Abstract
The study of P-element repression in Drosophila melanogaster led to the discovery of the telomeric Trans-Silencing Effect (TSE), a repression mechanism by which a transposon or a transgene inserted in subtelomeric heterochromatin (Telomeric Associated Sequence or TAS) has the capacity to repress in trans in the female germline, a homologous transposon, or transgene located in euchromatin. TSE shows variegation among egg chambers in ovaries when silencing is incomplete. Here, we report that TSE displays an epigenetic transmission through meiosis, which involves an extrachromosomal maternally transmitted factor. We show that this silencing is highly sensitive to mutations affecting both heterochromatin formation (Su(var)205 encoding Heterochromatin Protein 1 and Su(var)3–7) and the repeat-associated small interfering RNA (or rasiRNA) silencing pathway (aubergine, homeless, armitage, and piwi). In contrast, TSE is not sensitive to mutations affecting r2d2, which is involved in the small interfering RNA (or siRNA) silencing pathway, nor is it sensitive to a mutation in loquacious, which is involved in the micro RNA (or miRNA) silencing pathway. These results, taken together with the recent discovery of TAS homologous small RNAs associated to PIWI proteins, support the proposition that TSE involves a repeat-associated small interfering RNA pathway linked to heterochromatin formation, which was co-opted by the P element to establish repression of its own transposition after its recent invasion of the D. melanogaster genome. Therefore, the study of TSE provides insight into the genetic properties of a germline-specific small RNA silencing pathway.
Author Summary
The genome of the fruitfly was invaded in the last century by a mobile DNA element called the P element. After a transient period of genetic disorders due to P mobility, the P element established a repressive state for its transposition. We have shown that a major component of this repression comes from P copies inserted close to telomeres, the ends of linear chromosomes. One or two P copies inserted in subtelomeric heterochromatin (the DNA region highly compacted by protein complexes) can stabilize around 80 P copies. This finding allowed the discovery of a more general phenomenon called the “Trans-silencing effect” in which a transgene inserted in this subtelomeric heterochromatin represses, in the female germline, a homologous transgene, irrespective of the genetic location of the latter. We show that Trans-silencing requires not only the chromosomal copy of the telomeric silencer, but also a maternally transmitted factor whose influence can persist over generations. We have found that this epigenetic silencing is sensitive to mutations in genes involved in heterochromatin formation and in a recently discovered silencing pathway based on small RNAs. Trans-silencing thus provides a tool for mechanistic analysis of gene repression on the basis of chromatin changes combined with small RNA pathways in the germline.
Introduction
Repression of transposable elements (TEs) involves complex mechanisms that can be linked to either small RNA silencing pathways or chromatin structure modifications depending on the species and/or the TE family [1–6]. Drosophila species are particularly relevant to the study of these repression mechanisms since some families of TEs are recent invaders, allowing genetic analysis to be carried out on strains with or without these TEs [7,8]. In some cases, crossing these two types of strains induces hybrid dysgenesis, a syndrome of genetic abnormalities resulting from TE mobility [9,10]. In D. virilis, repression of hybrid dysgenesis has been correlated to RNA silencing since small RNAs of the retroelement Penelope, responsible for dysgenesis, were detected in nondysgenic embryos but not in dysgenic embryos [11]. In D. melanogaster, repression of retrotransposons can be established by noncoding fragments of the corresponding element (I factor [12,13], ZAM, and Idefix [14]) and can be in some cases (gypsy [15], mdg1 [16], copia [17], Het-A, TART [18,19], and ZAM, Idefix [C. Vaury and S. Desset, personal communication]) sensitive to mutations in genes from the Argonaute family involved in small RNA silencing pathways. In the same species, strong repression of the DNA P TE, by a cellular state that has been called “P cytotype” [10], can be established by one or two telomeric P elements inserted in heterochromatic “Telomeric Associated Sequences” (TAS) at the 1A cytological site corresponding to the left end of the X chromosome [20–24]. This includes repression of dysgenic sterility resulting from P transposition. We have previously shown that this P cytotype is sensitive to mutations affecting both Heterochromatin Protein 1 (HP1) [21] and the Argonaute family member AUBERGINE [25]. P repression corresponds to a new picture of TE repression shown, using an assay directly linked to transposition, to be affected by heterochromatin and small RNA silencing mutants.
In the course of the study of P cytotype, a new silencing phenomenon has been discovered. Indeed, a P-lacZ transgene or a single defective P element inserted in TAS can repress expression of euchromatic P-lacZ insertions in the female germline in trans, if a certain length of homology exists between telomeric and euchromatic insertions [23,26]. This homology-dependent silencing phenomenon has been termed Trans-Silencing Effect (TSE) [26]. Telomeric transgenes, but not centromeric transgenes, can be silencers and all euchromatic P-lacZ insertions tested can be targets [23,26]. TSE is restricted to the female germline (unpublished data) and has a maternal effect since repression occurs only when the telomeric transgene is maternally inherited [27]. Further, when TSE is not complete, variegating germline lacZ repression is observed from one egg chamber to another, suggesting a chromatin-based mechanism of repression [28]. Recently, an extensive analysis of small RNAs complexed with PIWI family proteins (AUBERGINE, PIWI, and AGO3) was performed in the Drosophila female germline [4]. The latter study showed that most of the RNA sequences associated to these proteins derive from TEs. TSE corresponds likely to such a situation.
Here, we analyze the genetic properties of TSE and show that it has an epigenetic transmission through meiosis, which involves an extrachromosomal maternally transmitted stimulating component. Further, in order to investigate the mechanism behind TSE, we performed a candidate gene analysis to identify genes whose mutations impair TSE. We found that TSE is strongly affected both by mutations in genes involved in heterochromatin formation and in the recently discovered small RNA silencing pathway called “repeat-associated small interfering RNAs” (rasiRNA) pathway [3,4,6,29]. In contrast, we show that TSE is not sensitive to genes specific to the classical RNA interference pathway linked to small interfering RNAs (siRNA) or to the micro RNA (miRNA) pathway. This suggests thus that TSE involves a rasiRNA pathway linked to heterochromatin formation and that such a mechanism, working in the germline, may underlie epigenetic transmission of repression through meiosis.
Results
Quantification of TSE
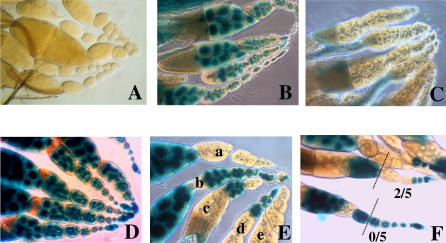
TSE was generated by combining telomeric transgene insertions (from the P-1152 line) as a silencer locus, with various euchromatic P-lacZ transgenes expressed in the germline as targets. Depending on the target, TSE can be almost total (Figure 1C) or intermediate (Figure 1E and 1F). When TSE is incomplete, variegation is observed since “on” and “off” lacZ expression is seen among egg chambers: egg chambers can show strong expression (dark blue) or no expression, but intermediate repression levels are not (or very rarely) found. In addition, a given ovary can present ovarioles showing all possible combinations of on or off egg chambers (Figure 1E). Simple quantification of TSE is thus possible by determining the percentage of repressed egg chambers (Figure 1F). We scored the number of repressed chambers among the first five egg chambers of a given ovariole for ten ovarioles chosen at random per ovary. For a given genotype, more than 1,000 egg chambers were classically counted (Table 1). This measure generally produces very reproducible results among replicate experiments allowing accurate quantification of TSE.
Figure 1. Phenotype and Quantification of TSE.
(A, B) Ovary β-galactosidase staining of P-lacZ enhancer-trap transgenes inserted in subtelomeric heterochromatin (line P-1152 [A]) and euchromatin (line BQ16 [B]).
(C) Complete repression of lacZ by TSE in the G1 females produced by the cross between P-1152 females and BQ16 males.
(D) Expression control of the euchromatic BC69 P-lacZ enhancer-trap.
(E, F) Incomplete TSE in the G1 females produced by the cross between P-1152 females and BC69 males. Variegation is observed inside ovaries that show individual ovarioles with both off (lacZ repressed) and on (lacZ not repressed) egg chambers (Ea, Ec, Ed) and ovarioles that are completely off (Ee) or completely on (Eb). (F) For the quantification of TSE, the number of repressed egg chambers among the first five egg chambers of a given ovariole is assayed. A total of ten ovarioles are scored per ovary for more than ten ovaries.
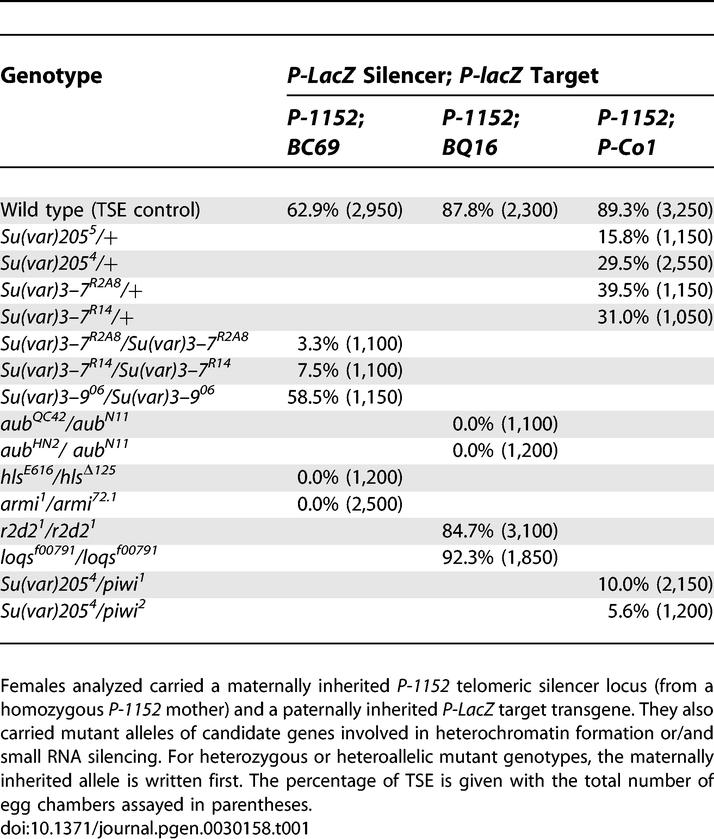
Table 1.
TSE Levels in Mutant Contexts
Transmission of TSE over Generations
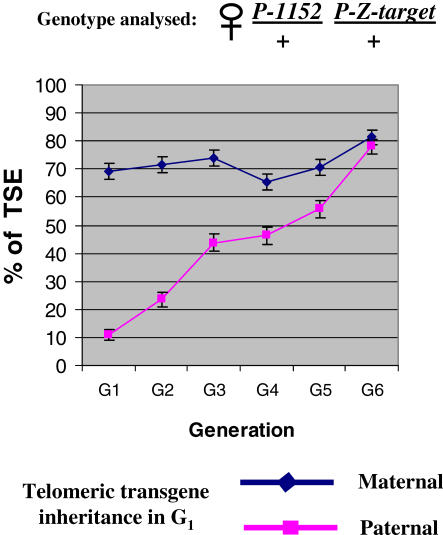
TSE was previously shown to have a maternal effect since strong repression occurs only in the progeny of crosses involving females carrying the telomeric silencer, whereas no or weak repression occurs if the telomeric silencer comes from the father [27,28]. Conversely, the parental origin of the target does not significantly affect TSE. We have thus tested whether TSE exhibits only a maternal effect (during one generation) or maternal inheritance over several generations as well. Two reciprocal G0 crosses were performed in which the telomeric silencer (P-1152) was maternally introduced (maternal lineage) or paternally introduced (paternal lineage) (Figure S1). Thus, the G1 females of the two lineages have the same genotype. These G1 females were backcrossed with males having the telomeric silencer and the target transgene in order to recover G2 females having one copy of the telomeric silencer locus and one copy of the target transgene. Such backcrosses were repeated for several generations in order to follow lineages with constant genotypes over generations. At each generation, the percentage of TSE was measured. Figure 2 shows that in the maternal lineage, strong TSE was detected in G1 (around 70%) and maintained over generations. By contrast, in G1 of the paternal lineage, the level of TSE was weak (around 10%), a result consistent with the maternal effect previously reported. However, in this paternal lineage, a strong level of repression was not observed in G2 females either (around 24%), despite the fact that G1 females were carrying a telomeric silencer. TSE thus shows a maternal inheritance. However, the level of TSE in G2 is greater than in G1 in the paternal lineage, and in succeeding generations TSE gradually increases. A total of six generations are necessary, however, to reach a repression level close to that of the maternal lineage. Thus, a memory of the initial maternal effect is observed over generations. In conclusion, TSE is characterized by maternal transmission that gets progressively reinforced over successive generations. It is therefore partially epigenetically transmitted through meiosis.
Figure 2. The Level of TSE Is Epigenetically Transmitted through Meiosis.
Two kinds of G1 females were generated that carry the telomeric silencer P-1152 and the P-otu-lacZ transgene (P-Co1) used as a TSE target (mating schemes are shown with the complete genotypes on Figure S1). In one lineage, P-1152 was maternally introduced, whereas it was paternally introduced in the other lineage. Because of the maternal effect of TSE, the first lineage (maternal lineage) corresponds to a TSE-positive context in G1, whereas the second (paternal lineage) corresponds to a TSE-negative context in G1. G1 females from the two lineages were crossed with P-1152; P-Z-target males to generate in both cases P-1152/ + ; P-Z-target/+ G2 females. Thus G2 females from the maternal and paternal lineages have the same genotype but differ by the cytoplasm inherited in G1. This (P-1152/ + ; P-Z-target/+) genotype was maintained over generations and TSE was measured at each generation. The evolution of TSE is shown in relation to succeeding generations. The blue and pink lines correspond to the lineages in which the P-1152 telomeric silencer locus was maternally and paternally inherited in G1, respectively. Confidence intervals of the percentages (α = 5%) are given.
TSE Requires an Extrachromosomally Transmitted Component
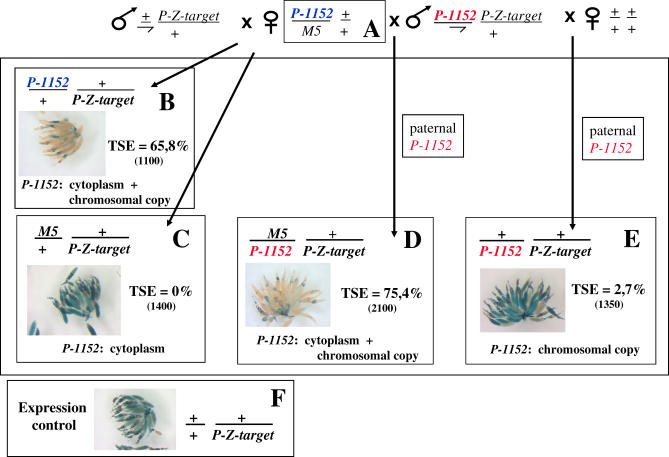
In the previous experiment, two main hypotheses can be proposed concerning the molecular basis of the difference inherited in G1 between the maternal and paternal lineages of TSE. First, maternally inherited telomeric transgenes may be imprinted while paternally inherited transgenes are not, and imprinting may be necessary for TSE to take place in the zygote. Second, a mother carrying telomeric transgenes may deposit an extrachromosomal factor in the oocyte, which is necessary for TSE to occur. This latter hypothesis was tested by using females hemizygous for the P-1152 telomeric silencer locus and a dominant genetic marker on the homologous chromosome to identify transmission of the chromosome carrying the telomeric silencer or the chromosome devoid of silencer (M5 balancer chromosome). Figure 3 shows that crossing these hemizygous females (“A” females) with males carrying a target transgene produced control G1 “B” females, which have inherited from their mother both the cytoplasm and a chromosomal copy of the telomeric silencer: in these females TSE is about 65%. However, sisters having inherited the M5 chromosome do not show any repression (“C” females, TSE = 0%). Thus, the cytoplasm of a P-1152 female without a chromosomal P-1152 copy cannot induce the TSE. Crossing P-1152 ; P-Z-target males with females devoid of telomeric silencer produces a weak repression in the progeny (2.7%) as shown by “E” females, a result consistent with the maternal effect of TSE reported previously [27]. Finally, crossing “A” females with males carrying a P-1152 telomeric silencer allows recovery of females having maternally inherited only a “P-1152” cytoplasm and paternally inherited a P-1152 chromosomal silencer. In that case, strong repression is observed (“D” females, 75% TSE). Thus the cytoplasmic component (incapable by itself of inducing TSE, as shown with “C” females) combined to a paternally inherited telomeric silencer can establish a strong TSE. Moreover, this repression is as strong as if the telomeric silencer was maternally inherited (“B” females). Consequently, the maternal effect of TSE cannot be attributed to a difference in imprinting between the maternally and paternally transmitted P-1152 telomeric silencers, but rather to an extrachromosomally transmitted factor likely deposited in the cytoplasm of the oocyte, which renders the telomeric silencers (paternally or maternally inherited) capable of establishing TSE.
Figure 3. TSE Involves a Strictly Maternally Transmitted Component.
“A” females, hemizygous for the P-1152 telomeric silencer locus, were established by crossing homozygous P-1152 females and males carrying the Muller-5 chromosome (M5) devoid of P sequence and marked by the semidominant Bar marker. These “A” females were crossed with males carrying the BQ16 euchromatic P-lacZ as target in order to recover the “B” and “C” females having inherited or not P-1152. “A” females were also crossed with P-1152; BQ16 males in order to recover the “D” females having inherited P-1152 from the father. P-1152; BQ16 males were also crossed with females devoid of P transgenes producing “E” females genotypically similar to “D” females, with the exception that they have inherited a naive cytoplasm, whereas, “D” females have inherited a P-1152 cytoplasm. “B–E” females were scored for TSE. “F” females show an expression control of the target.
Candidate Gene Analysis
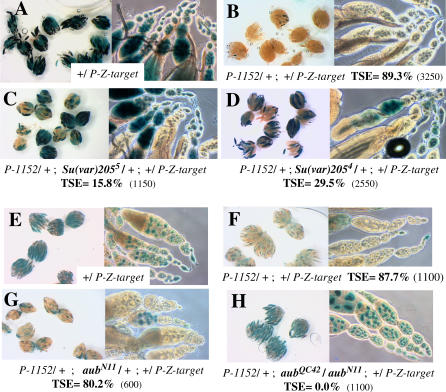
Given the variegating phenotype of TSE and the interaction observed above between a maternally transmitted factor and the chromosomal copy of the telomeric silencer, it seemed possible that heterochromatin formation and RNA silencing could be involved in this repression. A candidate gene analysis was thus performed to identify genes whose mutations affect TSE among genes encoding heterochromatin components and actors of the small RNA silencing pathways. For a given assay, a P-1152 telomeric silencer was combined with a P-lacZ target expressed in the female germline, in the absence (TSE positive control) or presence of mutant alleles of the candidate gene. The P-1152 silencer was inherited in each case from a homozygous P-1152 female. When tested in the heterozygous state, the mutant allele was maternally inherited. The first gene tested was Su(var)205 (Figure 4A–4D), which encodes HP1 [30]. HP1 is a multifunctional protein that binds at centromeres, telomeres, and some scattered sites on the chromosomal arms [31,32]. The TSE positive control produced almost 90% repression (Figure 4B), whereas one copy of the null allele Su(var)2055, corresponding to an almost completely amorphic allele, has a strong negative dominant effect on TSE (15.8% TSE remaining, Figure 4C). The allele Su(var)2054 encoding a truncated HP1 protein also behaves as a genetically null allele and also strongly impairs TSE (29.5% TSE remaining, Figure 4D). TSE is therefore sensitive to the dose of HP1. The same analysis was performed for SU(VAR)3–7, another nonhistone heterochromatin protein that binds at centromeres and telomeres and is a partner of HP1 [33,34]. SU(VAR)3–7 contains seven zinc finger motifs, which were shown to bind DNA in vitro [35,36]. Again a dose effect was observed since females heterozygous for the Su(var)3–7R2A8 null allele showed a reduced TSE (less than 40%, Table 1). Since Su(var)3–7 mutants can be homozygous viable, mutations in this gene were also tested at the homozygous state and an almost complete loss of TSE was observed (3.3% TSE remaining, Table 1). Such a result was also obtained with a second Su(var)3–7 mutant allele (Su(var)3–7R14, Table 1). Su(var)3–9 encodes a histone methyl transferase (HMT) responsible for the methylation of histone H3 on lysine 9 [37]. This protein is also a partner of HP1. The null allele of this gene tested here at the homozygous state had no significant effect on TSE (58.5% in the mutant versus 62.9% in the wild-type control, Table 1).
Figure 4. TSE Is Sensitive to Mutations in Genes Involved in Heterochromatin Formation and RNA Silencing.
In each case, the percentage of TSE is given with the total number of egg chambers counted in parentheses. Females analyzed carried a maternally inherited P-1152 telomeric silencer locus (from a homozygous P-1152 mother).
(A–D) Mutations affecting HP1. (A) Expression control of the P-otu-lacZ transgene (P-Co1) used as TSE target. (B) G1 females produced by the cross between P-1152 females and P-Co1 males. (C, D) Females having paternally inherited the P-Co1 target and maternally inherited the P-1152 telomeric silencer locus together with a Su(var)205 mutant allele.
(E–H) Mutations affecting AUBERGINE. (E) Expression control of the BQ16 target transgene. (F–H) These three types of females have maternally inherited a P-1152 telomeric silencer and paternally inherited the BQ16 target: (F) TSE positive control (aub+/aub+); (G) one maternally inherited mutant aub allele; (H) loss of function of aub in a heteroallelic mutant context.
We further tested genes involved in RNA silencing. Three primary RNA silencing pathways have been discovered so far: miRNA, siRNA (classically termed “RNA interference”), and rasiRNA pathways. These pathways may partially overlap, since they have certain actors in common, but they differ in terms of the biogenesis of short RNAs. miRNAs derive from hairpin RNAs and target numerous essential genes; siRNAs derive from double-strand RNAs and can serve as a defence mechanism against parasites such as viruses; and rasiRNAs derive from interactions between two complementary types of PIWI-interacting RNAs and have numerous targets but are more specific of TEs and heterochromatic sequences [3,4,6,29]. We tested three genes (aubergine [aub], homeless [hls], and armitage [armi]) involved in both the siRNA pathway [38,39] and the rasiRNA pathway in Drosophila ovaries [3,4,29]. Mutations of these genes can induce defaults in siRNA-guided cleavage or in the production of rasiRNAs depending on the target of the silencing [3,39,40]. These mutations are also responsible for disruption of embryonic axis specification linked to disturbance of microtubule polarisation, but this effect is mediated by a mechanism different from rasiRNA silencing itself [41]. The first gene tested is aub, an RNA binding protein and member of the Argonaute family (Figure 4E–4H) [42]. No significant dose effect of aub was detected (Figure 4G and unpublished data), but the two heteroallelic aub mutant genotypes tested completely abolish TSE (Figure 4H; Table 1). The second one is hls, which encodes an RNA helicase [43]. Again no dose effect was observed (unpublished data), but a heteroallelic hls mutant genotype completely abolishes TSE (Table 1). The third one is armi, which encodes a putative RNA helicase [44,45]. No dose effect was observed (unpublished data), but a complete loss of TSE was observed in a heteroallelic mutant context (Table 1). Finally piwi, another member of the Argonaute family [46] shown to be involved in the rasiRNA pathway [3,4,6,29,47] was tested. This gene is also necessary for germline stem-cell renewal [48] and was shown to be required for post-transcriptional transgene silencing (a phenomenon termed cosuppression)[49]. No dose effect on TSE was observed (unpublished data). Because piwi has deleterious effects on ovary structure at the homozygous state, we tested the effects of piwi mutant alleles at the heterozygous state in combination with a Su(var)205 mutant. In these combinations, piwi mutant alleles aggravate the negative effect of reduction of the dose of HP1: the single Su(var)205 mutant shows 29.5% TSE, whereas the double heterozygotes show 10.0% and 5.6% for piwi1 and piwi2 respectively (Table 1). These results indicate that the siRNA and/or the rasiRNA pathways are involved in TSE.
To determine which of these two pathways is responsible for TSE, several other mutations were tested. r2d2 is involved in the siRNA, but not the rasiRNA pathway [3,50,51]. R2D2 is a double-stranded RNA (dsRNA) binding protein involved in siRNA loading onto RISC [50]. In ovaries of females homozygous for the r2d21 null allele, the siRNA pathway is severely affected, as shown by a dsRNA-initiated RISC (or RNA-induced silencing complex) assay [52]. These ovaries show somewhat abnormal ovarioles (Figure S2), but quantification of TSE was nonetheless possible. The loss of function for r2d2 does not affect TSE (Table 1). A second protein DICER-2 (DCR-2), which combined with R2D2 is responsible for dsRNA cleavage, is also involved specifically in the siRNA pathway [53]. The loss of function of Dcr-2 is fertile. Unfortunately, we were not able to test the effect of a Dcr-2 mutant allele on TSE. Indeed, the control staining (in absence of a telomeric silencer) of the two different P-lacZ target transgenes that we tested in homozygous Dcr-2L811fsX mutants intriguingly showed almost no staining in ovaries, thus making it impossible to assay any target repression by a telomeric silencer in this context. Finally, a mutant allele of loquacious (loqs), a dsRNA binding protein involved in the miRNA pathway was tested. The loqsf00791 allele corresponds to a hypomorphic allele that alters miRNA-induced silencing in all tissues where it has been tested, including ovaries [54]. This allele is viable, allowing us to test homozygous mutant females. No effect on TSE was detected (Table 1).
In conclusion, TSE strongly depends on the function of two genes involved in heterochromatin formation and four genes involved in the rasiRNA silencing pathway but is not affected by mutations of genes involved in the siRNA or miRNA pathways.
Discussion
Our study of the regulatory mechanism of a TE that recently invaded the D. melanogaster genome allowed the discovery of a master site for establishing strong transposon repression in the germline; indeed, one or two P elements inserted in TASs are sufficient to repress the mobility of the whole P family [21]. TSE was further discovered [26], a mechanism by which a transgene inserted in subtelomeric heterochromatin has the capacity to repress a homologous transgene located in euchromatin. This phenomenon appears general since it can be induced by transgenes inserted at the telomere of the X chromosome as well as autosomal telomeres, and the targets can be located on all major chromosomes [26,28]. TSE corresponds thus to a useful tool to investigate the properties of the telomeric master site of P repression. Here, we show that TSE requires genes encoding proteins involved in heterochromatin formation (including HP1, a major component of heterochromatin) and proteins involved in the rasiRNA silencing pathway. Another TE master regulatory locus has been described in Drosophila close to the centromere of the X chromosome. It was first shown to regulate the gypsy retrotransposon and referred to as the flamenco locus [55]. This locus was further characterized as regulating together two other retrotransposons, ZAM and Idefix, and proposed as a repression center for multiple TEs, known as “COM” (Centre Organisateur de Mobilisation) [14]. Repression by this locus is based on RNA silencing, but differs from TSE in that repression of gypsy, ZAM, and Idefix occurs in the somatic follicle cells of the ovary. This point is important since in somatic follicle cells, AUBERGINE, a major actor of the rasiRNA pathway, is not expressed [4]. Conversely, in the germline, where TSE is active, all known components of the rasiRNA machinery (AUBERGINE, PIWI, and AGO3) are present. Consequently, TSE is particularly appropriate to investigate the genetic properties of a complete rasiRNA-based repression machinery.
What Is the Molecular Support of Epigenetic Transmission of TSE?
Here, we show that TSE exhibits a maternal memory that can be detected for six generations and has thus a partial epigenetic transmission through meiosis (Figure 2). The genetic analysis indicates that potentialization of the telomeric silencer (“potentialization step”) appears to be a prerequisite for repression of the target transgene by the telomeric silencer (“target repression step”). For the potentialization of the telomeric silencer, both an extrachromosomal maternally transmitted factor, produced by females carrying a telomeric silencer, and a chromosomal copy of the telomeric silencer are necessary (Figure 3). The molecular nature of this extrachromosomal maternally transmitted factor, responsible for the maternal effect, is not known at the moment but it may correspond to rasiRNAs since TAS homologous rasiRNAs have been reported (see Table S1 of [4]). We have attempted to identify short RNAs made by telomeric transgenes by northern blot on ovaries but did not detect them; this may be due to a low concentration of such short RNAs. We propose two nonmutually exclusive mechanisms. In the first, which derives from the “rasiRNA positive loop model” proposed by Brennecke et al. (“ping-pong model”) [4] and Gunawardane et al. [29], females deposit PIWI-interacting RNAs (piRNAs) in the oocyte produced by telomeric transgenes inserted in tandemly repeated TAS heterochromatic sequences. As proposed in their model, these RNAs would be primarily small antisense RNAs associated with PIWI or AUBERGINE. These piRNAs would interact in the embryo with sense RNAs produced by the telomeric transgenes and result in the production of small sense RNAs associated with AGO3, which in turn would promote the production of a new antisense piRNA, resulting in a positive feedback loop. Such a loop can explain the two-component interaction shown in Figure 3 (see below).
A second mechanism can be proposed for the potentialization step. Indeed, RNA silencing and heterochromatin formation have been shown to be connected since “short RNA-dependent heterochromatin formation” pathways have been described in several species (fission yeast [56], ciliates [57], plants [58], and Drosophila [59], for a review see [60–62]). According to this model, maternally transmitted small RNA molecules may modify the chromatin structure of the chromosomal copy of the (paternally or maternally transmitted) telomeric transgenes in the embryo and thereby confer to the telomeric transgene the capacity to be a silencer. Recently, RNA-dependent heterochromatin formation was observed at Drosophila telomeres since mutations of homeless were shown to cause an opening of chromatin at the level of telomeric retroelements in Drosophila ovaries (M. Klenov and V. Gvozdev, personal communication). Such a model is consistent with the results of our candidate gene analysis since mutations in two genes involved in heterochromatin formation (Su(var)205 and Su(var)3–7) strongly impair TSE. HP1 could play a role in heterochromatin formation, not only as a component of heterochromatin but also indirectly: indeed, in Schizosaccharomyces pombe CHP1 (a chromodomain protein) has been shown to be part of a complex that drives small RNAs to chromatin in order to modify histone H3 methylation [63]. Such a role for the HP1 chromodomain protein in a similar complex could also exist in Drosophila.
How can maternal memory of TSE over six generations be explained? Because of the modality of TSE transmission over several generations, it can be proposed that establishment of the hypothetical extrachromosomal component discussed above requires a number of generations to reach a sufficient concentration to establish strong repression. Under the “rasiRNA positive loop model” proposed by Brennecke et al. [4] and Gunawardane et al. [29], when males carrying a telomeric silencer are crossed with females bearing no silencer, the cytoplasm of the oocytes would lack piRNAs homologous to the transgene. However, synthesis of these RNAs would begin in these G1 females stimulating this pathway and increasing the piRNA concentration. At each generation, females would transmit to their daughters a higher concentration of rasiRNA molecules than at the previous generation. At generation six, a sufficient level would be reached to allow strong TSE. Under the “RNA-dependent heterochromatin formation” model, when males carrying a telomeric silencer are crossed with females bearing no silencer, again the cytoplasm of the oocytes would lack small RNAs homologous to the transgene. However, synthesis of these RNAs would begin in these G1 females stimulating heterochromatinization of the telomeric silencer. This change in the chromatin state would stimulate the production of rasiRNAs at the locus also establishing a positive loop, i.e., reciprocal stimulation between heterochromatin formation and production of small RNAs. Such a reciprocal positive loop between heterochromatin formation and small RNA silencing has been proposed for gene silencing in S. pombe [64,65]. Again, at each generation, females would receive from their mother and transmit to their daughters a higher concentration of RNA molecules than at the previous generation until a sufficient level is reached at generation six.
How Does the “Target Repression Step” Occur?
Repression of the target transgene by the telomeric silencer was shown to be homology dependent [23,26,28]. TSE however does not require homology at the level of the promoters of the telomeric and euchromatic insertions since a defective telomeric P element lacking the P promoter can repress a P-lacZ transgene in which lacZ is driven by a heat-shock promoter [23]. The TSE decision appears to be established at the cystoblast stage and stably maintained since the 15 deriving nurse cells are roughly identical in regard to their repression state. Another important point in understanding the repression mechanism is that the telomeric silencer can lack the lacZ gene, i.e., the gene repressed by TSE in the target transgene. For example, a P-white-yellow transgene [66] inserted at 1A can repress the euchromatic P-otu-lacZ-white (pCo) transgene used in this study (unpublished data). In this case, the homology between the telomeric and the euchromatic transgenes comes from the white transformation marker. This rules out the hypothesis that silencing of the target occurs via an interaction between rasiRNAs deriving from the telomeric silencer and the target transgene transcript; indeed, the targeted transcript in TSE corresponds to the lacZ sequence and no lacZ rasiRNAs can be produced by the silencer. Therefore, TSE would not occur by post-transcriptional gene silencing.
Target repression could thus occur via transcriptional gene silencing. This would fit with the variegated phenotype of repression observed. Such variegation strongly suggests that repressed targets undergo heterochromatinization. Two hypotheses can be proposed to explain such heterochromatinization. First, rasiRNAs produced by the telomeric silencer would interact with nascent transcripts on the target, and this interaction could provoke a local heterochromatinization of the target [64,65]. Heterochromatinization may then spread along the target as suggested by the repression of a P-otu-lacZ-white target by a telomeric P-white-yellow transgene. In this case heterochromatinization would start on the white sequence of the target and subsequently reach the lacZ sequence. HP1 and SU(VAR)3–7 could be involved in this spreading since centromeric heterochromatin spreading has been shown to be sensitive to the dose of these proteins [35,67]. An alternative hypothesis for target heterochromatinization is a DNA–DNA interaction between the telomeric and the euchromatic transgenes, leading to their pairing and to trans-heterochromatinization of the target following this pairing. Indeed, telomeric silencers are themselves in a heterochromatic state due to the heterochromatic nature of the TAS [68–71]. This is illustrated by a phenomenon referred to as telomeric position effect, in which a transgene inserted in TAS shows variegation in the eye for the transformation marker [72–75]. Again HP1 and SU(VAR)3–7 could play a role in this trans-heterochromatinization.
What Is the Role of the Telomere Cap in TSE?
Drosophila telomeres can be divided into three domains with respect to chromatin structure [71]: (1) the subtelomeric cluster of heterochromatic TAS repeats [68–70] in which the tested P elements or P transgenes are inserted; (2) distal to the TAS, a telomeric retrotransposon array tandemly repeated in the same orientation [76–80], the retrotransposon array is partially euchromatic [71], and transgenes located inside this domain do not show variegation for an eye marker [81]; and (3) at the extremity of the retrotransposon array, the telomere protein cap that prevents telomere fusion and regulates telomeric retrotransposon transcription and transposition [82–85]. Interestingly, HP1 (Su(var)205) and SU(VAR)3–7, both implicated in TSE, are part of the cap and therefore could be required for the telomeric locus to be a silencer. This hypothesis is supported by the analysis of the different Su(var)205 alleles. In addition to the two null alleles of Su(var)205 tested here, which showed a strong effect on TSE (Table 1), we also tested a deficiency and the Su(var)2052 allele corresponding to a mutation in the chromodomain (involved in the histone H3 K9-methylated binding), which leaves the capping activity intact [85,86]. The deletion of Su(var)205 as expected has a strong effect on TSE but Su(var)2052 has no effect (unpublished data). This suggests that HP1 effect can be mediated (at least partially) via its capping activity. Therefore, HP1 and SU(VAR)3–7 could affect the telomeric transgenes in TAS indirectly via such capping activity, for example, by affecting the localisation of the telomere inside the nucleus.
Finally, it could appear paradoxical that an amorphic mutant allele of the Su(var)3–9 HMT has no effect on TSE since RNA-dependent heterochromatin formation in most cases involves the histone H3 methylation transition from lysine 4 to lysine 9 to establish the link between short RNAs and formation of heterochromatin. However, this result can be explained by the fact that this HMT has been recently shown not to be responsible for histone H3 methylation on lysine 9 at Drosophila telomeres [85]; another unidentified HMT likely plays this role. We are therefore pursuing the candidate gene analysis in order to identify a HMT involved in TSE.
Conclusions
Telomeric TSE appears to be a complex repression mechanism that requires genes involved in heterochromatin formation and in RNA silencing. In Drosophila, interaction between small RNA silencing pathways and transcriptional repression was previously shown to exist in somatic tissues for cosuppression phenomena or for variegation phenotypes with the white marker in the eye [49,59]. TSE shows that interaction between RNA silencing and heterochromatin formation can also occur in the germline. This type of silencing appears to be the basic mechanism for P-element repression [28], although some P-encoded repressors encoded by euchromatic copies may contribute in some cases to P repression [10,87]. TSE has the same complex inheritance as the P cytotype [10,88,89]. The subtelomeric heterochromatin thus represents a piRNA producing “platform” [4] available for recent invaders of the genomes to establish their own repression. The study of TSE illustrates the genetic properties of such a platform.
Materials and Methods
Experimental conditions.
All crosses were performed at 25 °C and involved three to five couples in most cases. All ovary lacZ expression assays were carried out using X-gal overnight staining as described in Lemaitre et al. 1993 [90], except that ovaries were fixed for 6 min.
Transgenes and strains.
P-lacZ fusion enhancer trap transgenes (P-1152, BQ16, and BC69) contain an in-frame translational fusion of the Escherichia coli lacZ gene to the second exon of the P transposase gene and contain rosy+ as a transformation marker [91]. The P-1152 insertion comes from stock number 11152 of the Bloomington Stock Center (http://flystocks.bio.indiana.edu) and was mapped at the telomere of the X chromosome (site 1A); this stock was previously described to carry a single P-lacZ insertion in TAS [26]. However, in our number 11152 stock, we have mapped two P-lacZ insertions in the same TAS unit and in the same orientation that might have resulted from an unequal recombination event duplicating the P-lacZ transgene. P-1152 is homozygous viable and fertile. BQ16 is located at 64C in euchromatin of the third chromosome and is homozygous viable and fertile. BC69 is inserted on Chromosome 2 in the first exon of the vasa gene and results in a vasa loss of function; it is consequently homozygous female sterile. P-1152 shows no lacZ expression in the ovary (Figure 1A), whereas BQ16 and BC69 are strongly expressed in the nurse cells and in the oocyte (Figure 1B and 1D, respectively). P-Co1 is an insertion of the pCo transgene (P-otu-lacZ) on the third chromosome (87A-B), which is homozygous viable and fertile. β-galactosidase expression of the pCo transgene is driven by the otu promoter and is therefore strongly detected in both nurse cells and the mature oocyte [27]. This transgene contains a white gene as a transformation marker.
Lines carrying transgenes have M genetic backgrounds (devoid of P TEs), as well as the multimarked balancer stocks used in genetic experiments (yw; Cy; TM3Sb / T(2;3)apXa, M5; Cy / T(2;3)apXa and M5; TM3Sb/ T(2;3)apXa) and the strains carrying mutations used for the candidate gene analysis (see below).
Mutations used for the candidate gene analysis.
Su(var)205, aub, piwi, r2d2, and loqs genes are located on Chromosome 2, whereas Su(var)3–7, Su(var)3–9, hls (synonym of spindle-E), and armi are located on Chromosome 3. Loss of function is lethal in the case of Su(var)205 and loqs, female sterile in the case of aub, piwi, hls, r2d2, and armi, and is viable and fertile in the case of Su(var)3–9. Loss of function of Su(var)3–7 is viable when the mother is heterozygous mutant but lethal when the mother is homozygous mutant [92].
Su(var)2–505 was x-ray induced and corresponds to a null allele since it only encodes the first ten amino acids of the HP1 protein [67]. Su(var)2–504 was EMS induced and encodes a truncated HP1 protein, ∼85% of wild type size [67], missing a domain necessary for targeting of HP1 to the nucleus [93]. Lines carrying these mutations were kindly provided by Gunter Reuter.
Su(var)3–7R2A8 and Su(var)3–7R14 were generated by homologous recombination [92,94]. Su(var)3–7R14 produces a chimeric protein and Su(var)3–7R2A8 deletes most of the protein-coding sequence of the gene. These two alleles behave as genetic null mutations. Lines carrying these mutations were kindly provided by Marion Delattre.
Su(var)3–906 was generated by x-ray and corresponds to an amorphic allele [37,95]. Line carrying this mutation was kindly provided by Gunter Reuter.
Three strong mutant alleles of aub induced by EMS were used. All of them are homozygous female sterile. aubQC42 [96] comes from the Bloomington Stock Center (stock number 4968). aubHN2 [96] has an amino acid substitution. aubN11 [97] has a 154-bp deletion, resulting in a frameshift that is predicted to add 16 novel amino acids after residue 740 [98]. aubHN2 and aubN11 were kindly provided by Paul Macdonald.
piwi1 and piwi2 are P-element–induced mutations; piwi1 contains a P transgene inserted in the first exon [46]. The orientation of the transgene is opposite to that of the gene. piwi2 contains a P transgene inserted in exon 4 in the same orientation as the gene. In both cases, homozygous females are sterile. Lines carrying these mutations were kindly provided by Alain Bucheton.
hlsE616 (synonym of spn-E1) was generated by EMS [43,99]. hlsΔ125 (synonym of spn-EhlsΔ125) was generated by P-element excision resulting in deletion of coding sequences and adjacent sequences. Deletion may extend into other genes [43]. Lines carrying these mutations were kindly provided by Utpal Bhadra.
armi1 and armi72.1 are P-element–induced mutations; armi1 has an insertion of a P transgene in the 5′ UTR of the armitage gene [44]. armi72.1 derives from armi1 by an imprecise excision of the P element, resulting in a deletion of armi sequences in the 5′ untranslated region [39]. These two alleles are female sterile and come from the Bloomington Stock Center (stock numbers 8513 and 8544).
r2d21 results from a deletion induced by the remobilization of a P transgene located in 5′ of the r2d2 gene [50]. The 4.9-kb deletion removes the entire r2d2 open reading frame [52]. This allele is an amorphic allele and homozygous females are sterile due to abnormal ovaries. The siRNA pathway is severely affected as shown by a dsRNA-initiated RISC assay in ovaries of homozygous mutant females [52]. This line comes from the Bloomington Stock Center (stock number 8518).
loqsf00791 results from a piggyBac transgene insertion 57 bp upstream of the transcription start site of loqs [54], a gene necessary for miRNA-directed silencing. loqsf00791 is a hypomorphic allele that is homozygous viable. At the homozygous state, this allele has a strong effect on miRNA processing in ovaries. It also has a negative effect on siRNA silencing [54]. This allele comes from the Bloomington Stock Center (number 18371).
All the alleles described above are maintained over a balancer chromosome. Additional information about mutants and stocks are available at FlyBase (http://flybase.bio.indiana.edu).
Supporting Information
M5 indicates Muller5, a balancer of the X chromosome devoid of P sequence, Sb indicates Stubble, a dominant marker of the TM3 balancer Chromosome 3, and Xa means Xasta, a dominant marker of the attached Chromosomes 2 and 3 (T(2;3)apXa). In each case, the maternally inherited chromosomal complement is written above the bar. G1 females carrying the telomeric silencer P-1152 and the P-otu-lacZ transgene (P-Co1 located on Chromosome 3) used as a TSE target were generated by two reciprocal crosses; in the maternal lineage of TSE (left), P-1152 was maternally introduced, whereas it was paternally introduced in the paternal lineage (right). G1 females were crossed with P-1152; P-Co1 males to establish, in the two lineages, G2 females having the same genotype (P-1152/ M5; P-Co1/ Sb) but differing in the cytoplasm transmitted by the G0 females. The same procedure was repeated over six generations. At each generation, females were dissected and TSE was quantified in ovaries. The results are shown on Figure 2.
(51 KB PPT)
Ovaries from a homozygous r2d21 female that carries a P-1152 silencer locus and a BQ16 target transgene.
(2.4 MB PPT)
Accession Numbers
The National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov) and FlyBase (http://flybase.bio.indiana.edu) accession numbers for the genes described in this article are (respectively): armitage (CG11513, FBgn0041164); Su(var)205 (CG8409, FBgn0003607); aubergine (CG6137, FBgn0000146); piwi (CG6122, FBgn0004872); r2d2 (CG7138, FBgn0031951); loquacious (CG6866, FBgn0032515); Su(var)3–7 (CG8599, FBgn0003598); Su(var)3–9 (CG6476, FBgn0003600); and homeless (CG3158, FBgn0003483). For the P element, see FBgn0003055. For general properties of TAS, see FBgn0041614. For TAS data sequences, see LO3284, U58967, U58968, and U58969. Information about X-TAS piRNAs are in Table S1 of [4].
Acknowledgments
We thank Mikhail Klenov, Vladimir Gvozdev, Sophie Desset, and Chantal Vaury for personal communications. We thank Christophe Antoniewski, Alain Bucheton, Utpal Bhadra, Marion Delattre, Paul MacDonald Arno Muller, Gunter Reuter, and Mikiko Siomi for the gift of Drosophila lines. We thank the Bloomington Stock Center (http://flystocks.bio.indiana.edu) and FlyBase (http://flybase.bio.indiana.edu). We thank Sarah Ducamp, Julie Gabriel, Arnaud Suwalski, and Augustin de Vanssay for experimental help. We thank Valérie Delmarre and Paula Graca for technical assistance. We thank Anne-Marie Pret for her help in the preparation of the manuscript.
Abbreviations
- aub
aubergine
- Dcr-2
DICER-2
- dsRNA
double-stranded RNA
- hls
homeless
- HMT
histone methyl transferase
- HP1
Heterochromatin Protein 1
- miRNA
micro RNA
- piRNA
PIWI-interacting RNA
- rasiRNA
repeat-associated small interfering RNA
- siRNA
small interfering RNA
- TAS
Telomeric Associated Sequences
- TE
transposable element
- TSE
Trans-Silencing effect
Footnotes
A previous version of this article appeared as an Early Online Release on July 31, 2007 (doi:10.1371/journal.pgen.0030158.eor).
Author contributions. TJ, LT, ALT, CMS, and SR conceived and designed the experiments and performed the experiments. TJ, LT, ALT, CMS, DA, and SR analyzed the data. SR wrote the paper.
Funding. This work was supported by the Centre National de la Recherche Scientifique (Institut Jacques Monod-UMR 7592), by the Universités Pierre and Marie Curie-Paris 6 and Denis Diderot-Paris 7, by the Action Concertée Incitative–Biologie Cellulaire Moléculaire et Structurale program, and by the Association pour la Recherche sur le Cancer. TJ was supported by a graduate student fellowship from the French government (Ministère de l'Enseignement Supérieur et de la Recherche) and by the Association pour la Recherche sur le Cancer. ALT is supported by a postdoctoral position fellowship from the Fondation pour la Recherche Médicale.
Competing interests. The authors have declared that no competing interests exist.
References
- Lippman Z, May B, Yordan C, Singer T, Martienssen R. Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol. 2003;1:e67. doi: 10.1371/journal.pbio.0000067. doi: 10.1371/journal.pbio.0000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw NL, Fischer SE, Robert VJ, Thijssen KL, Fraser AG, et al. A genome-wide screen identifies 27 genes involved in transposon silencing in C. elegans. Curr Biol. 2003;13:1311–1316. doi: 10.1016/s0960-9822(03)00539-6. [DOI] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, et al. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, et al. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham PM, Kidwell MG, Rubin GM. The molecular basis of P-M hybrid dysgenesis: The role of the P element, a P-strain-specific transposon family. Cell. 1982;29:995–1004. doi: 10.1016/0092-8674(82)90463-9. [DOI] [PubMed] [Google Scholar]
- Bucheton A, Paro R, Sang HM, Pelisson A, Finnegan DJ. The molecular basis of I-R hybrid dysgenesis in Drosophila melanogaster: Identification, cloning, and properties of the I factor. Cell. 1984;38:153–163. doi: 10.1016/0092-8674(84)90536-1. [DOI] [PubMed] [Google Scholar]
- Kidwell MG, Kidwell JF, Sved JA. Hybrid dysgenesis in Drosophila melanogaster: A syndrome of aberrant traits including mutation, sterility, and male recombination. Genetics. 1977;86:813–833. doi: 10.1093/genetics/86.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels WR. In: P elements in Drosophila. Berg DE, Howe MM, editors. Washington, D. C.: American Society for Microbiology; 1989. pp. 437–484. [Google Scholar]
- Blumenstiel JP, Hartl DL. Evidence for maternally transmitted small interfering RNA in the repression of transposition in Drosophila virilis. Proc Natl Acad Sci U S A. 2005;102:15965–15970. doi: 10.1073/pnas.0508192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen S, Gassama MP, Heidmann T. Taming of transposable elements by homology-dependent gene silencing. Nat Genet. 1999;21:209–212. doi: 10.1038/5997. [DOI] [PubMed] [Google Scholar]
- Jensen S, Gassama MP, Dramard X, Heidmann T. Regulation of I-transposon activity in Drosophila: Evidence for cosuppression of nonhomologous transgenes and possible role of ancestral I-related pericentromeric elements. Genetics. 2002;162:1197–1209. doi: 10.1093/genetics/162.3.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desset S, Meignin C, Dastugue B, Vaury C. COM, a heterochromatic locus governing the control of independent endogenous retroviruses from Drosophila melanogaster. Genetics. 2003;164:501–509. doi: 10.1093/genetics/164.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarot E, Payen-Groschene G, Bucheton A, Pelisson A. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics. 2004;166:1313–1321. doi: 10.1534/genetics.166.3.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmykova AI, Klenov MS, Gvozdev VA. Argonaute protein PIWI controls mobilization of retrotransposons in the Drosophila male germline. Nucleic Acids Res. 2005;33:2052–2059. doi: 10.1093/nar/gki323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Klenov MS, Kalmykova AI, Stolyarenko AD, Kotelnikov RN, et al. The RNA interference proteins and Vasa locus are involved in the silencing of retrotransposons in the female germline of Drosophila melanogaster. RNA Biol. 2004;1:54–58. [PubMed] [Google Scholar]
- Savitsky M, Kwon D, Georgiev P, Kalmykova A, Gvozdev V. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes Dev. 2006;20:345–354. doi: 10.1101/gad.370206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Pane A, Schupbach T. cutoff and aubergine mutations result in retrotransposon upregulation and checkpoint activation in Drosophila. Curr Biol. 2007;17:637–642. doi: 10.1016/j.cub.2007.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronsseray S, Lehmann M, Anxolabehere D. The maternally inherited regulation of P elements in Drosophila melanogaster can be elicited by two P copies at cytological site 1A on the X chromosome. Genetics. 1991;129:501–512. doi: 10.1093/genetics/129.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronsseray S, Lehmann M, Nouaud D, Anxolabehere D. The regulatory properties of autonomous subtelomeric P elements are sensitive to a Suppressor of variegation in Drosophila melanogaster. Genetics. 1996;143:1663–1674. doi: 10.1093/genetics/143.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons MJ, Raymond JD, Niemi JB, Stuart JR, Merriman PJ. The P cytotype in Drosophila melanogaster: A maternally transmitted regulatory state of the germ line associated with telomeric P elements. Genetics. 2004;166:243–254. doi: 10.1534/genetics.166.1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin L, Lehmann M, Nouaud D, Izaabel H, Anxolabehere D, et al. P-Element repression in Drosophila melanogaster by a naturally occurring defective telomeric P copy. Genetics. 2000;155:1841–1854. doi: 10.1093/genetics/155.4.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart JR, Haley KJ, Swedzinski D, Lockner S, Kocian PE, et al. Telomeric P elements associated with cytotype regulation of the P transposon family in Drosophila melanogaster. Genetics. 2002;162:1641–1654. doi: 10.1093/genetics/162.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss D, Josse T, Anxolabehere D, Ronsseray S. Aubergine mutations in Drosophila melanogaster impair P cytotype determination by telomeric P elements inserted in heterochromatin. Mol Genet Genomics. 2004;272:336–343. doi: 10.1007/s00438-004-1061-1. [DOI] [PubMed] [Google Scholar]
- Roche SE, Rio DC. Trans-silencing by P elements inserted in subtelomeric heterochromatin involves the Drosophila Polycomb group gene, Enhancer of zeste. Genetics. 1998;149:1839–1855. doi: 10.1093/genetics/149.4.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronsseray S, Boivin A, Anxolabehere D. P-Element repression in Drosophila melanogaster by variegating clusters of P-lacZ-white transgenes. Genetics. 2001;159:1631–1642. doi: 10.1093/genetics/159.4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronsseray S, Josse T, Boivin A, Anxolabehere D. Telomeric transgenes and trans-silencing in Drosophila. Genetica. 2003;117:327–335. doi: 10.1023/a:1022929121828. [DOI] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, et al. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- James TC, Elgin SC. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol Cell Biol. 1986;6:3862–3872. doi: 10.1128/mcb.6.11.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James TC, Eissenberg JC, Craig C, Dietrich V, Hobson A, et al. Distribution patterns of HP1, a heterochromatin-associated nonhistone chromosomal protein of Drosophila. Eur J Cell Biol. 1989;50:170–180. [PubMed] [Google Scholar]
- Fanti L, Berloco M, Piacentini L, Pimpinelli S. Chromosomal distribution of heterochromatin protein 1 (HP1) in Drosophila: A cytological map of euchromatic HP1 binding sites. Genetica. 2003;117:135–147. doi: 10.1023/a:1022971407290. [DOI] [PubMed] [Google Scholar]
- Cleard F, Delattre M, Spierer P. SU(VAR)3–7, a Drosophila heterochromatin-associated protein and companion of HP1 in the genomic silencing of position-effect variegation. Embo J. 1997;16:5280–5288. doi: 10.1093/emboj/16.17.5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delattre M, Spierer A, Tonka CH, Spierer P. The genomic silencing of position-effect variegation in Drosophila melanogaster: Interaction between the heterochromatin-associated proteins Su(var)3–7 and HP1. J Cell Sci. 2000;113:4253–4261. doi: 10.1242/jcs.113.23.4253. [DOI] [PubMed] [Google Scholar]
- Reuter G, Giarre M, Farah J, Gausz J, Spierer A, et al. Dependence of position-effect variegation in Drosophila on dose of a gene encoding an unusual zinc-finger protein. Nature. 1990;344:219–223. doi: 10.1038/344219a0. [DOI] [PubMed] [Google Scholar]
- Cleard F, Spierer P. Position-effect variegation in Drosophila: The modifier Su(var)3–7 is a modular DNA-binding protein. EMBO Rep. 2001;2:1095–1100. doi: 10.1093/embo-reports/kve243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G, Ebert A, Krauss V, Fischer A, Hoffmann J, et al. Central role of Drosophila SU(VAR)3–9 in histone H3-K9 methylation and heterochromatic gene silencing. Embo J. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennerdell JR, Yamaguchi S, Carthew RW. RNAi is activated during Drosophila oocyte maturation in a manner dependent on aubergine and spindle-E. Genes Dev. 2002;16:1884–1889. doi: 10.1101/gad.990802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y, Du T, Haley B, Schwarz DS, Bennett R, et al. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell. 2004;116:831–841. doi: 10.1016/s0092-8674(04)00218-1. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Klenov MS, Vagin VV, Bantignies F, Cavalli G, et al. Dissection of a natural RNA silencing process in the Drosophila melanogaster germ line. Mol Cell Biol. 2004;24:6742–6750. doi: 10.1128/MCB.24.15.6742-6750.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C, Bratu DP, McGinnis-Schultz N, Koppetsch BS, Cook HA, et al. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev Cell. 2007;12:45–55. doi: 10.1016/j.devcel.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Palumbo G, Bozzetti MP, Tritto P, Pimpinelli S, et al. Genetic and molecular characterization of sting, a gene involved in crystal formation and meiotic drive in the male germ line of Drosophila melanogaster. Genetics. 1999;151:749–760. doi: 10.1093/genetics/151.2.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie DE, Berg CA. Homeless is required for RNA localization in Drosophila oogenesis and encodes a new member of the DE-H family of RNA-dependent ATPases. Genes Dev. 1995;9:2495–2508. doi: 10.1101/gad.9.20.2495. [DOI] [PubMed] [Google Scholar]
- Cook HA, Koppetsch BS, Wu J, Theurkauf WE. The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell. 2004;116:817–829. doi: 10.1016/s0092-8674(04)00250-8. [DOI] [PubMed] [Google Scholar]
- Dalmay T, Horsefield R, Braunstein TH, Baulcombe DC. SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. Embo J. 2001;20:2069–2078. doi: 10.1093/emboj/20.8.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, et al. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Szakmary A, Cox DN, Wang Z, Lin H. Regulatory relationship among piwi, pumilio, and bag-of-marbles in Drosophila germline stem cell self-renewal and differentiation. Curr Biol. 2005;15:171–178. doi: 10.1016/j.cub.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M, Bhadra U, Birchler JA. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol Cell. 2002;9:315–327. doi: 10.1016/s1097-2765(02)00440-9. [DOI] [PubMed] [Google Scholar]
- Liu Q, Rand TA, Kalidas S, Du F, Kim HE, et al. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Jiang F, Kalidas S, Smith D, Liu Q. Dicer-2 and R2D2 coordinately bind siRNA to promote assembly of the siRISC complexes. RNA. 2006;12:1514–1520. doi: 10.1261/rna.101606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, et al. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- Forstemann K, Tomari Y, Du T, Vagin VV, Denli AM, et al. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelisson A, Song SU, Prud'homme N, Smith PA, Bucheton A, et al. Gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissue-specific control of the Drosophila flamenco gene. Embo J. 1994;13:4401–4411. doi: 10.1002/j.1460-2075.1994.tb06760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, et al. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell. 2002;110:689–699. doi: 10.1016/s0092-8674(02)00909-1. [DOI] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M, Leibovitch BA, Gandhi SG, Rao M, Bhadra U, et al. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303:669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Allis CD. RNA meets chromatin. Genes Dev. 2005;19:1635–1655. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Cam H, Verdel A, Moazed D, Grewal SI. RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc Natl Acad Sci U S A. 2005;102:152–157. doi: 10.1073/pnas.0407641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel A, Moazed D. RNAi-directed assembly of heterochromatin in fission yeast. FEBS Lett. 2005;579:5872–5878. doi: 10.1016/j.febslet.2005.08.083. [DOI] [PubMed] [Google Scholar]
- Buhler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125:873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- Irvine DV, Zaratiegui M, Tolia NH, Goto DB, Chitwood DH, et al. Argonaute slicing is required for heterochromatic silencing and spreading. Science. 2006;313:1134–1137. doi: 10.1126/science.1128813. [DOI] [PubMed] [Google Scholar]
- Roseman RR, Johnson EA, Rodesch CK, Bjerke M, Nagoshi RN, et al. A P element containing suppressor of hairy-wing binding regions has novel properties for mutagenesis in Drosophila melanogaster. Genetics. 1995;141:1061–1074. doi: 10.1093/genetics/141.3.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg JC, Morris GD, Reuter G, Hartnett T. The heterochromatin-associated protein HP-1 is an essential protein in Drosophila with dosage-dependent effects on position-effect variegation. Genetics. 1992;131:345–352. doi: 10.1093/genetics/131.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis R, Hazelrigg T, Rubin GM. Effects of genomic position on the expression of transduced copies of the white gene of Drosophila. Science. 1985;229:558–561. doi: 10.1126/science.2992080. [DOI] [PubMed] [Google Scholar]
- Karpen GH, Spradling AC. Analysis of subtelomeric heterochromatin in the Drosophila minichromosome Dp1187 by single P element insertional mutagenesis. Genetics. 1992;132:737–753. doi: 10.1093/genetics/132.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter MF, Jang C, Kasravi B, Donath J, Mechler BM, et al. DNA organization and polymorphism of a wild-type Drosophila telomere region. Chromosoma. 1995;104:229–241. doi: 10.1007/BF00352254. [DOI] [PubMed] [Google Scholar]
- Andreyeva EN, Belyaeva ES, Semeshin VF, Pokholkova GV, Zhimulev IF. Three distinct chromatin domains in telomere ends of polytene chromosomes in Drosophila melanogaster Tel mutants. J Cell Sci. 2005;118:5465–5477. doi: 10.1242/jcs.02654. [DOI] [PubMed] [Google Scholar]
- Wallrath LL, Elgin SC. Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev. 1995;9:1263–1277. doi: 10.1101/gad.9.10.1263. [DOI] [PubMed] [Google Scholar]
- Cryderman DE, Morris EJ, Biessmann H, Elgin SC, Wallrath LL. Silencing at Drosophila telomeres: Nuclear organization and chromatin structure play critical roles. Embo J. 1999;18:3724–3735. doi: 10.1093/emboj/18.13.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubovsky MD, Konev AY, Walter MF, Biessmann H, Mason JM. Terminal retrotransposons activate a subtelomeric white transgene at the 2L telomere in Drosophila. Genetics. 2001;158:1111–1123. doi: 10.1093/genetics/158.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason JM, Konev AY, Golubovsky MD, Biessmann H. Cis- and trans-acting influences on telomeric position effect in Drosophila melanogaster detected with a subterminal transgene. Genetics. 2003;163:917–930. doi: 10.1093/genetics/163.3.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biessmann H, Champion LE, O'Hair M, Ikenaga K, Kasravi B, et al. Frequent transpositions of Drosophila melanogaster HeT-A transposable elements to receding chromosome ends. Embo J. 1992;11:4459–4469. doi: 10.1002/j.1460-2075.1992.tb05547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis RW, Ganesan R, Houtchens K, Tolar LA, Sheen FM. Transposons in place of telomeric repeats at a Drosophila telomere. Cell. 1993;75:1083–1093. doi: 10.1016/0092-8674(93)90318-k. [DOI] [PubMed] [Google Scholar]
- Sheen FM, Levis RW. Transposition of the LINE-like retrotransposon TART to Drosophila chromosome termini. Proc Natl Acad Sci U S A. 1994;91:12510–12514. doi: 10.1073/pnas.91.26.12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardue ML, DeBaryshe PG. Retrotransposons provide an evolutionarily robust non-telomerase mechanism to maintain telomeres. Annu Rev Genet. 2003;37:485–511. doi: 10.1146/annurev.genet.38.072902.093115. [DOI] [PubMed] [Google Scholar]
- Abad JP, De Pablos B, Osoegawa K, De Jong PJ, Martin-Gallardo A, et al. TAHRE, a novel telomeric retrotransposon from Drosophila melanogaster, reveals the origin of Drosophila telomeres. Mol Biol Evol. 2004;21:1620–1624. doi: 10.1093/molbev/msh180. [DOI] [PubMed] [Google Scholar]
- Biessmann H, Prasad S, Semeshin VF, Andreyeva EN, Nguyen Q, et al. Two distinct domains in Drosophila melanogaster telomeres. Genetics. 2005;171:1767–1777. doi: 10.1534/genetics.105.048827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biessmann H, Carter SB, Mason JM. Chromosome ends in Drosophila without telomeric DNA sequences. Proc Natl Acad Sci U S A. 1990;87:1758–1761. doi: 10.1073/pnas.87.5.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitsky M, Kravchuk O, Melnikova L, Georgiev P. Heterochromatin protein 1 is involved in control of telomere elongation in Drosophila melanogaster. Mol Cell Biol. 2002;22:3204–3218. doi: 10.1128/MCB.22.9.3204-3218.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenci G, Siriaco G, Raffa GD, Kellum R, Gatti M. The Drosophila HOAP protein is required for telomere capping. Nat Cell Biol. 2003;5:82–84. doi: 10.1038/ncb902. [DOI] [PubMed] [Google Scholar]
- Perrini B, Piacentini L, Fanti L, Altieri F, Chichiarelli S, et al. HP1 controls telomere capping, telomere elongation, and telomere silencing by two different mechanisms in Drosophila. Mol Cell. 2004;15:467–476. doi: 10.1016/j.molcel.2004.06.036. [DOI] [PubMed] [Google Scholar]
- Fanti L, Giovinazzo G, Berloco M, Pimpinelli S. The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol Cell. 1998;2:527–538. doi: 10.1016/s1097-2765(00)80152-5. [DOI] [PubMed] [Google Scholar]
- Rio DC. Regulation of Drosophila P element transposition. Trends Genet. 1991;7:282–287. doi: 10.1016/0168-9525(91)90309-E. [DOI] [PubMed] [Google Scholar]
- Ronsseray S, Lemaitre B, Coen D. Maternal inheritance of P cytotype in Drosophila melanogaster: A “pre-P cytotype” is strictly extra-chromosomally transmitted. Mol Gen Genet. 1993;241:115–123. doi: 10.1007/BF00280208. [DOI] [PubMed] [Google Scholar]
- Coen D, Lemaitre B, Delattre M, Quesneville H, Ronsseray S, et al. Drosophila P element: Transposition, regulation and evolution. Genetica. 1994;93:61–78. doi: 10.1007/BF01435240. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Ronsseray S, Coen D. Maternal repression of the P element promoter in the germline of Drosophila melanogaster: A model for the P cytotype. Genetics. 1993;135:149–160. doi: 10.1093/genetics/135.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Kane CJ, Gehring WJ. Detection in situ of genomic regulatory elements in Drosophila. Proc Natl Acad Sci U S A. 1987;84:9123–9127. doi: 10.1073/pnas.84.24.9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seum C, Pauli D, Delattre M, Jaquet Y, Spierer A, et al. Isolation of Su(var)3–7 mutations by homologous recombination in Drosophila melanogaster. Genetics. 2002;161:1125–1136. doi: 10.1093/genetics/161.3.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellum R, Raff JW, Alberts BM. Heterochromatin protein 1 distribution during development and during the cell cycle in Drosophila embryos. J Cell Sci. 1995;108:1407–1418. doi: 10.1242/jcs.108.4.1407. [DOI] [PubMed] [Google Scholar]
- Spierer A, Seum C, Delattre M, Spierer P. Loss of the modifiers of variegation Su(var)3–7 or HP1 impacts male X polytene chromosome morphology and dosage compensation. J Cell Sci. 2005;118:5047–5057. doi: 10.1242/jcs.02623. [DOI] [PubMed] [Google Scholar]
- Tschiersch B, Hofmann A, Krauss V, Dorn R, Korge G, et al. The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3–9 combines domains of antagonistic regulators of homeotic gene complexes. Embo J. 1994;13:3822–3831. doi: 10.1002/j.1460-2075.1994.tb06693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics. 1991;129:1119–1136. doi: 10.1093/genetics/129.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JE, Connell JE, Schlenker JD, Macdonald PM. Novel genetic screen for genes involved in posterior body patterning in Drosophila. Dev Genet. 1996;19:199–209. doi: 10.1002/(SICI)1520-6408(1996)19:3<199::AID-DVG3>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Harris AN, Macdonald PM. Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development. 2001;128:2823–2832. doi: 10.1242/dev.128.14.2823. [DOI] [PubMed] [Google Scholar]
- Tearle RG, Nusslein-Volhard C. Tubingen mutants and stock list. Drosoph Inf Serv. 1987;66:209–269. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
M5 indicates Muller5, a balancer of the X chromosome devoid of P sequence, Sb indicates Stubble, a dominant marker of the TM3 balancer Chromosome 3, and Xa means Xasta, a dominant marker of the attached Chromosomes 2 and 3 (T(2;3)apXa). In each case, the maternally inherited chromosomal complement is written above the bar. G1 females carrying the telomeric silencer P-1152 and the P-otu-lacZ transgene (P-Co1 located on Chromosome 3) used as a TSE target were generated by two reciprocal crosses; in the maternal lineage of TSE (left), P-1152 was maternally introduced, whereas it was paternally introduced in the paternal lineage (right). G1 females were crossed with P-1152; P-Co1 males to establish, in the two lineages, G2 females having the same genotype (P-1152/ M5; P-Co1/ Sb) but differing in the cytoplasm transmitted by the G0 females. The same procedure was repeated over six generations. At each generation, females were dissected and TSE was quantified in ovaries. The results are shown on Figure 2.
(51 KB PPT)
Ovaries from a homozygous r2d21 female that carries a P-1152 silencer locus and a BQ16 target transgene.
(2.4 MB PPT)