Abstract
Background
Spt7 is an integral component of the multi-subunit SAGA complex that is required for the expression of ~10% of yeast genes. Two forms of Spt7 have been identified, the second of which is truncated at its C-terminus and found in the SAGA-like (SLIK) complex.
Results
We have found that C-terminal processing of Spt7 to its SLIK form (Spt7SLIK) and to a distinct third form (Spt7Form3) occurs in the absence of the SAGA complex components Gcn5, Spt8, Ada1 and Spt20, the latter two of which are required for the integrity of the complex. In addition, N-terminally truncated derivatives of Spt7, including a derivative lacking the histone fold, are processed, indicating that the C-terminus of Spt7 is sufficient for processing and that processing does not require functional Spt7. Using galactose inducible Spt7 expression, we show that the three forms of Spt7 appear and disappear at approximately the same rate with full-length Spt7 not being chased into Spt7SLIK or Spt7Form3. Interestingly, reduced levels of Spt7SLIK and Spt7Form3 were observed in a strain lacking the SAGA component Ubp8, suggesting a regulatory role for Ubp8 in the truncation of Spt7.
Conclusion
We conclude that truncation of Spt7 occurs early in the biosynthesis of distinct Spt7 containing complexes rather than being a dynamic process linked to the action of the SAGA complex in transcriptional regulation.
Background
The multisubunit SAGA (Spt-Ada-Gcn5-Acetyltransferase) complex is a prototype for transcriptional coactivators that interface between DNA binding transcriptional regulators, the chromosomal template and the basal transcriptional machinery [1]. SAGA is a dynamic entity, with components existing in subcomplexes having related but distinct functions [2-6].
The Ada components of SAGA (ADA2, NGG1/ADA3, GCN5/ADA4) were first identified based on their involvement in regulated transcription in Saccharomyces cerevisiae [7-11]. SAGA possesses histone acetyltransferase (HAT) activity mediated by Gcn5 [2,12], which stimulates or represses transcription in a promoter specific fashion [13-17]. The structural core of SAGA is composed of a subset of the TBP-associated factors (TAFs) that are also found in TFIID [18,19]. Spt7, Ada1 and Spt20, members of the TBP-subclass of Spt (Suppressors of Ty insertions) [2,20-22] proteins, are located near the core of the complex and are required for its integrity, forming an interface between TAFs and other SAGA-specific components [19]. The additional Spt proteins, Spt3 and Spt8 are involved in direct interactions with TBP [3,22-25]. Tra1, a 437 kDa, phosphatidylinositol-3-kinase related protein [26,27] is found at the periphery of the complex and interacts with DNA-binding transcriptional regulators thus conferring promoter specificity to SAGA [28-32]. Through mass spectrometry analysis of purified SAGA complex, other components have been identified [33-36]. One of these proteins, Sus1, is associated with the nuclear pore mRNA export machinery [35] suggesting that SAGA may have a broader role in nuclear processes than previously envisioned.
Two forms of Spt7 have been described. The full-length protein of 1332 amino acid residues is found within the SAGA complex [2]. The second, a C-terminally truncated form likely arising from processing between residues 1125 and 1150 is found in the SLIK (SAGA-like) complex [3-6]. As suggested by the alternate name for this complex, SALSA (SAGA altered, Spt8 absent), SLIK also differs from SAGA in that it lacks Spt8 [3,4]. The absence of Spt8 can be attributed to the fact that it interacts with Spt7 in the region deleted in the truncated form [4,6]. The truncation of Spt7 and the subsequent loss of Spt8 may be a mechanism to create a complex that acts without effects mediated through TBP; for example, the derepression of the HIS3 promoter under starvation conditions [3,4]. A further difference between SAGA and SLIK is that Rtg2, which is required for the retrograde response pathway in yeast [37], is found specifically in SLIK [5]. Two other Rtg proteins, Rtg1 and Rtg3 are transcription factors required for the expression of retrograde target genes such as CIT2 [38]. Pray-Grant et al. [5] propose that SLIK facilitates transcriptional activation by Rtg1 and Rtg3 and perhaps also at a distinct set of promoters. However, the exact role of SLIK is unclear as the interpretation of transcriptional effects upon disruption of RTG2 is complicated since Rtg2 is required for nuclear import of Rtg1 and Rtg3 [39].
Two models for the relationship between the Spt7-containing complexes can be envisioned. In the first, processing at the C-terminus would occur as part of the biosynthesis of distinct SAGA and SLIK complexes. Alternatively, processing might occur during the normal functioning of SAGA, perhaps to irreversibly signal that a transcriptional event had occurred and/or enabling subsequent activity of the SLIK complex. In this study we performed experiments to differentiate between these models. We show that the conversion of Spt7 from its full-length SAGA-form (Spt7SAGA) to its truncated SLIK-form (Spt7SLIK) and a third C-terminally processed form (Spt7Form3) occurs in the absence of components of the complex including Ada1 and Spt20, which are required for the integrity of the complex. Processing to these forms occurs rapidly after its synthesis and in the absence of full-length Spt7. Taken together our results suggest that processing of Spt7 occurs in the biosynthesis of distinct Spt7-containing complexes.
Results and discussion
Truncated forms of Spt7 occur in the absence of fully functional SAGA complex
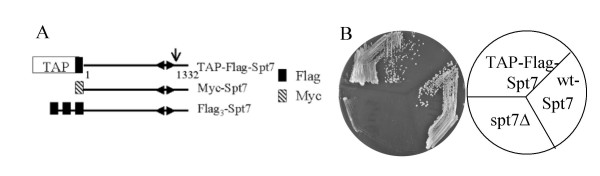
Two forms of Spt7 have been described that differ in their length [3-6]. Full-length Spt7 is 1332 amino acid residues and is found in the SAGA complex. This form, Spt7SAGA, migrates with an apparent molecular mass of approximately 200 kDa on SDS-PAGE. The SLIK complex contains a C-terminal truncation of Spt7 that results in an apparent molecular mass of approximately 180 kDa [4]. Based upon the mobility of this form and the differential function of Spt7 derivatives with C-terminal deletions, Spt7SLIK is predicted to contain a C-terminal truncation of approximately 200 amino acids [4,6]. To detect these forms of Spt7 we engineered a centromeric plasmid expressing a TAP and Flag-tagged version of Spt7 that could be identified after C-terminal truncation (YCpDed-TAP-Flag-SPT7, Figure 1a). We first addressed whether this molecule was functional by determining if it would complement a deletion of spt7 as assayed by growth on media depleted for inositol. As shown in Figure 1b, a strain lacking SPT7 is unable to grow on media lacking inositol. YCpDed-TAP-Flag-SPT7 allows for near complete restoration of growth.
Figure 1.
Epitope-tagged Spt7 molecules used in this study. A TAP-Flag-Spt7 contains N-terminal TAP and Flag tags fused in frame to the initiator Met of Spt7 (1332 amino acids) via a triple alanine linker encoded by a NotI site. Myc-Spt7 has a similar constructed N-terminal myc-epitope while Flag3-Spt7 contains 3 tandem copies of the Flag epitope. The position of the histone fold (amino acids 979–1045; [43]) is indicated with inverted arrows. An arrow indicates the approximate position of the cleavage site that generates Spt7SLIK. B. TAP-Flag-Spt7 is functional. Yeast strains BY4741 (SPT7), BY3218 (spt7Δ0) and BY3218 transformed with YCpDed-TAP-Flag-SPT7 were grown on a minimal plate depleted of inositol for 3 days at 30°C.
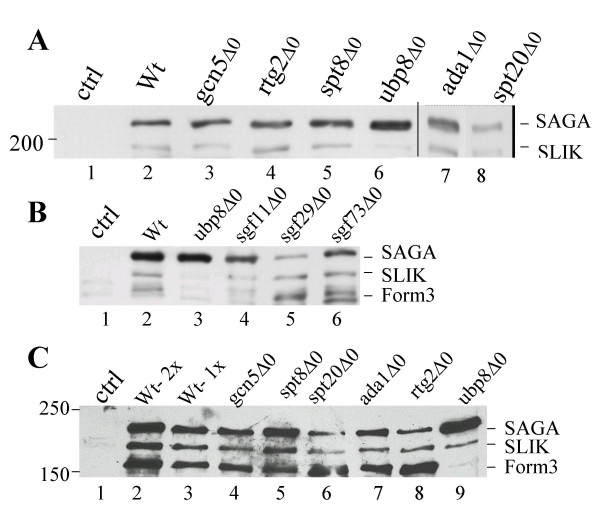
To determine if the components of the SAGA or SLIK complex influence the truncation of Spt7, we introduced YCpDed-TAP-Flag-SPT7 into yeast strain BY4741 (wild-type) and isogenic strains deleted for GCN5, RTG2, SPT8, UBP8, ADA1 and SPT20. Cells were grown in YPD media and protein extracts examined for Spt7 by Western blotting with anti-Flag antibody (Figure 2a). Two proteins with apparent molecular masses of approximately 220 and 200 kDa were present in wild-type cells containing YCpDed-TAP-Flag-SPT7 but not in control cells (compare lanes 1 and 2). The size of these molecules suggest that they represent the TAP-Flag tagged full-length (Spt7SAGA) and SLIK-forms (Spt7SLIK) of Spt7. The ratio of SAGA:SLIK forms of Spt7 was ~3:1 in the wild-type strain BY4741 (lane 2). This relative amount of Spt7SLIK was not reduced upon disruption of GCN5, RTG2, ADA1, SPT20 or SPT8. Since Spt20 and Ada1 are required for the integrity of the SAGA complex [6], the presence of Spt7SLIK in these deletion strains indicates that C-terminal processing can occur in the absence of the fully intact and functional complex. (We do note that there was an apparent decrease in the ratio of SAGA:SLIK forms in the spt20Δ0 strain and speculate that this may be due to differential stability of the forms in this strain background.) In addition, the wild-type level of Spt7SLIK seen in the spt8 deletion background suggests that the Spt8 interaction with Spt7 does not alter access of the putative processing enzyme(s) to the cleavage site. Furthermore, the lack of a requirement for Rtg2 indicates that the SLIK complex need not fully assemble to allow truncation of Spt7.
Figure 2.
Processed forms of Spt7 in yeast deletion strains. A. Wild-type (BY4741) or the indicated yeast deletion strains (gcn5Δ0, rtg2Δ0, spt8Δ0, ubp8Δ0, ada1Δ0, spt20Δ0; lanes 3–8, respectively) were transformed with YCpDed-TAP-Flag-SPT7, with the exception of lane 1 (ctrl) which was transformed with the control vector YCp-TAP-Flag. Cells were grown to an A600 = 1.0–1.5 in YP containing 2% glucose and crude cell lysates prepared by glass bead disruption. 50 μg of protein was separated by 5% SDS-PAGE and Western blotted with anti-Flag antibody. The mobility of a 200 kDa molecular weight marker is indicated on the left. Spt7SAGA and Spt7SLIK are marked. B. BY4741 (Wt, lane 2) and the indicated deletion strains (ubp8Δ0, lane 3; sgf11Δ0, lane 4; sgf29Δ0, lane 5; sgf73Δ0, lane 6) containing TAP-Flag-Spt7 were grown as above, crude yeast extracts separated by SDS-PAGE and Western blotted with anti-Flag-antibody. Lane one contains crude extract from the wild-type strain BY4741. Spt7SAGA, Spt7SLIK and Spt7Form3 are marked. C. Wild-type (BY4741 lanes 2 and 3) or the indicated yeast deletion strains, gcn5Δ0 (lane 4), spt8Δ0 (lane 5), spt20Δ0 (lane 6), ada1Δ0 (lane 7), rtg2Δ0 (lane 8), ubp8Δ0 (lane 9) containing YCpDed-TAP-Flag-SPT7 were grown in YP broth containing 2% glucose. Lane 1 (ctrl) contains BY4741 transformed with the control vector YCp-TAP-Flag. TAP-Flag-Spt7 was purified through affinity chromatography on IgG-Agarose and eluted after cleavage with TEV protease. Approximately equal amounts of Spt7 were separated by 5% SDS-PAGE and Western blotted with anti-Flag antibody. The mobility of 250 and 150 kDa molecular mass markers are indicated on the left. Lane 3 contains one half of the wild-type sample applied in lane 2 to facilitate quantitation.
Although still apparent, the amount of Spt7SLIK was reduced approximately three-fold in the strain deleted for the ubiquitin protease Ubp8 (Figure 2a, lane 6). We note that the ubp8Δ0 effect was found to vary from approximately two-fold to five-fold between different experiments (not shown). To determine whether the role of Ubp8 was the result of its involvement with SAGA, we examined the Spt7 forms in a strain deleted for Sgf11, a subunit of SAGA required for the association of Ubp8 with components of the complex [40-42]. As shown in Figure 2b, deletion of sgf11 partially reduced the level of Spt7SLIK. This effect was not to the same extent as deletion of Ubp8, perhaps because low levels of Ubp8 remain associated with SAGA in the absence of Sgf11 [40]. Nonetheless, while not eliminating other mechanisms, the result is consistent with a model whereby binding of Ubp8 to SAGA may alter the accessibility of the C-terminus of Spt7 to cleavage. Interestingly, another Flag-antibody reactive band, Spt7Form3, was evident in the sample lanes but not in the control. Since Spt7Form3 was somewhat obscured by a Flag antibody cross-reactive band, we analyzed the Spt7 forms in different strain backgrounds after first purifying TAP-Flag-Spt7 on IgG-agarose (Figure 2c). After purification a Flag-reactive molecule with an apparent mass approximately 20 kDa less than Spt7SLIK (also see Figure 3b) was revealed. The observed difference in mass of this molecule relative to intact Spt7 estimates an end-point at approximately amino acid residue 950. As was the case for Spt7SLIK, the level of this third form of Spt7 was comparable in each of the deletion strains (gcn5Δ0, spt8Δ0, spt20Δ0, ada1Δ0, rtg2Δ0) with the exception of ubp8Δ0 where it was reduced.
Figure 3.
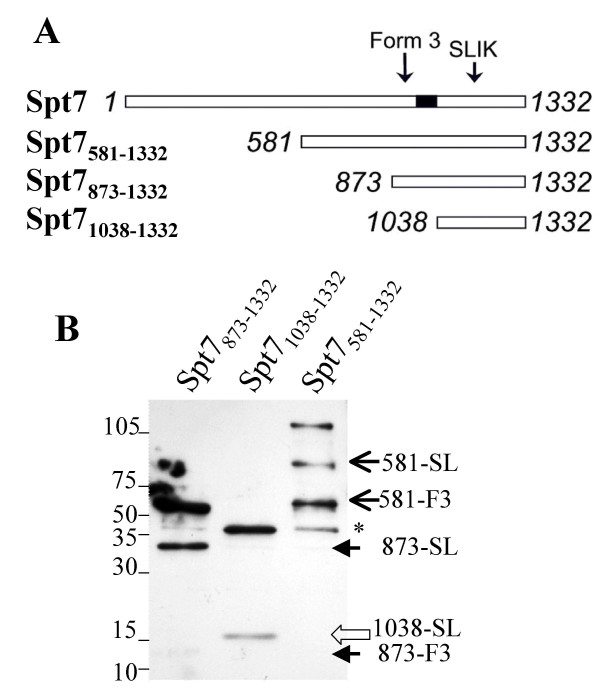
Formation of truncated forms of Spt7 in N-terminally deleted derivatives. A. N-terminally deleted derivatives of Spt7 were constructed containing amino acids 581–1332 (Spt7581–1332), 873–1332 (Spt7873–1332) and 1038–1332 (Spt71038–1332). Note that each molecule is N-terminally TAP and Flag tagged to allow purification then detection of processed fragments. The position of the histone fold is shown with a black box and the approximate position of the Spt7SLIK and Spt7Form3 processing sites by arrows. B. YCpDed-TAP-Flag- Spt7581–1332, Spt7873–1332 and Spt71038–1332 were expressed in BY3218 (spt7Δ0), tandem affinity purified and Spt7 forms detected by Western blotting with anti-Flag antibody after electrophoresis on an 8–15% gradient gel. Mobility of protein standards with the indicated molecular masses (kDa) is indicated on the left. Arrows on the right marked with 581, 873 and 1038 represent the mobility of truncated TAP-Flag- Spt7581–1332, Spt7873–1332 and Spt71038–1332, respectively. * marks an additional cleavage product seen with Spt7581–1332.
C-terminal processing of Spt7 occurs in the absence of its N-terminal amino acid residues
The finding that Spt7SLIK and Spt7Form3 were formed in the absence of components of SAGA, including Spt20 and Ada1, suggested that processing is not linked directly to the activity of the complex. If processing were part of the functional dynamics of SAGA, that is, occurring as a result of its activities in the regulation of transcription, then one would predict that it would require Spt7 in a functional form. We thus examined whether N-terminal deletion derivatives of Spt7, expressed in BY3218 (spt7Δ0) would be processed (Figure 3). Each molecule was N-terminally TAP and Flag tagged to allow purification and identification of Spt7 forms. The two longest derivatives (residues 580–1332 and 873–1332) contain the elements of the protein required for partial function [6]. By comparison, the shortest derivative (residues 1038–1332) lacks the histone fold, which is required for interaction with other components of the complex [43]. A protein reduced by approximately 20 kDa, an estimated mass consistent with a truncation at the Spt7SLIK processing site, was seen for each of the derivatives (denoted as SL). A further truncation, approximately 20 kDa smaller than Spt7SLIK and potentially corresponding to processing at the Spt7Form3 site (F3), was evident for Spt7581–1332 and Spt7873–1332 (though at a reduced level). Note that the third-form equivalent was not seen for Spt71037–1332, because its putative cleavage site is removed from this construct. This result suggests that the elements required for processing are intrinsic to the C-terminal end of Spt7 and that the N-terminus of the protein is not required. Furthermore, the appearance of a cleavage product for Spt71037–1332 suggests that processing can take place independently from SAGA and is thus not linked to the action of the complex. For Spt7581–1332, a fourth form was apparent just below Form 3 (marked by *). We have not characterized this form further and can not exclude that it is a degradation product.
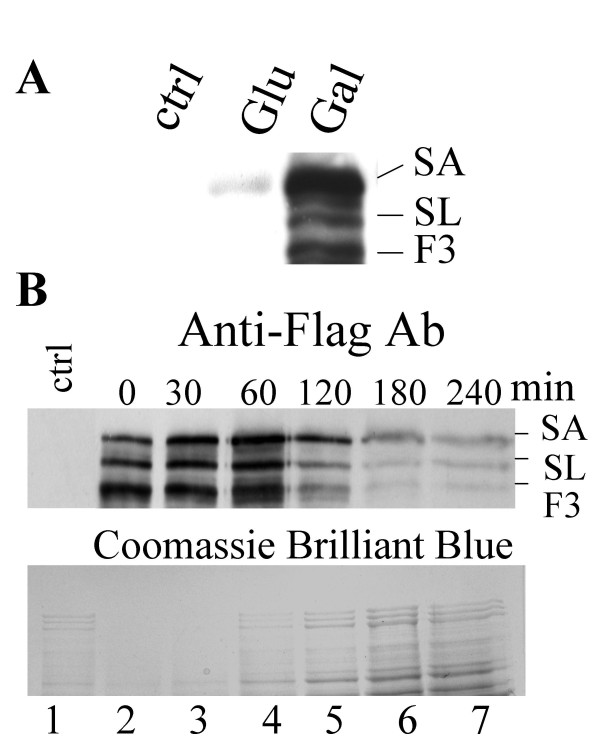
If the forms of Spt7 were created progressively from one another in response to the activity of the complex, one would predict that the longer forms would be chased into shorter forms. We analyzed the appearance and disappearance of the Spt7 forms upon inducing expression of a triple Flag-tagged Spt7 under control of the GAL10 promoter (GAL-Flag3-Spt7; see Figure 1). As shown in Figure 4a, cells containing GAL-Flag3-Spt7 rapidly induce the three Spt7 forms when shifted from raffinose to galactose containing media but not upon shifting to glucose containing media. Figure 4b is a time course of the appearance and disappearance of the Spt7 forms after induction in galactose containing media and subsequent shift to media containing glucose. Expression of the forms continued to increase after promoter shut-off until approximately the 60 minute time point. All three forms increased at approximately the same rate. After 60 minutes, the amounts of the Spt7 forms began to decay. This was in contrast to total protein, which continued to increase (comparing the Western blot versus the stained gel in Figure 4b). Similar to their appearance, the three forms disappeared at approximately the same rate. The longer forms were not chased into the shorter molecules, suggesting that Spt7SLIK and Spt7Form3 are produced early after the synthesis of Spt7SAGA and not sequentially as the result of the action of the complex. Combined with the results above, processing appears inherent to the C-terminus of the molecule and occurs independently from the action of the SAGA complex.
Figure 4.
Time course of the induction and decay of Spt7 forms. A. CY1811 containing YIplac211-GAL-Flag3-SPT7 was grown in minimal media containing 2% raffinose then diluted 1:5 into YP media containing 2% glucose (lane 2) or 2%galactose (lane 3) and grown for 30 minutes. Extracts were prepared by glass bead disruption and 100 μg separated by SDS-PAGE (5%). Flag3-Spt7 was detected by Western blotting with anti-Flag antibody. Lane one contains 100 μg of protein extract prepared after growth of BY4741 in galactose-containing media. B. CY1811 containing YIplac211-GAL – Flag3-SPT7 was grown in raffinose. Expression of Spt7 was induced by the addition of galactose then inhibited by the addition of YP media containing 2% glucose. Cells were grown for 90 minutes which was empirically determined as time 0. Additional equal volume samples were taken at time 0, 30, 60, 120 and 240 minutes (lanes 2–7). Protein extracts were prepared and equal volumes separated by SDS-Page (5%). Flag3-Spt7 was detected by Western blotting (top panel) or stained with Coomassie Brilliant Blue (bottom panel).
Further characterization of the 160 kDa form of Spt7
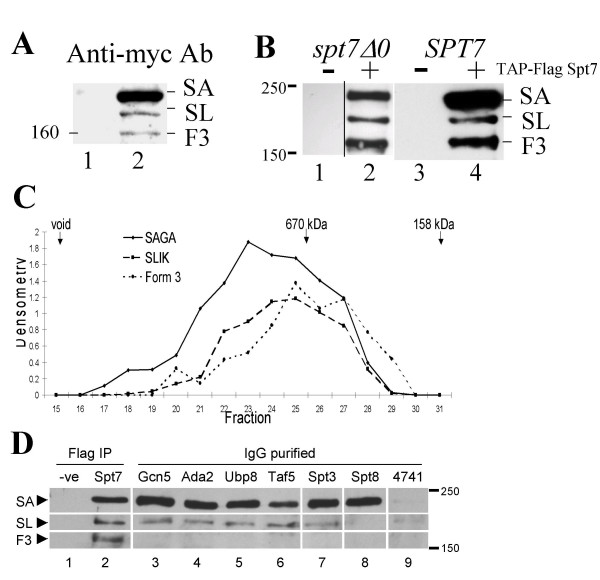
To eliminate the possibility that the appearance of Spt7Form3 was related to blotting with Flag antibody or to the TAP-tag, we expressed N-terminally myc-tagged Spt7, separated crude protein extract by SDS-PAGE and blotted with anti-myc antibody (Figure 5a). A band approximately 20 kDa smaller than myc-Spt7SAGA, likely myc-Spt7SLIK was present in wild-type cells containing YCp88-myc-SPT7 but not in control cells (compare lanes 1 and 2) as was the third-form at an apparent mass of ~160 kDa. To determine if Spt7Form3 was an artifact of the partial TAP purification or expression in cells also containing a wild-type copy of SPT7, we purified TAP-Flag-Spt7 from BY4741 (wild-type) and BY3281 (spt7Δ0) through both tandem affinity steps, IgG-Agarose and Calmodulin-Sepharose. As shown in Figure 5b, a ~160 kDa, Flag-detected protein was apparent after purification (lanes 2 and 4) and not in a mock purification (lanes 1 and 3) in both strain backgrounds.
Figure 5.
Characterization of Spt7 Form3. A. YCp88-myc was transformed into BY4741 (lane 1, ctrl) and YCp88-myc-SPT7 into BY4741 (lane 2). Crude protein extracts were prepared and Western blotted with anti-myc antibody. The mobility of a 160 kDa molecular weight marker is indicated. Spt7Form3 is marked as F3. B. BY4741 (wild-type, right panel) and BY3218 (spt7Δ0, left panel) were transformed with YCpDed-TAP-Flag (lanes 1 and 3) or YCpDed-TAP-Flag-SPT7 into (lanes 2 and 4). Extracts were prepared by grinding 1 litre of cells in liquid nitrogen, and subjected to TAP purification. 20 μl of the peak fractions were blotted with anti-Flag antibody. C. Gel filtration chromatography of Spt7 from crude yeast extracts. 8.5 mg of protein extract was prepared from yeast strain BY4741 containing YCpDed-TAP-Flag-SPT7 and applied to a Superose 6 HR10/30 column (Pharmacia). 500 μl fractions were collected, precipitated with 10% trichloroacetic acid, separated on a 5% SDS-polyacrylamide gel and Western blotted with anti-Flag antibody. Relative amounts of each of the three forms as determined by densitometry are shown as are the peak fraction for elution of the indicated molecular mass standards. D. Wild-type (4741; lane 9) or strains containing C-terminally TAP-tagged Gcn5, Ada2, Ubp8, Taf5, Spt3 and Spt8, expressing Flag-Spt7 (lanes 3–8) were grown in YP media containing 2% glucose to an OD600 = 2.0. Twenty-five milligrams of crude lysate was purified on IgG agarose. Bound protein was separated by 5% SDS-PAGE and Flag-Spt7 forms detected with anti-Flag antibody. To demonstrate the position of the three Spt7 forms lanes 1 and 2 are extracts of BY4741 transformed with YCpDed-Flag-SPT7 that have been purified on Sepharose 4B (-'ve) or anti-Flag antibody coupled to Sepharose (Spt7) then Western blotted with anti-Flag antibody. The positions of molecular mass markers, Spt7SAGA (SA), SPT7SLIK (SL) and Spt7Form3 (F3) are indicated.
To examine if Spt7Form3 is found within complexes, TAP-Flag-Spt7 was expressed in BY4741 and a crude protein extract applied to a Superose 6 HR10/30 gel filtration column. The elution profile of the different forms of Spt7 was examined by Western blotting with anti-Flag antibody and the relative amounts of the three forms of Spt7 determined by densitometry (Figure 5c). All three forms of Spt7 eluted in fractions corresponding to a high molecular mass, the largest being Spt7SAGA, followed by Spt7SLIK. Spt7Form3 eluted from the column with an average apparent molecular size slightly less than Spt7SLIK. To determine if Spt7Form3 was associated with components of the SAGA complex, extracts were prepared from strains containing N-terminally Flag-tagged Spt7 and TAP-tagged versions of Gcn5, Ada2, Ubp8, Taf5, Spt3 and Spt8. Complexes containing the TAP-tagged molecules were partially purified and the presence of the different forms of Spt7 analyzed by Western blotting with anti-Flag antibody (Figure 5d, lanes 3–9). Identification of the forms was established by immunoprecipitation of Flag-Spt7 from crude extracts with anti-Flag antibody (compare lanes 1 and 2). As expected from the interaction of Spt8 with the C-terminal end of Spt7 [4,6], Spt8 interacted only with full-length Spt7 (lane 8). The other SAGA components interacted with both Spt7SAGA and Spt7SLIK. Spt7Form3 did not co-purify with the SAGA components to the extent of either Spt7SAGA or Spt7SLIK.
The function of Spt7Form3 is unknown. The fact that Spt7Form3 appears as stable as the longer forms is consistent with it having a function(s), likely distinct from components of the SAGA complex. It is interesting to note that Spt7 has been found to associate with Ccl1, Rtf1 and Prp43, and that these interactions are not reciprocated by C-terminally TAP-tagged Spt7 or generally by other components of the SAGA complex (Saccharomyces Genome Database). In addition, we previously observed a possible SAGA-independent form of Spt7 by fractionation of crude extracts on a Mono Q column [27]. We also must consider an alternative model, whereby Spt7Form3 is an intermediate along a pathway of degradation. In this regard, we did observe an apparent increase in Spt7Form3 during TAP-purification.
Conclusion
In this report we show that processing of the C-terminus of Spt7 to the form found in the SLIK complex and to a novel third form does not require a functional SAGA complex. C-terminal truncation occurs in the absence of key components of SAGA and SLIK complexes. Spt20 and Ada1 are necessary for the integrity of a complete SAGA complex with their deletion resulting in severe transcriptional effects [6,22], yet processing occurs in their absence. In addition, since the truncated Spt7 molecules were present in a gcn5Δ0 strain, histone acetylation is not required for processing. Also suggesting that processing of Spt7 is independent of the function of SAGA, we found that a truncated derivative of Spt7 that lacks the histone fold is capable of being processed in a strain lacking full-length Spt7. Finally a time course revealing the appearance and disappearance of the Spt7 forms showed that Spt7SAGA is not chased into the shorter forms. We thus suggest that processing of Spt7 is an independent event in the biosynthesis of distinct Spt7 containing complexes rather than being linked to transcriptional regulation by the SAGA complex.
Processing of Spt7 showed partial dependence on the ubiquitin protease Ubp8. The role of Ubp8 is unclear but based on a partial effect upon deletion of Sgf11, it may require the association of Ubp8 with SAGA. We were intrigued by the possibility that the effect of Ubp8 may be related to our previous observation of the ubiquitylation of Spt7 dependent on the E3-ubiquitin ligase Tom1 [44]; however, because the relative amounts of the Spt7 forms appear unchanged in a tom1Δ0 strain (not shown) we have concluded that these observations are unrelated.
Our study has identified a third form of Spt7 with an apparent molecular mass of ~160 kDa. The identification of this form with three different tagged derivatives suggests that it is not an artifact of tagging; however, we can not fully exclude the possibility that N-terminal tagging results in its appearance. Gel-filtration experiments suggest that Spt7Form3 is found within a complex; albeit, distinct from SAGA or SLIK. As Spt7 was N-terminally tagged, the mobility of Spt7Form3 on SDS-PAGE suggests a molecule containing amino acid residues 1 to ~950. Determining the size of the released C-terminal fragment would facilitate mapping of the cleavage site; however, we have been unable to identify this molecule using C-terminally TAP-tagged Spt7.
Methods
Yeast strains
The wild-type yeast strains BY4741 (MATa hisΔ0 leu2Δ0 met1Δ0 ura3Δ0) and BY4742 (MATα hisΔ0 leu2Δ0 lys2Δ0 ura3Δ0) and the consortium constructed isogenic knockout strains [45] BY3218 (spt7Δ0), BY10809 (ubp8Δ0), BY17285 (gcn5Δ0), BY14619 (rtg2Δ0), BY12666 (spt8Δ0), BY14433 (sgf73Δ0), BY13418 (sgf29Δ0), BY2781 (sgf11Δ0), BY1038 (ada1Δ0) and BY17390 (spt20Δ0), and strains containing C-terminally TAP-tagged SAGA components were purchased from Open Biosystems or Research Genetics. CY1811 is a derivative of BY4741 that contains a YIplac211-GAL-Flag3-SPT7 integrated at the endogenous locus.
DNA constructs
YCpDed-TAP is a derivative of the LEU2 containing centromeric plasmid YCplac111 [46] that contains a DED1 promoter [9] driving expression of a TAP epitope. The DED1 promoter was synthesized by PCR using oligonucleotides 4149-1 and 4149-2 (see Table 1) and cloned as a PstI-BamHI fragment into YCplac111. The TAP epitope was subsequently cloned as a BamHI to SstI fragment into this molecule after its PCR using oligonucleotides 4149-4 and 4168-1 and pFA6a as the template (kindly provided by Kathy Gould). The coding sequence for the Flag-epitope was cloned into this molecule at the engineered NotI site using oligonucleotides 4213-1 and 4213-2 to give YCpDed-TAP-Flag. Epitope-tagged Spt7 was constructed through PCR amplification of two tandem parts of the 3999 base pair gene using oligonucleotides 4051-1 and 4051-2 for the 5'-segment and 4009-1 and 4009-2 for the 3'-segment, respectively. The two segments were ligated using the common internal SalI restriction site and cloned into YCpDed-TAP-Flag as a NotI-SstI fragment to generate YCpDed-TAP-Flag-SPT7. A molecule expressing Myc-tagged Spt7 was constructed by cloning a NotI-EcoRI fragment from YCpDed-Tap-Flag-SPT7 into the URA3 centromeric plasmid YCp88-myc [9] to generate YCp88-myc-SPT7. Single Flag-tagged Spt7 similarly contains a Met-Tyr-Lys-Asp4-Lys coding sequence inserted directly before the NotI site.
Table 1.
Oligonucleotides used in this study
| Oligo# | Sequence (5'-3') | Construct(s) |
| 4009-1 | GCGGCCGCTGGAAGAAAAGGATTGAATATGG | 3' fragment of SPT7 |
| 4009-2 | GAATTCTATTCAACTATTTAGCGCGCTC | |
| 4051-1 | GCGGCCGCAATGACTGAAAGAATACCAATAAAG | 5'fragment of SPT7 |
| 5051-2 | CCAAATCGTCTCTATCGTCATCC | |
| 4536-1 | GATGCGGCCGCGATGGAAGACGCTTCCGTG | 5' truncations of SPT7 |
| 4536-2 | GATCGGCCGCCGGTATCAACAGGCCAGAC | |
| 4213-1 | GGCCGACTACAAGGACGACGATGACAAGGC | N-terminal Flag epitope |
| 4213-2 | GGCCGCCTTGTCATCGTCGTCCTTGTAGTC | |
| 4149-4 | GAGCTCGCGGCCGCATAATCAAGTGCCCCGGAG | TAP-tag |
| 4168-1 | GGATCCATGAAAGCTGATGCGCAACAAATT | |
| 4356-1 | GATGCGGCCGCGATGGAAGACGCTTCGTG | DED1 promoter |
| 4356-2 | GATGCGGCCGCCGGTATCAACAGGCCAGAC | |
| 2764-1 | AACTGCAGTAATACGCTTAACTGCTC | GAL10 promoter |
| 2764-2 | CCCAAGCTTGACGTTAAAGTATAGAGGT | |
| 2821-1 | ACGAAGCTTACCATGGACTACAAGGACGACGAT | Met-Flag tag |
| 2821-2 | TTGAGCTCTGCGGCCGCCTTGTCATCGTCGTCC |
TAP-tagged derivatives of Spt7 with 752, 460 and 295 C-terminal amino acids were constructed by PCR using oligonucleotides 4009-1, 4536-1 and 4536-2 as 5' primer, and 4009-2 as 3' primer. Each was cloned into YCpDed-TAP-Flag as a NotI-EcoRI fragment to generate YCpDed-TAP-Flag-SPT7580–1332, YCpDed-TAP-Flag-SPT7872–1332 and YCpDed-TAP-Flag-SPT71038–1332.
The parent construct for plasmids expressing Flag3-Spt7 is pCB1450. This molecule contains a N-terminal Flag epitope (engineered with oligonucleotides 2821-1 and 2821-2) flanked upstream by the PstI site of the pUC polylinker and downstream by a NotI site. Genes to be expressed can be inserted as NotI-(SacI)EcoRI fragments. The polylinker HindIII site was deleted allowing insertion of promoter fragments as PstI-HindIII fragments. The GAL10 promoter was synthesized by PCR using oligonucleotides 2764-1 and 2764-2 and inserted into pCB1450. SPT7 from YCpDed-TAP-Flag-SPT7 was inserted as a NotI-EcoRI fragment and two additional Flag epitopes were inserted at the NotI site using oligonucleotides 4213-1 and 4213-2. For integration into yeast the GAL-Flag3-SPT7 module was inserted into YIplac211 [46]. Integration into the endogenous locus was possible after digestion with SalI.
TAP purification
Tandem affinity purification (TAP) was carried out as described [47] with minor modifications. Yeast cells were grown in YPD to an A600 = 1.5 and lysed by grinding in liquid nitrogen [48]. All further steps were performed at 4°C. Per liter of starting culture, cell extracts were suspended in ~5 ml of IPP150 (10 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.1% NP-40) with protease inhibitors (1 mM PMSF, 0.1 mM benzamidine hydrochloride, 2 μg/ml pepstatin A, 2 μg/ml leupeptin and 0.1 mg/ml trypsin inhibitor). The extract was cleared by centrifugation at 40,000 g for 1 h and incubated with 400 μl of IgG-Agarose (Sigma-Aldrich Canada Ltd) suspension for 2 h. After washing with 30 ml of IPP150 and 10 ml of TEV cleavage buffer (10 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.5 mM EDTA, 1 mM dithiothreitol, 0.1% NP-40), IgG beads were suspended in 1 ml of TEV cleavage buffer and incubated with ~100 U TEV protease for 2 h. 3 μl of 1 M CaCl2 was added to the TEV-cleaved eluent which was then diluted with 3 ml of calmodulin binding buffer (10 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1 mM magnesium acetate, 1 mM imidazole, 2 mM CaCl2, 1 mM beta-mercaptoethanol, 0.1% NP-40). For the second step of purification the protein was applied to 400 μl of Calmodulin-Sepharose (Stratagene Inc) and incubated for 2 h. After the calmodulin beads were washed with 30 ml of calmodulin binding buffer, bound protein was eluted with calmodulin elution buffer (10 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1 mM magnesium acetate, 1 mM imidazole, 20 mM EGTA, 1 mM beta-mercaptoethanol, 0.1% NP-40), in 200 μl fractions. 20 μl of the peak fractions were separated by 5% SDS-PAGE and subjected to Western blotting.
Western blotting
Yeast extracts were prepared by glass bead disruption [9] or grinding in liquid nitrogen [48]. Protein was separated by SDS polyacrylamide gel electrophoresis, transferred to PVDF membranes (Roche Applied Science) using a wet transfer system (Bio-Rad) in 48 mM Tris, 40 mM glycine, 0.0375% (6.5 mM) SDS and 20% methanol for 1 h at 100 V and detected using a 20% solution of Immobilon Western (Millipore Corp). Anti-Flag antibodies (M2, Sigma-Aldrich Canada Ltd) were used at a ratio of 1:4000; anti-myc antibody (Myc1-9E10 cell line) was used at a ratio of 1:2000. Densitometric scanning of films was performed using AlphaImager 3400 software (Alpha Innotech, Inc). Serial dilutions of samples were analyzed to provide standard curves for suitable film exposure.
Time course of the induction and decay of Spt7
A 10 ml culture of CY1811 was grown to an A600 ~3.0 in minimal media containing 0.6% casamino acids (Difco) and 2% raffinose. The culture was diluted with 10 ml of prewarmed media containing 2% galactose and grown for 30 minutes at 30°C. 80 ml of YP media containing 2% glucose was added and the cells grown for 90 minutes. This became time 0, and a 15 ml sample was taken. Cells were pelleted by centrifugation, washed in ice-cold water and frozen at -80°C. Additional samples were taken after 30, 60, 120 and 240 minutes. Yeast extracts were prepared by glass bead disruption in 20 mM Hepes-NaOH (pH 8.0), 240 mM NaCl containing protease inhibitors. Equal volumes were separated by SDS-PAGE (5%) and Western blotted with anti-Flag antibody.
Gel-filtration chromatography
Approximately 8.5 mg of whole cell extract from BY4741 containing YCpDed-TAP-Flag-SPT7 was prepared in 40 mM Tris-HCl (pH 7.7), 300 mM NaCl, 0.1% Nonidet P-40 and 10% glycerol, and subsequently applied to a Superose 6 HR10/30 column (Pharmacia Inc) at a flow rate of 0.3 ml/min. Protein from 250 μl aliquots of 500-μl selected fractions were precipitated with 10% trichloroacetic acid, acetone washed, resuspended in SDS loading buffer and separated on a 5% SDS-polyacrylamide gel.
Authors' contributions
SH carried out the analysis of Spt7 in yeast deletion strains, copurifications and mapping studies. GL performed analyses with Spt7 deletion constructs and biochemical fractionation of Spt7. AM performed functional analyses of Spt7 alleles. JG carried out the time course analysis of Spt7 synthesis and degradation. CB directed the experiments and wrote the manuscript.
Acknowledgments
Acknowledgements
We would like to thank Amy Tong and Drs. Brenda Andrews, Charlie Boone and Kathleen Gould for providing reagents as well as David Canton, David Litchfield, Meagan Davey, David Edgell, Kerri Kobryn and David Haniford for their discussions on this manuscript. This work was supported by a Canadian Institutes of Health Research operating grant to CB. SH is supported by a NSERC studentship, GL and AM by Western Graduate Research Scholarships.
Contributor Information
Stephen MT Hoke, Email: shoke@uwo.ca.
Gaoyang Liang, Email: gaoyang_liang@med.unc.edu.
A Irina Mutiu, Email: abulboac@uwo.ca.
Julie Genereaux, Email: jgenerea@uwo.ca.
Christopher J Brandl, Email: cbrandl@uwo.ca.
References
- Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/MMBR.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant PA, Duggan L, Côté J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, Berger SL, Workman JL. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- Belotserkovskaya R, Sterner DE, Deng M, Sayre MH, Lieberman PM, Berger SL. Inhibition of TATA-binding protein function by SAGA subunits Spt3 and Spt8 at Gcn4-activated promoters. Mol Cell Biol. 2000;20:634–647. doi: 10.1128/MCB.20.2.634-647.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Belotesrkovskaya R, Berger SL. SALSA, a variant of yeast SAGA, contains truncated Spt7, which correlates with activated transcription. Proc Natl Acad Sci USA. 2002;99:11622–11627. doi: 10.1073/pnas.182021199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pray-Grant MG, Schieltz D, McMahon SJ, Wood JM, Kennedy EL, Cook RG, Workman JL, Yates JR, 3rd, Grant PA. The novel SLIK histone acetylatransferase complex functions in the yeast retrograde response pathway. Mol Cell Biol. 2002;22:8774–8786. doi: 10.1128/MCB.22.24.8774-8786.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu PY, Winston F. Analysis of Spt7 function in the Saccharomyces cerevisiae SAGA coactivator complex. Mol Cell Biol. 2002;22:5367–5379. doi: 10.1128/MCB.22.15.5367-5379.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL, Pina B, Silverman N, Marcus GA, Agapite J, Regier JL, Triezenberg SJ, Guarente L. Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell. 1992;70:251–265. doi: 10.1016/0092-8674(92)90100-Q. [DOI] [PubMed] [Google Scholar]
- Georgakopoulos T, Thireos G. Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription. EMBO J. 1992;11:4145–4152. doi: 10.1002/j.1460-2075.1992.tb05507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandl CJ, Furlanetto AM, Martens JA, Hamilton KS. Characterization of NGG1, a novel yeast gene required for glucose repression of GAL4p-regulated transcription. EMBO J. 1993;12:5255–5265. doi: 10.1002/j.1460-2075.1993.tb06221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pina B, Berger S, Marcus GA, Silverman N, Agapite J, Guarente L. ADA3: a gene, identified by resistance to GAL4-VP16, with properties similar to and different from those of ADA2. Mol Cell Biol. 1993;13:5981–5989. doi: 10.1128/mcb.13.10.5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus GA, Silverman N, Berger SL, Horiuchi J, Guarente L. Functional similarity of physical association between GCN5 and ADA2: putative transcriptional adaptors. EMBO J. 1994;13:4807–4815. doi: 10.1002/j.1460-2075.1994.tb06806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: a transcriptional co-activator linking gene expression to histone acetylation. Cell. 1996;84:843–851. doi: 10.1016/S0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- Kuo MH, Zhou J, Jambeck P, Churchill ME, Allis CD. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utley RT, Ikeda K, Grant PA, Côté J, Steger DJ, Eberharter A, John S, Workman JL. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- Wang L, Liu L, Berger SL. Critical residues for histone acetylation by GCN5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes Dev. 1998;12:640–653. doi: 10.1101/gad.12.5.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci AR, Genereaux J, Brandl CJ. Components of the SAGA histone acetylatransferase complex are required for repressed transcription of ARG1 in rich medium. Mol Cell Biol. 2002;22:4033–4042. doi: 10.1128/MCB.22.12.4033-4042.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbaric S, Walker J, Schmid A, Svejstrup JQ, Horz W. Increasing the rate of chromatin remodeling and gene activation – a novel role for the histone. EMBO J. 2001;20:4944–4951. doi: 10.1093/emboj/20.17.4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant PA, Schieltz D, Pray-Grant MG, Steger DJ, Reese JC, Yates JR, 3rd, Workman JL. A subset of TAFIIs are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell. 1998;94:45–53. doi: 10.1016/S0092-8674(00)81220-9. [DOI] [PubMed] [Google Scholar]
- Wu PY, Ruhlmann C, Winston F, Schultz P. Molecular architecture of the S. cerevisiae SAGA complex. Mol Cell. 2004;15:199–208. doi: 10.1016/j.molcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Horiuchi J, Silverman N, Pina B, Marcus GA, Guarente L. ADA1, a novel component of the ADA/GCN5 complex has broader effects than GCN5, ADA2, or ADA3. Mol Cell Biol. 1997;17:3220–3228. doi: 10.1128/mcb.17.6.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SM, Winston F. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics. 1997;147:451–465. doi: 10.1093/genetics/147.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Grant PA, Roberts SM, Duggan LJ, Belotesrkovskaya R, Pacella LA, Winston F, Workman JL, Berger SL. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-Binding protein interaction. Mol Cell Biol. 1999;19:86–98. doi: 10.1128/mcb.19.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann DM, Arndt KM, Ricupero SL, Rodney JW, Winston F. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae . Genes Dev. 1992;6:1319–1331. doi: 10.1101/gad.6.7.1319. [DOI] [PubMed] [Google Scholar]
- Eisenmann DM, Chapon C, Roberts SM, Dollard C, Winston F. The Saccharomyces cerevisiae SPT8 gene encodes a very acidic protein that is functionally related to SPT3 and TATA-binding protein. Genetics. 1994;137:647–657. doi: 10.1093/genetics/137.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley AM, Rougeulle C, Winston F. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo . Genes Dev. 1999;13:2940–2945. doi: 10.1101/gad.13.22.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant PA, Schieltz D, Pray-Grant MG, Yates JR, 3rd, Workman JL. The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Mol Cell. 1998;2:863–867. doi: 10.1016/S1097-2765(00)80300-7. [DOI] [PubMed] [Google Scholar]
- Saleh A, Schieltz D, Ting N, McMahon SB, Litchfield DW, Yates JR, 3rd, Lees-Miller SP, Cole MD, Brandl CJ. Tra1 is a component of the yeast Ada-Spt transcriptional regulatory complexes. J Biol Chem. 1998;273:26559–26565. doi: 10.1074/jbc.273.41.26559. [DOI] [PubMed] [Google Scholar]
- Brown CE, Howe L, Sousa K, Alley SC, Carrozza MJ, Tan S, Workman JL. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science. 2001;292:2333–2337. doi: 10.1126/science.1060214. [DOI] [PubMed] [Google Scholar]
- Kulesza CA, van Buskirk HA, Cole MD, Reese JC, Smith MM, Engel DA. Adenovirus E1A requires the yeast SAGA histone acetyltransferase complex and associates with SAGA components Gcn5 and Tra1. Oncogene. 2002;21:1411–1422. doi: 10.1038/sj.onc.1205201. [DOI] [PubMed] [Google Scholar]
- Bhaumik SR, Raha T, Aiello DP, Green MR. In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev. 2004;18:333–343. doi: 10.1101/gad.1148404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishburn J, Mohibullah N, Hahn S. Function of a eukaryotic transcription activator during the transcription cycle. Mol Cell. 2005;18:369–378. doi: 10.1016/j.molcel.2005.03.029. [DOI] [PubMed] [Google Scholar]
- Reeves WM, Hahn S. Targets of the Gal4 Transcription Activator in Functional Transcription Complexes. Mol Cell Biol. 2005;25:9092–9102. doi: 10.1128/MCB.25.20.9092-9102.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SL, Jennings J, Canutescu A, Link AJ, Weil PA. Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol Cell Biol. 2002;22:4723–4738. doi: 10.1128/MCB.22.13.4723-4738.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell DA, Weaver CM, Jennings JL, McAfee KJ, He Y, Weil PA, Link AJ. Cluster analysis of mass spectrometry data reveals a novel component of SAGA. Mol Cell Biol. 2004;24:7249–7259. doi: 10.1128/MCB.24.16.7249-7259.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodirguez-Navarro S, Fischer T, Luo MJ, Antunez O, Brettschneider S, Lechner J, Perez-Ortin JE, Reed R, Hurt E. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell. 2004;116:75–86. doi: 10.1016/S0092-8674(03)01025-0. [DOI] [PubMed] [Google Scholar]
- Pray-Grant MG, Daniel JA, Schieltz D, Yates JR, 3rd, Grant PA. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- Liu Z, Butow RA. A transcriptional switch in the expression of yeast tricarboxylic acid cycle genes in response to a reduction or loss of respiratory function. Mol Cell Biol. 1999;19:6720–6728. doi: 10.1128/mcb.19.10.6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Rothermel B, Thornton J, Butow RA. A basic helix-loop-helix-leucine zipper transcription complex in yeast functions in a signaling pathway from mitochondria to the nucleus. Mol Cell Biol. 1997;17:1110–1117. doi: 10.1128/mcb.17.3.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekito T, Thornton J, Butow RA. Mitochondria-to-nuclear signaling is regulated by the subcellular localization of the transcription factors Rtg1p and Rtg3p. Mol Biol Cell. 2000;11:2103–2115. doi: 10.1091/mbc.11.6.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvarsdottir K, Krogan NJ, Emre NC, Wyce A, Thompson NJ, Emili A, Hughes TR, Greenblatt JF, Berger SL. H2B Ubiquitin Protease Ubp8 and Sgf11 constitute a discrete functional module within the Saccharomyces cerevisiae SAGA complex. Mol Cell Biol. 2005;25:1162–1172. doi: 10.1128/MCB.25.3.1162-1172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, Florens L, Swanson SK, Washburn MP, Workman JL. The deubiquitylation activity of Ubp8 is dependent upon Sgf11 and its association with the SAGA complex. Mol Cell Biol. 2005;25:1173–1182. doi: 10.1128/MCB.25.3.1173-1182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A, Stanojevic N, Duan Z, Sen PT, Bhaumik SR. Ubp8p, a histone deubiquitinase whose association with SAGA is mediated by Sgf11p, differentially regulates lysine 4 methylation of histone H3 in vivo. Mol Cell Biol. 2006;26:3339–3352. doi: 10.1128/MCB.26.9.3339-3352.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff YG, Sanders SL, Romier C, Kirschner D, Weil PA, Tora L, Davidson I. Histone folds mediate selective heterodimerization of yeast TAF(II)25 with TFIID components yTAF(II)47 and yTAF(II)65 and with SAGA component ySPT7. Mol Cell Biol. 2001;21:1841–1853. doi: 10.1128/MCB.21.5.1841-1853.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A, Collart M, Martens JA, Genereaux J, Allard S, Côté J, Brandl CJ. Tom1p, a yeast hect-domain protein which mediates transcriptional regulation through the yeast ADA/SAGA coactivator complexes. J Mol Biol. 1998;282:933–946. doi: 10.1006/jmbi.1998.2036. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Davis RW. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast E. coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Séraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- Saleh A, Lang V, Cook R, Brandl CJ. Identification of native complexes containing the yeast coactivator/repressor proteins NGG1/ADA3 and ADA2. J Biol Chem. 1997;272:5571–5578. doi: 10.1074/jbc.272.9.5571. [DOI] [PubMed] [Google Scholar]