiae) | 1.12 | 0.01 | 1827 | 265 | 2043 | 345 |
| AF000578_at | Rn.54977 | 85434 | Cdc5l | cell division cycle 5-like (S. pombe) | 1.10 | 0.00 | 228 | 18 | 252 | 31 |
| M95768_at | Rn.11199 | 81652 | Ctbs | chitobiase, di-N-acetyl- | 1.08 | 0.00 | 369 | 34 | 401 | 46 |
| rc_AI169005_at | Rn.4089 | 65160 | Clns1a | chloride channel, nucleotide-sensitive, 1A | 1.14 | 0.00 | 830 | 67 | 950 | 138 |
| rc_AA892559_at | Rn.6067 | 25707 | Cntf | ciliary neurotrophic factor | 1.11 | 0.00 | 234 | 25 | 260 | 38 |
| X92097_at | Rn.1022 | 65165 | Rnp24 | coated vesicle membrane protein | 1.10 | 0.00 | 421 | 49 | 462 | 49 |
| L20427_at | Rn.3824 | 29309 | Coq3 | coenzyme Q3 homolog, methyltransferase (yeast) | 1.07 | 0.01 | 664 | 56 | 709 | 82 |
| rc_AI178135_at | Rn.2765 | 29681 | C1qbp | complement component 1, q subcomponent binding protein | 1.09 | 0.00 | 944 | 94 | 1032 | 157 |
| X71127_at | Rn.6702 | 29687 | C1qb | complement component 1, q subcomponent, beta polypeptide | 1.08 | 0.01 | 1066 | 108 | 1155 | 180 |
| D88250_at | Rn.4037 | 192262 | C1s | complement component 1, s subcomponent | 1.07 | 0.00 | 118 | 11 | 128 | 15 |
| M29866_s_at | Rn.11378 | 24232 | C3 | complement component 3 | 1.19 | 0.00 | 147 | 19 | 176 | 27 |
| X52477_at | Rn.11378 | 24232 | C3 | complement component 3 | 1.12 | 0.00 | 280 | 26 | 316 | 40 |
| D87248_at | Rn.10644 | 27256 | Cntn6 | contactin 6 | 1.09 | 0.00 | 178 | 24 | 195 | 32 |
| X59737mRNA_g_at | Rn.155589 | 29593 | Ckmt1 | creatine kinase, mitochondrial 1, ubiquitous | 1.10 | 0.01 | 3684 | 630 | 4035 | 780 |
| M55534mRNA_s_at | Rn.98208 | 25420 | Cryab | crystallin, alpha B | 1.32 | 0.00 | 827 | 170 | 1092 | 245 |
| X60351cds_s_at | Rn.98208 | 25420 | Cryab | crystallin, alpha B | 1.29 | 0.00 | 1307 | 264 | 1677 | 347 |
| rc_AA893699_s_at | Rn.4157 | 362201 | Ccndbp1 | cyclin D-type binding-protein 1 | 1.11 | 0.00 | 976 | 92 | 1089 | 136 |
| rc_AA866477_at | Rn.2026 | 303393 | Cox7b | cytochrome c oxidase subunit VIIb | 1.09 | 0.01 | 2623 | 300 | 2848 | 379 |
| U40004_s_at | Rn.91314 | 313375 | Cyp2j9 | cytochrome P450, family 2, subfamily j, polypeptide 9 | 1.08 | 0.00 | 267 | 29 | 289 | 30 |
| U36992_at | Rn.53969 | 25429 | Cyp7b1 | cytochrome P450, family 7, subfamily b, polypeptide 1 | 1.12 | 0.00 | 493 | 88 | 560 | 125 |
| X59859_i_at | Rn.106103 | 29139 | Dcn | Decorin | 1.12 | 0.00 | 736 | 184 | 817 | 162 |
| Z12298cds_s_at | Rn.106103 | 29139 | Dcn | Decorin | 1.17 | 0.00 | 536 | 112 | 625 | 132 |
| D86041_at | Rn.7398 | 64157 | Ddah1 | Dimethylarginine dimethylaminohydrolase 1 | 1.21 | 0.00 | 997 | 148 | 1221 | 243 |
| rc_AI058941_s_at | Rn.7398 | 64157 | Ddah1 | Dimethylarginine dimethylaminohydrolase 1 | 1.10 | 0.00 | 206 | 22 | 229 | 37 |
| AJ011607_g_at | Rn.11846 | 301323 | Prim2 | DNA primase, p58 subunit | 1.06 | 0.00 | 433 | 36 | 459 | 34 |
| rc_AI170685_at | Rn.3904 | 84026 | Dnaja2 | DnaJ (Hsp40) homolog, subfamily A, member 2 | 1.11 | 0.01 | 1919 | 256 | 2131 | 322 |
| rc_AA799570_at | Rn.27927 | 300721 | Dnaja4 | DnaJ (Hsp40) homolog, subfamily A, member 4 | 1.10 | 0.00 | 1282 | 170 | 1418 | 239 |
| U15138_at | Rn.31981 | 81655 | Dncli2 | dynein, cytoplasmic, light intermediate polypeptide 2 | 1.09 | 0.01 | 4691 | 641 | 5078 | 587 |
| U08976_at | Rn.6148 | 64526 | Ech1 | enoyl coenzyme A hydratase 1, peroxisomal | 1.12 | 0.00 | 455 | 52 | 511 | 71 |
| M26125_at | Rn.3603 | 25315 | Ephx1 | epoxide hydrolase 1, microsomal | 1.09 | 0.01 | 552 | 76 | 606 | 92 |
| rc_AI031019_at | Rn.9181 | 64514 | Eif2b1 | eukaryotic translation initiation factor 2B, subunit 1 alpha | 1.19 | 0.00 | 352 | 25 | 417 | 44 |
| rc_AI031019_g_at | Rn.9181 | 64514 | Eif2b1 | eukaryotic translation initiation factor 2B, subunit 1 alpha | 1.23 | 0.00 | 258 | 15 | 319 | 44 |
| rc_AI230914_at | Rn.8873 | 64511 | Fntb | farnesyltransferase, CAAX box, beta | 1.17 | 0.00 | 779 | 60 | 912 | 86 |
| X73371_at | Rn.33323 | 289211 | Fcgr2b | Fc receptor, IgG, low affinity IIb | 1.11 | 0.00 | 83 | 6 | 93 | 13 |
| rc_AI169802_at | Rn.54447 | 25319 | Fth1 | ferritin, heavy polypeptide 1 | 1.26 | 0.00 | 4521 | 639 | 5668 | 729 |
| AJ007291_g_at | Rn.30105 | 117287 | Park7 | fertility protein SP22 | 1.15 | 0.00 | 2290 | 213 | 2642 | 331 |
| M84719_at | Rn.867 | 25256 | Fmo1 | flavin containing monooxygenase 1 | 1.13 | 0.01 | 278 | 77 | 325 | 123 |
| M59861_at | Rn.2328 | 64392 | Fthfd | Formyltetrahydrofolate dehydrogenase | 1.07 | 0.01 | 746 | 111 | 803 | 152 |
| L14684_at | Rn.10913 | 114017 | Gfm | G elongation factor | 1.18 | 0.00 | 234 | 28 | 275 | 40 |
| AF058795_at | Rn.118960 | 83633 | Gpr51 | G protein-coupled receptor 51 | 1.12 | 0.00 | 2365 | 558 | 2636 | 680 |
| X15466cds_at | Rn.114781 | 25450 | Gabrb1 | gamma-aminobutyric acid receptor, subunit beta 1 | 1.21 | 0.00 | 1653 | 291 | 2002 | 376 |
| AF028784mRNA#1_s_at | Rn.91512 | 24387 | Gfap | glial fibrillary acidic protein | 1.23 | 0.00 | 10243 | 2952 | 12547 | 3161 |
| AB008807_at | Rn.25166 | 114846 | Gsto1 | glutathione S-transferase omega 1 | 1.20 | 0.00 | 1386 | 171 | 1669 | 222 |
| X04229cds_s_at | Rn.93760 | 24423 | Gstm1 | glutathione S-transferase, mu 1 | 1.11 | 0.01 | 3563 | 725 | 3951 | 672 |
| E01415cds_s_at | Rn.6036 | 81869 | Gstm3 | glutathione S-transferase, mu type 3 | 1.15 | 0.00 | 2997 | 371 | 3469 | 448 |
| L10326_at | Rn.31 | 24896 | Gnas | GNAS complex locus | 1.10 | 0.00 | 2300 | 259 | 2538 | 434 |
| AF076619_at | Rn.30028 | 58844 | Grb14 | growth factor receptor bound protein 14 | 1.21 | 0.00 | 299 | 38 | 366 | 71 |
| X15705cds_at | Rn.112579 | 60460 | Hspa2 | heat shock protein 2 | 1.27 | 0.00 | 159 | 47 | 208 | 81 |
| D17711cds_s_at | Rn.11854 | 117282 | Hnrpk | heterogenous nuclear ribonucleoprotein K | 1.07 | 0.00 | 1766 | 194 | 1895 | 200 |
| U09551_at | Rn.89226 | 27080 | Hbp1 | high mobility group box transcription factor 1 | 1.07 | 0.01 | 179 | 15 | 191 | 18 |
| rc_AA892014_r_at | Rn.3516 | 114612 | Bat1a | HLA-B-associated transcript 1A | 1.06 | 0.01 | 212 | 13 | 224 | 18 |
| AF009656mRNA_s_at | Rn.47 | 24465 | Hprt | hypoxanthine guanine phosphoribosyl transferase | 1.15 | 0.00 | 1046 | 181 | 1188 | 158 |
| Y09507_at | Rn.10852 | 29560 | Hif1a | hypoxia inducible factor 1, alpha subunit | 1.10 | 0.00 | 471 | 48 | 517 | 57 |
| X96437mRNA_g_at | Rn.23638 | 294235 | Ler3 | immediate early response 3 | 1.11 | 0.00 | 240 | 28 | 267 | 42 |
| AF008554_at | Rn.43578 | 116967 | Iag2 | Implantation-associated protein | 1.18 | 0.00 | 189 | 21 | 225 | 42 |
| X56917_at | Rn.9877 | 81677 | Itpka | inositol 1,4,5-trisphosphate 3-kinase A | 1.08 | 0.00 | 2083 | 307 | 2247 | 403 |
| X83231_at | Rn.89576 | 50693 | Itih3 | inter-alpha trypsin inhibitor, heavy chain 3 | 1.16 | 0.00 | 1528 | 280 | 1778 | 379 |
| AJ222813_s_at | Rn.11118 | 29197 | Il18 | interleukin 18 | 1.15 | 0.00 | 203 | 32 | 233 | 34 |
| U77777_s_at | Rn.11118 | 29197 | Il18 | interleukin 18 | 1.17 | 0.00 | 91 | 17 | 107 | 27 |
| M58587_at | Rn.1716 | 24499 | Il6r | interleukin 6 receptor | 1.11 | 0.01 | 309 | 55 | 343 | 59 |
| rc_AA799581_at | Rn.17123 | 362418 | Irak2 | interleukin-1 receptor-associated kinase 2 | 1.08 | 0.00 | 632 | 57 | 679 | 67 |
| U20181_at | Rn.10132 | 64831 | Ireb2 | iron responsive element binding protein 2 | 1.06 | 0.01 | 361 | 29 | 383 | 38 |
| rc_AI231213_g_at | Rn.3022 | 83628 | Kai1 | kangai 1 | 1.11 | 0.00 | 909 | 128 | 1007 | 117 |
| rc_AI639470_g_at | Rn.125065 | 450225 | Krt10 | keratin 10 | 1.07 | 0.01 | 105 | 8 | 113 | 12 |
| D25224_at | Rn.999 | 29236 | Lamr1 | laminin receptor 1 (ribosomal protein SA) | 1.08 | 0.00 | 3716 | 323 | 4049 | 487 |
| rc_AI232268_at | Rn.10293 | 116565 | Lrpap1 | low density lipoprotein receptor-related protein associated protein 1 | 1.15 | 0.00 | 779 | 109 | 913 | 197 |
| Z11995cds_at | Rn.10293 | 116565 | Lrpap1 | low density lipoprotein receptor-related protein associated protein 1 | 1.08 | 0.00 | 223 | 23 | 242 | 25 |
| Z11995cds_g_at | Rn.10293 | 116565 | Lrpap1 | low density lipoprotein receptor-related protein associated protein 1 | 1.13 | 0.00 | 499 | 51 | 567 | 69 |
| rc_AA892775_at | Rn.2283 | 25211 | Lyz | Lysozyme | 1.54 | 0.00 | 1177 | 182 | 1910 | 700 |
| rc_AI639359_at | Rn.98599 | 58808 | Mpp4 | membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) | 1.08 | 0.00 | 78 | 8 | 83 | 8 |
| AF102854_at | Rn.42893 | 59322 | Cnksr2 | membrane-associated guanylate kinase-interacting protein | 1.08 | 0.00 | 696 | 87 | 752 | 104 |
| U05784_s_at | Rn.41412 | 64862 | Map1lc3b | Microtubule-associated proteins 1A/1B light chain 3 | 1.07 | 0.00 | 3095 | 215 | 3323 | 353 |
| M64301_g_at | Rn.88457 | 58840 | Mapk6 | mitogen-activated protein kinase 6 | 1.08 | 0.01 | 1053 | 118 | 1130 | 179 |
| rc_AI111401_s_at | Rn.27882 | 29688 | Minpp1 | multiple inositol polyphosphate histidine phosphatase 1 | 1.08 | 0.00 | 357 | 32 | 384 | 41 |
| L21995_s_at | Rn.9687 | 24558 | Mog | Myelin oligodendrocyte glycoprotein | 1.08 | 0.00 | 2345 | 453 | 2532 | 471 |
| M99485_at | Rn.9687 | 24558 | Mog | Myelin oligodendrocyte glycoprotein | 1.11 | 0.01 | 1041 | 317 | 1145 | 300 |
| D14688_s_at | Rn.103179 | 50685 | Mrlcb | myosin light chain, regulatory B | 1.10 | 0.00 | 4342 | 372 | 4767 | 479 |
| U88958_at | Rn.3546 | 83834 | Nrn1 | Neuritin | 1.10 | 0.00 | 4094 | 1045 | 4477 | 1119 |
| rc_AA943331_s_at | Rn.11271 | 24605 | Nras | neuroblastoma ras oncogene | 1.07 | 0.00 | 370 | 30 | 398 | 34 |
| Z12152_at | Rn.10971 | 24588 | Nef3 | neurofilament 3, medium | 1.16 | 0.00 | 1021 | 285 | 1163 | 300 |
| rc_AA818677_at | Rn.108194 | 24587 | Nefh | neurofilament, heavy polypeptide | 1.18 | 0.00 | 809 | 258 | 931 | 271 |
| X13804cds_at | Rn.108194 | 24587 | Nefh | neurofilament, heavy polypeptide | 1.11 | 0.00 | 514 | 98 | 561 | 91 |
| AF031880_at | Rn.18568 | 83613 | Nfl | neurofilament, light polypeptide | 1.08 | 0.01 | 4871 | 681 | 5219 | 747 |
| D82074_at | Rn.44289 | 29458 | Neurod1 | Neurogenic differentiation 1 | 1.14 | 0.00 | 124 | 23 | 143 | 36 |
| rc_AI072943_at | Rn.54707 | 266777 | Nptx1 | neuronal pentraxin 1 | 1.14 | 0.00 | 489 | 147 | 552 | 164 |
| Z11504cds_at | Rn.11642 | 29358 | Npy1r | neuropeptide Y receptor Y1 | 1.08 | 0.00 | 296 | 35 | 319 | 44 |
| U66274_at | Rn.10532 | 25340 | Npy5r | neuropeptide Y receptor Y5 | 1.11 | 0.00 | 72 | 10 | 79 | 8 |
| X97121_at | Rn.127792 | 64636 | Ntsr2 | neurotensin receptor 2 | 1.08 | 0.00 | 3808 | 326 | 4138 | 492 |
| E03082cds_s_at | Rn.9715 | 81737 | Ntf3 | neurotrophin 3 | 1.07 | 0.00 | 121 | 15 | 130 | 23 |
| U31203_at | Rn.10154 | 25495 | Nog | Noggin | 1.09 | 0.00 | 307 | 61 | 333 | 70 |
| U93197_at | Rn.9783 | 171116 | Opa1 | optic atrophy 1 homolog (human) | 1.12 | 0.00 | 296 | 33 | 333 | 51 |
| M93297cds_at | Rn.1430 | 64313 | Oat | Ornithine aminotransferase | 1.11 | 0.00 | 899 | 132 | 994 | 143 |
| X07944exon#1-12_s_at | Rn.874 | 24609 | Odc1 | Ornithine decarboxylase 1 | 1.06 | 0.01 | 817 | 91 | 865 | 72 |
| rc_AI175539_at | Rn.2005 | 25269 | Pvalb | Parvalbumin | 1.16 | 0.00 | 1271 | 527 | 1451 | 606 |
| AF002281_at | Rn.13361 | 114108 | Pdlim3 | PDZ and LIM domain 3 | 1.15 | 0.00 | 129 | 18 | 148 | 17 |
| M57728_g_at | Rn.11175 | 296588 | Pmpca | Peptidase (mitochondrial processing) alpha | 1.09 | 0.00 | 392 | 34 | 427 | 46 |
| D13907_at | Rn.841 | 64198 | Pmpcb | Peptidase (mitochondrial processing) beta | 1.10 | 0.00 | 466 | 48 | 515 | 67 |
| D13907_g_at | Rn.841 | 64198 | Pmpcb | Peptidase (mitochondrial processing) beta | 1.12 | 0.00 | 375 | 51 | 421 | 65 |
| U95920_at | Rn.98622 | 81740 | Pcm1 | pericentriolar material 1 | 1.30 | 0.00 | 534 | 83 | 693 | 105 |
| E03344cds_s_at | Rn.4065 | 29534 | Pxmp3 | Peroxisomal membrane protein 3 | 1.08 | 0.01 | 135 | 13 | 145 | 17 |
| rc_AA892797_at | Rn.108127 | 24644 | Pgk1 | Phosphoglycerate kinase 1 | 2.14 | 0.00 | 2205 | 508 | 4687 | 941 |
| U16655_at | Rn.37434 | 140693 | Plcd4 | phospholipase C, delta 4 | 1.12 | 0.00 | 377 | 37 | 425 | 56 |
| D45920_at | Rn.10684 | 84587 | Plcl1 | phospholipase C-like 1 | 1.14 | 0.00 | 524 | 106 | 595 | 134 |
| rc_AI232379_at | Rn.55127 | 25267 | Pdgfra | platelet derived growth factor receptor, alpha polypeptide | 1.13 | 0.00 | 230 | 43 | 265 | 76 |
| rc_AA899935_at | Rn.24751 | 64189 | Pafah1b2 | platelet-activating factor acetylhydrolase, isoform 1b, alpha2 subunit | 1.07 | 0.01 | 601 | 70 | 644 | 57 |
| AF022819_at | Rn.15693 | 59324 | Kcnk1 | potassium channel, subfamily K, member 1 | 1.08 | 0.00 | 1056 | 184 | 1135 | 258 |
| X62840mRNA_s_at | Rn.33095 | 25327 | Kcnc1 | potassium voltage-gated channel, Shaw-related subfamily, member 1 | 1.17 | 0.00 | 586 | 141 | 679 | 189 |
| AF033027_at | Rn.6525 | 64351 | Ykt6 | prenylated SNARE protein | 1.11 | 0.00 | 493 | 52 | 546 | 64 |
| D50093_s_at | Rn.3936 | 24686 | Prnp | prion protein | 1.10 | 0.01 | 4160 | 504 | 4561 | 570 |
| D90265_s_at | Rn.2668 | 29668 | Psma1 | proteasome (prosome, macropain) subunit, alpha type 1 | 1.09 | 0.00 | 868 | 92 | 951 | 121 |
| rc_AA891383_i_at | Rn.1276 | 29672 | Psma5 | proteasome (prosome, macropain) subunit, alpha type 5 | 1.55 | 0.00 | 127 | 26 | 196 | 33 |
| M86870_at | Rn.39305 | 116598 | Pdia4 | protein disulfide isomerase associated 4 | 1.13 | 0.00 | 377 | 34 | 425 | 39 |
| M18330_at | Rn.98279 | 170538 | Prkcd | protein kinase C, delta | 1.14 | 0.00 | 247 | 34 | 284 | 54 |
| M13707_at | Rn.9747 | 24681 | Prkcc | protein kinase C, gamma | 1.11 | 0.00 | 3374 | 846 | 3768 | 950 |
| AF062740_at | Rn.31799 | 54705 | Ppm2c | protein phosphatase 2C, magnesium dependent, catalytic subunit | 1.13 | 0.00 | 376 | 141 | 429 | 193 |
| AF062740_g_at | Rn.31799 | 54705 | Ppm2c | protein phosphatase 2C, magnesium dependent, catalytic subunit | 1.10 | 0.01 | 869 | 268 | 968 | 361 |
| L27843_s_at | Rn.9459 | 29463 | Ptp4a1 | protein tyrosine phosphatase 4a1 | 1.10 | 0.00 | 2226 | 208 | 2447 | 287 |
| U40790_at | Rn.10278 | 29645 | Ptprj | protein tyrosine phosphatase, receptor type, J | 1.28 | 0.00 | 925 | 106 | 1216 | 294 |
| rc_AI072770_s_at | Rn.4550 | 24943 | Plp | proteolipid protein | 1.14 | 0.00 | 6459 | 1959 | 7244 | 1900 |
| D10729_s_at | Rn.29244 | 24968 | Psmb8 | proteosome (prosome, macropain) subunit, beta type 8 | 1.08 | 0.00 | 339 | 36 | 367 | 45 |
| J03481mRNA_at | Rn.241 | 64192 | Qdpr | quinoid dihydropteridine reductase | 1.18 | 0.00 | 830 | 177 | 1001 | 285 |
| J03481mRNA_g_at | Rn.241 | 64192 | Qdpr | quinoid dihydropteridine reductase | 1.18 | 0.00 | 1944 | 383 | 2288 | 470 |
| M75153_at | Rn.1016 | 81830 | Rab11a | RAB11a, member RAS oncogene family | 1.12 | 0.00 | 821 | 101 | 925 | 185 |
| rc_AI169372_at | Rn.105901 | 304268 | Rasl11a | RAS-like family 11 member A | 1.13 | 0.00 | 309 | 49 | 349 | 58 |
| AF036548_at | Rn.3504 | 117183 | Rgc32 | response gene to complement 32 | 1.09 | 0.00 | 408 | 47 | 450 | 65 |
| AF036548_g_at | Rn.3504 | 117183 | Rgc32 | response gene to complement 32 | 1.14 | 0.00 | 1515 | 210 | 1730 | 273 |
| X52817cds_s_at | Rn.55126 | 116644 | Rtn1 | reticulon 1 | 1.14 | 0.00 | 6407 | 742 | 7327 | 749 |
| rc_AA900505_at | Rn.2042 | 64373 | Rhob | rhoB gene | 1.10 | 0.01 | 2569 | 427 | 2832 | 449 |
| X93352_at | Rn.2262 | 81729 | Rpl10a | ribosomal protein L10A | 1.12 | 0.00 | 3857 | 408 | 4323 | 651 |
| X13549_s_at | Rn.3359 | 81773 | Rps10 | ribosomal protein S10 | 1.10 | 0.00 | 4665 | 461 | 5131 | 681 |
| L23128_g_at | Rn.107070 | 24750 | RT1-N3 | RT1 class Ib gene, H2-TL-like, grc region (N3) | 1.22 | 0.00 | 564 | 52 | 693 | 83 |
| M15562_at | Rn.103146 | 294269 | RT1-Da | RT1 class II, locus Da | 1.17 | 0.00 | 308 | 47 | 368 | 96 |
| M15562_g_at | Rn.103146 | 294269 | RT1-Da | RT1 class II, locus Da | 1.20 | 0.00 | 324 | 106 | 405 | 175 |
| U75928UTR#1_s_at | Rn.98989 | 24791 | Sparc | secreted acidic cysteine rich glycoprotein | 1.19 | 0.00 | 2761 | 915 | 3269 | 935 |
| M63901_at | Rn.6173 | 25719 | Sgne1 | secretory granule neuroendocrine protein 1 | 1.10 | 0.00 | 1081 | 110 | 1185 | 121 |
| M63901_g_at | Rn.6173 | 25719 | Sgne1 | secretory granule neuroendocrine protein 1 | 1.09 | 0.00 | 4179 | 457 | 4561 | 570 |
| AB011530_at | Rn.30002 | 65047 | Slit1 | slit homolog 1 (Drosophila) | 1.17 | 0.00 | 616 | 65 | 728 | 106 |
| rc_AI145680_s_at | Rn.6085 | 25027 | Slc16a1 | solute carrier family 16 (monocarboxylic acid transporters), member 1 | 1.09 | 0.00 | 884 | 72 | 963 | 115 |
| Y16774_at | Rn.90036 | 64469 | Slc30a4 | solute carrier family 30 (zinc transporter), member 4 | 1.09 | 0.00 | 296 | 43 | 322 | 56 |
| rc_AI639256_at | Rn.40672 | 301255 | Spats1 | spermatogenesis associated, serine-rich 1 | 1.28 | 0.00 | 160 | 19 | 204 | 29 |
| U83883_at | Rn.36148 | 64635 | Snd1 | staphylococcal nuclease domain containing 1 | 1.13 | 0.00 | 973 | 123 | 1105 | 129 |
| M15114_g_at | Rn.83595 | 83792 | Scd2 | stearoyl-Coenzyme A desaturase 2 | 1.08 | 0.00 | 6769 | 655 | 7301 | 851 |
| X99337cds_s_at | Rn.37476 | 56064 | Sdfr1 | stromal cell derived factor receptor 1 | 1.10 | 0.00 | 1369 | 160 | 1507 | 212 |
| X99338cds_i_at | Rn.37476 | 56064 | Sdfr1 | stromal cell derived factor receptor 1 | 1.09 | 0.00 | 2601 | 296 | 2832 | 346 |
| M27925_at | Rn.506 | 29179 | Syn2 | synapsin II | 1.17 | 0.00 | 860 | 150 | 1014 | 211 |
| L10362_at | Rn.58137 | 117556 | Sv2b | synaptic vesicle glycoprotein 2b | 1.11 | 0.01 | 1702 | 358 | 1888 | 396 |
| rc_AI639484_at | Rn.58137 | 117556 | Sv2b | synaptic vesicle glycoprotein 2b | 1.09 | 0.00 | 3424 | 586 | 3738 | 811 |
| U40188_at | Rn.11270 | 171152 | Taf9l | TAF9-like RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa | 1.14 | 0.00 | 110 | 11 | 126 | 17 |
| rc_AA893860_at | Rn.22757 | 294810 | Tars | threonyl-tRNA synthetase | 1.08 | 0.00 | 655 | 73 | 710 | 87 |
| K01934mRNA#2_at | Rn.81140 | 25357 | Thrsp | thyroid hormone responsive protein | 1.09 | 0.00 | 362 | 66 | 394 | 75 |
| U09256_at | Rn.5950 | 64524 | Tkt | transketolase | 1.11 | 0.00 | 1117 | 94 | 1240 | 136 |
| L04760_at | Rn.6763 | 81002 | Trim23 | tripartite motif protein 23 | 1.07 | 0.01 | 415 | 40 | 446 | 57 |
| AB011679_at | Rn.2458 | 29214 | Tubb5 | tubulin, beta 5 | 1.16 | 0.00 | 336 | 57 | 387 | 43 |
| rc_AI103911_at | Rn.2603 | 291103 | Uqcrfs1 | ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 | 1.08 | 0.00 | 1213 | 97 | 1313 | 165 |
| U14746_at | Rn.11059 | 24874 | Vhl | von Hippel-Lindau syndrome homolog | 1.10 | 0.00 | 585 | 62 | 646 | 109 |
| rc_AA946044_s_at | Rn.4338 | 81515 | Lyn | Yamaguchi sarcoma viral (v-yes-1) oncogene homolog | 1.10 | 0.00 | 85 | 7 | 93 | 7 |
| U67082_at | Rn.10663 | 25165 | Znf386 | zinc finger protein 386 (Kruppel-like) | 1.15 | 0.00 | 201 | 22 | 232 | 31 |
S.D. = standard deviation; FDR = false discovery rate; iP = inbred alcohol-preferring rat; iNP = inbred alcohol-non-preferring rat; RMA = Robust multi-chip average.
Positive fold difference is iP/iNP; negative is iNP/iP.
Expression levels are those obtained after RMA background correction and scaling as indicated in Methods.
Table 5.
Significant fold differences in combined region analysis and in individual region analyses
| Probe set | Unigene | Entrez gene |
Chr. | Base pair location |
QTL | Gene symbol | Gene name | Comb. regions |
ACB | AMYG | FC | HIPP |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D00512_at | Rn.4054 | 25014 | 8 | -57,063,521 | Acat1 | acetyl-coenzyme A acetyltransferase 1 | -1.11 | |||||
| D90109_at | Rn.6215 | 25288 | 16 | -49,036,966 | Alc7 & 11 | Acsl1 | acyl-CoA synthetase long-chain family member 1 | 1.27 | ||||
| S56508_s_at | Rn.33697 | 117243 | 10 | 39,743,427 | Acsl6 | acyl-CoA synthetase long-chain family member 6 | 1.15 | |||||
| J00692_at | Rn.82732 | 29437 | 19 | -54,086,370 | Acta1 | actin, alpha 1, skeletal muscle | 1.10 | |||||
| Rc_AA891812_at | Rn.5788 | 24170 | 14 | -81,752,822 | Add1 | adducin 1 (alpha) | -1.09 | |||||
| X83715_at | Rn.5788 | 24170 | 14 | -81,752,822 | Add1 | adducin 1 (alpha) | -1.14 | |||||
| S47609_s_at | Rn.11180 | 25369 | 20 | -13,815,945 | Adora2a | adenosine A2a receptor | -1.08 | |||||
| M12112mRNA#3_s_at | Rn.6319 | 24179 | 19 | -54,743,436 | Agt | angiotensinogen | 1.20 | |||||
| Rc_AA943892_at | Rn.6319 | 24179 | 19 | -54,743,436 | Agt | angiotensinogen | 1.16 | |||||
| Rc_AI639111_at | Rn.93081 | 308923 | 1 | 16,332,862 | Ahi1 | Abelson helper integration site 1 | -1.46 | -1.54 | -1.35 | -1.48 | ||
| U48288_at | Rn.10557 | 25228 | 2 | 0 | Akap11 | A kinase (PRKA) anchor protein 11 | 1.24 | 1.50 | 1.32 | |||
| U67136_at | Rn.122003 | 171026 | 6 | 98,926,570 | Alc17 | Akap5 | A kinase (PRKA) anchor protein 5 | 1.23 | ||||
| M60322_g_at | Rn.107801 | 24192 | 4 | -61,921,567 | Alc14 & 16 | Akr1b4 | aldo-keto reductase family 1, member B4 (aldose reductase) | 1.19 | ||||
| D17309_at | Rn.25716 | 192242 | 4 | 65,249,037 | Alc14 & 16 | Akr1d1 | aldo-keto reductase family 1, member D1 | -1.09 | ||||
| D30041_at | Rn.87066 | 25233 | 1 | 82,764,343 | Akt2 | thymoma viral proto-oncogene 2 | -1.11 | |||||
| D86297_at | Rn.32517 | 25748 | X | 39,853,992 | Alas2 | aminolevulinic acid synthase 2 | -1.14 | |||||
| Rc_AA945321_at | Rn.34353 | 24186 | 14 | -19,126,964 | Alb | albumin | 1.06 | |||||
| AF001898_at | Rn.6132 | 24188 | 1 | 223,895,697 | Aldh1a1 | aldehyde dehydrogenase family 1, member A1 | 1.20 | |||||
| Rc_AA891631_at | Rn.116338 | 295365 | 2 | 203,715,385 | Alc15 | Amigo | amphoretin-induced gene and ORF | 1.11 | 1.22 | |||
| U90888_at | Rn.11106 | 25095 | 1 | 168,644,154 | Ampd3 | adenosine monophosphate deaminase 3 | 1.27 | 1.37 | ||||
| D42137exon_s_at | Rn.3318 | 25673 | 2 | -122,894,171 | Anxa5 | annexin A5 | 1.10 | |||||
| Rc_AI170212_s_at | Rn.11007 | 140667 | 16 | 73,688,647 | Ap3m2 | adaptor-related protein complex 3, mu 2 subunit | -1.12 | |||||
| AF029107_at | Rn.62687 | 83610 | 1 | 119,049,224 | Apba2 | amyloid beta (A4) precursor protein-binding, family A, member 2 | -1.13 | |||||
| L07114_at | Rn.10002 | 25383 | 4 | -159,278,107 | Alc18 | Apobec1 | apolipoprotein B editing complex 1 | -1.15 | ||||
| X55572_at | Rn.11339 | 25239 | 11 | 882,464 | Apod | apolipoprotein D | 1.25 | |||||
| AF026505_at | Rn.24612 | 114901 | 16 | -49,722,613 | Alc7 & 11 | Argbp2 | Arg/Abl-interacting protein ArgBP2 | 1.21 | ||||
| Rc_AA891194_s_at | Rn.24612 | 114901 | 16 | -49,722,613 | Alc7 & 11 | Argbp2 | Arg/Abl-interacting protein ArgBP2 | 1.22 | ||||
| Rc_AI639196_at | Rn.33757 | 66013 | X | -82,667,647 | Arhgef9 | Cdc42 guanine nucleotide exchange factor (GEF) 9 | -1.22 | |||||
| U07201_at | Rn.11172 | 25612 | 4 | -32,877,146 | Asns | asparagine synthetase | 1.19 | 1.25 | ||||
| D89514_at | Rn.15114 | 81643 | 9 | 70,823,725 | Atic | 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase | 1.25 | 1.32 | ||||
| J04024_at | Rn.2305 | 29693 | 12 | 35,129,987 | Alc6 & 10 | Atp2a2 | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 | -1.13 | ||||
| Rc_AA859645_at | Rn.53846 | 83526 | 3 | 118,444,840 | Atrn | attractin | 1.26 | 1.33 | ||||
| Rc_AA893602_at | Rn.14812 | 316051 | 8 | 122,981,038 | Azi2 | 5-azacytidine induced gene 2 | -1.23 | -1.28 | ||||
| Rc_AI170268_at | Rn.1868 | 24223 | 3 | 108,833,491 | B2m | beta-2 microglobulin | -1.18 | -1.31 | ||||
| Rc_AA892014_r_at | Rn.3516 | 114612 | 20 | -3,611,355 | Bat1a | HLA-B-associated transcript 1A | 1.06 | |||||
| U72350_at | Rn.10323 | 24888 | 3 | -143,034,710 | Bcl2l1 | Bcl2-like 1 | -1.07 | |||||
| U34963_s_at | Rn.10323 | 24888 | 3 | -143,034,710 | Bcl2l1 | Bcl2-like 1 | -1.11 | |||||
| AF065431_s_at | Rn.82709 | 64547 | 3 | 115,597,895 | Bcl2l11 | BCL2-like 11 (apoptosis facilitator) | -1.08 | |||||
| Rc_AA925846_s_at | Rn.89639 | 117271 | 12 | -39,383,652 | Alc10 | Bid3 | BH3 interacting (with BCL2 family) domain, apoptosis agonist | 1.09 | ||||
| U30831_at | Rn.31973 | 192189 | 1 | -176,738,853 | Bk | brain and kidney protein | 1.10 | |||||
| D63860_s_at | Rn.53974 | 25667 | 14 | -12,065,634 | Bmp3 | bone morphogenetic protein 3 | 1.06 | |||||
| AJ000347_g_at | Rn.8453 | 64473 | 13 | 101,540,167 | Bpnt1 | bisphosphate 3 -nucleotidase 1 | 1.13 | |||||
| M60921_g_at | Rn.27923 | 29619 | 13 | -47,041,028 | Btg2 | B-cell translocation gene 2, antiproliferative | 1.13 | 1.19 | ||||
| X71127_at | Rn.6702 | 29687 | 5 | -155,657,561 | Alc4 | C1qb | complement component 1, q subcomponent, beta polypeptide | 1.08 | ||||
| Rc_AI178135_at | Rn.2765 | 29681 | 10 | -57,895,408 | C1qbp | complement component 1, q subcomponent binding protein | 1.09 | |||||
| D88250_at | Rn.4037 | 192262 | 4 | -160,981,068 | Alc18 | C1s | complement component 1, s subcomponent | 1.07 | ||||
| M29866_s_at | Rn.11378 | 24232 | 9 | -64,513,131 | C3 | complement component 3 | 1.19 | |||||
| X52477_at | Rn.11378 | 24232 | 9 | -64,513,131 | C3 | complement component 3 | 1.12 | |||||
| U16802_at | Rn.88197 | 26989 | 15 | 13,859,285 | Cadps | Ca2+-dependent secretion activator | 1.08 | |||||
| M31178_g_at | Rn.3908 | 83839 | 5 | -198,836,347 | Alc4 | Calb1 | calbindin 1 | 1.16 | ||||
| X13933_s_at | Rn.4166 | 24242 | 6 | 124,481,414 | Alc17 | Calm1 | calmodulin 1 | 1.07 | ||||
| S83194_s_at | Rn.4851 | 60341 | 10 | 59,922,371 | Camkk1 | calcium/calmodulin-dependent protein kinase kinase 1, alpha | 1.12 | |||||
| rc_AI638943_at | Rn.43383 | 298657 | 5 | 0 | Alc4 | Car6 | carbonic anhydrase 6 | 1.09 | ||||
| U84410_s_at | Rn.10562 | 25402 | 16 | -48,944,301 | Alc7 & 11 | Casp3 | caspase 3, apoptosis related cysteine protease | 1.21 | ||||
| rc_AA926149_g_at | Rn.3001 | 24248 | 3 | 88,550,504 | Cat | catalase | 1.32 | 1.42 | ||||
| M11670_at | Rn.3001 | 24248 | 3 | 88,550,504 | Cat | catalase | 1.18 | |||||
| rc_AA893699_s_at | Rn.4157 | 362201 | 3 | 107,683,801 | Ccndbp1 | cyclin D-type binding-protein 1 | 1.11 | |||||
| U49062_at | Rn.6007 | 25145 | 20 | 47,531,055 | Cd24 | CD24 antigen | 1.18 | 1.32 | ||||
| U49062_g_at | Rn.6007 | 25145 | 20 | 47,531,055 | Cd24 | CD24 antigen | 1.09 | 1.24 | ||||
| X53517_at | Rn.2357 | 29185 | 1 | -95,788,071 | Cd37 | CD37 antigen | 1.08 | |||||
| D29646_at | Rn.11414 | 25668 | 14 | -72,322,869 | Cd38 | CD38 antigen | 1.13 | |||||
| X13016_at | Rn.3705 | 245962 | 13 | 87,873,428 | Cd48 | CD48 antigen | 1.18 | |||||
| rc_AA818025_g_at | Rn.1231 | 25407 | 3 | 89,140,026 | Cd59 | CD59 antigen | 1.10 | |||||
| X13044_at | Rn.33804 | 25599 | 18 | 56,828,742 | Cd74 | CD74 antigen (invariant polpypeptide of major histocompatibility class II antigen-associated) | 1.21 | |||||
| X13044_g_at | Rn.33804 | 25599 | 18 | 56,828,742 | Cd74 | CD74 antigen (invariant polpypeptide of major histocompatibility class II antigen-associated) | 1.18 | |||||
| X14254cds_g_at | Rn.33804 | 25599 | 18 | 56,828,742 | Cd74 | CD74 antigen (invariant polpypeptide of major histocompatibility class II antigen-associated) | 1.12 | |||||
| rc_AI176308_at | Rn.60067 | 64465 | 5 | 156,116,164 | Alc4 | Cdc42 | cell division cycle 42 homolog (S. cerevisiae) | 1.12 | ||||
| AF000578_at | Rn.54977 | 85434 | 9 | 11,162,952 | Cdc5l | cell division cycle 5-like (S. pombe) | 1.10 | |||||
| D83349_at | Rn.108785 | 29182 | 6 | -156,062,114 | Alc17 | Cdh22 | cadherin 22 | 1.43 | ||||
| L41275cds_s_at | Rn.10089 | 114851 | 20 | 7,376,551 | Cdkn1a | cyclin-dependent kinase inhibitor 1A | -1.11 | |||||
| AB009999_g_at | Rn.18983 | 81925 | 14 | -9,072,988 | Cds1 | CDP-diacylglycerol synthase 1 | 1.09 | |||||
| rc_AA875414_at | Rn.2912 | 83465 | 2 | 0 | Celsr2 | cadherin EGF LAG seven-pass G-type receptor 2 | 1.14 | 1.25 | ||||
| AF019974_at | Rn.11090 | 24259 | 3 | 120,477,308 | Chgb | chromogranin B | 1.25 | |||||
| X59737mRNA_g_at | Rn.155589 | 29593 | 3 | 108,058,997 | Ckmt1 | creatine kinase, mitochondrial 1, ubiquitous | 1.10 | |||||
| rc_AI169005_at | Rn.4089 | 65160 | 1 | 155,025,118 | Clns1a | chloride channel, nucleotide-sensitive, 1A | 1.14 | |||||
| AF102854_at | Rn.42893 | 59322 | X | 58,251,535 | Cnksr2 | membrane-associated guanylate kinase-interacting protein | 1.08 | |||||
| rc_AA944422_at | Rn.57635 | 54321 | 2 | 218,007,864 | Alc15 | Cnn3 | calponin 3, acidic | 1.16 | ||||
| rc_AA892559_at | Rn.6067 | 25707 | 1 | -216,001,097 | Cntf | ciliary neurotrophic factor | 1.11 | |||||
| D87248_at | Rn.10644 | 27256 | 4 | 140,322,847 | Alc14 & 16 | Cntn6 | contactin 6 | 1.09 | ||||
| L20427_at | Rn.3824 | 29309 | 5 | 36,640,896 | Alc4 | Coq3 | coenzyme Q3 homolog, methyltransferase (yeast) | 1.07 | ||||
| rc_AA866477_at | Rn.2026 | 303393 | X | 94,211,488 | Cox7b | cytochrome c oxidase subunit VIIb | 1.09 | |||||
| S66024_at | Rn.10251 | 25620 | 17 | 62,793,252 | Crem | cAMP responsive element modulator | 1.07 | |||||
| J02844_s_at | Rn.4896 | 83842 | 4 | -22,023,912 | Crot | carnitine O-octanoyltransferase | -1.12 | |||||
| U26033_at | Rn.4896 | 83842 | 4 | -22,023,912 | Crot | carnitine O-octanoyltransferase | -1.15 | |||||
| M55534mRNA_s_at | Rn.98208 | 25420 | 8 | 54,126,302 | Cryab | crystallin, alpha B | 1.32 | |||||
| X60351cds_s_at | Rn.98208 | 25420 | 8 | 54,126,302 | Cryab | crystallin, alpha B | 1.29 | |||||
| rc_AI013513_at | Rn.8046 | 64462 | 10 | -110,177,115 | Csnk1d | casein kinase 1, delta | -1.10 | -1.18 | ||||
| M95768_at | Rn.11199 | 81652 | 2 | 244,635,104 | Ctbs | chitobiase, di-N-acetyl- | 1.08 | |||||
| U40004_s_at | Rn.91314 | 313375 | 5 | -116,749,562 | Alc4 | Cyp2j9 | cytochrome P450, family 2, subfamily j, polypeptide 9 | 1.08 | ||||
| U36992_at | Rn.53969 | 25429 | 2 | 0 | Cyp7b1 | cytochrome P450, family 7, subfamily b, polypeptide 1 | 1.12 | |||||
| L33894_at | Rn.10058 | 79127 | 14 | -20,886,695 | Dck | deoxycytidine kinase | -1.12 | |||||
| Z12298cds_s_at | Rn.106103 | 29139 | 7 | 35,024,504 | Dcn | decorin | 1.17 | 1.29 | ||||
| X59859_i_at | Rn.106103 | 29139 | 7 | 35,024,504 | Dcn | decorin | 1.12 | |||||
| D86041_at | Rn.7398 | 64157 | 2 | 243,918,960 | Ddah1 | dimethylarginine dimethylaminohydrolase 1 | 1.21 | |||||
| rc_AI058941_s_at | Rn.7398 | 64157 | 2 | 243,918,960 | Ddah1 | dimethylarginine dimethylaminohydrolase 1 | 1.10 | |||||
| rc_AA894273_at | Rn.7398 | 64157 | 2 | 243,918,960 | Ddah1 | dimethylarginine dimethylaminohydrolase 1 | -1.19 | |||||
| AF016247_at | Rn.92730 | 83573 | 13 | 0 | Ddr2 | discoidin domain receptor family, member 2 | -1.18 | |||||
| rc_AI170685_at | Rn.3904 | 84026 | 19 | 22,858,527 | Dnaja2 | DnaJ (Hsp40) homolog, subfamily A, member 2 | 1.11 | |||||
| rc_AA799570_at | Rn.27927 | 300721 | 8 | 58,269,846 | Dnaja4 | DnaJ (Hsp40) homolog, subfamily A, member 4 | 1.10 | |||||
| U15138_at | Rn.31981 | 81655 | 19 | 469,162 | Dncli2 | dynein, cytoplasmic, light intermediate polypeptide 2 | 1.09 | |||||
| D85035_g_at | Rn.133148 | 81656 | 2 | 214,894,962 | Alc15 | Dpyd | dihydropyrimidine dehydrogenase | -1.11 | ||||
| Z46882cds_at | Rn.2889 | 25416 | 15 | 0 | Dpysl2 | dihydropyrimidinase-like 2 | -1.10 | |||||
| AF013144_at | Rn.10877 | 171109 | 1 | 259,965,278 | Dusp5 | dual specificity phosphatase 5 | -1.07 | |||||
| X94185cds_s_at | Rn.4313 | 116663 | 7 | 36,913,534 | Dusp6 | dual specificity phosphatase 6 | -1.28 | |||||
| U08976_at | Rn.6148 | 64526 | 1 | 84,010,830 | Ech1 | enoyl coenzyme A hydratase 1, peroxisomal | 1.12 | |||||
| D89730_g_at | Rn.8037 | 305604 | 14 | 109,759,971 | Efemp1 | epidermal growth factor-containing fibulin-like extracellular matrix protein 1 | -1.13 | |||||
| D89730_at | Rn.8037 | 305604 | 14 | 109,759,971 | Efemp1 | epidermal growth factor-containing fibulin-like extracellular matrix protein 1 | -1.15 | |||||
| rc_AA893770_g_at | Rn.144669 | 245963 | 3 | 4,760,573 | Egfl7 | EGF-like domain 7 | -1.07 | |||||
| rc_AI031019_g_at | Rn.9181 | 64514 | 12 | 33,001,710 | Alc6 & 10 | Eif2b1 | eukaryotic translation initiation factor 2B, subunit 1 alpha | 1.23 | 1.29 | |||
| rc_AI031019_at | Rn.9181 | 64514 | 12 | 33,001,710 | Alc6 & 10 | Eif2b1 | eukaryotic translation initiation factor 2B, subunit 1 alpha | 1.19 | 1.17 | |||
| X02610_g_at | Rn.4236 | 24333 | 5 | 167,404,823 | Alc4 | Eno1 | enolase 1, alpha | -1.11 | ||||
| X02610_at | Rn.4236 | 24333 | 5 | 167,404,823 | Alc4 | Eno1 | enolase 1, alpha | -1.15 | ||||
| X78689cds_at | Rn.24569 | 79208 | 14 | 0 | Epha5 | EphA5 | -1.11 | |||||
| X59290mRNA_at | Rn.62934 | 60589 | 5 | 0 | Alc4 | Epha8 | Eph receptor A8 | -1.08 | ||||
| M26125_at | Rn.3603 | 25315 | 13 | -96,911,908 | Ephx1 | epoxide hydrolase 1, microsomal | 1.09 | |||||
| U07619_at | Rn.9980 | 25584 | 2 | 218,334,111 | Alc15 | F3 | coagulation factor III | -1.19 | ||||
| X73371_at | Rn.33323 | 289211 | 13 | -86,823,198 | Fcgr2b | Fc receptor, IgG, low affinity IIb | 1.11 | |||||
| D12498_s_at | Rn.9797 | 79114 | 16 | -70,870,238 | Fgfr1 | Fibroblast growth factor receptor 1 | -1.11 | -1.21 | ||||
| M91599mRNA_at | Rn.24104 | 25114 | 17 | 0 | Fgfr4 | fibroblast growth factor receptor 4 | -1.09 | |||||
| M84719_at | Rn.867 | 25256 | 13 | -78,517,957 | Fmo1 | flavin containing monooxygenase 1 | 1.13 | |||||
| X05834_at | Rn.1604 | 25661 | 9 | -70,849,162 | Fn1 | fibronectin 1 | -1.34 | -1.32 | -1.55 | |||
| U82612cds_g_at | Rn.1604 | 25661 | 9 | -70,849,162 | Fn1 | fibronectin 1 | 1.15 | 1.25 | ||||
| rc_AA955600_at | Rn.1604 | 25661 | 9 | -70,849,162 | Fn1 | fibronectin 1 | -1.07 | |||||
| M28259cds_at | Rn.1604 | 25661 | 9 | -70,849,162 | Fn1 | fibronectin 1 | 1.13 | |||||
| L00191cds#1_s_at | Rn.1604 | 25661 | 9 | -70,849,162 | Fn1 | fibronectin 1 | -1.27 | |||||
| rc_AI230914_at | Rn.8873 | 64511 | 6 | 99,431,422 | Alc17 | Fntb | farnesyltransferase, CAAX box, beta | 1.17 | ||||
| L27421_at | Rn.62653 | 65153 | 3 | 10,200,569 | Freq | frequenin homolog (Drosophila) | -1.22 | |||||
| rc_AI169802_at | Rn.54447 | 25319 | 1 | 212,588,882 | Fth1 | ferritin, heavy polypeptide 1 | 1.26 | 1.29 | ||||
| M59861_at | Rn.2328 | 64392 | 0 | 0 | Fthfd | formyltetrahydrofolate dehydrogenase | 1.07 | |||||
| X15466cds_at | Rn.114781 | 25450 | 14 | -38,532,649 | Gabrb1 | gamma-aminobutyric acid receptor, subunit beta 1 | 1.21 | |||||
| rc_AI639378_at | Rn.40168 | 362760 | 6 | 0 | Alc17 | Galntl1 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 1 | -1.11 | ||||
| rc_AA859837_g_at | Rn.24783 | 83585 | 1 | -224,743,773 | Gda | guanine deaminase | -1.21 | |||||
| rc_AA859837_at | Rn.24783 | 83585 | 1 | -224,743,773 | Gda | guanine deaminase | -1.28 | |||||
| AF028784mRNA#1_s_at | Rn.91512 | 24387 | 10 | -92,074,249 | Gfap | glial fibrillary acidic protein | 1.23 | 1.44 | ||||
| L06986_at | Rn.10935 | 24388 | 14 | 2,597,302 | Gfi1 | growth factor independent 1 | -1.09 | |||||
| L14684_at | Rn.10913 | 114017 | 2 | 157,233,008 | Gfm | G elongation factor | 1.18 | 1.30 | ||||
| X55246_at | Rn.10109 | 25674 | 10 | -40,936,489 | Glra1 | glycine receptor, alpha 1 subunit | -1.11 | |||||
| M17526_g_at | Rn.90161 | 50664 | 19 | -11,476,908 | Gnao | guanine nucleotide binding protein, alpha o | -1.26 | |||||
| L10326_at | Rn.31 | 24896 | 3 | 0 | Gnas | GNAS complex locus | 1.10 | |||||
| L02896_at | Rn.7044 | 58920 | 0 | 0 | Gpc1 | glypican 1 | -1.12 | |||||
| AF058795_at | Rn.118960 | 83633 | 5 | -63,241,575 | Alc4 | Gpr51 | G protein-coupled receptor 51 | 1.12 | ||||
| AF076619_at | Rn.30028 | 58844 | 3 | -46,728,757 | Grb14 | growth factor receptor bound protein 14 | 1.21 | 1.32 | ||||
| M92076_at | Rn.41715 | 24416 | 4 | 0 | Grm3 | glutamate receptor, metabotropic 3 | -1.31 | |||||
| X96790_at | Rn.10409 | 81672 | 4 | 146,577,418 | Alc14 & 16 | Grm7 | glutamate receptor, metabotropic 7 | -1.08 | ||||
| X62660mRNA_g_at | Rn.57528 | 300850 | 8 | 0 | Gsta4 | glutathione S-transferase, alpha 4 | -1.81 | -1.52 | -1.78 | -1.75 | -2.07 | |
| X62660mRNA_at | Rn.57528 | 300850 | 8 | 0 | Gsta4 | glutathione S-transferase, alpha 4 | -1.29 | -1.32 | -1.36 | |||
| X04229cds_s_at | Rn.93760 | 24423 | 2 | -203,538,197 | Alc15 | Gstm1 | glutathione S-transferase, mu 1 | 1.11 | ||||
| E01415cds_s_at | Rn.6036 | 81869 | 2 | -203,454,119 | Alc15 | Gstm3 | glutathione S-transferase, mu type 3 | 1.15 | ||||
| AB008807_at | Rn.25166 | 114846 | 1 | 253,493,290 | Gsto1 | glutathione S-transferase omega 1 | 1.20 | |||||
| X56325mRNA_s_at | Rn.107334 | 25632 | 10 | -15,585,407 | Alc5 & 9 | Hba-a1 | hemoglobin alpha, adult chain 1 | -1.19 | ||||
| M94918mRNA_f_at | Rn.36966 | 24440 | 1 | -161,666,147 | Hbb | hemoglobin beta chain complex | -1.21 | |||||
| U09551_at | Rn.89226 | 27080 | 6 | -50,204,414 | Alc17 | Hbp1 | high mobility group box transcription factor 1 | 1.07 | ||||
| D13417_g_at | Rn.19727 | 29577 | 11 | -72,661,439 | Hes1 | hairy and enhancer of split 1 (Drosophila) | -1.08 | |||||
| Y09507_at | Rn.10852 | 29560 | 6 | 96,422,479 | Alc17 | Hif1a | hypoxia inducible factor 1, alpha subunit | 1.10 | ||||
| U89695_at | Rn.29981 | 84017 | 7 | -59,659,284 | Hmga2 | high mobility group AT-hook 2 | -1.07 | |||||
| X55286_g_at | Rn.9437 | 25675 | 2 | -27,047,872 | Hmgcr | 3-hydroxy-3-methylglutaryl-Coenzyme A reductase | -1.28 | |||||
| M33648_at | Rn.29594 | 24450 | 2 | 193,091,489 | Hmgcs2 | 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 | -1.07 | |||||
| M33648_g_at | Rn.29594 | 24450 | 2 | 193,091,489 | Hmgcs2 | 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 | -1.15 | |||||
| D17711cds_s_at | Rn.11854 | 117282 | 17 | 12,196,629 | Hnrpk | heterogenous nuclear ribonucleoprotein K | 1.07 | |||||
| U16845_at | Rn.10075 | 50864 | 8 | -28,587,115 | Hnt | neurotrimin | -1.43 | |||||
| AB007689_g_at | Rn.30014 | 29547 | 1 | -137,905,533 | Homer2 | homer homolog 2 (Drosophila) | -1.09 | |||||
| AF009656mRNA_s_at | Rn.47 | 24465 | X | 140,003,079 | Hprt | hypoxanthine guanine phosphoribosyl transferase | 1.15 | |||||
| X15705cds_at | Rn.112579 | 60460 | 6 | 99,003,760 | Alc17 | Hspa2 | heat shock protein 2 | 1.27 | ||||
| AF008554_at | Rn.43578 | 116967 | X | -94,168,934 | Iag2 | implantation-associated protein | 1.18 | 1.31 | ||||
| U77777_s_at | Rn.11118 | 29197 | 8 | 53,955,637 | Il18 | interleukin 18 | 1.17 | |||||
| AJ222813_s_at | Rn.11118 | 29197 | 8 | 53,955,637 | Il18 | interleukin 18 | 1.15 | |||||
| M58587_at | Rn.1716 | 24499 | 2 | -182,028,156 | Il6r | interleukin 6 receptor | 1.11 | |||||
| rc_AA799581_at | Rn.17123 | 362418 | 4 | 149,787,869 | Alc14 & 16 | Irak2 | interleukin-1 receptor-associated kinase 2 | 1.08 | 1.22 | |||
| U20181_at | Rn.10132 | 64831 | 8 | 58,452,371 | Ireb2 | iron responsive element binding protein 2 | 1.06 | |||||
| rc_AA893384_g_at | Rn.1499 | 292892 | 1 | 95,548,394 | Irf3 | interferon regulatory factor 3 | 1.06 | |||||
| X83231_at | Rn.89576 | 50693 | 16 | 6,341,044 | Alc7, 11, & 13 | Itih3 | inter-alpha trypsin inhibitor, heavy chain 3 | 1.16 | ||||
| X56917_at | Rn.9877 | 81677 | 3 | 106,153,909 | Itpka | inositol 1,4,5-trisphosphate 3-kinase A | 1.08 | |||||
| rc_AI231213_g_at | Rn.3022 | 83628 | 3 | -77,728,200 | Kai1 | kangai 1 | 1.11 | |||||
| rc_AI639313_at | Rn.87882 | 84009 | 11 | 68,205,034 | Kalrn | kalirin, RhoGEF kinase | 1.09 | |||||
| X62840mRNA_s_at | Rn.33095 | 25327 | 1 | 97,006,385 | Kcnc1 | potassium voltage-gated channel,Shaw-related subfamily, member 1 | 1.17 | |||||
| AJ007627_at | Rn.144567 | 27150 | 7 | 138,061,054 | Kcnh3 | potassium voltage-gated channel, subfamily H (eag-related), member 3 | -1.09 | |||||
| AF022819_at | Rn.15693 | 59324 | 19 | 56,221,480 | Kcnk1 | potassium channel, subfamily K, member 1 | 1.08 | |||||
| U93306_at | Rn.88869 | 25589 | 14 | 0 | Kdr | kinase insert domain protein receptor | 1.28 | 1.50 | ||||
| AF071204_at | Rn.44216 | 60427 | 7 | 37,734,427 | Kitl | kit ligand | 1.07 | |||||
| AF071204_g_at | Rn.44216 | 60427 | 7 | 37,734,427 | Kitl | kit ligand | -1.09 | |||||
| K02814_at | Rn.128333 | 24903 | 11 | 80,167,087 | Kng1 | kininogen 1 | 1.09 | |||||
| rc_AI639470_g_at | Rn.125065 | 450225 | 10 | -88,302,989 | Krt10 | keratin 10 | 1.07 | |||||
| D25224_at | Rn.999 | 29236 | 8 | 125,119,975 | Lamr1 | laminin receptor 1 (ribosomal protein SA) | 1.08 | |||||
| rc_AA892316_at | Rn.1057 | 155918 | 10 | 19,021,026 | Alc9 & 12 | Lcp2 | lymphocyte cytosolic protein 2 | 1.18 | ||||
| U07181_g_at | Rn.1785 | 24534 | 4 | 180,306,690 | Alc18 | Ldhb | lactate dehydrogenase B | 1.23 | ||||
| X96437mRNA_g_at | Rn.23638 | 294235 | 20 | -3,076,800 | Ler3 | immediate early response 3 | 1.11 | |||||
| AF090134_at | Rn.31766 | 85327 | 7 | 45,990,892 | Lin7a | lin-7 homolog a (C. elegans) | -1.16 | |||||
| AF090136_at | Rn.44269 | 60442 | 3 | 95,224,369 | Lin7c | lin-7 homolog C (C. elegans) | -1.35 | -1.51 | -1.27 | |||
| S66184_s_at | Rn.11372 | 24914 | 18 | -47,931,499 | Lox | lysyl oxidase | 1.22 | |||||
| rc_AI232268_at | Rn.10293 | 116565 | 14 | 0 | Lrpap1 | low density lipoprotein receptor-related protein associated protein 1 | 1.15 | |||||
| Z11995cds_g_at | Rn.10293 | 116565 | 14 | 0 | Lrpap1 | low density lipoprotein receptor-related protein associated protein 1 | 1.13 | |||||
| Z11995cds_at | Rn.10293 | 116565 | 14 | 0 | Lrpap1 | low density lipoprotein receptor-related protein associated protein 1 | 1.08 | |||||
| X76985_at | Rn.11404 | 59073 | 2 | -157,259,999 | Lxn | latexin | -1.19 | 1.34 | ||||
| rc_AA946044_s_at | Rn.4338 | 81515 | 5 | 16,933,105 | Alc4 | Lyn | Yamaguchi sarcoma viral (v-yes-1) oncogene homolog | 1.10 | ||||
| rc_AA892775_at | Rn.2283 | 25211 | 7 | -56,628,438 | Lyz | lysozyme | 1.54 | 2.11 | 1.81 | |||
| U05784_s_at | Rn.41412 | 64862 | 19 | 51,852,067 | Map1lc3b | microtubule-associated proteins 1A/1B light chain 3 | 1.07 | |||||
| M64301_g_at | Rn.88457 | 58840 | 8 | -80,231,782 | Mapk6 | mitogen-activated protein kinase 6 | 1.08 | |||||
| rc_AI227608_s_at | Rn.2455 | 29477 | 10 | 93,425,375 | Mapt | microtubule-associated protein tau | -1.29 | |||||
| AF095741_at | Rn.11802 | 171451 | 13 | -100,998,349 | Mg87 | Mg87 protein | -1.11 | |||||
| rc_AA892864_at | Rn.40396 | 29254 | 4 | 123,066,973 | Alc14 & 16 | Mgll | monoglyceride lipase | -1.11 | ||||
| rc_AI111401_s_at | Rn.27882 | 29688 | 1 | 0 | Minpp1 | multiple inositol polyphosphate histidine phosphatase 1 | 1.08 | |||||
| M15944_at | Rn.33598 | 24590 | 2 | 152,981,686 | Mme | membrane metallo endopeptidase | -1.05 | |||||
| M99485_at | Rn.9687 | 24558 | 20 | 1,601,730 | Mog | myelin oligodendrocyte glycoprotein | 1.11 | |||||
| L21995_s_at | Rn.9687 | 24558 | 20 | 1,601,730 | Mog | myelin oligodendrocyte glycoprotein | 1.08 | |||||
| rc_AI639359_at | Rn.98599 | 58808 | 9 | -57,843,928 | Mpp4 | membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) | 1.08 | |||||
| D14688_s_at | Rn.103179 | 50685 | 9 | -110,380,900 | Mrlcb | myosin light chain, regulatory B | 1.10 | |||||
| X06564_at | Rn.11283 | 24586 | 8 | -52,841,403 | Ncam1 | neural cell adhesion molecule 1 | -1.20 | |||||
| Z12152_at | Rn.10971 | 24588 | 15 | -47,714,153 | Nef3 | neurofilament 3, medium | 1.16 | |||||
| rc_AA818677_at | Rn.108194 | 24587 | 14 | -85,614,415 | Nefh | neurofilament, heavy polypeptide | 1.18 | |||||
| X13804cds_at | Rn.108194 | 24587 | 14 | -85,614,415 | Nefh | neurofilament, heavy polypeptide | 1.11 | |||||
| D82074_at | Rn.44289 | 29458 | 3 | -62,121,015 | Neurod1 | neurogenic differentiation 1 | 1.14 | |||||
| M82826_i_at | Rn.10686 | 24592 | 10 | 65,545,464 | Nf1 | neurofibromatosis 1 | -1.16 | |||||
| AF031880_at | Rn.18568 | 83613 | 15 | 47,652,082 | Nfl | neurofilament, light polypeptide | 1.08 | |||||
| U31203_at | Rn.10154 | 25495 | 10 | 0 | Nog | noggin | 1.09 | |||||
| rc_AI072943_at | Rn.54707 | 266777 | 10 | -108,933,164 | Nptx1 | neuronal pentraxin 1 | 1.14 | 1.32 | ||||
| M15880_at | Rn.9714 | 24604 | 4 | 78,314,142 | Alc14 & 16 | Npy | neuropeptide Y | -1.14 | ||||
| Z11504cds_at | Rn.11642 | 29358 | 16 | -24,747,181 | Alc7 & 11 | Npy1r | neuropeptide Y receptor Y1 | 1.08 | ||||
| U66274_at | Rn.10532 | 25340 | 16 | 24,765,172 | Alc7 & 11 | Npy5r | neuropeptide Y receptor Y5 | 1.11 | ||||
| rc_AA943331_s_at | Rn.11271 | 24605 | 2 | 198,255,369 | Nras | neuroblastoma ras oncogene | 1.07 | |||||
| U88958_at | Rn.3546 | 83834 | 0 | 0 | Nrn1 | neuritin | 1.10 | 1.19 | ||||
| L14851_at | Rn.10926 | 116508 | 6 | 112,277,485 | Alc17 | Nrxn3 | neurexin 3 | -1.11 | ||||
| V01543mRNA_at | Rn.2865 | 25247 | 14 | 77,934,337 | Nsg1 | neuron specific gene family member 1 | -1.07 | |||||
| E03082cds_s_at | Rn.9715 | 81737 | 4 | -162,680,717 | Alc18 | Ntf3 | neurotrophin 3 | 1.07 | ||||
| M86742cds_s_at | Rn.44225 | 25730 | 1 | 95,963,687 | Ntf5 | neurotrophin 5 | -1.06 | |||||
| X97121_at | Rn.127792 | 64636 | 0 | 0 | Ntsr2 | neurotensin receptor 2 | 1.08 | |||||
| rc_AI176253_at | Rn.133954 | 59319 | 3 | 0 | Nyw1 | ischemia related factor NYW-1 | -1.38 | |||||
| M93297cds_at | Rn.1430 | 64313 | 1 | -192,176,503 | Oat | ornithine aminotransferase | 1.11 | |||||
| X07944exon#1-12_s_at | Rn.874 | 24609 | 6 | 41,312,265 | Alc17 | Odc1 | ornithine decarboxylase 1 | 1.06 | ||||
| U93197_at | Rn.9783 | 171116 | 11 | -73,063,058 | Opa1 | optic atrophy 1 homolog (human) | 1.12 | |||||
| U47031_at | Rn.7176 | 29659 | 12 | -34,807,290 | Alc6 & 10 | P2rx4 | purinergic receptor P2X, ligand-gated ion channel 4 | -1.08 | ||||
| rc_AA899935_at | Rn.24751 | 64189 | 8 | -48,947,322 | Pafah1b2 | platelet-activating factor acetylhydrolase, isoform 1b, alpha2 subunit | 1.07 | |||||
| U35345_s_at | Rn.3840 | 29432 | 11 | 70,587,549 | Pak2 | p21 (CDKN1A)-activated kinase 2 | -1.20 | |||||
| AJ007291_g_at | Rn.30105 | 117287 | 5 | -168,060,336 | Alc4 | Park7 | fertility protein SP22 | 1.15 | ||||
| rc_AA891220_at | Rn.7264 | 297508 | 6 | 51,132,285 | Alc17 | Pbef1 | pre-B-cell colony enhancing factor 1 | 1.25 | ||||
| U95920_at | Rn.98622 | 81740 | 16 | 0 | Pcm1 | pericentriolar material 1 | 1.30 | 1.52 | ||||
| rc_AI232379_at | Rn.55127 | 25267 | 14 | 0 | Pdgfra | platelet derived growth factor receptor, alpha polypeptide | 1.13 | |||||
| M86870_at | Rn.39305 | 116598 | 4 | -76,205,527 | Alc14 & 16 | Pdia4 | protein disulfide isomerase associated 4 | 1.13 | ||||
| AF002281_at | Rn.13361 | 114108 | 16 | -49,637,142 | Alc7 & 11 | Pdlim3 | PDZ and LIM domain 3 | 1.15 | 1.28 | |||
| U77697_at | Rn.1878 | 29583 | 0 | -96,070,590 | Pecam | platelet/endothelial cell adhesion molecule | -1.14 | |||||
| D63673_at | Rn.10675 | 117265 | 9 | 10,081,699 | Pex6 | peroxisomal biogenesis factor 6 | -1.08 | |||||
| rc_AA892797_at | Rn.108127 | 24644 | X | 94,397,574 | Pgk1 | phosphoglycerate kinase 1 | 2.14 | 2.05 | 2.08 | 2.08 | 2.28 | |
| X97772_at | Rn.6872 | 58835 | 2 | -193,110,696 | Phgdh | 3-phosphoglycerate dehydrogenase | -1.33 | -1.36 | -1.42 | -1.33 | ||
| X97772_g_at | Rn.6872 | 58835 | 2 | -193,110,696 | Phgdh | 3-phosphoglycerate dehydrogenase | -1.31 | -1.28 | -1.39 | |||
| rc_AI008677_s_at | Rn.6872 | 58835 | 2 | -193,110,696 | Phgdh | 3-phosphoglycerate dehydrogenase | -1.27 | |||||
| M98826mRNA_g_at | Rn.10399 | 29353 | 12 | -27,893,003 | Alc10 | Phkg1 | phosphorylase kinase gamma 1 | -1.08 | ||||
| L14003UTR#1_f_at | Rn.54456 | 25046 | 13 | 43,799,887 | Pigr | polymeric immunoglobulin receptor | -1.07 | |||||
| U16655_at | Rn.37434 | 140693 | 9 | 74,025,487 | Plcd4 | phospholipase C, delta 4 | 1.12 | |||||
| D45920_at | Rn.10684 | 84587 | 9 | 54,335,647 | Plcl1 | phospholipase C-like 1 | 1.14 | |||||
| rc_AI072770_s_at | Rn.4550 | 24943 | X | 124,562,059 | Plp | proteolipid protein | 1.14 | |||||
| M57728_g_at | Rn.11175 | 296588 | 3 | 4,560,960 | Pmpca | peptidase (mitochondrial processing) alpha | 1.09 | |||||
| D13907_g_at | Rn.841 | 64198 | 4 | -8,746,903 | Pmpcb | peptidase (mitochondrial processing) beta | 1.12 | 1.26 | ||||
| D13907_at | Rn.841 | 64198 | 4 | -8,746,903 | Pmpcb | peptidase (mitochondrial processing) beta | 1.10 | |||||
| J02776_s_at | Rn.9346 | 29240 | 16 | 73,864,971 | Polb | polymerase (DNA directed), beta | -1.08 | |||||
| AF062740_at | Rn.31799 | 54705 | 5 | -26,234,515 | Alc4 | Ppm2c | protein phosphatase 2C, magnesium dependent, catalytic subunit | 1.13 | ||||
| AF062740_g_at | Rn.31799 | 54705 | 5 | -26,234,515 | Alc4 | Ppm2c | protein phosphatase 2C, magnesium dependent, catalytic subunit | 1.10 | ||||
| AJ011607_g_at | Rn.11846 | 301323 | 9 | -32,207,688 | Prim2 | DNA primase, p58 subunit | 1.06 | |||||
| AJ011607_at | Rn.11846 | 301323 | 9 | -32,207,688 | Prim2 | DNA primase, p58 subunit | -1.13 | |||||
| U42413_at | Rn.11089 | 25520 | 7 | -137,658,778 | Prkag1 | protein kinase, AMP-activated, gamma 1 noncatalytic subunit | -1.13 | |||||
| M13707_at | Rn.9747 | 24681 | 1 | -64,223,859 | Prkcc | protein kinase C, gamma | 1.11 | |||||
| M18330_at | Rn.98279 | 170538 | 16 | -5,954,214 | Alc7, 11, & 13 | Prkcd | protein kinase C, delta | 1.14 | ||||
| rc_AA799421_at | Rn.34966 | 29340 | 6 | 9,631,241 | Alc17 | Prkce | protein kinase C, epsilon | -1.29 | ||||
| D50093_s_at | Rn.3936 | 24686 | 3 | 119,581,709 | Prnp | prion protein | 1.10 | |||||
| X16554_at | Rn.9761 | 29562 | X | 128,323,278 | Prps1 | phosphoribosyl pyrophosphate synthetase 1 | 1.19 | |||||
| D90265_s_at | Rn.2668 | 29668 | 1 | -172,373,871 | Psma1 | proteasome (prosome, macropain) subunit, alpha type 1 | 1.09 | |||||
| rc_AA891383_i_at | Rn.1276 | 29672 | 2 | 203,787,813 | Alc15 | Psma5 | proteasome (prosome, macropain) subunit, alpha type 5 | 1.55 | 1.44 | 1.57 | 1.78 | |
| D10729_s_at | Rn.29244 | 24968 | 20 | -4,786,490 | Psmb8 | proteosome (prosome, macropain) subunit, beta type 8 | 1.08 | |||||
| L27843_s_at | Rn.9459 | 29463 | 9 | -29,618,696 | Ptp4a1 | protein tyrosine phosphatase 4a1 | 1.10 | |||||
| U40790_at | Rn.10278 | 29645 | 3 | -74,693,135 | Ptprj | protein tyrosine phosphatase, receptor type, J | 1.28 | |||||
| rc_AA874857_at | Rn.114933 | 498407 | 14 | -87,249,491 | pur-beta | transcription factor Pur-beta | -1.16 | |||||
| rc_AI175539_at | Rn.2005 | 25269 | 7 | -116,148,899 | Pvalb | parvalbumin | 1.16 | |||||
| E03344cds_s_at | Rn.4065 | 29534 | 2 | 98,306,852 | Pxmp3 | peroxisomal membrane protein 3 | 1.08 | |||||
| rc_AA800190_at | Rn.1518 | 25739 | 3 | 0 | Pygb | brain glycogen phosphorylase | 1.10 | |||||
| J03481mRNA_at | Rn.241 | 64192 | 14 | 70,744,388 | Qdpr | quinoid dihydropteridine reductase | 1.18 | |||||
| J03481mRNA_g_at | Rn.241 | 64192 | 14 | 70,744,388 | Qdpr | quinoid dihydropteridine reductase | 1.18 | |||||
| M75153_at | Rn.1016 | 81830 | 8 | -68,950,978 | Rab11a | RAB11a, member RAS oncogene family | 1.12 | |||||
| rc_AA894207_g_at | Rn.28921 | 246324 | 9 | -104,618,026 | Rab31 | RAB31, member RAS oncogene family | -1.18 | |||||
| rc_AA800305_at | Rn.44477 | 64633 | 9 | -1,492,865 | Rab5a | RAB5A, member RAS oncogene family | -1.15 | -1.29 | ||||
| AF072935_at | Rn.44477 | 64633 | 9 | -1,492,865 | Rab5a | RAB5A, member RAS oncogene family | -1.10 | |||||
| rc_AA900900_s_at | Rn.7107 | 84014 | 9 | -104,826,889 | Ralbp1 | ralA binding protein 1 | -1.11 | |||||
| rc_AI169372_at | Rn.105901 | 304268 | 12 | -8,735,920 | Rasl11a | RAS-like family 11 member A | 1.13 | |||||
| rc_AA893471_s_at | Rn.98353 | 24718 | 4 | 8,150,872 | Reln | reelin | -1.14 | |||||
| AF036548_g_at | Rn.3504 | 117183 | 15 | 0 | Rgc32 | response gene to complement 32 | 1.14 | |||||
| AF036548_at | Rn.3504 | 117183 | 15 | 0 | Rgc32 | response gene to complement 32 | 1.09 | |||||
| AF035151_s_at | Rn.9796 | 54292 | 14 | -81,365,932 | Rgs12 | regulator of G-protein signaling 12 | -1.07 | |||||
| AB015191_g_at | Rn.1943 | 60414 | 5 | 153,648,680 | Alc4 | Rhced | Rhesus blood group CE and D | -1.08 | ||||
| AB015191_at | Rn.1943 | 60414 | 5 | 153,648,680 | Alc4 | Rhced | Rhesus blood group CE and D | -1.09 | ||||
| rc_AA900505_at | Rn.2042 | 64373 | 6 | -32,055,314 | Alc17 | Rhob | rhoB gene | 1.10 | ||||
| X92097_at | Rn.1022 | 65165 | 12 | -33,028,678 | Alc6 & 10 | Rnp24 | coated vesicle membrane protein | 1.10 | ||||
| X93352_at | Rn.2262 | 81729 | 20 | 6,569,309 | Rpl10a | ribosomal protein L10A | 1.12 | |||||
| S71021_s_at | Rn.2660 | 117042 | 12 | -36,347,261 | Alc6 & 10 | Rpl6 | ribosomal protein L6 | 1.14 | ||||
| rc_AA799672_s_at | Rn.2660 | 117042 | 12 | -36,347,261 | Alc6 & 10 | Rpl6 | ribosomal protein L6 | 1.15 | ||||
| X13549_s_at | Rn.3359 | 81773 | 20 | -5,849,544 | Rps10 | ribosomal protein S10 | 1.10 | |||||
| L81138exon_i_at | Rn.2115 | 83789 | 10 | 13,975,349 | Alc5 & 9 | Rps2 | ribosomal protein S2 | -1.60 | -1.48 | -1.63 | -1.76 | -1.53 |
| L81136cds_f_at | Rn.2115 | 83789 | 10 | 13,975,349 | Alc5 & 9 | Rps2 | ribosomal protein S2 | -1.15 | 1.20 | |||
| M64795_f_at | - | 24735 | 20 | 0 | RT1@ | Major histocompatibility locus | -1.14 | |||||
| M11071_f_at | Rn.40130 | 24737 | 20 | 3,412,286 | RT1-Aw2 | RT1 class Ib, locus Aw2 | -1.38 | -1.37 | -1.41 | -1.41 | ||
| M10094_g_at | Rn.40130 | 24737 | 20 | 3,412,286 | RT1-Aw2 | RT1 class Ib, locus Aw2 | -1.20 | -1.36 | ||||
| M31018_f_at | Rn.40130 | 24737 | 20 | 3,412,286 | RT1-Aw2 | RT1 class Ib, locus Aw2 | -1.22 | -1.23 | ||||
| M24324_f_at | Rn.40130 | 24737 | 20 | 3,412,286 | RT1-Aw2 | RT1 class Ib, locus Aw2 | -1.09 | |||||
| AF074609mRNA_f_at | Rn.40130 | 24737 | 20 | 3,412,286 | RT1-Aw2 | RT1 class Ib, locus Aw2 | -1.14 | |||||
| U65217_i_at | Rn.33311 | 309622 | 20 | -4,730,786 | RT1-Bb | RT1 class II, locus Bb | 1.46 | 1.39 | ||||
| X56596_at | Rn.33311 | 309622 | 20 | -4,730,786 | RT1-Bb | RT1 class II, locus Bb | -1.16 | |||||
| M24026_f_at | Rn.145396 | 309600 | 20 | 147,139 | RT1-CE12 | RT1 class I, CE12 | 1.17 | |||||
| AF025308_f_at | Rn.160576 | 24977 | 20 | 3,552,628 | RT1-Cl | RT1 class Ib, locus Cl | -1.12 | |||||
| M15562_g_at | Rn.103146 | 294269 | 20 | 4,636,585 | RT1-Da | RT1 class II, locus Da | 1.20 | |||||
| M15562_at | Rn.103146 | 294269 | 20 | 4,636,585 | RT1-Da | RT1 class II, locus Da | 1.17 | |||||
| L23128_g_at | Rn.107070 | 24750 | 20 | 2,807,067 | RT1-N3 | RT1 class Ib gene, H2-TL-like, grc region (N3) | 1.22 | 1.26 | 1.35 | |||
| AF029240_g_at | Rn.143874 | 294228 | 20 | 0 | RT1-S3 | RT1 class Ib, locus S3 | -1.06 | |||||
| rc_AI235890_s_at | Rn.143874 | 294228 | 20 | 0 | RT1-S3 | RT1 class Ib, locus S3 | -1.21 | |||||
| X52817cds_s_at | Rn.55126 | 116644 | 6 | -94,400,519 | Alc17 | Rtn1 | reticulon 1 | 1.14 | ||||
| M15114_g_at | Rn.83595 | 83792 | 1 | 249,616,941 | Scd2 | stearoyl-Coenzyme A desaturase 2 | 1.08 | |||||
| Y00766_at | Rn.87394 | 25657 | 3 | -47,398,275 | Scn3a | sodium channel, voltage-gated, type III, alpha polypeptide | -1.36 | -1.38 | -1.44 | -1.39 | ||
| Y00766_g_at | Rn.87394 | 25657 | 3 | -47,398,275 | Scn3a | sodium channel, voltage-gated, type III, alpha polypeptide | -1.38 | 1.41 | ||||
| M62763complete_seq_at | Rn.31887 | 25541 | 5 | -129,158,061 | Alc4 | Scp2 | sterol carrier protein 2 | 1.25 | ||||
| X99337cds_s_at | Rn.37476 | 56064 | 8 | 62,408,946 | Sdfr1 | stromal cell derived factor receptor 1 | 1.10 | |||||
| X99338cds_i_at | Rn.37476 | 56064 | 8 | 62,408,946 | Sdfr1 | stromal cell derived factor receptor 1 | 1.09 | |||||
| rc_AA799700_at | Rn.100471 | 308993 | 1 | 0 | Sephs2 | selenophosphate synthetase 2 | -1.45 | -1.61 | -1.39 | -1.47 | ||
| M63901_at | Rn.6173 | 25719 | 3 | -99,525,619 | Sgne1 | secretory granule neuroendocrine protein 1 | 1.10 | |||||
| M63901_g_at | Rn.6173 | 25719 | 3 | -99,525,619 | Sgne1 | secretory granule neuroendocrine protein 1 | 1.09 | |||||
| AF009604_at | Rn.5909 | 81921 | 1 | 138,476,741 | Sh3gl3 | SH3 domain protein 2 C1 | 1.18 | |||||
| U90261UTR#1_g_at | Rn.107226 | 84357 | X | -56,129,005 | Sh3kbp1 | SH3-domain kinase binding protein 1 | 1.10 | |||||
| AB001453_at | Rn.59227 | 114858 | 17 | 0 | Shc3 | src homology 2 domain-containing transforming protein C3 | -1.28 | -1.29 | ||||
| rc_AA875411_s_at | Rn.37542 | 65041 | 1 | 159,205,528 | Skd3 | suppressor of K+ transport defect 3 | 1.06 | |||||
| rc_AA891445_at | Rn.37542 | 65041 | 1 | 159,205,528 | Skd3 | suppressor of K+ transport defect 3 | 1.11 | |||||
| rc_AI145680_s_at | Rn.6085 | 25027 | 2 | 199,823,090 | Slc16a1 | solute carrier family 16 (monocarboxylic acid transporters), member 1 | 1.09 | |||||
| U66723_s_at | Rn.10140 | 60423 | 3 | -109,256,103 | Slc28a2 | solute carrier family 28 (sodium-coupled nucleoside transporter), member 2 | -1.15 | -1.25 | ||||
| Y16774_at | Rn.90036 | 64469 | 3 | -109,549,289 | Slc30a4 | solute carrier family 30 (zinc transporter), member 4 | 1.09 | |||||
| AF004017_at | Rn.11114 | 84484 | 14 | -20,381,544 | Slc4a4 | solute carrier family 4, member 4 | -1.24 | |||||
| AJ001290cds_at | Rn.79242 | 114507 | 11 | 32,119,517 | Slc5a3 | solute carrier family 5 (inositol transporters), member 3 | -1.49 | -1.40 | -1.77 | -1.44 | -1.47 | |
| M96601_at | Rn.9968 | 29464 | 4 | 126,120,297 | Alc14 & 16 | Slc6a6 | solute carrier family 6 (neurotransmitter transporter, taurine), member 6 | -1.37 | ||||
| AB011530_at | Rn.30002 | 65047 | 1 | -246,928,385 | Slit1 | slit homolog 1 (Drosophila) | 1.17 | 1.32 | ||||
| AB011531_at | Rn.12298 | 83467 | 10 | 20,098,220 | Slit3 | slit homolog 3 (Drosophila) | -1.08 | |||||
| U83883_at | Rn.36148 | 64635 | 4 | 55,601,577 | Snd1 | staphylococcal nuclease domain containing 1 | 1.13 | |||||
| U75928UTR#1_s_at | Rn.98989 | 24791 | 10 | -40,823,352 | Sparc | secreted acidic cysteine rich glycoprotein | 1.19 | |||||
| rc_AI639256_at | Rn.40672 | 301255 | 9 | 11,125,212 | Spats1 | spermatogenesis associated, serine-rich 1 | 1.28 | 1.29 | ||||
| Y07534cds_s_at | Rn.91257 | 24794 | 6 | -128,383,141 | Alc17 | Spin2b | Serine protease inhibitor | -1.07 | ||||
| rc_AA800712_g_at | Rn.13142 | 361813 | 20 | -7,257,777 | Stk38 | serine/threonine kinase 38 | -1.17 | -1.37 | -1.21 | |||
| D12519_s_at | Rn.9943 | 116470 | 12 | -690,406 | Stx1a | syntaxin 1A (brain) | -1.10 | |||||
| L10362_at | Rn.58137 | 117556 | 1 | -130,761,497 | Sv2b | synaptic vesicle glycoprotein 2b | 1.11 | |||||
| rc_AI639484_at | Rn.58137 | 117556 | 1 | -130,761,497 | Sv2b | synaptic vesicle glycoprotein 2b | 1.09 | |||||
| M27925_at | Rn.506 | 29179 | 4 | 151,500,081 | Alc14 & 16 | Syn2 | synapsin II | 1.17 | ||||
| AF053938_s_at | Rn.9908 | 192117 | 20 | -32,816,659 | Syngap1 | synaptic Ras GTPase activating protein 1 homolog (rat) | -1.15 | |||||
| U85513_at | Rn.74259 | 60567 | 7 | -128,149,672 | Syt10 | synaptotagmin X | -1.07 | |||||
| U20105_at | Rn.48074 | 60565 | 2 | 198,770,093 | Syt6 | synaptotagmin 6 | -1.09 | |||||
| U20106_at | Rn.91884 | 59267 | 1 | 213,035,104 | Syt7 | synaptotagmin 7 | -1.13 | |||||
| U40188_at | Rn.11270 | 171152 | X | -94,415,515 | Taf9l | TAF9-like RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa | 1.14 | |||||
| M83107_at | Rn.34397 | 25123 | 8 | -48,910,973 | Tagln | transgelin | -1.17 | -1.22 | -1.23 | |||
| rc_AA893860_at | Rn.22757 | 294810 | 2 | -60,694,140 | Tars | threonyl-tRNA synthetase | 1.08 | |||||
| U09228_at | Rn.23354 | 84382 | 18 | 66,209,063 | Tcf4 | transcription factor 4 | -1.18 | |||||
| rc_AI639517_at | Rn.23354 | 84382 | 18 | 66,209,063 | Tcf4 | transcription factor 4 | -1.20 | |||||
| rc_AA893702_s_at | Rn.1829 | 64365 | 14 | -84,598,335 | Tcn2 | transcobalamin 2 | -1.07 | |||||
| K01934mRNA#2_at | Rn.81140 | 25357 | 1 | -154,725,746 | Thrsp | thyroid hormone responsive protein | 1.09 | 1.30 | ||||
| U88630_at | Rn.2398 | 81813 | 7 | -73,842,010 | Tieg | TGFB inducible early growth response | -1.07 | |||||
| U09256_at | Rn.5950 | 64524 | 16 | 5,908,756 | Alc7, 11, & 13 | Tkt | transketolase | 1.11 | ||||
| rc_AA818982_at | Rn.3364 | 25359 | 7 | -28,106,846 | Tmpo | thymopoietin | -1.16 | |||||
| rc_AI639532_at | Rn.43529 | 296369 | 3 | -155,713,170 | Tnnc2 | troponin C2, fast | -1.15 | |||||
| rc_AA899854_at | Rn.90996 | 360243 | 10 | -87,785,706 | Top2a | topoisomerase (DNA) 2 alpha | -1.09 | |||||
| rc_AI230748_at | Rn.36610 | 116646 | 15 | 56,768,246 | Tpt1 | tumor protein, translationally controlled 1 | -1.11 | |||||
| L04760_at | Rn.6763 | 81002 | 2 | 0 | Trim23 | tripartite motif protein 23 | 1.07 | |||||
| rc_AI169370_at | Rn.99661 | 64158 | 7 | -137,805,841 | Tuba1 | tubulin, alpha 1 | -1.09 | |||||
| AB011679_at | Rn.2458 | 29214 | 20 | 3,060,491 | Tubb5 | tubulin, beta 5 | 1.16 | |||||
| rc_AI103911_at | Rn.2603 | 291103 | 17 | 40,432,646 | Uqcrfs1 | ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 | 1.08 | |||||
| M32167_at | Rn.1923 | 83785 | 9 | 10,519,065 | Vegfa | vascular endothelial growth factor A | -1.09 | |||||
| M32167_g_at | Rn.1923 | 83785 | 9 | 10,519,065 | Vegfa | vascular endothelial growth factor A | -1.16 | |||||
| U14746_at | Rn.11059 | 24874 | 4 | 149,774,236 | Alc14 & 16 | Vhl | von Hippel-Lindau syndrome homolog | 1.10 | ||||
| rc_AI071435_at | Rn.51354 | 25218 | 20 | -5,074,730 | Vps52 | vacuolar protein sorting 52 (yeast) | -1.22 | |||||
| AF033027_at | Rn.6525 | 64351 | 14 | 86,638,972 | Ykt6 | prenylated SNARE protein | 1.11 | |||||
| U67082_at | Rn.10663 | 25165 | 6 | 143,317,934 | Alc17 | Znf386 | zinc finger protein 386 (Kruppel-like) | 1.15 |
Comb. regions = combined regions; Acb. = accumbens; Amyg. = amygdala; Fr. Ctx. = frontal cortext; Hipp. = hippocampus; CPU = caudate-putamen.
Positive fold differences are I/iNP; negative are iNP/iP.
Analysis for over-represented GO categories in the set of significant genes found with the overall analysis indicated that 17 GO categories were over-represented (P <.05). Six of the most neurobiologically interesting categories are axon guidance, gliogenesis, negative regulation of programmed cell death, regulation of programmed cell death, regulation of synapse structure function, and transmission of nerve impulse (Table 6); the full list can be viewed in Table 7. More genes demonstrate increased expression in the iP versus iNP rat comparison in the categories of regulation of cell death, negative regulation of cell death, regulation of synapse structure and function, and gliogenesis; whereas, more genes demonstrate decreased expression in the iP versus iNP in the category of axon guidance. Equal numbers of genes with increased and decreased expression in the iP versus iNP are present in the category of transmission of nerve impulse.
Table 6.
Biologically interesting, nonredundant, significantly over-represented GO biologic process categoriesa
| GO term | Fischer exact P value | Number of significant genes | Number of significant genes (iPO>NP) | Number of significant genes (iP<iNP) |
|---|---|---|---|---|
| Transmission of nerve impulse | .047 | 26 | 13 | 13 |
| Regulation of programmed cell death | .014 | 24 | 14 | 10 |
| Negative regulation of programmed cell death | .014 | 13 | 7 | 6 |
| Axon guidance | .017 | 7 | 2 | 5 |
| Regulation of synapse structure and function | .037 | 6 | 4 | 3 |
| Gliogenesis | .002 | 5 | 3 | 2 |
GO = Gene Ontology; iP = inbred alcohol-preferring rat; iNP = inbred alcoholenon-preferring rat; FDR = false discovery rate.
As determined by Fisher’s Exact test using the hypergeometric distribution (P <.05) on genes significant in the combined regions analysis (FDR < 0.10). Categories with >300 or <10 total genes in the chipset and those with <5 significant genes in a category were excluded. For the complete list of significant categories (P <.05), see Table 7.
Table 7.
All significant Gene Ontology (GO) biologic process categories
| GO ID | GO term | Fischer exact P | Sig genes | Total genes in chipset |
|---|---|---|---|---|
| GO:0008219 | Cell death | 0.033 | 28 | 288 |
| GO:0016265 | Death | 0.037 | 28 | 291 |
| GO:0019226 | Transmission of nerve impulse | 0.047 | 26 | 272 |
| GO:0007268 | Synaptic transmission | 0.041 | 25 | 256 |
| GO:0043067 | Regulation of programmed cell death | 0.014 | 24 | 221 |
| GO:0042981 | Regulation of apoptosis | 0.023 | 23 | 219 |
| GO:0043066 | Negative regulation of apoptosis | 0.014 | 13 | 97 |
| GO:0043069 | Negative regulation of programmed cell death | 0.014 | 13 | 97 |
| GO:0001568 | Blood vessel development | 0.038 | 10 | 78 |
| GO:0042060 | Wound healing | 0.028 | 9 | 64 |
| GO:0006959 | Humoral immune response | 0.028 | 9 | 64 |
| GO:0007411 | Axon guidance | 0.017 | 7 | 40 |
| GO:0019882 | Antigen presentation | 0.001 | 6 | 17 |
| GO:0046651 | Lymphocyte proliferation | 0.037 | 6 | 37 |
| GO:0050803 | Regulation of synapse structure and function | 0.037 | 6 | 37 |
| GO:0030333 | Antigen processing | 0.001 | 5 | 13 |
| GO:0042063 | Gliogenesis | 0.002 | 5 | 14 |
For the purposes of further discussion, the authors found it desirable to use a gene classification schema that would include genes found significant in individual regions and that would include functional neuroinformatic categories such as neuroplasticity. Such customized, neuroinformatic classifications of genes found significant in any of the individual regions, or in the combined analysis, were obtained by first assigning all genes to their appropriate GO biologic process categories, regardless of statistical significance of the category. Of the categories obtained, further selection was made based on the number of genes within category and degree of neuroinformatic interest. The resultant GO categories were synaptic transmission, regulation of action potential, positive regulation of transport, potassium ion transport, chloride transport, metal ion homeostasis (collapsed to form “neurotransmission,” Table 8); apoptosis, axon guidance, axonogenesis, cell migration, central nervous system development, cytoskeleton organization and biogenesis, gliogenesis, myelination, negative regulation of apoptosis, neuron differentiation, positive regulation of apoptosis, regulation of apoptosis, regulation of axonogenesis, regulation of neurogenesis, regulation of programmed cell death, and regulation of synapse structure and function (collapsed to form “neuroplasticity,” Table 9); and small GTPase mediated signal transduction, negative regulation of protein kinase activity, negative regulation of enzyme activity, transmembrane receptor protein tyrosine kinase signaling pathway (collapsed to form “intracellular messaging,” Table 10). Finally, the lists of genes thus classified were supplemented with manual curation based on a search of the literature. One last category was obtained by classifying genes as transcription factors based on results obtained with Genomatix Suite (Genomatix, Munich, Germany; Table 11).
Table 8.
Neurotransmission-related significant genesa
| Entrez gene | Gene symbol | Gene name | QTL | Combined regions fold (iP vs. iNP) | Individual regionb significant in (fold) |
|---|---|---|---|---|---|
| Decreased expression | |||||
| 140667 | Ap3m2 | adaptor-related protein complex 3, mu 2 subunit | -1.12 | ||
| 25369 | Adora2a | adenosine A2a receptor | -1.08 | ||
| 83610 | Apba2 | amyloid beta (A4) precursor protein-binding, family A, member 2 | -1.13 | ||
| 65153 | Freq | frequenin homolog (Drosophila) | -1.22 | ||
| 24416 | Grm3 | glutamate receptor, metabotropic 3 | -1.31 | ||
| 81672 | Grm7 | glutamate receptor, metabotropic 7 | Alc14 & 16 | -1.08 | |
| 25674 | Glra1 | glycine receptor, alpha 1 subunit | -1.11 | ||
| 50664 | Gnao | guanine nucleotide binding protein, alpha o | -1.26 | ||
| 29547 | Homer2 | homer homolog 2 (Drosophila) | -1.09 | ||
| 84009 | Kalrn | kalirin, RhoGEF kinase | -1.09 | ||
| 24903 | Kng1 | kininogen 1 | -1.09 | ||
| 116508 | Nrxn3 | neurexin 3 | Alc17 | -1.11 | |
| 24604 | Npy | neuropeptide Y | Alc14 & 16 | -1.14 | |
| 25730 | Ntf5 | neurotrophin 5 | -1.06 | ||
| 27150 | Kcnh3 | potassium voltage-gated channel, subfamily H (age-related), member 3 | -1.09 | ||
| 29659 | P2rx4 | purinergic receptor P2X, ligand-gated ion channel 4 | Alc6 & 10 | -1.08 | |
| 114858 | Shc3 | src homology 2 domain-containing transforming protein C3 | -1.28 | AMYG (e1.29), CPU (e1.39) | |
| 60567 | Syt10 | synaptotagmin X | -1.07 | ||
| 116470 | Stx1a | syntaxin 1A (brain) | -1.10 | ||
| Increased expression | |||||
| 295365 | Amigo | amphoretin-induced gene and ORF | Alc15 | 1.11 | AMYG (1.22) |
| 26989 | Cadps | Ca2+-dependent secretion activator | 1.08 | ||
| 65160 | Clns1a | chloride channel, nucleotide-sensitive, 1A | 1.14 | ||
| 24259 | Chgb | chromogranin B | AMYG (1.25) | ||
| 25707 | Cntf | ciliary neurotrophic factor | 1.11 | ||
| 24232 | C3 | complement component 3 | 1.19 | ||
| 289211 | Fcgr2b | Fc receptor, IgG, low affinity IIb | 1.11 | ||
| 25319 | Fth1 | ferritin, heavy polypeptide 1 | 1.26 | AMYG (1.29) | |
| 83633 | Gpr51 | G protein-coupled receptor 51 | Alc4 | 1.12 | |
| 25450 | Gabrb1 | gamma-aminobutyric acid receptor, subunit beta 1 | 1.21 | ||
| 64831 | Ireb2 | iron responsive element binding protein 2 | 1.06 | ||
| 24558 | Mog | myelin oligodendrocyte glycoprotein | 1.08 | ||
| 25340 | Npy5r | neuropeptide Y receptor Y5 | Alc7 & 11 | 1.11 | |
| 81737 | Ntf3 | neurotrophin 3 | Alc18 | 1.07 | |
| 59324 | Kcnk1 | potassium channel, subfamily K, member 1 | 1.08 | ||
| 25327 | Kcnc1 | potassium voltage-gated channel, Shaw-related subfamily, member 1 | 1.17 | ||
| 24686 | Prnp | prion protein | 1.10 | ||
| 24681 | Prkcc | protein kinase C, gamma | 1.11 | ||
| 116644 | Rtn1 | reticulon 1 | Alc17 | 1.14 | |
| 29179 | Syn2 | synapsin II | Alc14 & 16 | 1.17 | |
| 117556 | Sv2b | synaptic vesicle glycoprotein 2b | 1.11 | ||
iP = inbred alcohol-preferring rat; iNP = inbred alcoholenon-preferring rat.
Defined as categorized by a Gene Ontology category (synaptic transmission, regulation of action potential, positive regulation of transport, potassium ion transport, chloride transport, metal ion homeostasis) or found through manual curation to have neurotransmission-related function (Ap3m2, Clns1a, Chgb, Grm7, Glra1, Gnao, Homer2, Kalrn, Kng1, Kcnh3, P2rx4, Rtn1, Stx1a).
ACB = nucleus accumbens, AMYG = amygdala, FC = frontal cortex, HIPP = hippocampus, CPU = caudate-putamen.
Table 9.
Neuroplasticity-related significant genesa
| Entrez gene | Gene symbol | Gene name | QTL | Combined regions fold (iP vs. iNP) | Individual regionb significant in (fold) |
|---|---|---|---|---|---|
| Decreased expression | |||||
| 58835 | Phgdh | 3-phosphoglycerate dehydrogenase | -1.27 | CPU (e1.33) | |
| 25369 | Adora2a | adenosine A2a receptor | -1.08 | ||
| 24888 | Bcl2l1 | Bcl2-like 1 | -1.11 | ||
| 64547 | Bcl2l11 | BCL2-like 11 (apoptosis facilitator) | -1.08 | ||
| 29182 | Cdh22 | cadherin 22 | Alc17 | -1.43 | |
| 25402 | Casp3 | caspase 3, apoptosis related cysteine protease | Alc7 & 11 | ACB (e1.21) | |
| 114851 | Cdkn1a | cyclin-dependent kinase inhibitor 1A | -1.11 | ||
| 79208 | Epha5 | EphA5 | -1.11 | ||
| 79114 | Fgfr1 | Fibroblast growth factor receptor 1 | -1.11 | AMYG (e1.21) | |
| 25661 | Fn1 | fibronectin 1 | -1.27 | ||
| 65153 | Freq | frequenin homolog (Drosophila) | -1.22 | ||
| 24416 | Grm3 | glutamate receptor, metabotropic 3 | -1.31 | ||
| 25589 | Kdr | kinase insert domain protein receptor | -1.28 | AMYG (e1.50) | |
| 116508 | Nrxn3 | neurexin 3 | Alc17 | -1.11 | |
| 25730 | Ntf5 | neurotrophin 5 | -1.06 | ||
| 29432 | Pak2 | p21 (CDKN1A)-activated kinase 2 | NS | AMYG (e1.20) | |
| 29583 | Pecam | platelet/endothelial cell adhesion molecule | -1.14 | ||
| 29240 | Polb | polymerase (DNA directed), beta | -1.08 | ||
| 24718 | Reln | reelin | -1.14 | ||
| 84357 | Sh3kbp1 | SH3-domain kinase binding protein 1 | -1.1 | ||
| 83467 | Slit3 | slit homolog 3 (Drosophila) | -1.08 | ||
| 60567 | Syt10 | synaptotagmin X | -1.07 | ||
| 116646 | Tpt1 | tumor protein, translationally controlled 1 | -1.11 | ||
| 83785 | Vegfa | vascular endothelial growth factor Ac | -1.16 | ||
| Increased expression | |||||
| 29437 | Acta1 | actin, alpha 1, skeletal muscle | 1.1 | ||
| 24188 | Aldh1a1 | aldehyde dehydrogenase family 1, member A1 | 1.2 | ||
| 295365 | Amigo | amphoretin-induced gene and ORF | Alc15 | 1.11 | AMYG (1.22) |
| 24179 | Agt | angiotensinogen | 1.2 | CPU (1.30) | |
| 83526 | Atrn | attractin | 1.26 | AMYG (1.33) | |
| 117271 | Bid3 | BH3 interacting (with BCL2 family) domain, apoptosis agonist | Alc10 | 1.09 | |
| 25668 | Cd38 | CD38 antigen | 1.13 | ||
| 25599 | Cd74 | CD74 antigen (invariant polpypeptide of major histocompatibility class II antigen-associated) | 1.21 | ||
| 64465 | Cdc42 | cell division cycle 42 homolog (S. cerevisiae) | Alc4 | 1.12 | |
| 25707 | Cntf | ciliary neurotrophic factor | 1.11 | ||
| 192262 | C1s | complement component 1, s subcomponent | Alc18 | 1.07 | |
| 27256 | Cntn6 | contactin 6 | Alc14 & 16 | 1.09 | |
| 29139 | Dcn | decorinc | 1.17 | HIPP (1.29) | |
| 29560 | Hif1a | hypoxia inducible factor 1, alpha subunit | Alc17 | 1.1 | |
| 294235 | Ler3 | immediate early response 3 | 1.11 | ||
| 29197 | Il18 | interleukin 18 | 1.15 | ||
| 362418 | Irak2 | interleukin-1 receptor-associated kinase 2 | Alc14 & 16 | 1.08 | AMYG (1.22) |
| 24558 | Mog | myelin oligodendrocyte glycoprotein | 1.08 | ||
| 83834 | Nrn1 | neuritin | 1.1 | AMYG (1.19) | |
| 24588 | Nef3 | neurofilament 3, medium | 1.16 | ||
| 24587 | Nefh | neurofilament, heavy polypeptide | 1.18 | CPU (1.44) | |
| 83613 | Nfl | neurofilament, light polypeptide | 1.08 | ||
| 81737 | Ntf3 | neurotrophin 3 | Alc18 | 1.07 | |
| 171116 | Opa1 | optic atrophy 1 homolog (human) | 1.12 | ||
| 114108 | Pdlim3 | PDZ and LIM domain 3 | Alc7 & 11 | 1.15 | AMYG (1.28) |
| 29534 | Pxmp3 | peroxisomal membrane protein 3 | 1.08 | ||
| 170538 | Prkcd | protein kinase C, delta | Alc7, 11, & 13 | 1.14 | |
| 64373 | Rhob | rhoB gene | Alc17 | 1.1 | |
| 81921 | Sh3gl3 | SH3 domain protein 2 C1 | NS | AMYG (1.18) | |
| 65047 | Slit1 | slit homolog 1 (Drosophila) | 1.17 | HIPP (1.32) | |
| 29214 | Tubb5 | tubulin, beta 5 | 1.16 | ||
iP = inbred alcohol-preferring rat; iNP = inbred alcoholenon-preferring rat.
Defined as categorized by a Gene Ontology category (apoptosis, regulation of apoptosis, negative regulation of apoptosis, regulation of programmed cell death, positive regulation of apoptosis, central nervous system development, gliogenesis, myelination, axon guidance, axonogenesis, cell migration, regulation of axonogenesis, regulation of neurogenesis, regulation of synapse structure and function, cytoskeleton organization and biogenesis, neuron differentiation) or found through manual curation to have neuroplasticity-related function (Cdh22, Cdc42, Fgfr1, Vegfa, Vegfa, Dcn, Dcn, Epha5, Nrn1, Bid3, Casp3, Ler3, Pak2, Sh3kbp1).
ACB = nucleus accumbens, AMYG = amygdala, FC = frontal cortex, HIPP = hippocampus, CPU = caudate-putamen.
Gene represented by more than one significant probe set. Largest of the multiple fold changes is reported in column 5.
Table 10.
Intracellular messaging-related significant genesa
| Entrez gene | Gene symbol | Gene name | QTL | Combined regions fold (iP vs. iNP) | Individual regionb significant in (fold) |
|---|---|---|---|---|---|
| Decreased expression (iP vs. iNP) | |||||
| 66013 | Arhgef9 | Cdc42 guanine nucleotide exchange factor (GEF) 9 | -1.22 | ||
| 114851 | Cdkn1a | cyclin-dependent kinase inhibitor 1A | -1.11 | ||
| 171109 | Dusp5 | dual specificity phosphatase 5 | -1.07 | ||
| 116663 | Dusp6 | dual specificity phosphatase 6 | -1.28 | ||
| 25114 | Fgfr4 | fibroblast growth factor receptor 4 | -1.09 | ||
| 25661 | Fn1 | fibronectin 1 | -1.27 | ||
| 24416 | Grm3 | glutamate receptor, metabotropic 3 | -1.31 | ||
| 25589 | Kdr | kinase insert domain protein receptor | -1.28 | AMYG (-1.50) | |
| 24592 | Nf1 | neurofibromatosis 1 | -1.16 | ||
| 29353 | Phkg1 | phosphorylase kinase gamma 1 | Alc10 | -1.08 | |
| 29583 | Pecam | platelet/endothelial cell adhesion molecule | -1.14 | ||
| 29340 | Prkce | protein kinase C, epsilon | Alc17 | -1.29 | |
| 84014 | Ralbp1 | ralA binding protein 1 | -1.11 | ||
| 24718 | Reln | reelin | -1.14 | ||
| 24794 | Spin2b | Serine protease inhibitor | Alc17 | -1.07 | |
| 361813 | Stk38 | serine/threonine kinase 38 | -1.17 | ACB (e1.37), CPU (e1.21) | |
| 192117 | Syngap1 | synaptic Ras GTPase activating protein 1 homolog (rat) | -1.15 | ||
| Increased expression (iP vs. iNP) | |||||
| 25673 | Anxa5 | annexin A5 | 1.10 | ||
| 83839 | Calb1 | calbindin 1 | Alc4 | 1.16 | |
| 24242 | Calm1 | calmodulin 1 | Alc17 | 1.07 | |
| 25420 | Cryab | crystallin, alpha B | 1.32 | CPU (1.35) | |
| 83633 | Gpr51 | G protein-coupled receptor 51 | Alc4 | 1.12 | |
| 155918 | LCPU2 | lymphocyte cytosolic protein 2 | Alc9 & 12 | AMYG (1.18) | |
| 59322 | Cnksr2 | membrane-associated guanylate kinase-interacting protein | 1.08 | ||
| 58840 | Mapk6 | mitogen-activated protein kinase 6 | 1.08 | ||
| 24605 | Nras | neuroblastoma ras oncogene | 1.07 | ||
| 84587 | Plcl1 | phospholipase C-like 1 | 1.14 | ||
| 54705 | Ppm2c | protein phosphatase 2C, magnesium dependent, catalytic subunitc | Alc4 | 1.13 | |
| 29645 | Ptprj | protein tyrosine phosphatase, receptor type, J | 1.28 | ||
| 64373 | Rhob | rhoB gene | Alc17 | 1.10 | |
iP = inbred alcohol-preferring rat; iNP = inbred alcoholenon-preferring rat.
Defined as categorized by a Gene Ontology category (small GTPase mediated signal transduction, negative regulation of protein kinase activity, negative regulation of enzyme activity, transmembrane receptor protein tyrosine kinase signaling pathway) or were found through manual curation to have an intracellular messaging-related function (Anxa5, Calb1, Calm1, Dusp5, Dusp6, Cnksr2, Mapk6, Nf1, Plcl1, Phkg1, Prkce, Ppm2c, Ppm2c, Spin2b, Stk38, Syngap1).
ACB = nucleus accumbens, AMYG = amygdala, FC = frontal cortex, HIPP = hippocampus, CPU = caudate-putamen.
Gene represented by more than one significant probe set. Largest of the multiple fold changes is reported in column 5.
Table 11.
Transcription-related significant genesa
| Entrez gene | Gene symbol | Gene name | QTL | Combined regions fold (iP vs. iNP) | Individual regionb significant in (fold) |
|---|---|---|---|---|---|
| Decreased expression (iP vs. iNP) | |||||
| 24388 | Gfi1 | growth factor independent 1 | -1.09 | ||
| 29577 | Hes1 | hairy and enhancer of split 1 (Drosophila) | -1.08 | ||
| 84017 | Hmga2 | high mobility group AT-hook 2 | -1.07 | ||
| 292892 | Irf3 | interferon regulatory factor 3 | -1.06 | ||
| 246324 | Rab31 | RAB31, member RAS oncogene family | -1.18 | ||
| 81813 | Tieg | TGFB inducible early growth response | -1.07 | ||
| 84382 | Tcf4c | transcription factor 4 | -1.20 | ||
| 498407 | pur-beta | transcription factor Pur-beta | -1.16 | ||
| Increased expression (iP vs. iNP) | |||||
| 25620 | Crem | cAMP responsive element modulator | 1.07 | ||
| 362201 | Ccndbp1 | cyclin D-type binding-protein 1 | 1.11 | ||
| 300721 | Dnaja4 | DnaJ (Hsp40) homolog, subfamily A, member 4 | 1.10 | ||
| 27080 | Hbp1 | high mobility group box transcription factor 1 | Alc17 | 1.07 | |
| 29560 | Hif1a | hypoxia inducible factor 1, alpha subunit | Alc17 | 1.1 | |
| 29458 | Neurod1 | neurogenic differentiation 1 | 1.14 | ||
| 81830 | Rab11a | RAB11a, member RAS oncogene family | 1.12 | ||
| 304268 | Rasl11a | RAS-like family 11 member A | 1.13 | ||
| 64635 | Snd1 | staphylococcal nuclease domain containing 1 | 1.13 | ||
| 171152 | Taf9l | TAF9-like RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa | 1.14 | ||
| 24874 | Vhl | von Hippel-Lindau syndrome homolog | Alc14 & 16 | 1.10 | |
| 81515 | Lyn | Yamaguchi sarcoma viral (v-yes-1) oncogene homolog | Alc4 | 1.10 | |
| 25165 | Znf386 | zinc finger protein 386 (Kruppel-like) | Alc17 | 1.15 | CPU (1.22) |
iP = inbred alcohol-preferring rat; iNP = inbred alcoholenon-preferring rat.
Defined as transcription factors by Genomatix Suite (Crem, Gfi1, Hbp1, Hif1a, Irf3, Klf10, Neurod1, Taf9l, Tcf4) or found through manual curation to have functions or putative functions related to the regulation of transcription.
ACB = nucleus accumbens, AMYG = amygdala, FC = frontal cortex, HIPP = hippocampus, CPU = caudate-putamen.
Gene represented by more than one significant probe set. Largest of the multiple fold changes is reported in column 5.
3.1. qRT-PCR confirmation
qRT-PCR measurements were conducted on genes to verify differences observed with microarray hybridization. Genes were selected on the basis of significant differential expression in at least one brain region and reasonable fold changes (Table 12). GSTa4 and Scn1a are not differentially expressed using the RMA analysis presented here, but were with an earlier analysis using MAS5 (unpublished observation). The direction of the expression differences using qRT-PCR was the same as in the microarray data for all genes and regions tested. For the Akap11 gene, the magnitude of the change of the qRT-PCR data for all five regions was far greater than that observed in the microarray data (fold change in CPU was 785, and in HIPP 1,900), which probably reflects technical differences between the two assays. Expression levels of Akap11 in iNP were near back-ground levels in the array experiment; qRT-PCR assays allow one to measure much lower levels of RNA with more accuracy in that range (background is much lower), which allows one to better estimate the fold change.
Table 12.
Quantitative RT-PCR confirmation of microarray fold change (iP vs. iNP) data
| Frontal cortex |
Caudate-putamen |
Nucleus accumbens |
||||
|---|---|---|---|---|---|---|
| Gene name | Microarray | qRT-PCR | Microarray | qRT-PCR | Microarray | qRT-PCR |
| Akap11 | 1.04 | 1053a | 1.23 | 786a | 1.5b | 386a |
| Scn1a | 1.03 | 1.19 | 1.23 | 1.54 | 1.07 | 1.16 |
| Fth1 | 1.30 | 1.12 | 1.22 | 1.20a | 1.27 | 1.14 |
| GSTa4 | -1.75b | -1.99a | -2.00b | -2.13a | -1.52b | -2.94a |
| Scn3a | -1.39b | -1.62a | -1.46b | -1.47 | -1.38b | -1.04 |
iP = inbred alcohol-preferring rat; iNP = inbred alcoholenon-preferring rat; qRT-PCR = real-time PCR; FDR = false discovery rate.
Statistically significant (P <.05).
Statistically significant (FDR < 0.10).
4. Discussion
The major findings of this study were that (1) there was a greater degree of between-region differential expression than within-region, between-strain differential expression (Fig. 1); (2) there was within-region, between-strain differential expression in all five regions examined, with many of these genes classifiable as being related to neurotransmission (Table 8), neuroplasticity (Table 9), intracellular messaging (Table 10), and regulation of transcription (Table 11), with the AMYG as the region demonstrating the greatest number of genes differentially expressed; (3) considerable statistical power could be obtained by minimizing the effect of variance by examining the main effect of strain in a linear model that contained all five brain regions and that with this approach over 300 genes demonstrated differential expression; and (4) in this main effect of strain model, the significant genes represented 17 over-represented GO categories, for example, axon guidance, gliogenesis, negative regulation of programmed cell death, regulation of programmed cell death, regulation of synapse structure function, and transmission of nerve impulse (Table 6).
Inbred P and NP rats were used in the present study in an attempt to reduce the biological variability in the gene expression data. However, there are two potential weaknesses with this approach, in that the particular inbred strains used in the present study may not necessarily have the same characteristics as the parent foundation lines of selectively bred P and NP rats; and, in that inbreeding randomly fixes genes that are not relevant to the phenotype of alcohol preference, which can result in some irrelevant differences in gene expression levels. Although preliminary findings indicate that similar differences are found for the inbred strains and parent lines in sweet preference, anxiety, spontaneous motor activity, and the development of rapid tolerance (Stewart et al., 2004), as well as alcohol drinking (Bell et al., 2004), extrapolating the results to the selectively bred lines should be done cautiously.
4.1. Within-region analysis
Principal components analysis (Fig. 1) revealed that the first two principal components resolve the differences between arrays based on brain region, but not on strain, indicating that most of the differences in gene expression between arrays in the present experiment are accounted for by brain region and not by strain. This finding is in agreement with observations made by others that regional differences in gene expression in the adult mouse brain are greatly influenced by embryological origins (Zapala et al., 2005) and that, across a range of behavioral phenotypes, the number of between-brain region differences of expression can be greater than within-region differences (Nadler et al., 2006; Pavlidis & Noble, 2001).
The AMYG had the greatest number of differences in gene expression between the iP and iNP strains (Tables 1 and 2), which may be a result of the multiple nuclei within this region that mediate different behavioral functions. Of the 54 differentially expressed genes in this region, the biologic process most represented is neuroplasticity, in which 8 genes have been implicated (Table 9). Co-citation analysis indicated a relationship between two pairs of these genes. Fgfr1 and Pak2 both show lower expression in the iP rat than iNP rat, and are involved in the Ras/ERK/PAK2 pathway that governs neuronal differentiation (Shin et al., 2002). Neuritin and attractin, both of which show higher expression in the iP than iNP rat, have been identified as part of a cluster of genes that are up-regulated during the recovery phase of spinal cord injury (Di Giovanni et al., 2004). A 36% reduction in expression of Phgdh in the AMYG of the iP rat compared to the iNP rat has the potential to alter cell survival, neuritogenesis, and voltage propagation (Furuya et al., 2000) in this region. The expression of sodium channel, voltage-gated, type III, alpha polypeptide (Scn3a) was significantly lower in the AMYG of the iP than iNP line, and was generally lower in the other regions of the iP rats. Widespread differences in the expression of Scn3a between the iP and iNP lines could contribute to differences in the overall neuronal excitability in these animals (Hains et al., 2003) and potentially alter responses of the two lines to alcohol.
Thirteen of the 54 differentially expressed genes in the AMYG were located within established alcohol QTLs (Bice et al., 1998; Carr et al., 1998, 2003; Foroud et al., 2002, 2003; Radcliffe et al., 2004; Terenina-Rigaldie et al., 2003) (Table 5). The pathway involving caspase 3 (lower expression in iP rats) and interleukin-1 receptor-associated kinase 2 (higher expression in iP rats) is involved in the age-related decline in function of hippocampal cells (Lynch & Lynch, 2002). Amphoretin-induced gene and ORF (Amigo), higher in the AMYG of the iP than iNP rat, is a transmembrane protein that modulates neurite out-growth (Kuja-Panula et al., 2003), and thus has the potential to mediate neuroplastic responses to alcohol.
The CPU demonstrated the second highest number of differentially expressed genes (Table 1). Src homology 2 domain-containing transforming protein C3 (Shc3) had lower expression in the iP than in the iNP. Although classified by GO as having a function in neurotransmission (Table 6), Shc3 is perhaps more appropriately thought of as playing a role in the regulation of neuronal development and synaptic function (Liu & Meakin, 2002). In the CPU, 8 of the 24 differentially expressed genes were located within established alcohol QTLs (Bice et al., 1998; Carr et al., 1998, 2003; Foroud et al., 2002, 2003; Radcliffe et al., 2004; Terenina-Rigaldie et al., 2003) (Table 5). Among these 8 genes, several are involved in protein formation, that is, ribosomal protein S2 heat shock protein 2, and zinc finger protein 386 (Kruppel-like). Some of the differences in the CPU between the iP and iNP rats may reflect differences in response to the low-dose stimulating (Rodd et al., 2004; Waller et al., 1986) and high-dose motor impairing (Lumeng et al., 1982) effects of alcohol exhibited by the parent lines.
The HIPP demonstrated significant differential expression of 21 genes (Table 1). Slit homolog 1 (Drosophila), higher in the iP line, has been implicated in processes related to neuroplasticity (Table 9), specifically in axon guidance (Chang et al., 2006; Long et al., 2004; Thompson et al., 2006). One of the 21 differentially expressed genes in the HIPP, ribosomal protein S2, was located within established alcohol QTLs (Bice et al., 1998; Carr et al., 1998, 2003; Foroud et al., 2002, 2003; Radcliffe et al., 2004; Terenina-Rigaldie et al., 2003) (Table 5). Some of these differences in the HIPP between iP and iNP rats may be associated with the differences in the development and/or persistence of tolerance to the motor impairing effects of alcohol observed for the parent lines (Bell et al., 2001; Gatto et al., 1987a, 1987b; Waller et al., 1983).
In a previous study that jointly analyzed the HIPP data here and data from different inbred iP and iNP strains, many more differences were observed in the HIPP between iP and iNP rats, most likely due to the higher power of that study (a total of 10 arrays used for each group) (Edenberg et al., 2005). Nine of the 21 genes reported here (indicated in Table 2 with footnote “b”) were found in the previous study; the absence of a greater overlap is likely due to differences in the study design discussed above, differences in the preprocessing of data (MAS5 was used in the previous study, whereas RMA was used in the present study), and the unavoidable fact that the true FDR of the two studies, while estimated to be low, was almost certainly not zero.
The ACB demonstrated significant differential expression of 16 genes (Table 1). Catalase, expressed higher in the iP line, is the major enzyme metabolizing ethanol to acetaldehyde in the CNS (Aragon et al., 1991, 1992; Cohen et al., 1980; Zimatkin and Lindros, 1996). Gill et al. (1996) have also reported higher catalase activity in the CNS of P rats compared to NP rats. Two of the 15 differentially expressed genes in the ACB were located within established alcohol QTLs (Bice et al., 1998; Carr et al., 1998, 2003; Foroud et al., 2002, 2003; Radcliffe et al., 2004; Terenina-Rigaldie et al., 2003), that is, ribosomal protein S2 and proteasome (prosome, macropain) subunit, alpha type 5 (Table 5). Some of the differences in the ACB could be related to the disparate alcohol drinking characteristics of the iP and iNP strains.
The FC demonstrated significant differential expression of eight genes (Table 1). The one gene that is differentially expressed in the FC but in no other region is glial fibrillary acidic protein (Gfap). This gene is abundantly expressed in glial cells of the central nervous system and can be used as a marker for glial cell density and health. It is decreased in the FC of human adult alcoholics and more so in alcoholics with depressive symptoms (Miguel-Hidalgo et al., 2002). Humans with a history of chronic alcoholism have been found to have substantial glial pathology in the FC irrespective of current alcohol use or liver pathology (Cullen & Halliday, 1994). It is possible that higher Gfap expression in the FC of the iP line confers protective benefits. The FC has only two significant genes that map to established alcohol QTLs (Bice et al., 1998; Carr et al., 1998, 2003; Foroud et al., 2002, 2003; Radcliffe et al., 2004; Terenina-Rigaldie et al., 2003) (Table 5), ribosomal protein S2 and proteasome (prosome, macropain) subunit, alpha type 5.
4.2. Overall strain differences
Within the 296 genes found significant in the overall analysis (Table 4), 17 GO categories were over-represented (P <.05, Table 7). Six categories associated with neuronal function included axon guidance, gliogenesis, negative regulation of programmed cell death, regulation of programmed cell death, regulation of synapse structure function, and transmission of nerve impulse (Table 6).
To guide determination of relationships between and within genes of the significant GO categories listed in Tables 8-11, a co-citation network graph was constructed. Of the 63 genes present in the combined lists (Tables 8-11), 24 genes are present in a co-citation graph with a threshold for inclusion of four co-citations per link (Fig. 2). The expression of Cdkn1a and Bcl2l1 was lower in the iP than iNP strain; both are involved in regulation of apoptosis, suggesting the reduced expression of these genes may have a positive impact on neuronal function. Ntf3 and Ntf5 code for proteins classified as neurotrophic factors, that is, they function to promote both neuroplasticity and neuro-protection (Filus & Rybakowski, 2005). There is evidence that these two neurotrophic factors might be involved in maintaining functional tolerance to ethanol (Szabo & Hoffman, 1995).
Fig. 2.
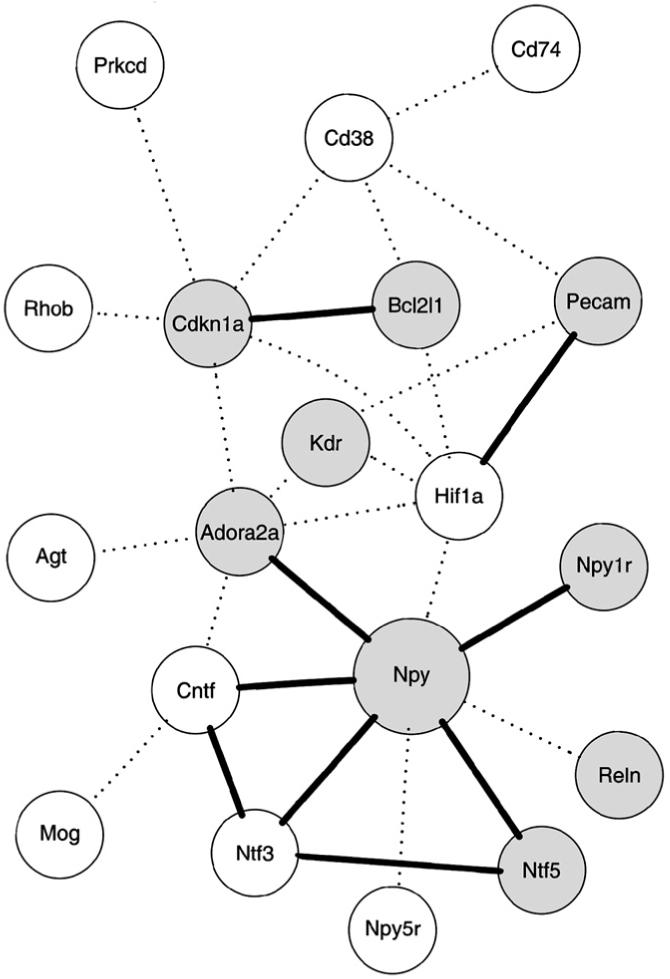
Co-citation network graph constructed with modified BioConductor software and manual curation (see Methods for details). Grey filled circles denote genes that have decreased expression in the inbred alcohol-preferring rat, unfilled circles denote genes that have increased expression. Those links supported by 16 or greater co-citations are depicted with bold solid lines, whereas those with less than 16 are depicted with dotted lines. The circle for Npy has been enlarged for emphasis. Genes Slit1, Slit3, C1s, B2m, and Il18 also met the minimum criteria but were only peripherally related or not related to the interactions shown in the figure. Therefore, for the sake of clarity, these genes were not included.
Cntf codes for a neurotrophic factor; however, unlike the Ntfs, this factor has been shown to play a positive role in myelinogenesis (Stankoff et al., 2002). Cntf and Ntf3 have both been shown to play a positive role in psychostimulant-induced behavioral sensitization via their participation in DA-mediated neuronal plasticity (Pierce & Bari, 2001). The higher expression of both of these factors in the iP than iNP strain could be a factor contributing to the reinforcing properties of ethanol observed for the P but not NP line (Gatto et al., 1994). The differences in expression of Ntf and Cntf between iP and iNP rats might indicate that the neuronal system in general might be more geared toward undergoing positive neuronal changes in response to environmental manipulations.
The lower expression of Npy in the iP than iNP strain may be the result of the combined effects of higher expression of Cnft and lower expression of Ntf5 in the iP rats. Cnft has been shown to decrease the expression of Npy (Rao et al., 1992) and the Cnft analogue CNTF (A×15) has been shown to exert a delayed effect in suppressing the expression of Npy in calorie restricted, diet-induced, obese rats (Bluher et al., 2004). Ntf5, but not Ntf3 has been shown to stimulate the expression of Npy in the rat neocortex (Wirth et al., 1998). The findings indicating lower expression of Npy in the iP than iNP rats are in general agreement with reports on tissue levels of NPY reported for the selectively bred P and NP lines (Ehlers et al., 1998; Hwang et al., 2004). A brief survey of other genes connected to Npy reveals that the products of Reln and Npy5r also interact with Npy. Reln, secreted by GABAergic interneurons, is involved in the control of neuronal migration and Npy immunopositive cells have Reln receptors (Pesold et al., 1999). A decrease in the expression of Reln in the iP line is consistent with the decreased expression of Npy in this line. The higher expression of Npy5r in the iP than iNP strain may reflect a compensatory response of having lower expression levels of Npy. The Npy5r antagonist L-152,804 has recently been shown to reduce ethanol self-administration and reinforcement in the iP rat (Schroeder et al., 2003, 2005). The higher expression of Npy5r may be a factor contributing to the high alcohol drinking behavior of the iP rats.
A review of Table 8 indicates that the metabotropic glutamate, glycine, and GABA receptors may also have roles in contributing to the disparate alcohol drinking behaviors of the iP and iNP rats. Table 9 lists many genes that may mediate differences in neuronal development and neuroplasticity between the iP and iNP strains, for example, neurexin 3 is an axonal receptor of neuroligan and this complex can control excitatory and inhibitory synapse formation (Dean et al., 2003; Hussain & Sheng, 2005). Table 10 lists many genes that are different between the strains and may play pivotal roles in intracellular messaging, for example, the expression of Prkce (protein kinase C, epsilon) is almost 30% lower in the iP than iNP strain. This relationship between alcohol drinking and Prkce levels is opposite to that found in Prkce knockout mice, which demonstrated decreased preference for ethanol and increased sensitivity to the behavioral effects of ethanol (Choi et al., 2002; Hodge et al., 1999). These apparently contradictory findings between the inbred rat strains and the mutant mice may be due to species differences and/or to compensatory changes in the knockout mouse. Finally, Table 11 lists transcription factors that were different between the iP and iNP line in the overall analysis. Too little is known about the role of transcription factors in the development of specific neural structures and cell types to speculate on the effects of these differences.
Among the top 20 or so candidates identified in the convergent functional genomics analysis of Rodd et al. (2006), there were seven genes (Fn1, Syn2, Agt, Lyz, Nrn1, Aldh1a1, and Prkce) that were also identified in the overall analysis in the present study as being significantly different between iP and iNP rats (Table 4). These genes should be considered as potential candidates contributing to high alcohol drinking behavior.
In summary, through careful analysis of differential expression of the genome of the iP and iNP rat strains, we have seen that the biggest difference is not between lines, but between regions. We have also seen that, at least in the alcoholnaïve state, the AMYG demonstrates the greatest number of differences in gene expression of the five regions examined, although all five regions demonstrate at least some genes with significant differential expression (FDR < 0.10). Taken together, the individual region and combined region analyses indicate that the expressions of genes involved in biologic networks of neurotransmitters, intracellular messengers, neuroplastins, neurotrophins, and transcription factors may all contribute to behavioral and neurobiological differences between the iP and iNP rats and their parent stocks. A network involving Npy and related genes has the most supporting evidence from both the data of this experiment and a co-citation bioinformatic analysis.
Acknowledgments
This study was supported in part by AA07611, AA07462, AA13521 [INIA], and AA16652 [INIA]. Microarrays were analyzed using the facilities of the Center for Medical Genomics at Indiana University School of Medicine, which is supported in part by the Indiana Genomics Initiative (INGEN®) of Indiana University. INGEN is supported in part by the Lilly Endowment Inc.
References
- Aragon CM, Rogan F, Amit Z. Ethanol metabolism in rat brain homogenates by a catalase-H2O2 system. Biochem Pharmacol. 1992;44(1):93–98. doi: 10.1016/0006-2952(92)90042-h. [DOI] [PubMed] [Google Scholar]
- Aragon CM, Stotland LM, Amit Z. Studies on ethanol-brain catalase interaction: evidence for central ethanol oxidation. Alcohol Clin Exp Res. 1991;15(2):165–169. doi: 10.1111/j.1530-0277.1991.tb01848.x. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, Suzek TO, Troup DB, Wilhite SE, Ngau WC, Ledoux P, Rudnev D, Lash AE, Fujibuchi W, Edgar R. NCBI GEO: mining millions of expression profiles—database and tools. Nucleic Acids Res. 2005;33:D562–D566. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell RL, Rodd ZA, Boutwell CL, Murphy JM, Lumeng L, Li T-K, McBride WJ. Repeated daily scheduled access results in binge-like ethanol consumption in adult inbred P rats. [Abstract] Alcohol Clin Exp Res. 2004;28(Suppl):143A. [Google Scholar]
- Bell RL, Stewart RB, Woods JE, 2nd, Lumeng L, Li TK, Murphy JM, McBride WJ. Responsivity and development of tolerance to the motor impairing effects of moderate doses of ethanol in alcohol-preferring (P) and -nonpreferring (NP) rat lines. Alcohol Clin Exp Res. 2001;25(5):644–650. [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc(B) 1995;57:289–290. [Google Scholar]
- Bice P, Foroud T, Bo R, Castelluccio P, Lumeng L, Li TK, Carr LG. Genomic screen for QTLs underlying alcohol consumption in the P and NP rat lines. Mamm Genome. 1998;9:949–955. doi: 10.1007/s003359900905. [DOI] [PubMed] [Google Scholar]
- Bluher S, Moschos S, Bullen J, Jr., Kokkotou E, Maratos-Flier E, Wiegand SJ, Sleeman MW, Mantzoros CS. Ciliary neurotrophic factorA×15 alters energy homeostasis, decreases body weight, and improves metabolic control in diet-induced obese and UCP1-DTA mice. Diabetes. 2004;53:2787–2796. doi: 10.2337/diabetes.53.11.2787. [DOI] [PubMed] [Google Scholar]
- Bonci A, Bernardi G, Grillner P, Mercuri NB. The dopamine-containing neuron: maestro or simple musician in the orchestra of addiction? Trends Pharmacol Sci. 2003;24:172–177. doi: 10.1016/S0165-6147(03)00068-3. [DOI] [PubMed] [Google Scholar]
- Carr LG, Foroud T, Bice P, Gobbett T, Ivashina J, Edenberg H, Lumeng L, Li TK. A quantitative trait locus for alcohol consumption in selectively bred rat lines. Alcohol Clin Exp Res. 1998;22:884–887. [PubMed] [Google Scholar]
- Carr LG, Habegger K, Spence J, Ritchotte A, Liu L, Lumeng L, Li TK, Foroud T. Analyses of quantitative trait loci contributing to alcohol preference in HAD1/LAD1 and HAD2/LAD2 rats. Alcohol Clin Exp Res. 2003;27:1710–1717. doi: 10.1097/01.ALC.0000097161.51093.71. [DOI] [PubMed] [Google Scholar]
- Chang C, Adler CE, Krause M, Clark SG, Gertler FB, Tessier-Lavigne M, Bargmann CI. MIG-10/lamellipodin and AGE-1/PI3K promote axon guidance and outgrowth in response to slit and netrin. Curr Biol. 2006;16:854–862. doi: 10.1016/j.cub.2006.03.083. [DOI] [PubMed] [Google Scholar]
- Choi DS, Wang D, Dadgar J, Chang WS, Messing RO. Conditional rescue of protein kinase C epsilon regulates ethanol preference and hypnotic sensitivity in adult mice. J Neurosci. 2002;22:9905–9911. doi: 10.1523/JNEUROSCI.22-22-09905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicero TJ. A critique of animal analogues of alcoholism. In: Majchrowicz E, Noble EP, editors. Biochemistry and Pharmacology of Alcohol. Vol. 2. Plenium Press; New York: 1979. pp. 533–560. [Google Scholar]
- Cohen G, Sinet PM, Heikkila R. Ethanol oxidation by rat brain in vivo. Alcohol Clin Exp Res. 1980;4(4):366–370. doi: 10.1111/j.1530-0277.1980.tb04833.x. [DOI] [PubMed] [Google Scholar]
- Cullen KM, Halliday GM. Chronic alcoholics have substantial glial pathology in the forebrain and diencephalon. Alcohol Alcohol. 1994;(Suppl 2):253–257. [PubMed] [Google Scholar]
- Dean C, Scholl FG, Choih J, DeMaria S, Berger J, Isacoff E, Scheiffele P. Neurexin mediates the assembly of presynaptic terminals. Nat Neurosci. 2003;6:708–716. doi: 10.1038/nn1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giovanni S, Faden AI, Yakovlev A, Duke-Cohan JS, Finn T, Thouin M, Knoblach S, De Biase A, Bregman BS, Hoffman EP. Neuronal plasticity after spinal cord injury: identification of a gene cluster driving neurite outgrowth. FASEB J. 2004;19(1):153–154. doi: 10.1096/fj.04-2694fje. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Strother WN, McClintick JN, Tian H, Stephans M, Jerome RE, Lumeng L, Li T-K, McBride WJ. Gene expression in the hippocampus of inbred alcohol-preferring and -non-preferring rats. Genes Brain Behav. 2005;4:20–30. doi: 10.1111/j.1601-183X.2004.00091.x. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene expression omni-bus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers CL, Chaplin RI, Wall TL, Lumeng L, Li TK, Owens MJ, Nemeroff CB. Corticotropin releasing factor (CRF): studies in alcohol preferring and non-preferring rats. Psycho-pharmacology (Berl) 1992;106:359–364. doi: 10.1007/BF02245418. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Li TK, Lumeng L, Hwang BH, Somes C, Jimenez P, Mathe AA. Neuropeptide Y levels in ethanol-naive alcohol-preferring and nonpreferring rats and in Wistar rats after ethanol exposure. Alcohol Clin Exp Res. 1998;22:1778–1782. [PubMed] [Google Scholar]
- Ehlers CL, Somes C, Li TK, Lumeng L, Kinkead B, Owens MJ, Nemeroff CB. Neurontensin studies in alcohol naive, preferring and non-preferring rats. Neuroscience. 1999;93:227–236. doi: 10.1016/s0306-4522(99)00113-x. [DOI] [PubMed] [Google Scholar]
- Filus JF, Rybakowski J. Neurotrophic factors and their role in the pathogenesis of affective disorders. Psychiatr Pol. 2005;39:883–897. [PubMed] [Google Scholar]
- Foroud T, Bice P, Castelluccio P, Bo R, Ritchotte A, Stewart R, Lumeng L, Li TK, Carr L. Mapping of QTL influencing saccharin consumption in the selectively bred alcohol-preferring and -nonpreferring rat lines. Behav Genet. 2002;32:57–67. doi: 10.1023/a:1014459912935. [DOI] [PubMed] [Google Scholar]
- Foroud T, Ritchotte A, Spence J, Liu L, Lumeng L, Li TK, Carr LG. Confirmation of alcohol preference quantitative trait loci in the replicate high alcohol drinking and low alcohol drinking rat lines. Psychiatr Genet. 2003;13:155–161. doi: 10.1097/00041444-200309000-00004. [DOI] [PubMed] [Google Scholar]
- Furuya S, Tabata T, Mitoma J, Yamada K, Yamasaki M, Makino A, Yamamoto T, Watanabe M, Kano M, Hirabayashi Y. L-serine and glycine serve as major astroglia-derived trophic factors for cerebellar Purkinje neurons. Proc Natl Acad Sci U S A. 2000;97:11528–11533. doi: 10.1073/pnas.200364497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatto GJ, McBride WJ, Murphy JM, Lumeng L, Li TK. Ethanol self-infusion into the ventral tegmental area by alcohol-preferring rats. Alcohol. 1994;11:557–564. doi: 10.1016/0741-8329(94)90083-3. [DOI] [PubMed] [Google Scholar]
- Gatto GJ, Murphy JM, Waller MB, McBride WJ, Lumeng L, Li TK. Chronic ethanol tolerance through free-choice drinking in the P line of alcohol-preferring rats. Pharmacol Biochem Behav. 1987a;28:111–115. doi: 10.1016/0091-3057(87)90021-9. [DOI] [PubMed] [Google Scholar]
- Gatto GJ, Murphy JM, Waller MB, McBride WJ, Lumeng L, Li TK. Persistence of tolerance to a single dose of ethanol in the selectively-bred alcohol-preferring P rat. Pharmacol Biochem Behav. 1987b;28:105–110. doi: 10.1016/0091-3057(87)90020-7. [DOI] [PubMed] [Google Scholar]
- Gentleman R. Using GO for statistical analyses; Proceedings of COMPSTAT 2004, Symposium; 2004.pp. 171–180. [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bio-conductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry J. R package version 1.12.1. 2006. Rgraphviz: provides plotting capabilities for R graph objects. [Google Scholar]
- Gill K, Amit Z, Smith BR. The regulation of alcohol consumption in rats: the role of alcohol-metabolizing enzymes-catalase and aldehyde dehydrogenase. Alcohol. 1996;13:347–353. doi: 10.1016/0741-8329(96)00006-7. [DOI] [PubMed] [Google Scholar]
- Hains BC, Klein JP, Saab CY, Craner MJ, Black JA, Waxman SG. Upregulation of sodium channel Nav1.3 and functional involvement in neuronal hyperexcitability associated with central neuropathic pain after spinal cord injury. J Neurosci. 2003;23:8881–8892. doi: 10.1523/JNEUROSCI.23-26-08881.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, Richter J, Rubin GM, Blake JA, Bult C, Dolan M, Drabkin H, Eppig JT, Hill DP, Ni L, Ringwald M, Balakrishnan R, Cherry JM, Christie KR, Costanzo MC, Dwight SS, Engel S, Fisk DG, Hirschman JE, Hong EL, Nash RS, Sethuraman A, Theesfeld CL, Botstein D, Dolinski K, Feierbach B, Berardini T, Mundodi S, Rhee SY, Apweiler R, Barrell D, Camon E, Dimmer E, Lee V, Chisholm R, Gaudet P, Kibbe W, Kishore R, Schwarz EM, Sternberg P, Gwinn M, Hannick L, Wortman J, Berriman M, Wood V, de laCruz N, Tonellato P, Jaiswal P, Seigfried T, White R. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge CW, Mehmert KK, Kelley SP, McMahon T, Haywood A, Olive MF, Wang D, Sanchez-Perez AM, Messing RO. Supersensitivity to allosteric GABA(A) receptor modulators and alcohol in mice lacking PKCepsilon. Nat Neurosci. 1999;2:997–1002. doi: 10.1038/14795. [DOI] [PubMed] [Google Scholar]
- Hussain NK, Sheng M. Neuroscience. Making synapses: a balancing act. Science. 2005;307:1207–1208. doi: 10.1126/science.1110011. [DOI] [PubMed] [Google Scholar]
- Hwang BH, Suzuki R, Lumeng L, Li TK, McBride WJ. Innate differences in neuropeptide Y (NPY) mRNA expression in discrete brain regions between alcohol-preferring (P) and -nonpreferring (NP) rats: a significantly low level of NPY mRNA in dentate gyrus of the hippocampus and absence of NPY mRNA in the medial habenular nucleus of P rats. Neuropeptides. 2004;38:359–368. doi: 10.1016/j.npep.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Institute of Laboratory Animal Resources. Commission on Life Sciences. National Research Council . Guide for the Care and Use of Laboratory Animals. National Academy Press; Washington, DC: 1996. [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Kerns RT, Ravindranathan A, Hassan S, Cage MP, York T, Sikela JM, Williams RW, Miles MF. Ethanol-responsive brain region expression networks: implications for behavioral responses to acute ethanol in DBA/2J versus C57BL/6J mice. J Neurosci. 2005;25:2255–2266. doi: 10.1523/JNEUROSCI.4372-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuja-Panula J, Kiiltomaki M, Yamashiro T, Rouhiainen A, Rauvala H. AMIGO, a transmembrane protein implicated in axon tract development, defines a novel protein family with leucinerich repeats. J Cell Biol. 2003;160:963–973. doi: 10.1083/jcb.200209074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang T, Spence J, Liu L, Strother WN, Chang HW, Ellison JA, Lumeng L, Li TK, Foroud T, Carr LG. Alpha-synuclein maps to a quantitative trait locus for alcohol preference and is differentially expressed in alcohol-preferring and -nonpreferring rats. Proc Natl Acad Sci U S A. 2003;100:4690–4695. doi: 10.1073/pnas.0737182100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SM, McConnell P, Johnson KF, Shoemaker J. MedlineR: an open source library in R for Medline literature data mining. Bioinformatics. 2004;20:3659–3661. doi: 10.1093/bioinformatics/bth404. [DOI] [PubMed] [Google Scholar]
- Liu H-Y, Meakin SO. ShcB and ShcC activation by the Trk family of receptor tyrosine kinases. J Biol Chem. 2002;277:26046–26056. doi: 10.1074/jbc.M111659200. [DOI] [PubMed] [Google Scholar]
- Long H, Sabatier C, Ma L, Plump A, Yuan W, Ornitz DM, Tamada A, Murakami F, Goodman CS, Tessier-Lavigne M. Conserved roles for Slit and Robo proteins in midline commissural axon guidance. Neuron. 2004;42:213–223. doi: 10.1016/s0896-6273(04)00179-5. [DOI] [PubMed] [Google Scholar]
- Lumeng L, Hawkins DT, Li T-K. New strains of rats with alcohol preference and nonpreference. In: Thurman RG, Williamson JR, Drott H, Chance B, editors. Alcohol and Aldehyde Metabolizing Systems. Vol. 3. Academic Press; New York: 1977. pp. 537–544. [Google Scholar]
- Lumeng L, Waller MB, McBride WJ, Li TK. Different sensitivities to ethanol in alcohol-preferring and -nonpreferring rats. Pharmacol Biochem Behav. 1982;16:125–130. doi: 10.1016/0091-3057(82)90023-5. [DOI] [PubMed] [Google Scholar]
- Lynch AM, Lynch MA. The age-related increase in IL-1 type I receptor in rat hippocampus is coupled with an increase in caspase-3 activation. Eur J Neurosci. 2002;15:1779–1788. doi: 10.1046/j.1460-9568.2002.02012.x. [DOI] [PubMed] [Google Scholar]
- Maldonado R. The neurobiology of addiction. J Neural Transm Suppl. 2003;66:1–14. doi: 10.1007/978-3-7091-0541-2_1. [DOI] [PubMed] [Google Scholar]
- McBride WJ, Chernet E, Dyr W, Lumeng L, Li T-K. Densities of dopamine D2 receptors are reduced in CNS regions of alcohol-preferring P rats. Alcohol. 1993a;10:387–390. doi: 10.1016/0741-8329(93)90025-j. [DOI] [PubMed] [Google Scholar]
- McBride WJ, Chernet E, McKinzie DL, Lumeng L, Li TK. Quantitative autoradiography of mu-opioid receptors in the CNS of alcohol-naive alcohol-preferring P and -nonpreferring NP rats. Alcohol. 1998;16:317–323. doi: 10.1016/s0741-8329(98)00021-4. [DOI] [PubMed] [Google Scholar]
- McBride WJ, Chernet E, Russell RN, Wong DT, Guan XM, Lumeng L, Li TK. Regional CNS densities of monoamine receptors in alcohol-naive alcohol-preferring P and -nonpreferring NP rats. Alcohol. 1997;14:141–148. doi: 10.1016/s0741-8329(96)00117-6. [DOI] [PubMed] [Google Scholar]
- McBride WJ, Guan X-M, Chernet E, Lumeng L, Li T-K. Regional serotonin-1A receptors in the CNS of alcohol-preferring and -nonpreferring rats. Pharmacol Biochem Behav. 1994;49:7–12. doi: 10.1016/0091-3057(94)90449-9. [DOI] [PubMed] [Google Scholar]
- McBride WJ, Li T-K. Animal models of alcoholism: neurobiology of high alcohol-drinking behavior in rodents. Crit Rev Neurobiol. 1998;12:339–369. doi: 10.1615/critrevneurobiol.v12.i4.40. [DOI] [PubMed] [Google Scholar]
- McBride WJ, Murphy JM, Gatto GJ, Levy AD, Yoshimoto K, Lumeng L, Li T-K. CNS mechanisms of alcohol self-administration. Alcohol Alcohol. 1993b;2:463–467. [PubMed] [Google Scholar]
- Miguel-Hidalgo JJ, Wei J, Andrew M, Overholser JC, Jurjus G, Stockmeier CA, Rajkowska G. Glia pathology in the pre-frontal cortex in alcohol dependence with and without depressive symptoms. Biol Psychiatry. 2002;52:1121–1133. doi: 10.1016/s0006-3223(02)01439-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan MK, Ponomarev I, Hitzemann RJ, Belknap JK, Tabakoff B, Harris RA, Crabbe JC, Blednov YA, Grahame NJ, Phillips TJ, Finn DA, Hoffman PL, Iyer VR, Koob GF, Bergeson SE. Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proc Natl Acad Sci U S A. 2006;103:6368–6373. doi: 10.1073/pnas.0510188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy JM, Stewart RB, Bell RL, Badia-Elder NE, Carr LG, McBride WJ, Lumeng L, Li T-K. Phenotypic and genotypic characterization of the Indiana University rat lines selectively bred for high and low alcohol preference. Behav Genet. 2002;32:363–388. doi: 10.1023/a:1020266306135. [DOI] [PubMed] [Google Scholar]
- Nadler JJ, Zou F, Huang H, Moy SS, Lauder JM, Crawley JN, Threadgill DW, Wright FA, Magnuson TR. Large scale gene expression differences across brain regions and inbred strain correlate with behavioral phenotype. Genetics. 2006 doi: 10.1534/genetics.106.061481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlidis P, Noble WS. Analysis of strain and regional variation in gene expression in mouse brain. Genome Biol. 2001;2:10. doi: 10.1186/gb-2001-2-10-research0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Watson C. The Rat Brain in Stereotaxic Coordinates. 4th ed. Academic Press; New York: 1998. [Google Scholar]
- Pesold C, Liu WS, Guidotti A, Costa E, Caruncho HJ. Cortical bitufted, horizontal, and Martinotti cells preferentially express and secrete reelin into perineuronal nets, nonsynaptically modulating gene expression. Proc Natl Acad Sci U S A. 1999;96:3217–3222. doi: 10.1073/pnas.96.6.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce RC, Bari AA. The role of neurotrophic factors in psychostimulant-induced behavioral and neuronal plasticity. Rev Neurosci. 2001;12:95–110. doi: 10.1515/revneuro.2001.12.2.95. [DOI] [PubMed] [Google Scholar]
- R Development Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2006. [Google Scholar]
- Radcliffe RA, Erwin VG, Draski L, Hoffmann S, Edwards J, Deng XS, Bludeau P, Fay T, Lundquist K, Asperi W, Deitrich RA. Quantitative trait Loci mapping for ethanol sensitivity and neurotensin receptor density in an F2 intercross derived from inbred high and low alcohol sensitivity selectively bred rat lines. Alcohol Clin Exp Res. 2004;28:1796–1804. doi: 10.1097/01.alc.0000148106.71801.d7. [DOI] [PubMed] [Google Scholar]
- Rao et al., 1992Rao MS, Tyrrell S, Landis SC, Patterson PH.Effects of ciliary neurotrophic factor (CNTF) and depolarization on neuropeptide expression in cultured sympathetic neurons Dev Biol 1992150281–293. [DOI] [PubMed] [Google Scholar]
- Rodd ZA, Bell RL, McKinzie DL, Webster AA, Murphy JM, Lumeng L, Li TK, McBride WJ. Low-dose stimulatory effects of ethanol during adolescence in rat lines selectively bred for high alcohol intake. Alcohol Clin Exp Res. 2004;28:535–543. doi: 10.1097/01.alc.0000122107.08417.d0. [DOI] [PubMed] [Google Scholar]
- Rodd ZA, Bertsh BA, Strother WN, Le-Niculescu H, Balaraman Y, Hayden E, Jerome RE, Edenberg HJ, McBride WJ, Niculescu AB. Candidate genes, pathways and mechanisms for alcoholism: an expanded convergent functional genomics approach. Pharmacogenomics J. 2006 doi: 10.1038/sj.tpj.6500420. [Advance online 2006].
- Scholtens D, von Heydebreck A. Analysis of differential gene expression studies. In: Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer; New York, NY: 2005. pp. 230–235. [Google Scholar]
- Schroeder JP, Iller KA, Hodge CW. Neuropeptide-Y Y5 receptors modulate the onset and maintenance of operant ethanol self-administration. Alcohol Clin Exp Res. 2003;27:1912–1920. doi: 10.1097/01.ALC.0000098873.80433.BA. [DOI] [PubMed] [Google Scholar]
- Schroeder JP, Overstreet DH, Hodge CW. The neuropeptide-Y Y5 receptor antagonist L-152,804 decreases alcohol self-administration in inbred alcohol-preferring (iP) rats. Alcohol. 2005;36:179–186. doi: 10.1016/j.alcohol.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Shin E-Y, Shin K-S, Lee C-S, Woo K-N, Quan S-H, Soung N-K, Kim YG, Cha CI, Kim S-R, Park D, Bokoch GM, Kim E-G. Phosphorylation of p85 beta PIX, a Rac/Cdc42-specific guanine nucleotide exchange factor, via the Ras/ERK/PAK2 pathway is required for basic fibroblast growth factor-induced neurite outgrowth. J Biol Chem. 2002;277:44417–44430. doi: 10.1074/jbc.M203754200. [DOI] [PubMed] [Google Scholar]
- Slawecki CJ, Jimenez-Vasquez P, Mathe AA, Ehlers CL. Substance P and neurokinin levels are decreased in the cortex and hypothalamus of alcohol-preferring (P) rats. J Stud Alcohol. 2001;62:736–740. doi: 10.15288/jsa.2001.62.736. [DOI] [PubMed] [Google Scholar]
- Smith DG, Learn JE, McBride WJ, Lumeng L, Li TK, Murphy JM. Alcohol-naive alcohol-preferring (P) rats exhibit higher local cerebral glucose utilization than alcohol-nonpreferring (NP) and Wistar rats. Alcohol Clin Exp Res. 2001;25:1309–1316. [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:1. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Stankoff B, Aigrot MS, Noel F, Wattilliaux A, Zalc B, Lubetzki C. Ciliary neurotrophic factor (CNTF) enhances myelin formation: a novel role for CNTF and CNTF-related molecules. J Neurosci. 2002;22:9221–9227. doi: 10.1523/JNEUROSCI.22-21-09221.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart RB, Badia-Elder NE, Stogsdill TR, Loholdt JM, Smith RJ, Murphy JM, Lumeng L. Behavioral pheno-typing of inbred alcohol preferring and nonpreferring (iP, iNP) rat strains. Alcohol Clin Exp Res. 2004;28(Suppl):89A. [Google Scholar]
- Strother WN, Chernet EJ, Lumeng L, Li TK, McBride WJ. Regional central nervous system densities of delta-opioid receptors in alcohol-preferring P, alcohol-nonpreferring NP, and unselected Wistar rats. Alcohol. 2001;25:31–38. doi: 10.1016/s0741-8329(01)00162-8. [DOI] [PubMed] [Google Scholar]
- Strother WN, Lumeng L, Li T-K, McBride WJ. Acute ethanol effects on cerebral glucose utilization in adult alcohol-preferring P and alcohol nonpreferring NP rats. Alcohol. 2005;35:119–128. doi: 10.1016/j.alcohol.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Szabo G, Hoffman PL. Brain-derived neurotrophic factor, neurotrophin-3 and neurotrophin-4/5 maintain functional tolerance to ethanol. Eur J Pharmacol. 1995;287:35–41. doi: 10.1016/0014-2999(95)00466-3. [DOI] [PubMed] [Google Scholar]
- Terenina-Rigaldie E, Moisan MP, Colas A, Beauge F, Shah KV, Jones BC, Mormede P. Genetics of behaviour: phenotypic and molecular study of rats derived from high- and low-alcohol consuming lines. Pharmacogenetics. 2003;13:543–554. doi: 10.1097/01.fpc.0000054120.14659.8c. [DOI] [PubMed] [Google Scholar]
- Thompson H, Barker D, Camand O, Erskine L. Slits contribute to the guidance of retinal ganglion cell axons in the mammalian optic tract. Dev Biol. 2006;296:476–484. doi: 10.1016/j.ydbio.2006.06.017. [DOI] [PubMed] [Google Scholar]
- Waller MB, McBride WJ, Lumeng L, Li TK. Initial sensitivity and acute tolerance to ethanol in the P and NP lines of rats. Pharmacol Biochem Behav. 1983;19:683–686. doi: 10.1016/0091-3057(83)90345-3. [DOI] [PubMed] [Google Scholar]
- Waller MB, Murphy JM, McBride WJ, Lumeng L, Li TK. Effect of low dose ethanol on spontaneous motor activity in alcohol-preferring and -nonpreferring lines of rats. Pharmacol Biochem Behav. 1986;24:617–623. doi: 10.1016/0091-3057(86)90567-8. [DOI] [PubMed] [Google Scholar]
- Wirth MJ, Gorba T, Wahle P. Epigenetic factors regulate the NPY expression in rat cortical neurons. Regul Pept. 1998;75-76:283–292. doi: 10.1016/s0167-0115(98)00080-9. [DOI] [PubMed] [Google Scholar]
- Witzmann FA, Li J, Strother WN, McBride WJ, Hunter L, Crabb DW, Lumeng L, Li TK. Innate differences in protein expression in the nucleus accumbens and hippocampus of inbred alcohol-preferring and -nonpreferring rats. Proteomics. 2003;3:1335–1344. doi: 10.1002/pmic.200300453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong DT, Reid LR, Li TK, Lumeng L. Greater abundance of serotonin1A receptor in some brain areas of alcohol-preferring (P) rats compared to nonpreferring (NP) rats. Pharmacol Biochem Behav. 1993;46:173–177. doi: 10.1016/0091-3057(93)90337-s. [DOI] [PubMed] [Google Scholar]
- Worst TJ, Tan JC, Robertson DJ, Freeman WM, Hyytia P, Kiianmaa K, Vrana KE. Transcriptome analysis of frontal cortex in alcohol-preferring and nonpreferring rats. J Neurosci Res. 2005;80:529–538. doi: 10.1002/jnr.20496. [DOI] [PubMed] [Google Scholar]
- Xu Y, Ehringer M, Yang F, Sikela JM. Comparison of global brain gene expression profiles between inbred long-sleep and inbred short-sleep mice by high-density gene array hybridization. Alcohol Clin Exp Res. 2001;25:810–818. [PubMed] [Google Scholar]
- Zapala MA, Hovatta I, Ellison JA, Wodicka L, Del Rio JA, Tennant R, Tynan W, Broide RS, Helton R, Stoveken BS, Winrow C, Lockhart DJ, Reilly JF, Young WG, Bloom FE, Lockhart DJ, Barlow C. Adult mouse brain gene expression patterns bear an embryologic imprint. Proc Natl Acad Sci U S A. 2005;102:10357–10362. doi: 10.1073/pnas.0503357102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou FC, Bledsoe S, Lumeng L, Li TK. Immunostained serotonergic fibers are decreased in selected brain regions of alcohol-preferring rats. Alcohol. 1991;8:425–431. doi: 10.1016/s0741-8329(91)90034-t. [DOI] [PubMed] [Google Scholar]
- Zhou FC, Bledsoe S, Lumeng L, Li TK. Reduced serotonergic immunoreactive fibers in the forebrain of alcohol-preferring rats. Alcohol Clin Exp Res. 1994a;18:571–579. doi: 10.1111/j.1530-0277.1994.tb00912.x. [DOI] [PubMed] [Google Scholar]
- Zhou FC, Pu CF, Murphy J, Lumeng L, Li TK. Serotonergic neurons in the alcohol-preferring rats. Alcohol. 1994b;11:397–403. doi: 10.1016/0741-8329(94)90024-8. [DOI] [PubMed] [Google Scholar]
- Zhou FC, Zhang JK, Lumeng L, Li TK. Mesolimbic dopamine system in alcohol-preferring rats. Alcohol. 1995;12:403–412. doi: 10.1016/0741-8329(95)00010-o. [DOI] [PubMed] [Google Scholar]
- Zimatkin SM, Lindros KO. Distribution of catalase in rat brain: aminergic neurons as possible targets for ethanol effects. Alcohol Alcohol. 1996;31(2):167–174. doi: 10.1093/oxfordjournals.alcalc.a008128. [DOI] [PubMed] [Google Scholar]


