Abstract
Co-ordinated regulation of gene expression is required for the transmission and survival of Borrelia burgdorferi in different hosts. The sigma factor RpoS (σS), as regulated by RpoN (σ54), has been shown to regulate key virulence factors (e.g. OspC) required for these processes. As important, multiple signals (e.g. temperature, pH, cell density, oxygen) have been shown to increase the expression of σS-dependent genes; however, little is known about the signal transduction mechanisms that modulate the expression of rpoS. In this report we show that: (i) rpoS has a σ54-dependent promoter that requires Rrp2 to activate transcription; (ii) Rrp2Δ123, a constitutively active form of Rrp2, activated σ54-dependent transcription of rpoS/P-lacZ reporter constructs in Escherichia coli; (iii) quantitative reverse transcription polymerase chain reaction (QRT-PCR) experiments with reporter cat constructs in B. burgdorferi indicated that Rrp2 activated transcription of rpoS in an enhancer-independent fashion; and finally, (iv) rpoN is required for cell density- and temperature-dependent expression of rpoS in B. burgdorferi, but histidine kinase Hk2, encoded by the gene immediately upstream of rrp2, is not essential. Based on these findings, a model for regulation of rpoS has been proposed which provides mechanisms for multiple signalling pathways to modulate the expression of the σS regulon in B. burgdorferi.
Introduction
Borrelia burgdorferi, the Lyme disease agent, is maintained in nature in two very distinct environments, Ixodes ticks and mammalian hosts (Burgdorfer et al., 1982; Steere et al., 1983). As B. burgdorferi moves from one host to another, its ability to adapt and survive is attributed to its capacity to sense changes in temperature, pH, cell density, oxygen and/or exposure to host factors and alter gene expression accordingly (Schwan et al., 1995; Stevenson et al., 1995; Indest et al., 1997; Ramamoorthy and Philipp, 1998; Carroll et al., 1999; Indest and Philipp, 2000; Schwan and Piesman, 2000; Ojaimi et al., 2003; Seshu et al., 2004). Previous reports have demonstrated that central to the regulation of these adaptive responses are σS (encoded by rpoS) and σ54 (encoded by rpoN, also known as ntrA). In addition, σ54-dependent expression of σS is responsible for the expression of key virulence factors [e.g. outer surface protein C (OspC), OspA and decorin binding protein A (DbpA)] required for infectivity and transmission during the infective cycle (Elias et al., 2000; Yang et al., 2000; Hübner et al., 2001; Caimano et al., 2004; Fisher et al., 2005).
σ54-dependent transcription is a tightly regulated process requiring an activator protein that stimulates the isomerization of the closed σ54-RNAP (RNA polymerase) holoenzyme/promoter complex to the open complex in a reaction that requires ATP hydrolysis by the activator (Sasse-Dwight and Gralla, 1988; Popham et al., 1989). In general, σ54-dependent activators are modular in structure consisting of an N-terminal regulatory domain, a central ATPase domain and a C-terminal DNA binding domain (North et al., 1996; Ogura and Wilkinson, 2001; Xu and Hoover, 2001). These activators bind enhancer-like sequences upstream of their target promoters and then contact the closed complexes through DNA looping (Su et al., 1990). In B. burgdorferi, the two-component response regulator Rrp2 (BB0763) is a σ54-dependent activator that controls rpoS expression (Yang et al., 2003a). Attempts to disrupt rrp2 have been unsuccessful so far, but a single amino-acid change within the ATPase domain of Rrp2 eliminates σS-dependent lipoprotein expression by disrupting the cascade involved in the σ54-dependent expression of rpoS (Yang et al., 2003a). Because both B. burgdorferi rpoN and rpoS mutants are viable (Elias et al., 2000; Hübner et al., 2001; Fisher et al., 2005), the inability to generate Rrp2-deficient mutants suggests that Rrp2 has an essential function in B. burgdorferi that is separate from its role in activating transcription of rpoS (Yang et al., 2003a).
The gene immediately upstream of rrp2, designated hk2 (BB0764), is predicted to encode a histidine kinase through which sensory information is channelled to activate Rrp2 (Yang et al., 2003a). Hk2 is thought to donate phosphate to Rrp2 to stimulate its activity; however, the specific signals that trigger the autophosphorylation of Hk2 and influence its interactions with Rrp2 have not been identified. Another intriguing aspect of the σ54–σS regulatory pathway is that multiple signals (e.g. temperature, pH and cell density) seem to simulate the expression of the σS-dependent genes, suggesting that additional systems influence the transcriptional control of rpoS.
In this article, we report the identification of the rpoS transcriptional start site downstream of a σ54-dependent promoter and reveal characteristics of Rrp2 that potentially explain how this protein plays a more complex role in transcriptional regulation of rpoS. In addition, we constructed an hk2 mutant and assessed the involvement of this putative histidine kinase with regard to rpoS and ospC expression. Based upon our findings in the present study, we put forward a model which may explain how multiple signalling pathways control the expression of σS and its regulon in B. burgdorferi.
Results
Confirmation of the σ54-dependent rpoS promoter
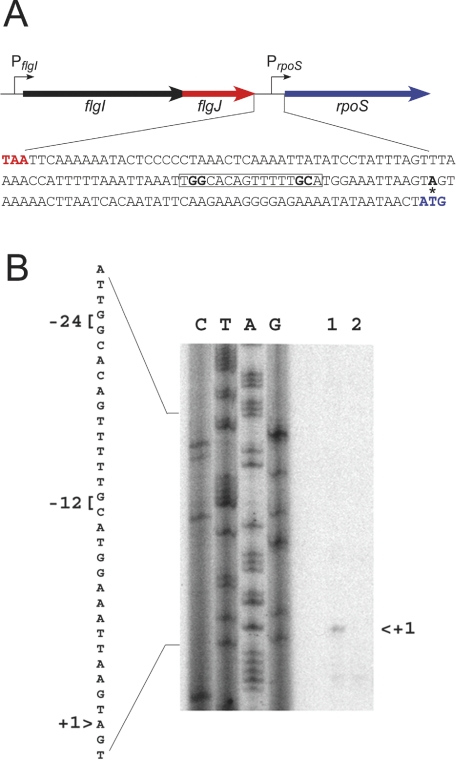
Several studies have shown that B. burgdorferi rpoS expression is regulated by σ54 (Hübner et al., 2001; Yang et al., 2003a,b; Fisher et al., 2005). In B. burgdorferi B31, a potential σ54-dependent promoter is located 62 bp upstream of the putative rpoS start codon (Fig. 1A) (Buck et al., 2000; Hübner et al., 2001). Primer extension reactions using RNA isolated from B. burgdorferi strain B31-A (high-passage) cells grown to high density (2 × 108 cells ml−1) revealed an extension product that terminated 14 bp downstream of the GC doublet of the putative σ54-dependent promoter sequence (Fig. 1B). B. burgdorferi strain B31-ARpoN is a derivative of B31-A in which rpoN is disrupted with a kanamycin-resistance cassette. RNA isolated from B31-ARpoN cells grown to high density (2 × 108 cells ml−1) failed to yield an extension product from the σ54-dependent promoter (Fig. 1B). Taken together, these observations strongly suggest that the sequence containing a predicted −24/−12 region upstream of rpoS is a σ54-dependent promoter. These results are consistent with those recently reported by both Smith et al. (2007) and Lybecker and Samuels (2007), which employed 5′-RACE analysis to identify the identical transcriptional start site in the infectious clone B. burgdorferi BbAH130 and low-passage strain 297, respectively.
Fig. 1. Primer extension analysis of rpoS transcripts.
A. Schematic of the B. burgdorferi rpoS region indicating flgI (black arrow), flgJ (red arrow) and rpoS (blue arrow); the approximate locations of putative promoters (small bent black arrows) are indicated. The gene names are as annotated in the B. burgdorferi B31 genome sequence (TIGR and/or ERGO, Integrated Genomics). The DNA sequence in the region between flgJ and rpoS is shown. The stop codon (TAA) for flgJ is indicated in red, and the start codon (ATG) for rpoS is indicated in blue. The σ54-dependent promoter for rpoS is highlighted with a box around it, and the conserved −24/−12 GG/GC doublets are shown in bold. The transcriptional start site for this promoter is indicated in bold with an asterisk below it.
B. Primer extension reactions were performed with RNA extracted from B. burgdorferi strains B31-A (1) and B31-ARpoN (2) at 2 × 108 cells ml−1. The sequencing ladder was generated with the rpoS-4 primer. The −24/−12 doublets are indicated, and the +1 site is indicated by an arrowhead (<) denoting the 5′ end of the transcript.
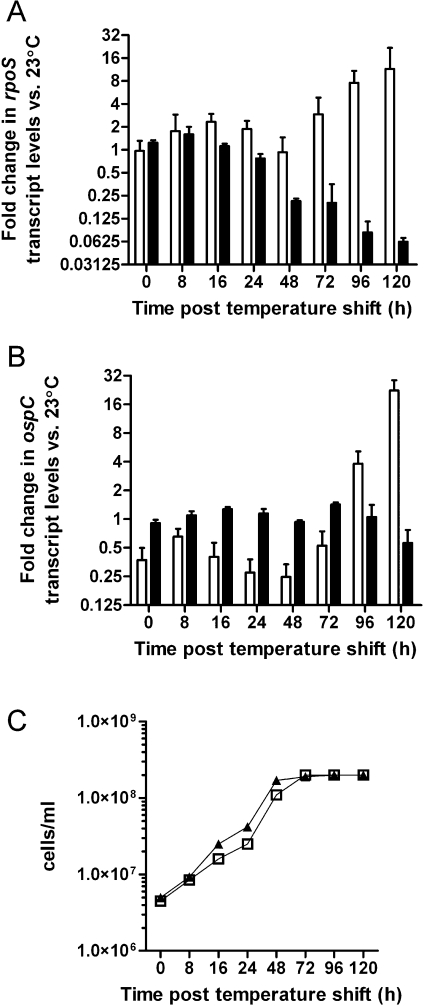
Although the σ54-dependent transcriptional start site for rpoS maps to the same location in both high- and low-passage B. burgdorferi strains, previous studies have demonstrated that the RpoN/RpoS pathway may be defective in high-passage isolates (Masuzawa et al., 1994; Elias et al., 2000; Schwan and Piesman, 2000; Lybecker and Samuels, 2007). To determine whether this pathway is functioning properly in high-passage B. burgdorferi strain B31-A, we conducted a side-by-side comparison of rpoS and ospC transcript levels in B31-A (high-passage) and B31-A3 (low-passage) at various time points following a temperature shift from 23°C to 34°C. Results of quantitative reverse transcription polymerase chain reaction (QRT-PCR) experiments demonstrated an increase in rpoS and ospC expression following an increase in growth temperature, indicating that the RpoN/RpoS pathway functions as expected in strain B31-A3 (Fig. 2). However, in B31-A, rpoS and ospC transcript levels initially remained unchanged following a temperature shift but decreased as cell density increased, indicating that temperature regulation was defective in this high-passage strain (Fig. 2). Based upon these results, all further experiments in this study were conducted with low-passage B. burgdorferi strain B31-A3 and its derivatives.
Fig. 2.
Quantitative RT-PCR analysis of rpoS and ospC transcripts following an increase in growth temperature from 23°C to 34°C. RNA was extracted from B. burgdorferi strains B31-A3 (low-passage, white bars) and B31-A (high-passage, black bars) grown at 23°C, and at various time points following a temperature shift to 34°C. Levels of transcripts were measured with specific primer/probe sets using Taqman, and values have been normalized to the internal control, flaB. Data presented represents averages of three assays performed in quadruplicate. Fold changes are expressed relative to spirochetes grown at 23°C. Error bars represent standard deviation.
A. QRT-PCR analysis of rpoS following a temperature shift.
B. QRT-PCR analysis of ospC following a temperature shift.
C. Growth curves of B31-A3 (white squares) and B31-A (black triangles) following a temperature shift from 23 to 34°C.
Purified Rrp2 and Rrp2Δ123 fail to bind DNA sequences immediately upstream of B. burgdorferi rpoS
Purification of recombinant Rrp2 from Escherichia coli for in vitro studies was complicated by poor expression and insolubility. Additionally, initial attempts to purify recombinant Rrp2 resulted in proteins with incorrect amino-acid sequences as determined by N-terminal sequencing (data not shown). Assessment of the rrp2 codon usage using GCUA Graphical Codon Usage Analyzer (http://gcua.schoedl.de/) revealed that approximately 38% of the codons in this open reading frame (ORF) were rare in E. coli, which likely accounted for the poor expression of Rrp2 in E. coli (Boylan et al., 2006). As an alternative, a codon-optimized version of rrp2, designated rrp2op, was synthesized. Because efficient binding of σ54-dependent activators may require phosphorylation of the N-terminal receiver domain (Lee et al., 1994; 2003), a truncated version of rrp2op was polymerase chain reaction (PCR) amplified by deleting the first 369 nucleotides (corresponding to 123 codons). Both the full-length and truncated genes were cloned into pBAD-TOPO and overexpressed in E. coli. The expression of the truncated gene was expected to result in a constitutively active form of Rrp2 (designated Rrp2Δ123) consisting of the central ATPase and DNA binding domains. Rrp2 and Rrp2Δ123were purified to homogeneity from the soluble fraction as assessed by SDS-PAGE using heparin–sepharose affinity chromatography (data not shown). The majority of Rrp2 and Rrp2Δ123 eluted from the heparin column in the 200–300 mM KCl fractions (data not shown). Both forms of Rrp2 were used in DNA mobility-shift assays.
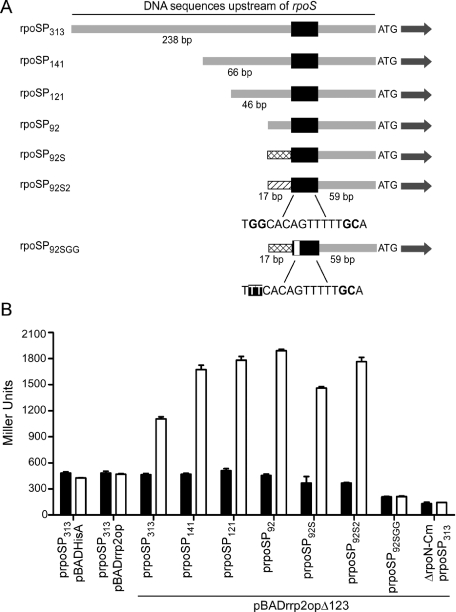
To identify a potential enhancer Rrp2 binding sequence upstream of rpoS, two DNA fragments, rpoSP313 (313 bp) and rpoSP141 (141 bp) (Fig. 3A), were amplified by PCR for use in gel mobility-shift assays. Each fragment consisted of the σ54-dependent rpoS promoter along with 59 bp of downstream DNA and either 238 or 66 bp of upstream DNA respectively. Because most σ54-dependent activators bind within 200 bp of their target promoters, the rpoSP313 fragment was predicted to contain enough upstream DNA sequence to include a potential enhancer. Despite numerous attempts, under a variety of binding conditions, no binding of Rrp2 or Rrp2Δ123 to either rpoSP313 or rpoSP141 was observed. These observations suggested that there is no high-affinity Rrp2 binding site within the 238 bp upstream of the rpoS promoter. Alternatively, it is possible that the correct conditions for binding of Rrp2 or Rrp2Δ123 were not satisfied in the in vitro assay.
Fig. 3.
A. Schematic representation of the promoter regions upstream of B. burgdorferi rpoS used for mobility-shift DNA binding assays and transcriptional fusion analyses. The labels on the left indicate the names of the rpoS promoter regions cloned into the promoter-less lacZ or cat reporter plasmids. The start of rpoS is indicated by an ATG codon. The grey lines to the left of the ATG represent the DNA upstream of rpoS. The black boxes indicate the location of the σ54-dependent promoter; the exact sequence of this promoter is shown below with the characteristic −12/−24 GC/GG bases shown in bold. The number of base pairs shown on either side of the black boxes specifies the length of the DNA fragment on either side of the σ54-dependent promoter (e.g. 238 bp upstream of the −24 or 59 bp downstream of the −12). The hatched lines shown on rpoSP92S and rpoSP92S2 indicate the DNA sequences that were randomly scrambled; exact sequences for these scrambled regions are shown in Table 2. The rpoSP92SGG fragment is identical to the rpoSP92S; however, it harbours a mutation in the σ54-dependent promoter at the −24 position (GG to TT) as indicated in white. B. Transcription from the rpoS promoter region is activated by Rrp2Δ123. β-Galactosidase (LacZ) activity was measured in E. coli Top 10 cells harbouring reporter constructs and expression vectors. lacZ-fusion plasmids harbouring varying-length DNA fragments encompassing the B. burgdorferi rpoS promoter region are indicated, along with either pBADHisA (vector control), pBADrrp2op (expresses full-length Rrp2), or pBADrrp2opΔ123 (expresses N-terminally truncated Rrp2, designated Rrp2Δ123). E. coliΔrpoN-Cm, an rpoN mutant, is indicated. Assays were performed after 6 h growth in LB media without (black bars) or with 0.002% l-arabinose (white bars) to induce protein expression. Error bars represent standard deviation.
Rrp2Δ123 functions with E. coli σ54-holoenzyme to activate transcription from the rpoS promoter
Due to our inability to generate an rrp2 mutant in B. burgdorferi, we decided to use E. coli as a surrogate system to assess Rrp2-dependent activation of transcription from the B. burgdorferi rpoS promoter region. Rrp2 activity was examined in E. coli using a reporter construct in which the B. burgdorferi rpoSσ54-dependent promoter, along with 59 bp of downstream DNA sequence and 238 bp of upstream DNA sequence (rpoSP313 fragment), was introduced upstream of a promoter-less lacZ in plasmid pPBMB101. This reporter plasmid, designated prpoSP313, was introduced into an E. coli Top 10 strain that expressed full-length Rrp2 or Rrp2Δ123. Because the levels of σ54 in E. coli remain relatively constant throughout growth phases (Jishage et al., 1996), we chose to assess promoter activity at 6 h following the addition of arabinose to allow for sufficient Rrp2 expression as confirmed by immunoblot analysis (data not shown). Full-length Rrp2 did not activate transcription from the rpoSP-lacZ reporter gene above background levels (i.e. in the absence of any Rrp2 protein) even when expression of the activator was induced with arabinose (Fig. 3B). This observation was expected because Rrp2 presumably must be phosphorylated to be active.
Induction of Rrp2Δ123 expression with arabinose, in contrast, stimulated expression of the rpoSP-lacZ reporter construct, prpoSP313 (Fig. 3B). A set of rpoSP-lacZ reporter genes with varying lengths of DNA (17–238 bp) upstream of the rpoS promoter were constructed and examined to determine whether RrpΔ123-mediated transcriptional activation from the rpoS promoter in E. coli required an upstream enhancer (Fig. 3A). Induction of Rrp2Δ123 expression with arabinose led to approximately a 3- to 4-fold increase in LacZ activity for all rpoSP–lacZ fusions harbouring an intact σ54-dependent promoter compared with no inducer (Fig. 3B). Similarly, two reporter constructs, prpoS92S and prpoS92S2, that included 17 bp of randomly scrambled DNA sequence immediately of upstream of the σ54-dependent promoter (Fig. 3A) also demonstrated approximately a fourfold increase in LacZ activity in the presence of Rrp2Δ123 (Fig. 3B). In contrast, the prpoS92SGG plasmid, in which the σ54-dependent promoter was mutated at the −24 position (GG to TT), showed no increase in LacZ activity in the presence of inducer (Fig. 3B), indicating that the promoter must be intact for transcriptional activation. Similarly, the levels of LacZ activity from the prpoSP313 reporter plasmid in an E. coli rpoN mutant did not increase with the addition of arabinose, indicating that the Rrp2Δ123-mediated transcriptional activation was dependent on σ54 (Fig. 3B). A recent report by Smith et al. (2007) demonstrating that E. coli RpoN is capable of binding upstream of the B. burgdorferi rpoS gene supported our data indicating that Rrp2Δ123-mediated transcriptional activation in our reporter system was dependent on E. coliσ54. Expression from the bosRPO-lacZ, was not affected by Rrp2Δ123 (data not shown) as expected, because transcription of bosR is not dependent on σ54 (Fisher et al., 2005). Taken together, these data suggest that the 238 bp of DNA sequence upstream of the σ54-dependent rpoS promoter does not harbour an enhancer binding sequence recognized by Rrp2Δ123 for transcriptional activation in E. coli, which is consistent with the results of the mobility-shift DNA binding assays.
Rrp2 does not require upstream DNA sequences for transcriptional activation from the rpoS promoter in B. burgdorferi
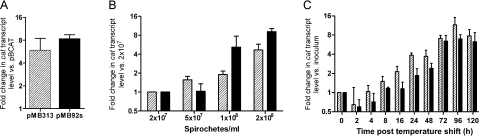
Previous studies have demonstrated that when produced in excess, σ54-activator proteins (e.g. NtrC or N-terminally truncated DctD) can lead to activation of transcription from solution (Lee et al., 1994; North and Kustu, 1997). Therefore, to confirm that overexpression of RrpΔ123 was not responsible for the observed activation of the rpoS promoter constructs in E. coli, two reporter plasmids were constructed fusing the rpoSσ54-dependent promoter and either 238 bp (pMB313) or 17 bp (pMB92S) of upstream DNA to a promoter-less cat gene in plasmid pBCAT, and were transformed into B. burgdorferi. This allowed for assessment of Rrp2-mediated transcriptional activation of rpoS in the presence of wild-type levels of Rrp2. Levels of cat expression from the two reporter constructs in B. burgdorferi B31-A3 (2 × 108 cells ml−1) were monitored by QRT-PCR and compared with that of pBCAT. Results indicated a 7-fold increase in cat expression in the strain harbouring pMB313 and a 9-fold increase in cat expression in the strain harbouring pMB92S compared with the same strain harbouring the pBCAT vector (Fig. 4A). Further experiments monitored the levels of cat expression from the two reporter plasmids as B. burgdorferi cell density increased or following a temperature shift from 23°C to 34°C. QRT-PCR results demonstrated that cat transcript levels increased 6- to 8-fold as spirochete density increased (Fig. 4B). Similarly, temperature-shift experiments showed that cat transcript levels increased as much as 8-fold following an increase in growth temperature (Fig. 4C) Taken together, these data indicated that, as observed with Rrp2Δ123 in E. coli, the 238 bp of DNA sequence immediately upstream of the rpoSσ54-dependent promoter is not required for activation of rpoS transcription by Rrp2 in B. burgdorferi.
Fig. 4. Transcript levels of cat in B. burgdorferi B31-A3 as measured by QRT-PCR. All values have been normalized to the internal control, flaB. Error bars represent standard deviation.
A. cat transcripts levels were measured in B. burgdorferi A3 harbouring cat reporter plasmids pMB313 (rpoSP313 fragment), pMB92S (rposP92S fragment) and pBCAT (vector control) at a cell density of 2 × 108 cells ml−1. Fold changes are relative to the vector control strain.
B. cat transcripts levels were measured in B. burgdorferi B31-A3 harbouring cat reporter plasmids pMB313 (hatched bars) and pMB92S (black bars) at varying cell densities. Fold changes are relative to the 2 × 107 spirochetes ml−1 culture.
C. cat transcripts levels were measured in B. burgdorferi B31-A3 harbouring cat reporter plasmids pMB313 (hatched bars) and pMB92S (black bars) following an increase in growth temperature from 23°C to 34°C. Fold changes are relative to the inoculums used at t = 0 h.
Construction of a B. burgdorferi hk2 mutant
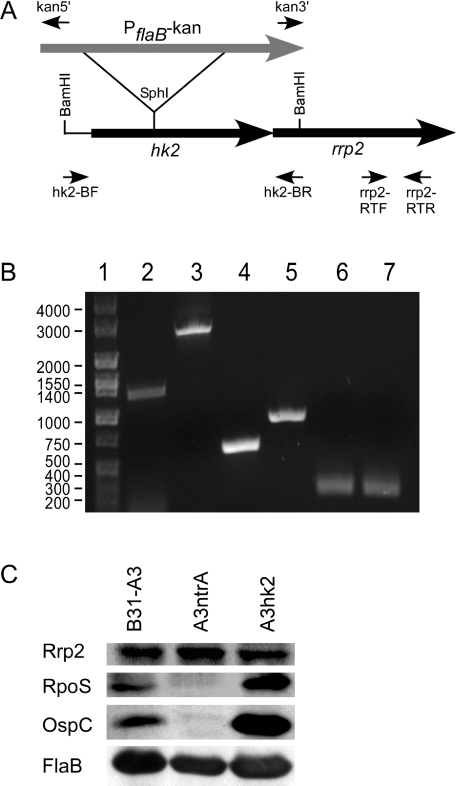
The gene immediately upstream of rrp2, hk2, encodes the putative cognate histidine kinase for Rrp2. Consistent with this prediction, purified Hk2 can autophosphorylate and subsequently donate phosphate to Rrp2, although this phospho-transfer is inefficient in vitro (unpublished data). To determine the influence of Hk2 on rpoS expression, an hk2 mutant in B. burgdorferi low-passage strain B31-A3 (designated as strain A3hk2) was generated by disrupting the gene with a kanamycin-resistance cassette (Fig. 5A). The disruption of hk2 in A3hk2 was confirmed by PCR (Fig. 5B). The kanamycin-resistance cassette was inserted into hk2 so that its promoter (PflaB) would allow for rrp2 expression, and the orientation of this insertion was confirmed by PCR (Fig. 5B). Subsequently, reverse transcription polymerase chain reaction (RT-PCR) using the rrp2-RTF and rrp2-RTR primer pair confirmed the presence of rrp2 transcript in both B31-A3 and A3hk2 (Fig. 5B). The levels of Rrp2 in B31-A3 and A3hk2 were equivalent as assessed by immunoblot analysis (Fig. 5C), confirming that insertion of the kanamycin-resistance cassette into hk2 was not polar on rrp2. B. burgdorferi A3ntrA, an rpoN mutant, was also shown to express similar levels of Rrp2. The plasmid content of A3hk2 was assessed and, in comparison with the parent strain, was observed to be missing lp21 and lp36.
Fig. 5. Construction of a B. burgdorferi hk2 mutant.
A. Schematic representation for inactivation of hk2 in B31-A3. hk2 and rrp2 are represented by black arrows as labelled. A DNA fragment harbouring hk2 was PCR amplified using hk2-BF and hk2-BR primers and insertionally disrupted at a unique SphI site with a kanamycin cassette (grey arrow) as described in the Experimental procedures section. Primers are denoted by short black arrows.
B. Agarose gel patterns of PCR products for B31-A3 (lane 2) and A3hk2 (lane 3) using the hk2-BF and hk2-BR primer pair. Disruption of hk2 by the kanamycin cassette resulted in an increased size PCR product (compare lanes 2 and 3). PCR products for the hk2-BF and kan5′ primer pair (lane 4), and the hk2-BR and kan3′ primer pair (lane 5), confirmed the orientation of the kanamycin cassette with respect to hk2 and rrp2 as diagrammed in panel A. RT-PCR analysis with the rrp2-RTF and rrp2-RTR primer pair confirmed the presence of rrp2 transcript in both B31-A3 (lane 6) and A3hk2 (lane 7). Lane 1 contains DNA markers with the sizes indicated to the left.
C. Immunoblot analysis of B31-A3, A3ntrA and A3hk2 grown to high cell density (2 × 108 cells ml−1 + 24 h). Whole-cell lysates of B. burgdorferi strains equivalent to ∼108 cells were separated on a 12% Tris-glycine gel, immobilized on a nitrocellulose membrane and probed with antiserum specific for the antigens indicated on the left. FlaB serves as a loading control to demonstrate equivalent protein amounts between samples.
Because Hk2 is predicted to phosphorylate Rrp2, which subsequently governs σ54-dependent RpoS and OspC production, the expression of these proteins was assessed in A3hk2. Spirochetes were grown to high density (2 × 108 cells ml−1), and immunoblotting was used to determine levels of RpoS and OspC compared with the parent strain B31-A3 and A3ntrA (negative control). Results showed that A3hk2 appeared to express increased levels of both RpoS and OspC compared with B31-A3, while neither protein was detected in A3ntrA (Fig. 5C). These data indicated that expression of these two proteins was σ54-dependent, but not Hk2-dependent.
rpoS mRNA and RpoS levels in B. burgdorferi cultures indicate that rpoN is required, but hk2 is not essential, for maximal expression of rpoS at high cell densities
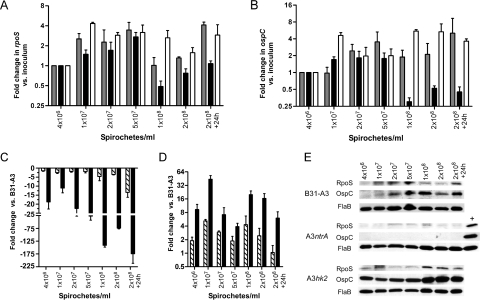
Previous studies have demonstrated that B. burgdorferi cell density influences the expression of RpoS-controlled genes, including ospC (Indest et al., 1997; Ramamoorthy and Philipp, 1998; Caimano et al., 2004). In order to examine the role of an hk2 mutation on the transcription of rpoS and ospC, levels of both rpoS and ospC transcripts were monitored using QRT-PCR in B. burgdorferi strains B31-A3, A3ntrA and A3hk2 as cultures of these strains transitioned from lower density (4 × 106 cells ml−1) to higher density (2 × 108 cells ml−1). A3ntrA was used as a control to confirm that σ54 was required for expression of rpoS when cells reach high density (2 × 108 cells ml−1) in low-passage B. burgdorferi (Fisher et al., 2005). Additionally, RpoS and OspC levels were determined using immunoblot analysis of whole-cell lysates corresponding to each time point at which transcript was measured.
Cultures were inoculated in 1 l of fresh Barbour–Stoenner–Kelly (BSKII) medium at 4 × 106 cells ml−1, and aliquots were harvested as the spirochete density increased. The RNA was purified and the levels of rpoS and ospC transcripts were measured, normalized to flaB as an internal control and compared with the levels in the initial inoculums. Interestingly, rpoS transcript levels increased 2- to 4-fold in all three B. burgdorferi strains in samples harvested at 1 × 107, 2 × 107 and 5 × 107 cells ml−1 in comparison with the inoculums (Fig. 6A). Levels of rpoS transcript decreased in all three strains as cell density increased to 1–2 × 108 cells ml−1, and then increased markedly in the B31-A3 and A3hk2, but not in A3ntrA, as the cultures reached higher densities (2 × 108 cells ml−1 + 24 h). Consistent with these results, the ospC transcript levels followed similar trends in all three strains (Fig. 6B). Analysis of the rpoS and ospC transcript levels in A3ntrA compared with B31-A3 as cell density increased revealed roughly a 2- to 15-fold decrease in rpoS and a 10- to 200-fold decrease in ospC transcripts (Fig. 6C). These results are consistent with previous findings demonstrating decreased rpoS transcript and protein levels in strains lacking σ54 (Hübner et al., 2001; Fisher et al., 2005; Smith et al., 2007). In contrast, comparison of rpoS and ospC transcripts in A3hk2 to B31-A3 demonstrated elevated rpoS (2- to 5-fold) and ospC (4- to 44-fold) transcript levels in A3hk2 at increased cell densities with the exception that at 2 × 108 cells ml−1 + 24 h, the rpoS transcript level was approximately the same in both strains (Fig. 6D).
Fig. 6. Quantitative RT-PCR analysis of rpoS and ospC transcripts and immunoblot analysis of RpoS and OspC as cell density increases.
RNA was extracted from B. burgdorferi strains B31-A3 (grey bars), A3ntrA (black bars) and A3hk2 (white bars) as spirochete density increased and transcripts were quantified using specific primers and probes with the Taqman system. Values have been normalized to the internal control, flaB. Data presented represents averages of three assays performed in quadruplicate. Error bars represent standard deviation.
A. QRT-PCR analysis of rpoS as cell density increased. Fold changes are expressed relative to the initial inoculum.
B. QRT-PCR analysis of ospC as cell density increased. Fold changes are expressed relative to the initial inoculum.
C. QRT-PCR analysis of rpoS (hatched bars) and ospC (black bars) transcripts in A3ntrA relative to B31-A3. Fold changes are expressed compared with B31-A3 at corresponding cell densities.
D. QRT-PCR analysis of rpoS (hatched bars) and ospC (black bars) transcripts in A3hk2 relative to B31-A3. Fold changes are expressed compared to the B31-A3 at corresponding cell densities.
E. Immunoblot analysis of RpoS and OspC levels in B. burgdorferi strains B31-A3, A3ntrA and A3hk2 as cell density increased. Whole-cell lysates of B. burgdorferi strains equivalent to approximately 8 × 107−1 × 108 cells were separated on 12% Tris-glycine gels, immobilized on nitrocellulose membranes and probed with antiserum specific for the antigens indicated on the left. FlaB serves as a loading control to demonstrate equivalent protein amounts between samples. Cell densities are indicated at the top of each lane, and positive controls for the A3ntrA samples are indicated by a plus sign (+).
In order to determine protein levels, immunoblot analyses were used to examine RpoS and OspC expression in the three B. burgdorferi strains as cultures transitioned from lower to higher cell density. In comparison with the initial inoculums, the RpoS levels in B31-A3 and A3hk2 initially increased, then decreased, and then increased again as cultures reached maximum cell density (Fig. 6E). In contrast, while RpoS was shown to be virtually undetectable in A3ntrA at low and high cell densities, trace amounts appeared to be detectable at moderate cell density (2 × 107−1 × 108 cells ml−1). OspC was undetectable at all cell densities in A3ntrA (Fig. 6E). Taken together, these results indicate that although rpoN is required for rpoS expression as cell density increases, it appears that hk2 is not essential.
Influence of hk2 and rpoN mutations on rpoS and ospC mRNA and protein levels in B. burgdorferi cultures following an increase in growth temperature
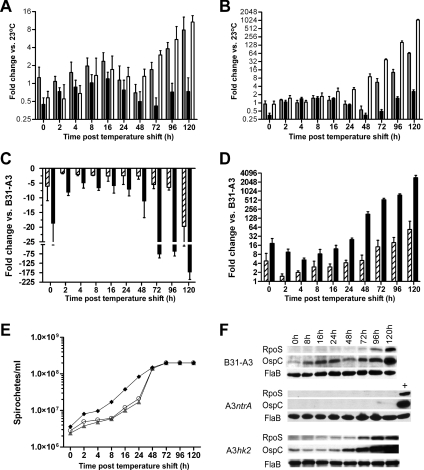
A previous study has shown that B. burgdorferi strain 297 rpoS transcript levels increase 2-fold in response to a temperature shift from 23°C to 37°C, 6-fold upon entry into stationary phase, and as much as 40- to 70-fold following several days growth at 37°C (Caimano et al., 2004). In order to examine the role of an hk2 mutation on the transcription of rpoS and ospC following a shift in growth temperature from 23°C to 34°C, both rpoS and ospC transcript levels were monitored using QRT-PCR in B. burgdorferi B31-A3, A3ntrA and A3hk2. At various time points following temperature elevation, spirochetes were harvested, RNA was purified, and the levels of rpoS and ospC transcripts were subsequently determined and compared with transcript levels from cultures grown at 23°C. As shown in Fig. 7A, when compared with cultures maintained at 23°C, both B31-A3 and A3hk2 showed increased rpoS transcript levels (approximately 2-fold) at 16 h post temperature shift. By 120 h post temperature shift, rpoS transcript levels increased 7- to 10-fold in B31-A3 and A3hk2. Similarly, ospC transcript levels increased approximately 8-fold in B31-A3 and A3hk2 by 72 and 48 h post temperature shift respectively, and continued to increase at the time points of 96 and 120 h (Fig. 7B). Interestingly, the observed increase in ospC transcript in A3hk2 (1000-fold) was much greater than that observed in B31-A3 (64-fold) at 120 h post temperature shift. As expected, both the rpoS and ospC transcripts in A3ntrA remained at similar levels to a culture at 23°C for the duration of the experiment (Fig. 7A and B). Analysis of the rpoS and ospC transcript levels in A3ntrA compared with B31-A3 following a temperature shift revealed a 2- to 20-fold decrease in rpoS and a 5- to 200-fold decrease in ospC transcripts (Fig. 7C). In contrast, comparison of rpoS and ospC transcripts in A3hk2 versus B31-A3 demonstrated markedly increased rpoS (2- to 60-fold) and ospC (4- to 2000-fold) transcript levels in A3hk2 (Fig. 7D). Growth curves relating cell densities with time points post temperature shift are shown (Fig. 7E).
Fig. 7.
Quantitative RT-PCR analysis of rpoS and ospC transcripts and immunoblot analysis of RpoS and OspC following an increase in growth temperature from 23°C to 34°C. RNA was extracted from B. burgdorferi strains B31-A3 (grey bars), A3ntrA (black bars) and A3hk2 (white bars) grown at 23°C and following a temperature shift to 34°C, and transcripts were quantified using specific primers and probes with the Taqman system. Values have been normalized to the internal control, flaB. Data presented represents averages of three assays performed in quadruplicate. Error bars represent standard deviation.
A. QRT-PCR analysis of rpoS following a temperature shift. Fold changes are expressed relative to spirochetes grown at 23°C.
B. QRT-PCR analysis of ospC following a temperature shift. Fold changes are expressed relative to spirochetes grown at 23°C.
C. QRT-PCR analysis of rpoS (hatched bars) and ospC (black bars) transcripts in A3ntrA relative to B31-A3. Fold changes are expressed compared with the B31-A3 at corresponding time points.
D. QRT-PCR analysis of rpoS (hatched bars) and ospC (black bars) transcripts in A3hk2 relative to B31-A3. Fold changes are expressed compared with the B31-A3 at corresponding time points.
E. Growth curves of B31-A3 (grey triangles), A3ntrA (black diamonds) and A3hk2 (open circles) following a temperature shift from 23°C to 34°C.
F. Immunoblot analysis of RpoS and OspC levels in B. burgdorferi strains B31-A3, A3ntrA and A3hk2 following an increase in growth temperature from 23°C to 34°C. Whole-cell lysates of B. burgdorferi strains equivalent to approximately 8 × 107−1 × 108 cells were separated on 12% Tris-glycine gels, immobilized on nitrocellulose membranes and probed with antiserum specific for the antigens indicated on the left. FlaB serves as a loading control to demonstrate equivalent protein amounts between samples. Time points are indicated at the top of each lane, and positive controls for the A3ntrA samples are indicated by a plus sign (+).
Immunoblot analyses were employed to examine RpoS and OspC expression in the three B. burgdorferi strains following an increase in growth temperature. Both RpoS and OspC levels in B31-A3 were initially almost undetectable but increased with time as cultures were incubated at 34°C, and both proteins were clearly evident by the 120 h post-temperature shift (Fig. 7F). A3hk2 demonstrated a similar pattern of RpoS and OspC expression to B31-A3 in that both proteins increased following the temperature shift; however, the levels of protein expressed by A3hk2 appeared to be slightly higher. As expected, RpoS and OspC were virtually undetectable in A3ntrA at all time points (Fig. 7F). Overall, these QRT-PCR and immunoblotting data support previous studies indicating that rpoN is required for rpoS expression following a temperature shift from 23°C to 34°C, and demonstrate that hk2 does not appear to be required for this adaptive response to occur.
Rrp2 requires rpoN, but not hk2 for transcriptional activation from the rpoS promoter in B. burgdorferi
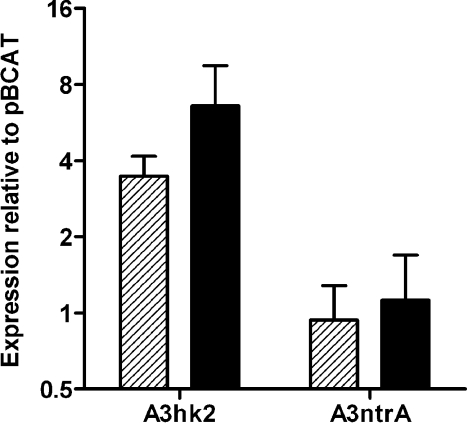
To investigate whether the rpoSP-cat expression was dependent on σ54 and Hk2, both pMB313 and pMB92S reporter plasmids were transformed into B. burgdorferi A3ntrA and A3hk2. QRT-PCR was employed to determine the levels of cat transcript when cells reached high density (2 × 108 cells ml−1) from strains containing pMB313 and pMB92S compared with strains harbouring vector alone. Results showed that cat transcript levels in A3hk2 increased approximately 4- to 8-fold compared with the vector only control (Fig. 8). In contrast, no appreciable increase in cat transcript levels was evident in A3ntrA consistent with decreased rpoS transcript levels in this mutant. The results obtained for A3hk2 are similar to the results observed for wild-type B31-A3 (Fig. 4A), and suggest that hk2 is not absolutely required for transcriptional activation from the σ54-dependent rpoS promoter.
Fig. 8.
Transcript levels of cat in B. burgdorferi A3ntrA and A3hk2 as measured by QRT-PCR. cat transcripts levels were measured in B. burgdorferi A3hk2 and A3ntrA harbouring plasmids pMB313 (hatched bars) and pMB92S (black bars). Fold changes are relative to strains harbouring pBCAT. All values have been normalized to the internal control, flaB. Data presented represents averages of three assays performed in quadruplicate. Error bars represent standard deviation.
Discussion
Signal transduction pathways are important mechanisms for bacteria to sense, respond and adapt to changing environmental stimuli. In the complex life cycle of B. burgdorferi, the σ54–σS regulatory cascade plays a critical role in the differential expression of a variety of lipoproteins associated with host adaptation and virulence (Elias et al., 2000; Hübner et al., 2001; Yang et al., 2003a,b; 2005; Fisher et al., 2005). It has been previously demonstrated that the two-component response regulator Rrp2 serves as the activator for this regulatory pathway (Yang et al., 2003a). In the present study, we confirmed transcription of rpoS from its σ54-dependent promoter and showed that Rrp2 activates rpoS transcription from this promoter.
Despite the fact that the σ54-dependent promoter of B. burgdorferi rpoS is a near-perfect match to the consensus sequence and would, therefore, be expected to have a high affinity for σ54-RNAP holoenzyme, QRT-PCR assays indicate that rpoS transcript levels are relatively low in B. burgdorferi, ranging from ∼1% to 4% of those for flaB. Consistent with previous reports, our results show that rpoS mRNA levels increase only 4- to 5-fold in B. burgdorferi as cultures transition from lower to higher cell density when grown at 34°C, and only about 2-fold immediately following a temperature shift from 23°C to 34°C. Moreover, even 5 days (120 h) post temperature shift when spirochetes had reached maximal cell density, we observed only a 7- to 10-fold increase in rpoS transcript levels. We postulate that the modest level of transcription initiating from the σ54-dependent rpoS promoter was because Rrp2 does not bind a nearby enhancer to activate transcription from this promoter. Two important findings from this study support this hypothesis. First, we observed that neither purified Rrp2 nor Rrp2Δ123 bound to DNA fragments up to 238 bp upstream and 59 bp downstream of the rpoS promoter in mobility-shift DNA binding assays using a variety of conditions. Second, in vivo investigations employing promoter–reporter gene fusion assays in both E. coli and B. burgdorferi revealed that neither DNA sequences upstream nor downstream of the rpoS promoter were required for transcriptional activation by Rrp2.
The predicted domain structure of Rrp2 indicates that it contains an amino-terminal receiver domain, a central ATPase domain and a carboxy-terminal domain with a potential helix–turn–helix DNA binding motif (Yang et al., 2003a). Most σ54-dependent activators contain a carboxy-terminal DNA binding domain that is responsible for enhancer recognition; examples include NtrC, NtC1 and DctD (Reitzer and Magasanik, 1986; Lee and Hoover, 1995; Doucleff et al., 2005). However, there are σ54-dependent activator proteins that lack a DNA binding domain, including Helicobacter pylori FlgR and Chlamydia trachomatis CtcC (Koo and Stephens, 2003; Brahmachary et al., 2004). In the case of FlgR, it has been shown that this activator does not require an upstream enhancer, but rather appears to contact σ54-holoenzyme directly to activate transcription (Brahmachary et al., 2004). Although Rrp2 does not appear to bind to a specific enhancer near the rpoS promoter to activate transcription, it may bind to low-affinity sites or contact the closed complex at the rpoS promoter directly from solution to activate transcription. Rrp2 did bind to heparin–sepharose during purification procedures, suggesting that its DNA binding domain is functional and further supporting the idea that Rrp2 displays non-sequence specific DNA binding.
Efforts to isolate B. burgdorferi rrp2 mutants have been unsuccessful. Yang and colleagues (2003a) have also reported on the inability to inactivate rrp2, but were able to replace the wild-type rrp2 allele with a mutant allele that presumably prevented Rrp2 from activating transcription while not have interfering with other activities of the protein such as DNA binding. Our inability to inactivate rrp2 does not appear to be due to polar effects on downstream genes because rrp2 is predicted to be the last gene within the putative rpoS operon. We postulate that Rrp2 has an additional, essential role in the cell that is unrelated to its function in activating transcription from rpoS or any other σ54-dependent gene. One vital function that Rrp2 may play in the cell is to repress one or more genes whose unregulated expression would otherwise compromise the viability of the cell. Studies are ongoing to identify specific Rrp2 binding sites within the B. burgdorferi genome that might help shed light on a potential repressor role for Rrp2.
Analysis of rpoS and cat transcript levels in B. burgdorferi rpoN and hk2 mutant strains revealed that σ54 is required for rpoS expression as previously reported, but that hk2 does not appear to be involved in cell density- or temperature-dependent regulation. When compared with both wild-type and the rpoN mutant, expression of rpoS mRNA was higher in the hk2 mutant, a similar trend was observed with cat transcript levels. We propose that Hk2 is dispensable for expression of rpoS because Rrp2 may be able to accept phosphate from another protein histidine kinase or a low-molecular-weight phosphate donor, such as acetyl phosphate, in the cell as reported for other systems (Lukat et al., 1992; Da Re et al., 1999; Yamamoto et al., 2005). It is unclear exactly what signal(s) lead to activation of Rrp2 or what cellular factors interact with Rrp2, but the ability of Rrp2 to accept phosphate from alternative donors could provide multiple sources of signal input and greater flexibility in regulating the σ54 and σS regulons in B. burgdorferi. Interestingly, rpoS transcript levels in the hk2 mutant were slightly higher than wild-type levels. This may be due to altered expression of rrp2 in the hk2 mutant. Alternatively, some histidine kinases stimulate the removal of phosphate from their cognate phosphorylated response regulators (Keener and Kustu, 1988). If Hk2 has such activity, then Rrp2-phosphate may accumulate to higher levels in the hk2 mutant, which could account for increased rpoS transcript levels in the mutant.
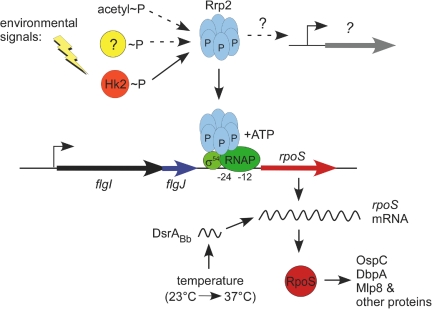
Based upon our present results in combination with previously published studies, we propose a model for the regulation of rpoS in B. burgdorferi (Fig. 9). In brief, signals are sensed by Hk2 or other histidine kinases (e.g. BB0420, annotated as a sensory transduction protein kinase; BB0363, annotated as a sensory box/GGDEF family protein, ERGO genome database, Integrated Genomics), triggering autophosphorylation and subsequent transfer of a phosphate to the N-terminal receiver domain of Rrp2. Alternately, Rrp2 may accept a phosphate from a low-molecular-weight phospho-donor(s) (e.g. acetyl phosphate). After activation, Rrp2 interacts with σ54-RNAP holoenzyme bound to the −12/−24 promoter region upstream of rpoS and activates transcription leading to expression of σS-controlled genes (e.g. ospC, dpbA, mlp8). If correct, this model would permit multiple signals to be channelled through Rrp2 to activate the σ54–σS regulatory network.
Fig. 9.
Proposed model for the regulation of rpoS in B. burgdorferi. Environmental signals are sensed by Hk2 (orange circle) leading to autophosphorylation and subsequent phosphotransfer to the receiver domain of Rrp2 (blue ovals). Alternately, other sensor kinases (yellow circle with question mark)(e.g. BB0420 or BB0363) or small phosphate donors, such as acetyl-phosphate (acetyl∼P), could activate Rrp2, which, in turn, would activate the transcription of rpoS (red arrow) from the σ54-dependent promoter (indicated by −24/−12). RNAP is represented as a green oval, σ54 is shown as small green circle, and RpoS is represented as a red circle. The genes upstream of rpoS are shown as follows: flgI (black arrow) and flgJ (blue arrow). It is possible that Rrp2 could interact with other genes of unknown function (grey arrow with question mark above). Putative promoters are indicated as small bent black arrows. A small RNA molecule (DsrABb) is shown as a wavy line and is proposed to regulate the efficiency of rpoS mRNA (long wavy line) translation in response to temperature (Lybecker and Samuels, 2007). The described model suggests that multiple signals could affect the regulation of the σS regulon.
Experimental procedures
Strains, plasmids and culture conditions
The strains and plasmids used in this study are shown in Table 1. E. coli strains were grown in Luria–Bertani (LB) broth or on LB agar at 37°C. B. burgdorferi strain B31-A3 and derivatives were grown in modified BSKII medium pH 7.6 (Barbour et al., 1984) or on BSKII agar plates at 34°C unless otherwise stated. Spirochete numbers were determined by dark field microscopy. When necessary, antibiotics were added to the media at the following concentrations: for E. coli, ampicillin, 100 μg ml−1; chloramphenicol, 25 μg ml−1; kanamycin, 25 μg ml−1; gentamicin, 10 μg ml−1; and for B. burgdorferi, rifampicin, 10 μg ml−1; amphotericin B, 7.5 μg ml−1; kanamycin, 200 μg ml−1; gentamicin, 40 μg ml−1. All chemicals were purchased from Sigma (St Louis, MO) unless otherwise stated.
Table 1.
Strains and plasmids used in this study.
| Strain/Plasmid | Description/relevant featuresa | Reference |
|---|---|---|
| Strain | ||
| E. coli strains | ||
| Top 10 | General cloning strain | Invitrogen |
| BW25113 | Red recombinase mutagenesis strain | Datsenko and Wanner (2000) |
| ΔrpoN-Cm | BW25113 rpoN deletion strain; CmR | This study |
| B. burgdorferi strains | ||
| B31-A | Clone A, high-passage, avirulent | Rosa et al. (1996) |
| B31-ARpoN | rpoN mutant of B31-A, KmR | This study |
| B31-A3 | Wild-type clone A3, low-passage, virulent | Elias et al. (2002) |
| A3ntrA | rpoN mutant of B31-A3, KmR | Fisher et al. (2005) |
| A3hk2 | hk2 mutant of B31-A3, KmR | This study |
| Plasmids | ||
| E. coli plasmids | ||
| pLysE | Constitutively expresses T7 lysozyme; CmR | Invitrogen |
| pPBMB101 | Promoter-less lacZ reporter vector, p15A ori; KmR | P.J. Brett (unpublished) |
| pBAD-TOPO | Arabinose-inducible expression vector, pBR322 ori; ApR | Invitrogen |
| prrp2op | pUC57::rrp2op; ApR | GenScript Corporation |
| pBADrrp2op | pBAD-TOPO::rrp2op; ApR | This study |
| pBADrrp2opΔ123 | pBAD-TOPO::rrp2opΔ123; ApR | This study |
| prpoSP313 | pPBMB101::rpoSP313; KmR | This study |
| prpoSP141 | pPBMB101::rpoSP141; KmR | This study |
| prpoSP121 | pPBMB101::rpoSP121; KmR | This study |
| prpoSP92 | pPBMB101::rpoSP92; KmR | This study |
| prpoSP92S | pPBMB101::rpoSP92S; KmR | This study |
| prpoSP92S2 | pPBMB101::rpoSP92S2; KmR | This study |
| prpoSP92SGG | pPBMB101::rpoSP92SGG; KmR | This study |
| pbosRPO | pPBMB101::bosRPO; KmR | This study |
| pJLB12a | PflaB-kan cassette | Bono et al. (2000) |
| pKD3 | CmR cassette for construction of ΔrpoN-Cm | Datsenko and Wanner (2000) |
| pPCRscriptCamSK + | Cloning vector, pBC SK(+) derivative, pUC ori, CmR | Stratagene |
| B. burgdorferi plasmids | ||
| pBSV2G | Borrelia shuttle vector; GmR | Elias et al. (2003) |
| pBCAT | pBSV2G::′cat; GmR | This study |
| pMB313 | pBCAT::rpoSP313; GmR | This study |
| pMB92S | pBCAT::rpoSP92S; GmR | This study |
| pHk2 | pPCRScriptCamSK + ::BB0764 ORF; CmR | This study |
| pSKHk2 | pHk2::PflaB-kan; CmR KmR | This study |
Antibiotic resistance: Ap, ampicillin; Cm, chloramphenicol; Gm, gentamicin; Km, kanamycin.
For rpoS and ospC transcript and protein-level quantification experiments, B. burgdorferi starter cultures were grown at 34°C from glycerol stocks. These cultures were diluted to ∼105 cells ml−1 in 200 ml of BSKII and incubated at 34°C until a density of ∼2–4 × 107 cells ml−1 was reached. Cultures were used to inoculate 1 l of BSKII at a density of ∼4 × 106 cells ml−1 and incubation continued at 34°C. At 8–12 h intervals, spirochetes were enumerated and aliquots were harvested for RNA isolation or protein analysis.
Temperature-shift experiments were performed based on previously described methods (Ojaimi et al., 2003; Caimano et al., 2004). Briefly, B. burgdorferi strains were inoculated from glycerol stocks, grown to mid-log phase at 34°C, diluted to ∼105 cells ml−1 and grown at 23°C to a density of ∼5 × 107 cells ml−1. Spirochetes were then diluted to ∼5 × 105 cells ml−1 in larger culture volumes and grown at 23°C to a density of 1–3 × 107 cells ml−1 (mid-log phase), at which time cells were transferred to pre-warmed BSKII at a density of 2–4 × 106 cells ml−1 and further incubated at 34°C. Cells were harvested for RNA isolation at t = 0, 2, 4, 8, 16, 24, 48, 72, 96 and 120 h, and for protein analysis at t = 0, 8, 16, 24, 48, 72, 96 and 120 h. Cultures were maintained at 23°C and allowed to continue to 7 × 107−1 × 108, at which time cells were harvested for RNA analysis. The doubling times of the spirochetes grown at 23°C and 34°C were 32–36 h and 8–10 h respectively.
Primer extension analysis
Total RNA was isolated from a B. burgdorferi B31-A and B31-ARpoN (2 × 108 cells ml−1) using TRI-Reagent (Sigma) as described by the manufacturer. A total of 1 μg of RNA was used as template for primer extension reactions with the Primer Extension System AMV (Promega, Madison, WI). A sequence ladder was generated with the fmol Cycle Sequencing System (Promega) using 1 μg chromosomal DNA isolated from B. burgdorferi B31-A. For primer extension and sequencing reactions, primers rpoS-4 and up-PE were labelled at the 5′-end with 32P[γ]-ATP (3000 Ci mmole−1) (ICN, Costa Mesa, CA) using T4 polynucleotide kinase (Promega). Reaction products were separated by electrophoresis on a 6% polyacrylamide, 7 M urea gel. Reaction products were visualized by autoradiography.
SDS-PAGE and immunoblots
For protein analysis, B. burgdorferi whole-cell lysates were prepared by washing spirochete pellets twice with Haley's buffer (20 mM HEPES, pH 7.6, 50 mM NaCl), followed by lysis with 1× Tris-glycine SDS sample buffer (Invitrogen). For immunoblotting, B. burgdorferi lysates equivalent to 8 × 107−1 × 108 cells were separated on 12% Tris-glycine polyacrylamide gels (Invitrogen) and electrophoretically transferred to nitrocellulose membranes (0.45 μm pore size, Invitrogen). Immunoblot analyses were performed at room temperature with incubations as follows: membranes were blocked with 5% skim milk in TBS (100 mM Tris-HCl, pH 7.5, 150 mM NaCl) for 30–60 min, followed by application of the primary antibody diluted in TBS-T (TBS, 0.05% Tween 20) for 1 h, then application of the secondary antibody diluted in TBS-T for 1 h. Membranes were washed three times with TBS-T following each incubation step. Blots were visualized with ECL™ Plus Western Blotting Detection Reagents (GE Healthcare) as per the manufacturer's instructions. Primary antibodies included anti-RpoS polyclonal antiserum (1/1200), anti-OspC polyclonal antiserum (1/1000), anti-Rrp2 polyclonal antiserum (1/1000) and anti-FlaB monoclonal antibody MCA9724 (1/1000). Secondary antibodies were either anti-rabbit IgG-HRP conjugate (1/5000) or anti-mouse IgG-HRP conjugate (1/5000). When appropriate, nitrocellulose membranes were stripped with Restore™ Western Blot Stripping Buffer (Pierce) and washed with TBS-T prior to reprobing.
Escherichia coli lysates and purified Rrp2 proteins were separated on 4–20% Tris-glycine polyacrylamide gels (ISC Bioexpress, Kaysville, UT) and stained with Coomassie brilliant blue or analysed by Western blot as described above.
Cloning and expression of rrp2op and rrp2opΔ123
For expression in E. coli, a codon-optimized version of B. burgdorferi rrp2 (designated rrp2op) was synthesized and cloned into pUC57 by GenScript Corporation (Piscataway, NJ). Full-length rrp2op (1356 bp) was PCR amplified from plasmid prrp2op using the Rrp2op-F and Rrp2op-R primer pair. A truncated version of rrp2 (987 bp) lacking the N-terminal 369 bp, designated rrp2opΔ123, was similarly PCR amplified using the Rrp2opΔ123-F and Rrp2op-R primer pair. The PCR products were individually cloned into the pBAD-TOPO vector (Invitrogen) to place the rrp2 alleles under control of the araBAD promoter/operator. DNA sequencing was used to confirm that the rrp2 gene sequences in the resulting plasmids, pBADrrp2op and pBADrrp2opΔ123, were correct.
Purification of Rrp2 and Rrp2Δ123
Rrp2 and Rrp2Δ123 were overexpressed in E. coli Top 10 (pLysE) cells. Bacteria were grown in 500 ml of LB medium with appropriate antibiotics at 37°C with aeration. When cells reached an OD600 of 0.6, expression of protein was induced using 0.02% l-arabinose for 18 h. Cells were harvested by centrifugation, resuspended in buffer A (20 mM Tris pH 8, 50 mM KCl, 5% glycerol, 0.5 mM DTT) and lysed by sonication. Insoluble material was removed by centrifugation, and the supernatant was applied to a Heparin Sepharose™6 Fast Flow (GE Healthcare, Uppsala, Sweden) column. The protein was eluted with a stepwise gradient of KCl (200 mM, 300 mM, 400 mM, 500 mM, 750 mM and 1 M KCl). Fractions were analysed by SDS-PAGE, and those containing Rrp2 or Rrp2Δ123 were pooled, concentrated and stored in buffer A at 4°C. Protein concentrations were determined using a BCA™ protein assay kit (Pierce Biotechnology, Rockford, IL). Purified protein was used in mobility-shift DNA binding assays.
Polymerase chain reaction, RT-PCR, QRT-PCR and DNA mobility-shift assays
Polymerase chain reactions were performed using the Expand High Fidelity PCR System (Roche Applied Science, Indianapolis, IN) as per the manufacturer's instructions. Genomic DNA used for PCRs was isolated from bacterial strains using the Wizard genomic DNA purification kit (Promega). PCR primers are shown in Table 2. All primers were obtained from Integrated DNA Technologies (Coralville, IA). DNA sequencing was performed by ACGT (Wheeling, IL).
Table 2.
Polymerase chain reaction (PCR), PE, RT-PCR and QRT-PCR primers and probes used in this study.
| Primer name | Primer sequence (5′→ 3′)a–d |
|---|---|
| PCR and PE primers | |
| rpoS-4 | TATGTTTAAATCAGCTTAATTTATTTC |
| up-PE | TAGATTTTGTGAATTAATTTTGGTTTCC |
| Rrp2op-F | ATGAGCAAAATTCTGGTGGCGG |
| Rrp2op-R | TTAATCAATATTATATTCGATAA |
| Rrp2opΔ123-F | GATAATGAAAATGCGAATCTGGAAAATATTCTG |
| rpoSP313-FB | AAGCGGATCCTTGAAAGCATGAAAAAA |
| rpoSP-RXhoI | CCGCTCGAGTATTATATTTTCTCCCCTT |
| rpoSP141-FB | AAGCGGATCCCAAAAAATACTCCCCCT |
| rpoSP121-FB | AAGCGGATCCCTCAAAATTATATCCTATTTAG |
| rpoSP92-FB | AAGCGGATCCCCATTTTTAAATTAAATTGGCAC |
| rpoSP92S-FB | AAGCGGATCCCACATTATTATAATATATGGCACAGTTTTTGCATG |
| rpoSP92S2-FB | AAGCGGATCCACCTATAAATTATATTATGGCACAGTTTTTGCATG |
| rpoSP92SGG-FB | AAGCGGATCCCACATTATTATAATATATTTCACAGTTTTTGCATG |
| rpoSP92-RXb | GTACTCTAGATATTATATTTTCTCCCCTT |
| rpoSP92S-FH | GATCAAGCTTCACATTATTATAATATATGGCACAGTTTTTGCATG |
| rpoSP313-FH | GATCAAGCTTCAAAAAATACTCCCCCT |
| bosRPO-FB | AAGCGGATCCAATTGAAAAAATAAATTCTAAGAAATGG |
| bosRPO-RX | CCGCTCGAGATGATTATACCTTTTTTGTTTAAATTAAAG |
| EcrpoN-F | ATGAAGCAAGGTTTGCAACTCAGGCTTAGCCAACAACTGGGTGTAGGCTGGAGCTGCTTC |
| EcrpoN-R | TCAAACGAGTTGTTTACGCTGGTTTGACGGCGGAATGGATCATATGAATATCCTCCTTA |
| c1 | TTATACGCAAGGCGACAAGG |
| yhbG-430 | GCCGTGTAGAAATTGCCCGC |
| hk2SphF | GTGGCATGCAGTGTTACAACCAATTAACC |
| hk2SphR | GTGGCATGCAATCTCTGATGTTACATTGC |
| hk2-BF | GTGGGATCCATCGCCCCTATAATCAAAAT |
| hk2-BR | GTGGGATCCAGCTTCTTCTTCGTCACTA |
| kan5′ | GCAATGTAACATCAGAGATT |
| kan3′ | GGTTAATTGGTTGTAACACT |
| RT-PCR, QRT-PCR | |
| primers and probes | |
| rrp2-RTF | GGCTGCAACAAACAAAAACATT |
| rrp2-RTR | TTTCATTGCATCATTAGAAAGAGTT |
| flaB-586F | AATCTTTTCTCTGGTGAGGGAGCT |
| flaB-657R | TCCTTCCTGTTGAACACCCTCT |
| flaB-611T | FAMc-AAACTGCTCAGGCTGCACCGGTTC-TAMRAd |
| cat-284F | ACAAGGGTGAACACTATCCCATATC |
| cat-256R | GAATGCTCATCCGGAATTACG |
| cat-310T | FAMc-CCAGCTCACCGTCTTTCATTGCCA-TAMRAd |
| rpoS-215F | AAGAAGGCAACTTGGGATTAATAAGA |
| rpoS-308R | TGCTTAATCCAAAATGATGCATAAG |
| rpoS-242T | FAMc-CTGCTGAAAAATATGACCCGAATAAAAATACCAAATT-TAMRAd |
| ospCRT-F1 | ACGGATTCTAATGCGGTTTTACTT |
| ospCRT-R1 | CAATAGCTTTAGCAGCAATTTCATCT |
| ospCRT-P | FAMc-ATGACAGCAACGCTTCAACCTCTTTCACAG-TAMRAd |
Linker sequences are underlined.
b. Scrambled sequences are italicized.
FAM = 6-carboxyflourescein.
TAMRA = 5-carboxytetramethylrhodamin
Borrelia burgdorferi RNA was extracted using TRI-Reagent (Sigma) as described by the manufacturer. RNA was treated with DNase I and further purified using the RNeasy miniprep kit (Qiagen). cDNA synthesis reactions for RT-PCR and QRT-PCR were performed using SuperScript III (Invitrogen) to synthesize first-strand cDNA following the manufacturer's instructions. RT-PCR primers specific for rrp2 are shown in Table 2. QRT-PCR primers and probes specific for cat, rpoS, ospC and flaB were designed using Primer Express 1.0 and are shown in Table 2. Reactions were performed in a total volume of 20 μl using TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, CA), 1–2 ng of first-strand cDNA, 300 nM forward and reverse primers, and 250 nM probe. All reactions were carried out on the ABI PRISM 7900HT Sequence Detection System (Applied BioSystems) using a PCR cycle of 2 min at 50°C, 10 min at 95°C, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. Each transcript was normalized by comparison with the constant, internal control flaB (Fisher et al., 2005). Three individual assays were performed in quadruplicate.
The rpoS promoter/operator (P/O) region was amplified by PCR using the rpoSP313-FB and rpoSP-RXhoI primer pair, generating a 313 bp DNA fragment, or using the rpoSP141-FB and rpoSP-RXhoI primer pair, generating a 141 bp DNA fragment. Mobility-shift DNA binding reactions were performed essentially as previously described (Porter et al., 1993; Boylan et al., 2003; 2006), or with the Sigma Mobility Shift Optimization Assay kit (Sigma) using purified Rrp2 or Rrp2Δ123 (concentrations ranging from 100 nM to 2 μM) and approximately 20 000 cpm of [γ-32P]-ATP-labelled target sequence. Varying concentrations of the components in the binding buffer were also used in attempts to optimize binding conditions. Purified BosR protein and a labelled DNA fragment bearing the napA-P/O were employed as a positive control (Boylan et al., 2003).
Escherichia coli‘lacZ reporter system
The B. burgdorferi rpoS-P/O regions (P313, P141, P121, P92, P92S P92S2 and P92SGG) diagrammed in Fig. 3A were PCR amplified from strain B. burgdorferi B31-A3 genomic DNA using one of the following forward primers rpoSP313-FB, rpoSP141-FB, rpoSP121-FB, rpoSP92-FB, rpoSP92S-FB, rpoSP92S2-FB, rpoSP92GG-FB and the reverse primer rpoSP-RXhoI. The B. burgdorferi rpoS-P/O regions were cloned upstream of a promoter-less lacZ into the BamHI/XhoI sites of pPBMB101 and transformed into the appropriate E. coli strain. As a control, the B. burgdorferi bosR-P/O region was similarly amplified using the bosRPO-FB and bosRPO-RX primer pair and cloned into the BamHI/XhoI sites of pPBMB101.
An E. coli rpoN mutant (ΔrpoN-Cm) was constructed in strain BW25113 via red recombinase mutagenesis (Datsenko and Wanner, 2000) using the EcrpoN-F and EcrpoN-R primer pair. Disruption of rpoN was confirmed by PCR using the c1 and yhbG-430 primer pair. The rpoS-lacZ reporter plasmids (pPBMB101 derivatives) and arabinose-inducible expression constructs pBADrrp2op or pBADrrp2opΔ123 were cotransformed into E. coli Top 10 cells or ΔrpoN-Cm cells, and were grown for 6 h at 37°C in the presence or absence of 0.002% l-arabinose. β-Galactosidase (LacZ) assays were performed by the method of Miller (1972) 2–3 times in triplicate.
Generating a B. burgdorferi hk2 mutant
To inactivate hk2, BB0764 was amplified by PCR from B. burgdorferi B31-A3 with primers hk2-BF and hk2-BR. The resulting PCR product was digested with BamHI to produce 1.3 kb fragment containing hk2, and was cloned into the BamHI restriction site of pPCRScriptCamSK + (Stratagene, La Jolla, CA), generating the plasmid pHk2. A kanamycin-resistance cassette driven by the B. burgdorferi flaB promoter region was amplified from pJLB12a (Bono et al., 2000) using primers hk2-SphF and hk2-SphR, which introduced SphI restriction sites on the ends. The PCR product and pHk2 were digested with SphI and ligated together to generate pSKHk2. The resulting plasmid was transformed into low-passage B. burgdorferi strain B31-A3, as described by Samuels (1995), and kanamycin-resistant colonies were analysed by PCR using primers hk2-BF and hk2-BR. To assess whether rrp2 was transcribed in the resulting mutant strain (A3hk2), total RNA was isolated, and RT-PCR was performed using the rrp2-RTF and rrp2-RTR primer pair. The plasmid profile of A3hk2 was assessed as previously described (Purser and Norris, 2000; Elias et al., 2002).
Borrelia burgdorferi rpoS promoter–cat reporter constructs
A B. burgdorferi reporter plasmid was constructed by amplifying a promoter-less chloramphenicol acetyltransferase (cat) gene from pPCRScriptCamSK + (Stratagene) and cloning it into the Borrelia shuttle vector, pBSV2G. The resulting plasmid was designated pBCAT. The B. burgdorferi rpoS-P/O regions (P313 and P92S) were PCR amplified using either the rpoS313-FH or the rpoS92S-FH primer in combination with the rpoSP92-RXb primer, and then cloned into the HindIII/XbaI sites of the B. burgdorferi reporter construct pBCAT. All inserts were confirmed by DNA sequencing. The plasmids pMB313, pMB92S and pBCAT (control) were subsequently transformed into B. burgdorferi B31-A3, A3ntrA and A3hk2. For QRT-PCR experiments, RNA was isolated from strains harbouring cat reporter constructs that were grown to appropriate cell densities or at various time points following a temperature shift.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Institute of Allergy and Infectious Diseases. We would like to thank Dan Sturdevant for Taqman primer and probe design, and Jonathan Warawa for help with Taqman procedures and data analysis. We would also like to thank Patricia Rosa for providing B. burgdorferi strains and plasmid pBSV2G, Tom Schwan for providing anti-FlaB and anti-OspC antibodies, Jon Skare for providing anti-RpoS antibodies, and Meghan Lybecker and Scott Samuels for helpful discussions. We are grateful to Gary Hettrick and Anita Mora for graphics support.
References
- Barbour AG, Tessier SL, Hayes SF. Variation in a major surface protein of Lyme disease spirochetes. Infect Immun. 1984;45:94–100. doi: 10.1128/iai.45.1.94-100.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono JL, Elias AF, Kupko JJ, Stevenson B, Tilly K, Rosa P. Efficient targeted mutagenesis in Borrelia burgdorferi. J Bacteriol. 2000;182:2445–2452. doi: 10.1128/jb.182.9.2445-2452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylan JA, Posey JE, Gherardini FC. Borrelia oxidative stress response regulator, BosR: a distinctive Zn-dependent transcriptional activator. Proc Natl Acad Sci USA. 2003;100:11684–11689. doi: 10.1073/pnas.2032956100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylan JA, Hummel CS, Benoit S, Garcia-Lara J, Treglown-Downey J, Crane EJ, 3rd, Gherardini FC. Borrelia burgdorferi bb0728 encodes a coenzyme A disulphide reductase whose function suggests a role in intracellular redox and the oxidative stress response. Mol Microbiol. 2006;59:475–486. doi: 10.1111/j.1365-2958.2005.04963.x. [DOI] [PubMed] [Google Scholar]
- Brahmachary P, Dashti MG, Olson JW, Hoover TR. Helicobacter pylori FlgR is an enhancer-independent activator of sigma54-RNA polymerase holoenzyme. J Bacteriol. 2004;186:4535–4542. doi: 10.1128/JB.186.14.4535-4542.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck M, Gallegos M-T, Studholme DJ, Guo Y, Gralla JD. The bacterial enhancer-dependent σ54 (σN) transcription factor. J Bacteriol. 2000;182:4129–4136. doi: 10.1128/jb.182.15.4129-4136.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgdorfer W, Barbour AG, Hayes SF, Benach JL, Grunwaldt E, Davis JP. Lyme disease – a tick borne spirochetosis? Science. 1982;216:1317–1319. doi: 10.1126/science.7043737. [DOI] [PubMed] [Google Scholar]
- Caimano MJ, Eggers CH, Hazlett KR, Radolf JD. RpoS is not central to the general stress response in Borrelia burgdorferi but does control expression of one or more essential virulence determinants. Infect Immun. 2004;72:6433–6445. doi: 10.1128/IAI.72.11.6433-6445.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JA, Garon CF, Schwan TG. Effects of environmental pH on membrane proteins in Borrelia burgdorferi. Infect Immun. 1999;67:3181–3187. doi: 10.1128/iai.67.7.3181-3187.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Re SS, Deville-Bonne D, Tolstykh T, Veron M, Stock JB. Kinetics of CheY phosphorylation by small molecule phosphodonors. FEBS Lett. 1999;457:323–326. doi: 10.1016/s0014-5793(99)01057-1. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucleff M, Chen B, Maris AE, Wemmer DE, Kondrashkina E, Nixon BT. Negative regulation of AAA + ATPase assembly by two component receiver domains: a transcription activation mechanism that is conserved in mesophilic and extremely hyperthermophilic bacteria. J Mol Biol. 2005;353:242–255. doi: 10.1016/j.jmb.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Elias A, Bono JL, Carroll JA, Stewart P, Tilly K, Rosa P. Altered stationary phase response in a Borrelia Burgdorferi rpoS mutant. J Bacteriol. 2000;178:2909–2918. doi: 10.1128/jb.182.10.2909-2918.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias AF, Stewart PE, Grimm D, Caimano MJ, Eggers CH, Tilly K, et al. Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infect Immun. 2002;70:2139–2150. doi: 10.1128/IAI.70.4.2139-2150.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias AF, Bono JL, Kupko JJ, Stewart PE, Krum JG, Rosa PA. New antibiotic resistance cassettes suitable for genetic studies in Borrelia burgdorferi. J Mol Microbiol Biotechnol. 2003;6:29–40. doi: 10.1159/000073406. [DOI] [PubMed] [Google Scholar]
- Fisher MA, Grimm D, Henion AK, Elias AF, Stewart PE, Rosa PA, Gherardini FC. Borrelia burgdorferiσ54 is required for mammalian infection and vector transmission but not for tick colonization. Proc Natl Acad Sci USA. 2005;102:5162–5167. doi: 10.1073/pnas.0408536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübner A, Yang X, Nolen DM, Popova TG, Cabello FC, Norgard MV. Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc Natl Acad Sci USA. 2001;98:12724–12729. doi: 10.1073/pnas.231442498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indest KJ, Philipp MT. DNA-binding proteins possibly involved in regulation of the post-logarithmic-phase expression of lipoprotein P35 in Borrelia burgdorferi. J Bacteriol. 2000;182:522–525. doi: 10.1128/jb.182.2.522-525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indest KJ, Ramamoorthy R, Sole M, Gilmore RD, Johnson BJB, Philipp M. Cell-density-dependent expression of Borrelia burgdorferi lipoprotein in vitro. Infect Immun. 1997;65:1165–1171. doi: 10.1128/iai.65.4.1165-1171.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jishage M, Iwata A, Ueda S, Ishihama A. Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of four species of sigma subunit under various growth conditions. J Bacteriol. 1996;178:5447–5451. doi: 10.1128/jb.178.18.5447-5451.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keener J, Kustu S. Protein kinase and phosphoprotein phosphatase activities of nitrogen regulatory proteins NTRB and NTRC of enteric bacteria: roles of the conserved amino-terminal domain of NTRC. Proc Natl Acad Sci USA. 1988;85:4976–4980. doi: 10.1073/pnas.85.14.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo IC, Stephens RS. A developmentally regulated two-component signal transduction system in Chlamydia. J Biol Chem. 2003;278:17314–17319. doi: 10.1074/jbc.M212170200. [DOI] [PubMed] [Google Scholar]
- Lee JH, Hoover TR. Protein crosslinking studies suggest that Rhizobium meliloti C4-dicarboxylic acid transport protein D, a sigma 54-dependent transcriptional activator, interacts with sigma 54 and the beta subunit of RNA polymerase. Proc Natl Acad Sci USA. 1995;92:9702–9706. doi: 10.1073/pnas.92.21.9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Scholl D, Nixon BT, Hoover TR. Constitutive ATP hydrolysis and transcriptional activation by a stable, truncated form of Rhizobium meliloti DCTD, a σ54-dependent transcriptional activator. J Biol Chem. 1994;269:20401–20409. [PubMed] [Google Scholar]
- Lee SY, De La Torre A, Yan D, Kustu S, Nixon BT, Wemmer DE. Regulation of the transcriptional activator NtrC1: structural studies of the regulatory and AAA+ ATPase domains. Genes Dev. 2003;17:2552–2563. doi: 10.1101/gad.1125603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukat GS, McCleary WR, Stock AM, Stock JB. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc Natl Acad Sci USA. 1992;89:718–722. doi: 10.1073/pnas.89.2.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lybecker MC, Samuels DS. Temperature-induced regulation of RpoS by a small RNA in Borrelia burgdorferi. Mol Microbiol. 2007;64:1075–1089. doi: 10.1111/j.1365-2958.2007.05716.x. [DOI] [PubMed] [Google Scholar]
- Masuzawa T, Kurita T, Kawabata H, Yanagihara Y. Relationship between infectivity and OspC expression in Lyme disease Borrelia. FEMS Microbiol Lett. 1994;123:319–324. doi: 10.1111/j.1574-6968.1994.tb07242.x. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- North AK, Kustu S. Mutant forms of the enhancer-binding protein NtrC can activate transcription from solution. J Mol Biol. 1997;267:17–36. doi: 10.1006/jmbi.1996.0838. [DOI] [PubMed] [Google Scholar]
- North AK, Weiss DS, Suzuki H, Flashner Y, Kustu S. Repressor forms of the enhancer-binding protein NtrC: some fail in coupling ATP hydrolysis to open complex formation by σ54-holoenzyme. J Mol Biol. 1996;260:317–331. doi: 10.1006/jmbi.1996.0403. [DOI] [PubMed] [Google Scholar]
- Ogura T, Wilkinson AJ. AAA+ superfamily ATPases: common structure – diverse function. Genes Cells. 2001;6:575–597. doi: 10.1046/j.1365-2443.2001.00447.x. [DOI] [PubMed] [Google Scholar]
- Ojaimi C, Brooks C, Casjens S, Rosa P, Elias A, Barbour A, et al. Profiling of temperature-induced changes in Borrelia burgdorferi gene expression by using whole genome arrays. Infect Immun. 2003;71:1689–1705. doi: 10.1128/IAI.71.4.1689-1705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popham D, Szeto D, Keener J, Kustu S. Function of a bacterial activator protein that binds to transcriptional enhancers. Science. 1989;243:629–635. doi: 10.1126/science.2563595. [DOI] [PubMed] [Google Scholar]
- Porter SC, North AK, Wedel AB, Kustu S. Oligomerization of NTRC at the glnA enhancer is required for transcriptional activation. Genes Dev. 1993;7:2258–2272. doi: 10.1101/gad.7.11.2258. [DOI] [PubMed] [Google Scholar]
- Purser JE, Norris SJ. Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc Natl Acad Sci USA. 2000;97:13865–13870. doi: 10.1073/pnas.97.25.13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamoorthy R, Philipp MT. Differential expression of Borrelia burgdorferi proteins during growth in vitro. Infect Immun. 1998;66:5119–5124. doi: 10.1128/iai.66.11.5119-5124.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzer LJ, Magasanik B. Transcription of glnA in E. coli is stimulated by activator bound to sites far from the promoter. Cell. 1986;45:785–792. doi: 10.1016/0092-8674(86)90553-2. [DOI] [PubMed] [Google Scholar]
- Rosa P, Samuels DS, Hogan D, Stevenson B, Casjens S, Tilly K. Directed insertion of a selectable marker into a circular plasmid of Borrelia burgdorferi. J Bacteriol. 1996;178:5946–5953. doi: 10.1128/jb.178.20.5946-5953.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels DS. Electroporation of the spirochete Borrelia burgdorferi. Methods Mol Biol. 1995;47:253–259. doi: 10.1385/0-89603-310-4:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasse-Dwight S, Gralla JD. Probing the Escherichia coli glnALG upstream activation mechanism in vivo. Proc Natl Acad Sci USA. 1988;85:8934–8938. doi: 10.1073/pnas.85.23.8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwan TG, Piesman J. Temporal changes in outer surface proteins A and C of the lyme disease-associated spirochete, Borrelia burgdorferi, during the chain of infection in ticks and mice. J Clin Microbiol. 2000;38:382–388. doi: 10.1128/jcm.38.1.382-388.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwan TG, Piesman J, Golde WT, Dolan MC, Rosa PA. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc Natl Acad Sci USA. 1995;92:2909–2913. doi: 10.1073/pnas.92.7.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshu J, Boylan JA, Gherardini FC, Skare JT. Dissolved oxygen levels alter gene expression and antigen profiles in Borrelia burgdorferi. Infect Immun. 2004;72:1580–1586. doi: 10.1128/IAI.72.3.1580-1586.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AH, Blevins JS, Bachlani GN, Yang XF, Norgard MV. Evidence that RpoS (σS) in Borrelia burgdorferi is controlled directly by RpoN (σ54/σN) J Bacteriol. 2007;189:2139–2144. doi: 10.1128/JB.01653-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steere AC, Bartenhagen NH, Craft JE, Hutchinson GJ, Newman JH, Rahn DW, et al. The early clinical manifestations of Lyme disease. Ann Intern Med. 1983;99:76–82. doi: 10.7326/0003-4819-99-1-76. [DOI] [PubMed] [Google Scholar]
- Stevenson B, Schwan TG, Rosa PA. Temperature-related differential expression of antigens in the Lyme disease spirochete, Borrelia burgdorferi. Infect Immun. 1995;63:4535–4539. doi: 10.1128/iai.63.11.4535-4539.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W, Porter S, Kustu S, Echols H. DNA-looping and enhancer activity: association between DNA-bound NTRC activator and RNA polymerase at the bacterial glnA promoter. Proc Natl Acad Sci USA. 1990;87:5504–5508. doi: 10.1073/pnas.87.14.5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Hoover TR. Transcriptional regulation at a distance in bacteria. Curr Opin Microbiol. 2001;4:138–144. doi: 10.1016/s1369-5274(00)00179-x. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Hirao K, Oshima T, Aiba H, Utsumi R, Ishihama A. Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli. J Biol Chem. 2005;280:1448–1456. doi: 10.1074/jbc.M410104200. [DOI] [PubMed] [Google Scholar]
- Yang X, Goldberg MS, Popova TG, Schoeler GB, Wikel SK, Hagman KE, Norgard MV. Interdependence of environmental factors influencing reciprocal patterns of gene expression in virulent Borrelia burgdorferi. Mol Microbiol. 2000;37:1470–1479. doi: 10.1046/j.1365-2958.2000.02104.x. [DOI] [PubMed] [Google Scholar]
- Yang XF, Alani SM, Norgard MV. The response regulator Rrp2 is essential for the expression of major membrane lipoproteins in Borrelia burgdorferi. Proc Natl Acad Sci USA. 2003a;100:11001–11006. doi: 10.1073/pnas.1834315100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XF, Hubner A, Popova TG, Hagman KE, Norgard MV. Regulation of expression of the paralogous Mlp family in Borrelia burgdorferi. Infect Immun. 2003b;71:5012–5020. doi: 10.1128/IAI.71.9.5012-5020.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XF, Lybecker MC, Pal U, Alani SM, Blevins J, Revel AT, et al. Analysis of the ospC regulatory element controlled by the RpoN-RpoS regulatory pathway in Borrelia burgdorferi. J Bacteriol. 2005;187:4822–4829. doi: 10.1128/JB.187.14.4822-4829.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]