Abstract
To identify novel antiapoptotic proteins encoded by DNA viruses, we searched viral genomes for proteins that might interfere with Fas and TNFR1 apoptotic signaling pathways. We report here that equine herpesvirus type 2 E8 protein and molluscum contagiosum virus MC159 protein both show sequence similarity to the death effector domains (DEDs) of the Fas/TNFR1 signaling components FADD and caspase-8. Yeast two-hybrid analysis revealed that E8 protein interacted with the caspase-8 prodomain whereas MC159 protein interacted with FADD. Furthermore, expression of either E8 protein or MC159 protein protected cells from Fas- and TNFR1-induced apoptosis indicating that certain herpesviruses and poxviruses use DED-mediated interactions to interfere with apoptotic signaling pathways. These findings identify a novel control point exploited by viruses to regulate Fas- and TNFR1-mediated apoptosis.
Keywords: FADD, caspase, tumor necrosis factor, molluscum contagiosum virus, equine herpesvirus type 2
Apoptosis, or programmed cell death, is an intrinsic biochemical pathway that plays an essential role in normal development and tissue homeostasis (1). Functioning as central effector molecules of mammalian apoptosis are a growing family of aspartate-specific cysteine proteases termed caspases (2, 3). These proteases are synthesized as proenzymes that must first undergo proteolytic processing to become active enzymatic complexes. Recent studies suggest that the extensive N-terminal prodomains associated with certain caspases function to directly link proenzyme processing and activation to specific apoptotic signaling pathways (4–6). Upon binding to their respective ligands, Fas and TNFR1 receptors trigger apoptosis by recruiting the cytosolic adaptor molecule FADD (MORT1) to the plasma membrane (7–10). FADD contains a death effector domain (DED) that binds to the N-terminal prodomain of caspase-8 (FLICE, MACH, Mch5), which itself contains two DEDs. Although it is uncertain how the binding of FADD to the caspase-8 prodomain results in the processing and activation of the protease, interactions between the DED of FADD and one or both prodomain DEDs may change the conformation of pro-caspase-8 and allow the proenzyme to be activated by autocleavage (4, 5). Thus, DEDs function as protein-binding domains that directly link both Fas and TNFR1 apoptotic signaling pathways to the activation of caspase-8. The recent observation of DEDs in the prodomain of caspase-10 (Mch4) suggests that these regulatory domains play a role in the processing and activation of other caspases (6).
Apoptosis is an effective antiviral defense mechanism used by the host to eliminate virus-infected cells and can be triggered by activated cytotoxic T cells, proinflammatory cytokines such as tumor necrosis factor (TNF), and by viral disruption of cellular metabolism and cell cycle regulation (11). To counter the host apoptotic response, many DNA viruses encode proteins that interfere with key regulatory steps in apoptotic signaling pathways. Antiapoptotic proteins encoded by viruses include Bcl-2 homologs (e.g., adenovirus E1B 19K) that function by an unknown mechanism to prevent activation of apoptotic caspases (12–14), as well as inhibitors of the caspases themselves (e.g., cowpox CrmA and baculovirus P35) (15–18). Infection of cells with mutant viruses lacking these genes often results in premature apoptotic death and reduced yields of progeny virus, indicating that antiapoptotic proteins are necessary for efficient virus replication (19–21). In addition, suppression of apoptotic cell death is important for viral persistence (22) and may be required for the establishment of certain latent infections (23). Thus, understanding the mechanisms that viruses use to inhibit apoptosis should provide further insight into apoptotic signaling pathways as well as virus replication and persistence.
To identify novel viral proteins that might interfere with apoptosis, we studied two DNA viruses that do not encode any known antiapoptotic proteins such as Bcl-2 homologs or direct inhibitors of caspase activity. Here we show that equine herpesvirus type 2 (EHV-2) E8 is a DED-containing protein that binds to the caspase-8 prodomain and blocks Fas- and TNFR1-induced apoptosis. Furthermore, we show that the MC159 protein encoded by molluscum contagiosum virus (MCV), a member of the poxvirus family, is a DED-containing antiapoptotic protein that binds to FADD. These findings suggest that the E8 and MC159 proteins function during virus infection to evade the host apoptotic response and identify novel sites for viral intervention in Fas and TNFR1 apoptotic signaling pathways.
MATERIALS AND METHODS
Expression Vectors.
EHV-2 E8 (24) and MCV 159L (25) open reading frames were amplified by PCR from viral DNAs and cloned into the pCI expression plasmid (Promega) to construct pCI-E8 and pCI-MC159. Baculovirus ORF P35 was subcloned from pPRM-35K-ORF (26) into pCI to construct pCI-P35. pCI-mut Fas expressing a truncated Fas (from patient 5) was described previously (27).
Cell Death Assays.
HeLa cells were cotransfected with 350 ng of pCMVβ-gal vector (Stratagene) and 700 ng of either pCI, pCI-E8, pCI-MC159, or pCI-P35 expression vectors using LipofectAMINE (Life Technologies, Gaithersburg, MD). Cells were treated 28 h after transfection with either 500 ng/ml anti-human Fas mAb CH11 (Oncor) and 5 μg/ml cycloheximide or 20 ng/ml recombinant (Escherichia coli) human TNF-α (Boehringer Mannheim) and 15 μg/ml cycloheximide for 16 h. Control cells received cycloheximide treatment alone. Cells were fixed and stained for β-galactosidase (β-gal) expression as described (28). The total number of flat, blue-staining cells in five random fields of view were counted. Protection from Fas- and TNF-induced apoptosis was measured as the ratio of the number of flat, blue-staining cells in treated to control wells.
MCF7 cells were cotransfected with 100 ng of pCMVβ-gal vector and 1 μg of either pCI, pCI-E8, pCI-MC159, or pCI-P35 expression vectors using Lipofectin (Life Technologies). Cells were treated 24 h after transfection with either 150 ng/ml anti-human Fas mAb CH11 and 1 μg cycloheximide (control cells received cycloheximide treatment alone) or 40 ng/ml recombinant human TNF-α for 12 h. For UV-irradiation treatment of cells, 6-well dishes were placed on a UV transilluminator (FisherBiotech) for 5 min and then incubated for 12 h. Cells were then fixed and stained for β-gal expression. The total number of flat and round blue-staining cells in five to seven fields of view were counted. Protection from Fas-, TNF-, and UV-induced apoptosis was measured as the ratio of flat, blue-staining cells to the total number of blue cells in treated to control wells.
Jurkat cells were electroporated with the murine class I expression vector H-2 Ld-pSRα and either pCI, pCI-E8, PCI-MC159, or pCI-mutant Fas (mFas) at a ratio of 3:1 (20 μg DNA total) and incubated for 16 h as described (27). Viable cells were isolated on Ficoll (Pharmacia) gradients and treated with 30 ng/ml anti-human Fas mAb CH11 in triplicate wells for 7 h; parallel control cultures received no antibody treatment. Transfected viable cells staining negative for annexin-V (an early marker of apoptosis) and positive for H-2 Ld were counted by flow cytometry. Percentage protection from Fas-induced apoptosis was calculated as the ratio of transfected viable cells in treated to control cultures.
Yeast Two-Hybrid Analysis.
Caspase-8 (DED-A, residues 3–80; DED-B, residues 133–209; DED A+B, residues 3–209) (6), murine FADD, E8, and E8 DED (residues 93–171) were subcloned into yeast two-hybrid vectors. A bait vector (LexA fusion protein), a prey vector (B42 fusion protein), and the β-gal expression plasmid p18–34 were transformed together into the yeast strain EGY191 (29). For filter assays, colonies were patched onto Sc, -ura, -trp, -his plates containing 2% galactose. Cells were lifted in duplicate using nitrocellulose filters (Schleicher & Schuell) and lysed in liquid nitrogen. For the detection of β-gal, each filter was placed in Z buffer (60 mM Na2HPO4/40 mM NaH2PO4/10 mM KCl/1 mM MgSO4/50 mM 2-mercaptoethanol) containing 1.25 mg/ml 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal). Four colonies from each transformation were assayed. For liquid assays, cells were grown in Sc, -ura, -trp, -his media containing 2% raffinose to an OD600 of 0.2 and induced with a 2% final concentration of galactose for 6 h. Cells (1.5 ml) were pelleted and resuspended in 300 μl Z buffer. Cells (100 μl) were lysed in 0.7 ml Z buffer containing 35 units of Lyticase (Boehringer Mannheim) for 1 h. To assay β-gal activity, 160 μl o-nitrophenyl β-d-galactoside (4 mg/ml) was added to the reaction, and the absorbance was measured at OD420 and expressed as Miller units. Three colonies from each transformation were assayed in duplicate.
RESULTS
E8 and MC159 Are DED-Containing Viral Proteins.
The EHV-2 and MCV genomes do not encode homologs of known antiapoptotic proteins such as Bcl-2 or inhibitors of caspase activity (24, 25). We therefore performed a computer search of the EHV-2 and MCV genomes to identify homologs of cellular proteins involved in apoptosis. Both EHV-2 E8 and MCV MC159 proteins showed homology to Fas/TNFR1 signaling proteins FADD and caspase-8 (Fig. 1). The C terminus of the 171-aa E8 protein shows a striking similarity to cellular DEDs. Residues 93–171 share 31% identity (42% similarity) with the DED of FADD (residues 4–81) and 23 and 20% identity (38 and 32% similarity) with the two DEDs present in the caspase-8 prodomain (DED-A, residues 3–80 and DED-B, residues 133–209, respectively). A short region from the amino-half of E8 (residues 58–72) also shows homology to the C terminus of cellular DEDs (data not shown). The 241-aa MC159 protein is also a viral DED-containing protein. MC159 contains two tandemly arranged DEDs (DED-A, residues 1–79 and DED-B, residues 97–175), each of which are homologous to the DEDs of FADD and caspase-8. The overall level of similarity between the viral DEDs and that of FADD or caspase-8 is comparable to the levels of similarity that exist between cellular DED-containing family members. Thus, E8 and MC159 proteins are viral members of the DED-containing family of apoptotic signaling proteins.
Figure 1.
EHV-2 E8 and MCV MC159 are DED-containing proteins. E8 and MC159 DEDs are aligned with the DEDs of FADD and caspase-8. Solid black and gray shading indicate identical and conserved residues, respectively. Dots indicate gaps in the sequence to allow optimal alignment.
E8 and MC159 Proteins Interact with the Caspase-8 Prodomain and FADD, Respectively.
Because DEDs are thought to mediate binding between DED-containing cellular proteins, and binding of FADD to the caspase-8 prodomain has been demonstrated previously by yeast two-hybrid analysis (4), we examined the ability of E8 and MC159 to interact with these cellular proteins. Yeast two-hybrid interactions were assessed by an X-Gal filter blue/white assay and by a quantitative liquid β-gal assay (Table 1). Full-length E8 protein (residues 1–171) and its C-terminal DED (residues 93–171) each interacted with the caspase-8 prodomain (DED A+B, residues 3–209). Moreover, the isolated E8 DED strongly interacted with DED-B (residues 133–209) of the caspase-8 prodomain, demonstrating that E8 DED is sufficient to mediate protein interactions with an isolated cellular DED. No interaction was observed between E8 DED and DED-A (residues 3–80). In addition, E8 DED did not interact with FADD, even though both FADD and the prodomain of caspase-8 were expressed at comparable levels in the assay (data not shown). Analysis of MC159 binding revealed a striking difference in binding specificity. Unlike E8, MC159 interacted strongly with FADD and showed little or no binding to the prodomain of caspase-8. These findings demonstrate that although viral DED-containing proteins share similar sequences, they show marked differences in their ability to interact with the two cellular DED family members, FADD and caspase-8.
Table 1.
Interactions between viral and cellular DED-containing proteins by yeast two-hybrid analysis
| DNA-binding hybrid | Activation hybrid | Filter assay (intensity of blue color) | Liquid assay (β-gal activity, Miller units) |
|---|---|---|---|
| LexA-caspase-8 DED A + B | B42-E8 | +++ | 163 ± 2.6 |
| LexA-caspase-8 DED A + B | B42-E8 DED | ++ | 16.3 ± 2.9 |
| LexA-caspase-8 DED A + B | B42 | — | 7.5 ± 0.8 |
| LexA-E8 DED | B42-caspase-8 DED A + B | ++ | 44 ± 8.0 |
| LexA-E8 DED | B42-caspase-8 DED B | +++ | 162 ± 44 |
| LexA-E8 DED | B42-caspase-8 DED A | — | 21 ± 2.0 |
| LexA-E8 DED | B42 | — | 21 ± 3.3 |
| LexA-E8 DED | B42-FADD | — | 15 ± 0.5 |
| LexA-E8 DED | B42 | — | 14 ± 1.5 |
| LexA-MC159 | B42-FADD | ++++ | 6890 ± 1627 |
| LexA-MC159 | B42-caspase-8 DED A + B | — | 31 ± 0.1 |
| LexA-MC159 | B42-caspase-8 DED B | + | 61 ± 8.9 |
| LexA-MC159 | B42-caspase-8 DED A | — | 39 ± 19 |
| LexA-MC159 | B42 | — | 27 ± 1.0 |
Data represent four independent experiments. ++++, Intense blue color; +++, strong blue color; ++, medium blue color; +, light blue color; —, white.
E8 and MC159 Proteins Block Fas- and TNFR1-Induced Apoptosis.
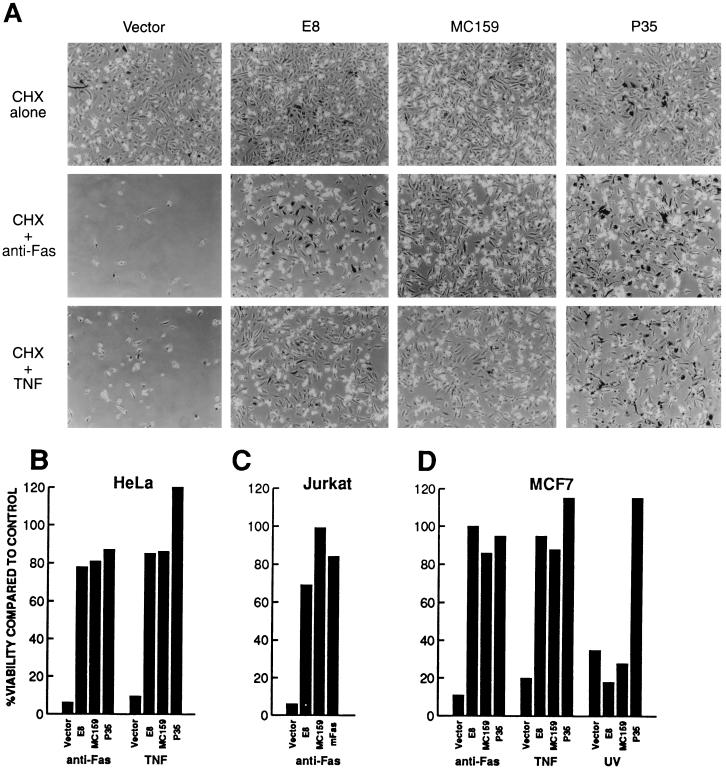
Since E8 and MC159 bound to the caspase-8 prodomain and FADD, respectively, we postulated that the DED-containing viral proteins might function to block Fas- and TNFR1-induced apoptosis. To determine whether E8 and MC159 have antiapoptotic activity, HeLa cells were cotransfected with plasmids expressing β-gal and either E8 or MC159 and then treated 28 h after transfection with either anti-Fas antibody or TNF in the presence of cycloheximide. Whereas treated cells that were cotransfected with the control vector and plasmid expressing β-gal underwent apoptosis with rounding up, membrane blebbing, and lifting off from the plate, many of the treated cells that were cotransfected with either E8 or MC159 and β-gal remained flat and attached to the plate (Fig. 2A). E8 and MC159 protected HeLa cells to a similar degree as baculovirus P35, which has been shown previously to be an effective inhibitor of Fas- and TNF-induced apoptosis (30). Comparison of the number of viable, blue-staining flat cells in treated wells with those in untreated control wells showed high levels (>75%) of protection from Fas- or TNF-induced apoptosis for E8, MC159, or baculovirus P35 proteins, whereas the vector alone afforded little or no (<10%) protection (Fig. 2B).
Figure 2.
E8 and MC159 block Fas- and TNFR1-induced apoptosis. HeLa, Jurkat, and MCF7 cells were transfected with the indicated plasmids and treated with either anti-Fas antibody, TNF, or UV-irradiation. (A) HeLa cells were fixed and stained for β-gal expression following treatment and then viewed by phase-contrast microscopy. (B–D) The percentage of viable cells was determined by measuring the number of cells surviving after treatment with various apoptotic inducers compared with the number of cells receiving no treatment.
E8 and MC159 also effectively blocked Fas- and TNFR1-induced apoptosis in two other cell lines, Jurkat T cells and MCF7 breast carcinoma cells (Fig. 2 C and D). E8 and MC159 protected Jurkat cells to nearly the same level as a truncated Fas mutant, which acts as a dominant-negative inhibitor of Fas-induced apoptosis (27). In MCF7 cells, E8 and MC159 blocked Fas- and TNFR1-induced apoptosis nearly as well as P35. However, in striking contrast to P35, E8 and MC159 did not block UV-induced apoptotic death. Therefore, the antiapoptotic activity of E8 and MC159 appears to be restricted to blocking Fas and TNFR1 death signals.
DISCUSSION
We have identified herpesvirus and poxvirus proteins that specifically block Fas and TNFR1 signaling pathways. TNF and Fas previously have been shown to be important for lysis of virus-infected cells. TNF is directly cytotoxic to cells infected with both DNA and RNA viruses (31, 32), and overexpression of the cytokine by a recombinant vaccinia virus leads to rapid virus clearance from infected mice with little or no cytopathic effects (33). Although the role of Fas in the elimination of virus-infected cells is less well studied than TNF, Fas-induced apoptosis has been shown in vitro to interfere with the replication of herpes simplex virus (34). In addition, Fas and perforin lytic pathways are major mechanisms of virus-specific T cell-mediated cytotoxicity (35). Thus, the ability of E8 and MC159 proteins to inhibit Fas and TNFR1 apoptotic signaling pathways may provide a selective advantage for EHV-2 and MCV replication in their respective hosts.
EHV-2 belongs to the gammaherpesvirus subfamily (36). These viruses establish latent infections in lymphocytes and usually persist for the lifetime of the host. Inhibition of apoptosis by gammaherpesviruses is thought to be important because all known members of this subfamily that have been sequenced, except for EHV-2, encode Bcl-2 homologs (24, 37). Because the EHV-2 E8 protein blocks Fas- and TNFR1-induced apoptosis, it may have a role analogous to the viral Bcl-2 homologs in blocking the host apoptotic response and preventing the premature destruction of virus-infected cells. Although little is known of EHV-2 infection in the horse, E8-mediated interference with Fas and TNFR1 signaling pathways in both lymphocytes and epithelial cells may be critical for the chronic, frequently asymptomatic infection caused by the virus (38).
Poxviruses encode members of the serpin family including SPI-1 and the caspase inhibitor SPI-2 (e.g., cowpox CrmA) that interfere with Fas- and TNFR1-induced apoptosis (39, 40). In addition, several poxviruses encode soluble TNF receptors that interfere with activation of the TNFR1 apoptotic signaling pathway by direct binding to TNF (41, 42). Surprisingly, MCV does not encode homologs of either of these types of apoptotic inhibitors (25). Infection of humans with MCV results in the formation of hyperplastic cutaneous lesions that can persist for months to years and typically show no inflammatory reaction (43). Our finding that MC159 blocks Fas and TNFR1 signaling pathways in two epithelial cell lines suggests that this protein may play an important role in the persistent infection of epithelial cells by MCV. Interestingly, MCV encodes another DED-containing protein called MC160 that has homology to both MC159 and cellular DEDs. Experiments are in progress to determine whether MC160 has antiapoptotic activity.
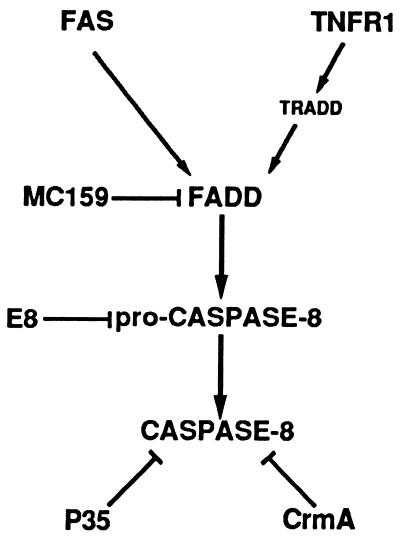
Our findings identify FADD and pro-caspase-8 as targets for viral intervention in Fas and TNFR1 signaling pathways. Caspase-8 and FADD are also involved in apoptosis mediated by the DR3 receptor (44), suggesting that E8 and MC159 might also block this apoptotic signaling pathway. The binding of DED-containing E8 and MC159 proteins to the prodomain of caspase-8 and FADD, respectively, is consistent with a model of apoptotic suppression that involves direct interaction with the cellular DED-containing proteins that mediate Fas and TNFR1 death signals (Fig. 3). The binding of FADD to the caspase-8 prodomain has been suggested to result in the processing and activation of the proenzyme to an active heterodimeric enzyme complex (4, 5). We propose that the binding of E8 to pro-caspase-8 or MC159 to FADD blocks Fas- and TNFR1-induced apoptosis by interfering with the ability of pro-caspase-8 to bind to FADD. The mechanism used by E8 and MC159 to block apoptosis is therefore different from that used by the cowpox CrmA (15, 45, 46) and baculovirus P35 (16–18) proteins, which inhibit Fas- and TNFR1-induced death by directly inhibiting active caspase-8 (ref. 47; data not shown). These findings demonstrate that DED-containing proteins can function as negative regulators of both Fas and TNFR1 signaling pathways and identify the DED-mediated pro-caspase-8/FADD interaction as a novel site of apoptotic regulation.
Figure 3.
Model for E8 and MC159 inhibition of Fas and TNFR1 signaling pathways. Fas and TNFR1 cell surface receptors induce apoptosis through the binding of FADD to the prodomain of caspase-8. E8 and MC159 proteins bind to the caspase-8 prodomain and FADD, respectively, and may block Fas- and TNFR1-induced apoptosis by interfering with the ability of pro-caspase-8 to bind to FADD. In contrast, CrmA and P35 antiapoptotic proteins directly inhibit caspase-8 enzymatic activity.
Acknowledgments
We thank A. Davison for EHV-2 DNA, V. Dixit for the MCF7 cell line, P. Friesen for plasmid pPRM-35K-ORF, E. Golemis for yeast strain EGY191, J. Sisler for oligonucleotide synthesis and DNA sequencing, C. Zacharchuk for plasmid H-2 Ld-pSRα, and S. Straus for critical reading of the manuscript. D.M. is a Howard Hughes Medical Institute–National Institutes of Health Scholar.
Footnotes
Abbreviations: DED, death effector domain; EHV-2, equine herpesvirus type 2; MCV, molluscum contagiosum virus; TNF, tumor necrosis factor; β-gal, β-galactosidase; X-Gal, 5-bromo-4-chloro-3-indolyl β-d-galactoside.
References
- 1.Vaux D L, Strasser A. Proc Natl Acad Sci USA. 1996;93:2239–2244. doi: 10.1073/pnas.93.6.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henkart P A. Immunology. 1996;4:195–201. doi: 10.1016/s1074-7613(00)80428-8. [DOI] [PubMed] [Google Scholar]
- 3.Alnemri E S, Livingston D J, Nicholson D W, Salvesen G, Thornberry N A, Wong W W, Yuan J. Cell. 1996;87:171. doi: 10.1016/s0092-8674(00)81334-3. [DOI] [PubMed] [Google Scholar]
- 4.Boldin M P, Goncharov T M, Goltsev Y V, Wallach D. Cell. 1996;85:803–815. doi: 10.1016/s0092-8674(00)81265-9. [DOI] [PubMed] [Google Scholar]
- 5.Muzio M, Chinnaiyan A M, Kischkel F C, O’Rourke K, Shevchenko A, Ni J, Scaffidi C, Bretz J D, Zhang M, Gentz R, Mann M, Krammer P H, Peter M E, Dixit V M. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 6.Fernandes-Alnemri T, Armstrong R C, Krebs J, Srinivasula S M, Wang L, Bullrich F, Fritz L C, Trapani J A, Tomaselli K J, Litwack G, Alnemri E S. Proc Natl Acad Sci USA. 1996;93:7464–7469. doi: 10.1073/pnas.93.15.7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boldin M P, Varfolomeev E E, Pancer Z, Mett I L, Camonis J H, Wallach D. J Biol Chem. 1995;270:7795–7798. doi: 10.1074/jbc.270.14.7795. [DOI] [PubMed] [Google Scholar]
- 8.Chinnaiyan A M, O’Rourke K, Tewari M, Dixit V M. Cell. 1995;81:505–512. doi: 10.1016/0092-8674(95)90071-3. [DOI] [PubMed] [Google Scholar]
- 9.Kischkel F C, Hellbardt S, Behrmann I, Germer M, Pawlita M, Krammer P H, Peter M E. EMBO J. 1995;14:5579–5588. doi: 10.1002/j.1460-2075.1995.tb00245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chinnaiyan A M, Tepper C G, Seldin M F, O’Rourke K, Kischkel F C, Hellbardt S, Krammer P H, Peter M E, Dixit V M. J Biol Chem. 1996;271:4961–4965. doi: 10.1074/jbc.271.9.4961. [DOI] [PubMed] [Google Scholar]
- 11.Shen Y, Shenk T E. Curr Opin Genet Dev. 1995;5:105–111. doi: 10.1016/s0959-437x(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 12.Boulakia C A, Chen G, Ng F W H, Teodoro J G, Branton P E, Nicholson D W, Poirier G G, Shore G C. Oncogene. 1996;12:529–535. [PubMed] [Google Scholar]
- 13.Chinnaiyan A M, Orth K, O’Rourke K, Duan H, Poirier G G, Dixit V M. J Biol Chem. 1996;271:4573–4576. doi: 10.1074/jbc.271.9.4573. [DOI] [PubMed] [Google Scholar]
- 14.Armstrong R C, Aja T, Xiang J, Gaur S, Krebs J F, Hoang K, Bai X, Korsmeyer S J, Karanewsky D S, Fritz L C, Tomaselli K J. J Biol Chem. 1996;271:16850–16855. doi: 10.1074/jbc.271.28.16850. [DOI] [PubMed] [Google Scholar]
- 15.Ray C A, Black R A, Kronheim S R, Greenstreet T A, Sleath P R, Salvesen G S, Pickup D J. Cell. 1992;69:597–604. doi: 10.1016/0092-8674(92)90223-y. [DOI] [PubMed] [Google Scholar]
- 16.Bump N J, Hackett M, Hugunin M, Seshagriri S, Brady K, Chen P, Ferenz C, Franklin S, Ghayur T, Licari L P, Mankovich J, Shi L, Greenberg A H, Miller L K, Wong W W. Science. 1995;269:1885–1888. doi: 10.1126/science.7569933. [DOI] [PubMed] [Google Scholar]
- 17.Xue D, Horvitz H R. Nature (London) 1995;377:248–251. doi: 10.1038/377248a0. [DOI] [PubMed] [Google Scholar]
- 18.Bertin J, Mendrysa S M, LaCount D J, Gaur S, Krebs J F, Armstrong R C, Tomaselli K J, Friesen P D. J Virol. 1996;70:6251–6259. doi: 10.1128/jvi.70.9.6251-6259.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pilder S, Logan J, Shenk T. J Virol. 1984;52:664–671. doi: 10.1128/jvi.52.2.664-671.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hershberger P A, Dickson J A, Friesen P D. J Virol. 1992;66:5525–5533. doi: 10.1128/jvi.66.9.5525-5533.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brooks M A, Ali A N, Turner P C, Moyer R W. J Virol. 1995;69:7688–7698. doi: 10.1128/jvi.69.12.7688-7698.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levine B, Huang Q, Isaacs J T, Reed J C, Griffin D E, Hardwick J M. Nature (London) 1993;361:739–742. doi: 10.1038/361739a0. [DOI] [PubMed] [Google Scholar]
- 23.Gregory C D, Dive C, Henderson S, Smith C A, Williams G T, Gordon J, Rickinson A B. Nature (London) 1991;349:612–614. doi: 10.1038/349612a0. [DOI] [PubMed] [Google Scholar]
- 24.Telford E A R, Watson M S, Aird H C, Perry J, Davison A J. J Mol Biol. 1995;249:520–528. doi: 10.1006/jmbi.1995.0314. [DOI] [PubMed] [Google Scholar]
- 25.Senkevich T G, Bugert J J, Sisler J R, Koonin E V, Darai G, Moss B. Science. 1996;273:813–816. doi: 10.1126/science.273.5276.813. [DOI] [PubMed] [Google Scholar]
- 26.Hershberger P A, LaCount D J, Friesen P D. J Virol. 1994;68:3467–3477. doi: 10.1128/jvi.68.6.3467-3477.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fisher G H, Rosenberg F J, Straus S E, Dale J K, Middelton L A, Lin A Y, Strober W, Lenardo M J, Puck J M. Cell. 1995;81:935–946. doi: 10.1016/0092-8674(95)90013-6. [DOI] [PubMed] [Google Scholar]
- 28.Uren A G, Pakusch M, Hawkins C J, Puls K L, Vaux D L. Proc Natl Acad Sci USA. 1996;93:4974–4978. doi: 10.1073/pnas.93.10.4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gyuris J, Golemis E, Chertkov H, Brent R. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 30.Beidler D R, Tewari M, Friesen P D, Poirier G G, Dixit V M. J Biol Chem. 1995;270:16526–16528. doi: 10.1074/jbc.270.28.16526. [DOI] [PubMed] [Google Scholar]
- 31.Mestan J, Digel W, Mittnacht S, Hillen H, Blohm D, Moller A, Jacobsen H, Kirchner H. Nature (London) 1986;323:816–819. doi: 10.1038/323816a0. [DOI] [PubMed] [Google Scholar]
- 32.Wong G H W, Goeddel D V. Nature (London) 1986;323:819–822. doi: 10.1038/323819a0. [DOI] [PubMed] [Google Scholar]
- 33.Sambhi S K, Kohonen-Corish M R J, Ramshaw I A. Proc Natl Acad Sci USA. 1991;88:4025–4029. doi: 10.1073/pnas.88.9.4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sieg S, Yildirim Z, Smith D, Kayagaki N, Yagita H, Huang Y, Kaplan D. J Virol. 1996;70:8747–8751. doi: 10.1128/jvi.70.12.8747-8751.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kagi D, Vignaux F, Ledermann B, Burki K, Depraetere V, Nagata S, Hengartner H, Goldstein P. Science. 1994;265:528–530. doi: 10.1126/science.7518614. [DOI] [PubMed] [Google Scholar]
- 36.Telford E A R, Studdert M J, Agius C T, Watson M S, Aird H C, Davison A J. Virology. 1993;195:492–499. doi: 10.1006/viro.1993.1400. [DOI] [PubMed] [Google Scholar]
- 37.Smith C A. Trends Cell Biol. 1995;5:344. doi: 10.1016/s0962-8924(00)89061-3. [DOI] [PubMed] [Google Scholar]
- 38.Agius C T, Studdert M. Adv Virus Res. 1994;44:357–379. doi: 10.1016/s0065-3527(08)60333-4. [DOI] [PubMed] [Google Scholar]
- 39.Tewari M, Dixit V M. J Biol Chem. 1995;270:3255–3260. doi: 10.1074/jbc.270.7.3255. [DOI] [PubMed] [Google Scholar]
- 40.Macen J L, Garner R S, Musy P Y, Brooks M A, Turner P C, Moyer R W, McFadden G, Bleackley R C. Proc Natl Acad Sci USA. 1996;93:9108–9113. doi: 10.1073/pnas.93.17.9108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith C A, Davis T, Wignall J M, Din W S, Farrah T, Upton C, McFadden G, Goodwin R G. Biochem Biophys Res Commun. 1991;176:335–342. doi: 10.1016/0006-291x(91)90929-2. [DOI] [PubMed] [Google Scholar]
- 42.Smith C A, Hu F-Q, Davis Smith T, Richards C L, Smolak P, Goodwin R G, Pickup D J. Virology. 1996;223:132–147. doi: 10.1006/viro.1996.0462. [DOI] [PubMed] [Google Scholar]
- 43.Gottlieb S L, Myskowski P L. Int J Dermatol. 1994;33:453–461. doi: 10.1111/j.1365-4362.1994.tb02853.x. [DOI] [PubMed] [Google Scholar]
- 44.Chinnaiyan A M, O’Rourke K, Yu G-L, Lyons R H, Garg M, Duan D R, Xing L, Gentz R, Ni J, Dixit V M. Science. 1996;274:990–992. doi: 10.1126/science.274.5289.990. [DOI] [PubMed] [Google Scholar]
- 45.Komiyama T, Ray C A, Pickup D J, Howard A D, Thornberry N A, Peterson E P, Salvesen G. J Biol Chem. 1994;269:19331–19337. [PubMed] [Google Scholar]
- 46.Tewari M, Quan L T, O’Rourke K, Desnoyers S, Zeng Z, Beidler D R, Poirier G G, Salvesen G S, Dixit V M. Cell. 1995;81:801–809. doi: 10.1016/0092-8674(95)90541-3. [DOI] [PubMed] [Google Scholar]
- 47.Srinivasula S M, Ahmad M, Fernandes-Alnemri T, Litwack G, Alnemri E S. Proc Natl Acad Sci USA. 1996;93:14486–14491. doi: 10.1073/pnas.93.25.14486. [DOI] [PMC free article] [PubMed] [Google Scholar]