Abstract
The harmfulness of 8-oxo-7,8-dihydro-2′-deoxyguanosine (8oxodG) damage resides on its dual coding potential, as it can pair with the correct dCMP (dC) or the incorrect dAMP (dA). Here, we investigate the translesional synthesis ability of family B ϕ29 DNA polymerase on 8oxodG-containing templates. We show that this polymerase preferentially inserts dC opposite 8oxodG, its 3′–5′ exonuclease activity acting indistinctly on both dA or dC primer terminus. In addition, ϕ29 DNA polymerase shows a favoured extension of the 8oxodG/dA pair, but with an efficiency much lower than that of the canonical dG/dC pair. Additionally, we have analysed the role of the invariant tyrosine from motif B of family B DNA polymerases in translesional synthesis past 8oxodG, replacing the corresponding ϕ29 DNA polymerase Tyr390 by Phe or Ser. The lack of the aromatic portion in mutant Y390S led to a lost of discrimination against dA insertion opposite 8oxodG. On the contrary, the absence of the hydroxyl group in the Y390F mutant precluded the favoured extension of 8oxodG:dA base pair with respect to 8oxodG:dC. Based on the results obtained, we propose that this Tyr residue contributes to dictate nucleotide insertion and extension preferences during translesion synthesis past 8oxodG by family B replicases.
INTRODUCTION
The high fidelity displayed by replicases from families A, B and C outcomes from 104 to 106-fold polymerase preference for inserting correct rather than incorrect nucleotides (1). Such selectivity has traditionally been considered to rely on the base-pair shape and size, as the result of the geometric restraints imposed by the polymerase active site to tolerate equivalent Watson–Crick base pairs, ruling out those differing from this geometry (1–3). Additionally, recent studies performed with non-natural nucleotides also suggest a role for the π–π stacking interactions between the aromatic rings of the incoming dNTP and amino acid residues for an efficient polymerization at least in family B DNA polymerases (4). Polymerases contact DNA at positions in which both, topology and chemistry are identical among the four canonical base pairs. Thus, interactions occur mainly with the base, sugar and triphosphate moieties of the incoming nucleotide, with the nucleotide placed immediately 5′ with respect to the templating nucleotide and, most importantly, with the minor groove of the nascent base pair (1,3). In this sense, structural studies of DNA polymerase complexes have revealed the presence of conserved residues at their catalytic sites that make contacts with the purine N3 and pyrimidine O2 atoms that act as hydrogen-bond acceptors and are positioned at similar locations in the minor groove (1,5–8). Incorrect insertion of a nucleotide will move such H-acceptors out of position, breaking interactions with the polymerase and diminishing the DNA-binding stability at the polymerization site. This will result in an increased chance of the primer DNA to be switched to the 3′-5′ exonuclease site of the replicase to have the incorrect nucleotide removed (8–10).
Lesions in the genomes arise by their continuous exposure to the action of toxic environmental agents such as ultraviolet and ionizing radiation, genotoxic chemicals, by-products of the normal metabolism like the reactive oxygen species (ROS), in addition to spontaneous breaks of chemical bonds (11). Most organisms code for a number of enzymes to repair such damages before replication fork meeting them since replicative DNA polymerases stall when they come across one of these lesions as they cannot form a proper and catalytically competent ternary complex with the damaged nucleotide at their catalytic sites. One exception is the specially deleterious lesion 8-oxo-7,8,-dihydro-2′-deoxyguanosine (8oxodG), produced by ROS inside the cell (12) as, if it escapes the repair machinery, it can be used by the replicase as template or as incoming nucleotide. The harmfulness of this lesion resides in its dual coding potential as it can pair with both cytosine and adenine during DNA synthesis, in the latter case leading to G to T transversions (13,14). Structural studies of both 8oxodG:dC and 8oxodG:dA base pairs showed that 8oxodG adopts the classical anti conformation opposite dC, pairing in a Watson–Crick fashion, whereas its glycosidic bond adopts the syn orientation to form a Hoogsteen base pair with dA (15–19).
Structural modelling of catalytically competent complexes of RB69 and T7 DNA polymerases have suggested that the preferential insertion of dC opposite such a lesion is accomplished by a handicapped dA incorporation since the O8 atom of the 8oxodG(syn):dA mispair sterically clashes with specific residues at the corresponding active sites (19,20). However, notwithstanding their high insertion fidelity and preferential dC insertion during the bypass of 8oxodG-containing templates, replicases can misincorporate dA with a moderately high efficiency (19–24) due to the fact that such a base pair establishes appropriate hydrogen-bond interactions with the minor groove sensing residues at the catalytic site (19).
Bacteriophage ϕ29 DNA polymerase is a protein-primed DNA-dependent replicase belonging to the eukaryotic-type family of DNA polymerases (family B). Like many other replicases, it possesses, within a single polypeptide chain, both 5′–3′ polymerization and 3′–5′ exonuclease activities. It displays a high intrinsic nucleotide insertion discrimination [104 to 106 (25)], which is further improved 100-fold through proofreading by the exonuclease domain (26). ϕ29 DNA polymerase displays two unique characteristics compared with most replicases. First, a DNA polymerase molecule replicates the entire genome processively without the assistance of processivity factors (27), in contrast to most replicases that require accessory proteins to clamp the enzyme to the DNA (28–31). Second, ϕ29 DNA polymerase couples processive DNA polymerization to strand displacement. This ability allows the enzyme to replicate the ϕ29 double-strand genome without the need for a helicase (27).
In this article, we study the ability of ϕ29 DNA polymerase to perform translesional synthesis past 8oxodG by assaying the nucleotide insertion opposite the lesion and further extension steps, as well as its capacity to proofread the formed pairs, this issue being of importance, as this enzyme is currently used for isothermal rolling circle amplification and whole genome amplification (32,33). Structural models mentioned above, together with multiple sequence alignments of DNA polymerases (34), as well as the availability of the crystallographic structure of ϕ29 DNA polymerase (35), have led us to analyse the role in translesional synthesis past 8oxodG of the invariant Tyr residue of the highly conserved B motif of family B DNA polymerases by means of substitutions at the corresponding ϕ29 DNA polymerase residue Tyr390. The results obtained allow us to propose a principal and dual role for this residue as one of the key determinants that dictate nucleotide insertion and extension preferences during translesion synthesis past 8oxodG by family B replicases.
MATERIALS AND METHODS
Nucleotides and DNAs
Unlabelled nucleotides were purchased from Amersham Pharmacia Biochemicals. [γ-32P]ATP (3000 Ci/mmol) was obtained from Amersham Pharmacia. Oligonucleotides Pber (5′CTGCAGCTGATGCGC), Pber-2 (5′CTGCAGCTGATGC), PberA (5′CTGCAGCTGATGCGCA), PberC (5′CTGCAGCTGATGCGCC) and sp1 (5′GATCACAGTGAGTAC) were 5′-labelled with [γ-32P]ATP and phage T4 polynucleotide kinase and purified electrophoretically on 8 M urea–20% polyacrylamide gels. Labelled Pber, Pber-2, PberA and PberC were hybridized to the 34-mer 8oxodG containing oligonucleotide 5′GTACCCGGGGATCCGTAC(8oxodG)GCGCATCAGCTGCAG, and labelled sp1 to sp1cC+5 (5′TCTATCGTACTCACTGTGATC) and to sp1cA+5 (5′TCTATAGTACTCACTGTGATC). Hybridizations were performed in the presence of 0.2 M NaCl and 50 mM Tris-HCl (pH 7.5), resulting in primer/template structures. Oligonucleotides were obtained from Invitrogen.
Proteins
Phage T4 polynucleotide kinase was obtained from New England Biolabs. ϕ29 DNA polymerase variants at Tyr390 residue Y390F and Y390S were constructed by J. Saturno in an exonuclease deficient background (D12A/D66A) (hereafter PolY390FExo− and PolY390SExo−, respectively (unpublished data) and further purified from Escherichia coli BL21(DE3) cells harbouring the corresponding recombinant plasmid (36).
Translesion synthesis past 8oxodG by ϕ29 DNA polymerase
The hybrid molecules Pber-2/T4 and Pber-2/8oxodG, containing the unmodified dG and an 8oxodG lesion at the +3 position of the template, respectively, can be used both as substrate for the 3′–5′ exonuclease activity and for DNA-dependent DNA polymerization. The incubation mixture contained, in a final volume of 12.5 μl, 50 mM Tris-HCl (pH 7.5), 1 mM dithiothreitol, 4% glycerol, 0.1 mg/ml bovine serum albumin (BSA), 10 mM MgCl2, 1.2 nM of either 5′-labelled Pber-2/T4 or Pber-2/8oxodG, 24 nM of either wild-type or mutant D12A/D66A (Exo−) DNA polymerase, and the indicated concentration of the four dNTPs. After incubation for 5 min at 25°C, the reaction was stopped by adding EDTA up to 10 mM. Samples were analysed by 8 M urea–20% PAGE and autoradiography. Polymerization or 3′–5′ exonuclease activity are detected as an increase or decrease, respectively, in the size (13-mer) of the 5′-labelled Pber-2 primer.
3′–5′ exonuclease activity of wild-type ϕ29 DNA polymerase
The incubation mixture contained, in 12.5 µl, 50 mM Tris-HCI (pH 7.5), 10 mM MgCl2, 1 mM dithiothreitol, 4% glycerol, 0.1 mg/ml BSA, 6 nM of wild-type ϕ29 DNA polymerase and 1.2 nM of either 5′-labelled PberC/T4, PberA/8oxodG or PberC/8oxodG double-stranded DNA (dsDNA) substrate. Samples were incubated at 25°C for the indicated times and quenched by adding EDTA up to 10 mM. Reactions were analysed by 8 M urea–20% PAGE and autoradiography.
Insertion of dC and dA opposite 8oxodG
The incubation mixture contained, in a final volume of 12.5 μl, 50 mM Tris-HCl (pH 7.5), 1 mM dithiothreitol, 4% glycerol, 0.1 mg/ml BSA and 10 mM MgCl2. As substrate, 1.2 nM of the 5′-labelled hybrid molecule Pber/8oxodG dsDNA was used. The amount of DNA polymerase added (12, 30 and 18 nM of PolwtExo−, PolY390FExo− and PolY390SExo− mutants, respectively) was adjusted to obtain linear conditions. Samples were incubated for 15 s at 25°C, in the presence of the indicated concentrations of either dCTP or dATP, and quenched by adding 3 μl of gel loading buffer. Reactions were analysed by electrophoresis in 8 M urea–20% PAGE and quantified using a Molecular Dynamics PhosphorImager. Formation of the extended product was plotted against dNTP concentration. Apparent values for Michaelis–Menten constant Km and Vmax for incorporation opposite 8oxodG were obtained by least-squares nonlinear regression to a rectangular hyperbola using Kaleidagraph 3.6.4 software. kcat was calculated by dividing the Vmax by the enzyme concentration. Catalytic efficiency was obtained by dividing kcat by Km. Discrimination factor (f ins) for the nucleotide insertion opposite 8oxodG was obtained by dividing the catalytic efficiency for the incorporation of dCTP by the corresponding one for the insertion of dATP. Assays were performed at least three times to guarantee reproducibility.
Extension of DNA synthesis past a 8oxodG lesion
As substrate, 1.2 nM of the 5′-labelled hybrid molecules PberA/8oxodG and PberC/8oxodG were used to analyse extension of 8oxodG:dA and 8oxodG:dC base pairs, respectively. Assay was carried out under the same experimental conditions as described above, but in the presence of the indicated concentrations of dGTP, the next correct nucleotide to be inserted. Extension parameters were obtained as mentioned. Discrimination factor (fext) for the extension of DNA synthesis past the 8oxodG lesion was obtained by dividing the catalytic efficiency for the extension of 8oxodG:dA by the corresponding one for the extension of 8oxodG:dC. Assays were performed at least three times to guarantee reproducibility.
Measurement of DNA polymerase dissociation rates
Reactions were carried out essentially as described (37,38).The incubation mixture contained, in a final volume of 12.5 μl, 50 mM Tris-HCl (pH 7.5), 1 mM dithiothreitol, 4% glycerol, 0.1 mg/ml BSA, 10 mM MgCl2, 1.2 nM of the 5′-labelled hybrid molecule PberA/8oxodG or PberC/8oxodG and 25 nM of either PolwtExo− or 75 nM of PolY390FExo− mutant. After 5 min at 25°C to reach the binding equilibrium, samples were mixed with a 1000-fold excess of unlabelled substrate to trap dissociated DNA polymerase molecules. After the delay times indicated, 100 μM dGTP was added. Reactions were quenched by adding 3 μl of gel loading buffer 15 s after addition of dGTP. For 0 min delay time point, dGTP was included in the solution with the trapping DNA. Control reaction of the effectiveness of the trap was performed by incubating the DNA polymerase with the labelled and trapping DNA simultaneously. Reactions were analysed by electrophoresis in 8 M urea–20% PAGE and quantified using a Molecular Dynamics PhosphorImager. The fraction of primer molecules extended (y) by the saturating concentration of dGTP was plotted against the delay time (t). These data were fit to the single exponential y = Ae−kt + B to obtain the rate constant for dissociation (k). Assays were performed at least three times to guarantee reproducibility.
8oxodGTP incorporation opposite dC and dA
The incubation mixture contained, in a final volume of 12.5 μl, 50 mM Tris-HCl (pH 7.5), 1 mM dithiothreitol, 4% glycerol, 0.1 mg/ml BSA, 10 mM MgCl2 and 12 nM of PolwtExo−. As substrate, 1.2 nM of the 5′-labelled hybrid molecule sp1/sp1cC+5 or sp1/sp1cA+5 was used. Samples were incubated at 25°C for 15 s in the presence of the indicated concentrations of 8oxodGTP and quenched by adding 3 μl of gel loading buffer. Reactions were analysed and quantified as described above.
Extension of dC:8oxodG and dA:8oxodG base pairs
The incubation mixture contained, in a final volume of 12.5 μl, 50 mM Tris-HCl (pH 7.5), 1 mM dithiothreitol, 4% glycerol, 0.1 mg/ml BSA, 10 mM MgCl2 and 12 nM of PolwtExo−. As substrate, 1.2 nM of the 5′-labelled hybrid molecule sp1/sp1cC+5 or sp1/sp1cA+5 was used. Samples were incubated at 25°C, in the presence of either 100 nM dGTP or 64 μM 8oxodGTP to obtain the dC:dG, dC:8oxodG and dA:8oxodG base pairs poised for extension by the indicated dATP concentrations. Samples were incubated at 25°C for 15 s and quenched by adding 3 μl of gel loading buffer. Reactions were analysed by electrophoresis in 8 M urea–20% PAGE.
RESULTS AND DISCUSSION
Translesion synthesis past 8oxodG by ϕ29 DNA polymerase
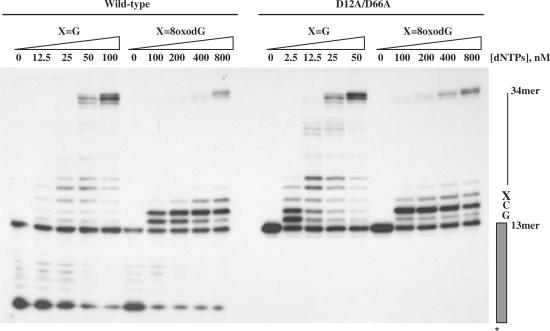
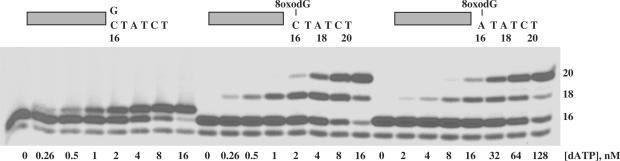
ϕ29 DNA polymerase is the most processive replicase known, being able to incorporate, without the assistance of processivity factors, more than 70 000 nt during a single encounter with an undamaged DNA template (27). Considering the presence of ROS derived from normal cellular metabolism that react with and modify DNA, is predictable that ϕ29 DNA polymerase has to deal with such DNA lesions during replication. Here, we have analysed the efficiency of translesion synthesis past one of the most abundant DNA lesions caused by ROS, the 8oxodG. To test ϕ29 DNA polymerase ability to carry out nucleotide insertion and further extension opposite such a lesion, primer extension reactions were conducted in the presence of increasing concentrations of the four dNTPs (see the Materials and Methods section). The 5′-labelled primer molecule was annealed to templates containing at the +3 position with respect to the 3′-end of primer terminus either the natural dG (X=G) or 8oxodG (X=8oxodG). As shown in Figure 1, in the absence of nucleotides, the wild-type enzyme digests the primer strand by means of its 3′–5′ exonuclease activity and requires 50–100 nM dNTPs to get a net elongation of the primer strand with the unmodified template, yielding full-length (34-mer) products. In contrast, at those high dNTPs concentrations (100 nM), the polymerase stalls at the +1 and +2 positions, that is, prior to the 8oxodG lesion, showing nucleotide insertion opposite damage as a limiting step. As dNTP concentration increases, a band at the +3 position appears just opposite 8oxodG, indicating that extension of the primer terminus paired to 8oxodG is also impaired, needing an 800 nM dNTP concentration to come across the damage. In parallel, we tested the behaviour of the exonuclease deficient ϕ29 DNA polymerase mutant D12A/D66A on these molecules to study the effect of the 3′–5′ exonuclease activity during translesion synthesis past 8oxodG (Figure 1). The absence of competition by the 3′–5′ exonuclease activity allows mutant polymerase to insert and further elongate nucleotides at dNTPs concentrations lower than those of the wild-type enzyme with the unmodified template. Interestingly, the mutant polymerase was able to insert nucleotides opposite 8oxodG at 100 nM dNTPs, as manifested by the presence of a band at the +3 position. To get full-length products with the damaged DNA, both mutant and wild-type DNA polymerases required a dNTP concentration 16-fold higher than that needed with the undamaged template. From this, it could be inferred that the 3′–5′ exonuclease activity of the wild-type enzyme exerts a similar pressure with both substrates, suggesting that during translesional synthesis DNA polymerase could proofread the primer terminus paired with 8oxodG.
Figure 1.
Translesion synthesis past 8oxodG by ϕ29 DNA polymerase. The assay was carried out as described in the Materials and Methods section. Twenty-four nanomolar of either wild-type or mutant D12A/D66A (Exo−) DNA polymerase were incubated with 32P-labelled primer/template DNA hybrid molecules, containing at the +3 position of the template either dG (X=G) or 8oxodG (X=8oxodG), and in the presence of the indicated concentration of the four dNTPs. After incubation for 5 min at 25°C, samples were analysed by 8 M urea–20% PAGE and autoradiography. Polymerization or 3′–5′ exonuclease activity are detected as an increase or decrease, respectively, in the size (13-mer) of the 5′-labelled primer.
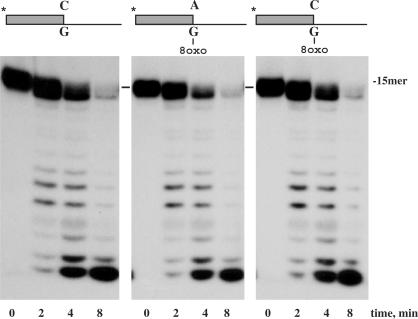
To carry out a comparative analysis of the capacity of the wild-type DNA polymerase to excise 3′ terminal dA or dC paired to 8oxodG versus dC opposite the unmodified dG, we performed a 3′–5′ exonuclease assay using as substrates the hybrid molecules depicted on top of Figure 2 (see also Materials and Methods section). As shown in Figure 2, the wild-type DNA polymerase was able to degrade the three substrates with the same efficiency, irrespective of their terminus base pair. The inherent potential deleteriousness of the 8oxodG(syn):dA mispair is a consequence of not being identified as aberrant by the polymerase since, in such orientation, the O8 group mimics the O2 group of a thymidine, establishing a proper hydrogen-bond contact with the minor groove sensing residues at the catalytic site (19). This fact allows stable mispair formation at the polymerization site, avoiding its exonucleolytic correction. In addition, DNA polymerases contain residues that make interactions through the minor groove of the DNA at post-replication positions that contribute to binding affinity and to the ability to sense base-pair geometry (3). The misinsertion and further extension of nucleotides provoke changes in the geometry of the DNA with the subsequent lost of several of these contacts, causing DNA polymerase to stall and proofread the misinserted nucleotides (9). Protrusion of the Hoogsteen base pair 8oxodG(syn):dA into the major groove does not alter the overall structure of the DNA, contributing to avoid its exonucleolytic proofread.
Figure 2.
3′–5′ Exonuclease activity of wild-type ϕ29 DNA polymerase on dA- or dC- containing primer-terminus paired to 8oxodG. The assay was carried out in the conditions described in the Materials and Methods section, using 1.2 nM of the hybrid molecule depicted on top of the figure and in the presence of 6 nM of wild-type ϕ29 DNA polymerase. After incubation at the indicated times at 25°C, degradation of the labelled DNA was analysed by electrophoresis in 8 M urea–20% PAGE and autoradiography.
Error-free synthesis opposite 8oxodG depends on the invariant Tyrosine residue of motif B
As most DNA polymerases assayed in their capacity to insert nucleotides opposite 8oxodG [see (39) and references therein], ϕ29 DNA polymerase is able to catalyse mainly the insertion of both dC and dA (data not shown). Crystallization of bacteriophage T7 (family A) (19) and RB69 (family B) (20) DNA polymerases complexed with a nascent 8oxodG(anti):dCTP base pair have given light about the structural rationale for the efficient bypass of 8oxodG, in contrast to other lesions as abasic sites and thymine dimers that completely stall the replication machinery until the lesion is either repaired or bypassed. In both structures, the unfavourable steric interaction between the O8 group and O4 oxygen of the 8oxodG in its non-mutagenic (anti) conformation is alleviated by a kinking of the phophodiester backbone of the template strand at the catalytic site. In T7 DNA polymerase, there is an additional contact between the ε amino group of residue Lys536 and the O8 group of 8oxodG steadying the anti conformation for base pairing with dCTP (19,24). On the other hand, modelling of a nascent 8oxodG(syn):dATP base pair in the active site of both DNA polymerases allowed Brieba et al. (19) and Freisinger et al. (20) to predict steric and electrostatic clashes between the O8 group and the side chain of residues located at the fingers subdomain, as responsible for the discrimination against the 8oxodG in its syn (mutagenic) conformation. In spite of the presence of polymerase-dependent specific residues, in both DNA polymerases the Tyr residue of the highly conserved motif B (Tyr567 and Tyr530 in RB69 and T7 DNA polymerases, respectively) (40) is proposed to play a role in discrimination during nucleotide insertion opposite an 8oxodG site.
To study the role of such Tyr residue in 8oxodG translesion synthesis, we have analysed the insertion and elongation capacities opposite 8oxodG-containing templates of ϕ29 DNA polymerase variants at the homologous Tyr390 residue, in an exonuclease deficient background (D12A/D66A). PolY390FExo− and PolY390SExo− mutants, allowed us to evaluate the importance of the hydroxyl and aromatic groups, respectively, in both translesional phases by comparing their activities with the wild-type enzyme (PolwtExo−).
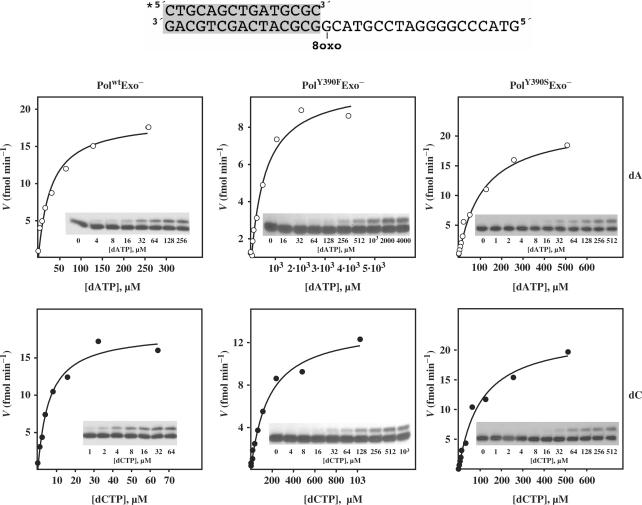
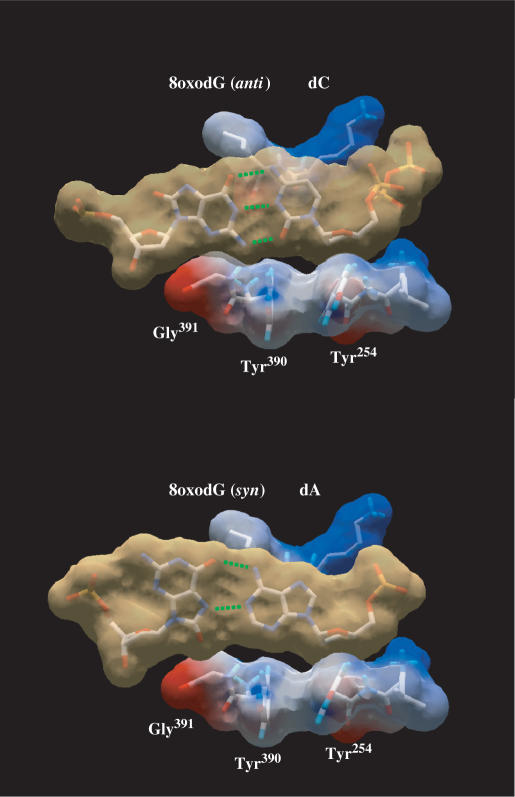
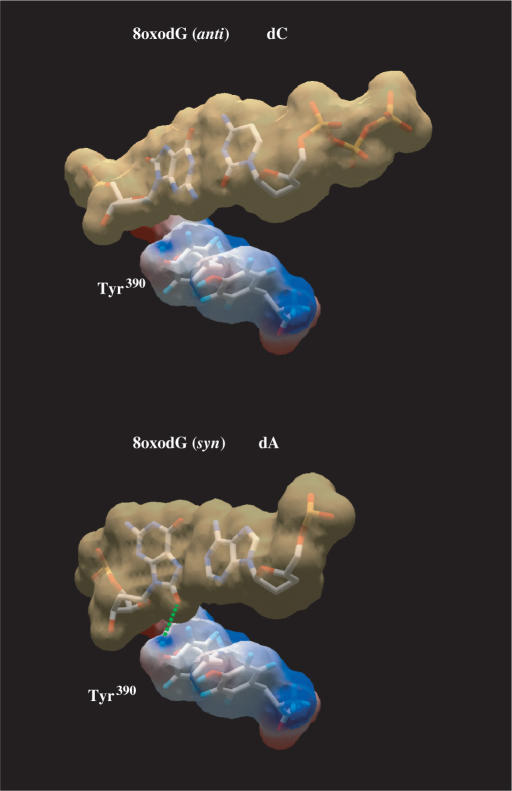
To analyse the ability of ϕ29 DNA polymerase mutants to carry out nucleotide insertion opposite 8oxodG, primer extension reactions were conducted in the presence of increasing concentrations of the corresponding dNTP (see Materials and Methods section). A 5′-labelled primer molecule (15-mer) was annealed to a template (34-mer) containing 8oxodG at the nascent position (see Materials and Methods section). As it can be observed in Figure 3 and Table 1, mutant PolwtExo− displayed a clear preference to incorporate dC with respect to dA, mainly due to the lower Km for dC, showing a discrimination factor (fins in Table 1) of about 5, a moderately high insertion fidelity in comparison with other polymerases belonging to families A, B, X, Y and reverse transcriptase (RT) (13,20,21,23,24,41–50), in which the dC:dA insertion ratio ranges from 0.075 for HIV-1 RT (21) to 91 for Dpo4 DNA polymerase (51) [compiled in (39)]. The incorporation of dA cannot be attributed to a decreased insertion fidelity of PolwtExo−, as it discriminates 3 × 105-fold against dA insertion opposite the undamaged dG (data not shown). Under those conditions, and consistent with their reduced efficiency in incorporating nucleotides opposite undamaged templates due to a reduced dNTP-binding capacity (52,53), PolY390FExo− and PolY390SExo− mutants were also less active than PolwtExo− in inserting both dA and dC at 8oxodG (Figure 3 and Table 1). Thus, correct dC insertion by PolY390FExo− and PolY390SExo− mutants is reduced to two and one orders of magnitude relative to the wild-type polymerase, respectively. These data are in good agreement with the comparative efficiencies displayed by these two mutants during incorporation of dNTPs on undamaged substrates (53), precluding any additional incorporation defect due to the presence of 8oxodG at the templating position. However, their discrimination factors clearly changed. Whereas PolY390FExo− mutant discriminates against dA insertion in a similar fashion as PolwtExo− (fins∼ 4), PolY390SExo− mutant displays nearly identical insertion efficiency for both nucleotides (fins∼ 1). Such behaviour is not a direct consequence of the diminished insertion fidelity showed by this mutant, as it has a discrimination factor against erroneous nucleotide insertion opposite a non-damaged nucleotide of 3 × 102 (52,53). This result clearly reflects the importance of the aromatic moiety of ϕ29 DNA polymerase residue Tyr390 in discriminating against the syn orientation (error-prone) of the purine base of the 8oxodG. Freisinger et al. modelled a nascent 8oxodG:dATP base pair at the catalytic active site of RB69 DNA polymerase (20). Such a model led them to propose that the O8 group of 8oxodG(syn) would clash with the molecular surface formed by residues Tyr567 and Gly568, allowing the canonical and non-mutagenic anti conformation to be preferentially placed (20). The DNA and incoming nucleotide from the structure of the RB69 ternary complex (8) can be homology modelled onto the structure of the apo ϕ29 DNA polymerase by aligning the palm subdomains of both ϕ29 and RB69 DNA polymerases, the position of the modelled DNA and incoming nucleotide being consistent with the placement of the ϕ29 DNA polymerase catalytic carboxylates, steric gate residues that distinguish ribo- from deoxyribonucleotides, and residues involved in binding DNA (35). This fact allowed us also to model a nascent 8oxodG:dATP in the corresponding active site of ϕ29 DNA polymerase (Figure 4) in which ϕ29 DNA polymerase residues Tyr390 and Gly391 are placed at the same location and orientation that the homologous RB69 DNA polymerase residues Tyr567 and Gly568, mentioned above. Based on that, the absence of discrimination showed by PolY390SExo− mutant could reflect that the lack of the aromatic group avoids the steric contacts with the O8 of the 8oxodG(syn) either by removing a direct interaction with Tyr390 or by tolerating a subtle shift of Gly391 backbone to accommodate the O8 atom, or both. Interestingly, recent studies have involved the homologous Tyr766 residue of E. coli DNA polymerase I in preventing incorrect nucleotide insertion across from the dG:N-2-aminofluorene adduct, as mutant Y766S was significantly less selective for correct nucleotide incorporation than the wild-type enzyme (54).
Figure 3.
Nucleotide selection during insertion opposite 8oxodG. Here, 12, 30 and 18 nM of PolwtExo−, PolY390FExo− and PolY390SExo− mutants, respectively, were incubated at 25°C for 15 s with 1.2 nM of the hybrid molecule depicted on top of the figure and in the presence of the indicated concentrations of either dATP or dCTP (see Materials and Methods section). Formation of the extended product was plotted against dNTP concentration. Inset, electrophoretic analysis of the primer extension experiments at the corresponding nucleotide concentration indicated below. Each data set was fit by least-squares nonlinear regression to a rectangular hyperbola.
Table 1.
Insertion of dCTP and dATP opposite 8oxodG
| Mutant polymerase | dA insertion | dC insertion | |||||
|---|---|---|---|---|---|---|---|
| Km (μM) | Kcat (min−1) | (Kcat/Km)(μM−1min−1) | Km (μM) | Kcat (min−1) | (Kcat/Km) (μM−1min−1) | fins | |
| PolwtExo− | 28.05 ± 6.2 | 0.124 | 4.42 × 10−3 | 5.8 ± 1.2 | 0.122 | 2.1 × 10−2 | 4.8 |
| PolY390FExo− | 457.6 ± 122.1 | 0.027 | 5.9 × 10−5 | 168.2 ± 33.3 | 0.036 | 2.14 × 10−4 | 3.6 |
| PolY390SExo− | 116.7 ± 16.4 | 0.098 | 8.4 × 10−4 | 104.1 ± 18 | 0.101 | 9.7 × 10−4 | 1.1 |
(Kcat/Km) stands for the catalytic efficiency displayed by mutant polymerases for the insertion of the indicated nucleotide. fins stands for insertion discrimination factor obtained by dividing catalytic efficiency for the insertion of dC by that for the insertion of dA.
Figure 4.
Modelling of nascent 8oxodG(anti):dC and 8oxodG(syn):dA base pairs in the polymerase active site. Modelling an 8oxodG(syn):dA base pair shows the steric hindrance between the O8 atom and the surface formed by ϕ29 DNA polymerase residues Tyr390 and Gly391, according to (20). van der Waals surfaces are shown for base pairs (light brown) and for protein side chains (coloured according to their electrostaticity). 8oxodG(anti):dC [coordinates obtained from PDB1 Q9Y (20)], and 8oxodG(syn):dA [PDB1 TK8 (19)] base pairs were first modelled into RB69 DNA polymerase active site (PDB1 IG9). Superposition of RB69 and ϕ29 (PDB1 XHX) DNA polymerase active sites allowed us to model both base pairs at the active site of the latter. Green dotted lines represent the potential hydrogen bonds formed between the nucleotides.
Tyrosine residue of motif B dictates the extension preferences of nucleotides base-paired to 8oxodG lesion
To study the next step after insertion opposite 8oxodG, 16/34-mer hybrid molecules, with the primer 3′-end containing either dC or dA paired with 8oxodG at the template (see Materials and Methods section), were used as substrate to analyse single nucleotide incorporation of dG, the next correct nucleotide following the 8oxodG:dC/8oxodG:dA base-pair formation. As seen in Table 2 (see also Supplementary Figure S1), PolwtExo− mutant displayed a 14-fold favoured extension of the 8oxodG:dA base pair (see in Table 2 discrimination factor, fext). Interestingly, PolY390FExo− mutant lost discrimination ability to elongate the base pairs under study, as it showed a 5-fold preference to extend the 8oxodG:dA pair. In contrast, PolY390SExo− mutant recovered a nearly wild-type phenotype, exhibiting a 12-fold preference in the extension of such a base pair (see Table 2). These results were pointing to the importance of the hydroxyl group of ϕ29 DNA polymerase residue Tyr390 in the establishment of contacts with the syn conformation of 8oxodG that allow the pair 8oxodG:dA to be preferentially extended.
Table 2.
Extension of DNA synthesis past an 8oxodG lesion
| Mutant polymerase | 8oxodG:dA | 8oxodG:dC | |||||
|---|---|---|---|---|---|---|---|
| Km (μM) | Kcat (min−1) | (Kcat/Km) (μM−1min−1) | Km (μM) | Kcat (min−1) | (Kcat/Km) (μM−1min−1) | fext | |
| PolwtExo− | 0.09 ± 0.01 | 0.15 | 1.70 | 1.08 ± 0.1 | 0.13 | 0.12 | 14.2 |
| PolY390FExo− | 2.86 ± 0.26 | 0.07 | 2.5 × 10−2 | 10.12 ± 1.73 | 0.10 | 5 × 10−3 | 5 |
| PolY390SExo− | 0.56 ± 0.05 | 0.13 | 0.23 | 3.85 ± 0.25 | 0.11 | 0.02 | 11.5 |
(Kcat/Km) stands for the catalytic efficiency displayed by mutant polymerases for extension of either 8oxodG:dA or 8oxodG:dA base pair. fext stands for extension discrimination factor obtained by dividing catalytic efficiency for the extension of 8oxodG:dA base pair by that for the extension of 8oxodG:dC base pair.
The structural basis for the poor extension efficiency of the 8oxodG:dC base pair have been highlighted by modelling this pair, poised for extension, at the polymerization site of T7 and RB69 DNA polymerases (19,20). Thus, the anti conformation of 8oxodG would promote local perturbations of the DNA backbone to alleviate steric clashes between the O8 group of 8oxodG and its 5′ phosphate group, leading to a subtle shift in the sugar moiety of 8oxodG which would decrease the catalytic efficiency of the elongation of this base pair (19). In addition, avoidance of those steric clashes would be accounted by a slight movement of the newly replicated duplex towards the DNA major groove with the direct consequence of a diminished binding stability at the polymerase active site (20). To elucidate differences in the proper stabilization at the polymerase active site of both DNA substrates, koff assays with ϕ29 PolwtExo− were carried out by measuring the DNA-binding stability of the enzyme to DNA hybrid molecules containing either a dA- or dC- 3′-end primer terminus paired to 8oxodG (see Materials and Methods section and Supplementary Figure S2). Interestingly, the 8oxodG:dA base pair (koff = 0.23 ± 0.01 min−1) was bound 3-fold more stably to the PolwtExo− polymerization site than the 8oxodG:dC pair (koff = 0.70 ± 0.02 min−1). Thus, our results support the original proposal of a diminished binding stability of the template 8oxodG(syn) when is forming a base pair poised for extension (20).
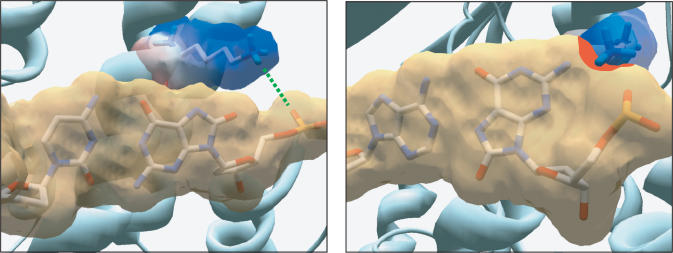
RB69 DNA polymerase Tyr567 forms part of the minor groove mismatch sensing mechanism (8) by means of making, through a water molecule, hydrogen bonds to the sugar moiety and the acceptors located at the N3 of a purine or O2 of a pyrimidine of the ultimate template base (8). Modelling of 8oxodG(syn):dA pair ready for extension predicted that the O8 group could substitute this water molecule and make a hydrogen bond with the hydroxyl group of Tyr567, in an analogous manner as with the N3 of a non-modified dG (20) (Figure 5), not detecting this mispair as being aberrant. In fact, the minor groove surface of this mispair is identical to the normal dT:dA base pair (19). On the contrary, the steric clashes between the O8 of 8oxodG(anti) and sugar moiety could difficult the proper formation of the water-mediated hydrogen bond between the Tyr residue and the N3 of 8oxodG in the 8oxodG:dC pair. This elegant model proposed by Freisinger et al. (20) to explain the structural determinants for insertion and further extension opposite 8oxodG is substantiated by the biochemical results presented here. Thus, the absence of the hydroxyl group in PolY390FExo− mutant would preclude stabilization of the 8oxodG(syn) through such hydrogen bond, diminishing the discrimination for extending both base pairs. In this sense, relative stabilization of the 8oxodG:dA base pair (koff = 0.32 ± 0.02 min−1) with respect to 8oxodG:dC (koff = 0.58 ± 0.03 min−1) at the polymerization site of PolY390FExo− mutant decreased in comparison with PolwtExo− (Supplementary Figure S2), while PolY390SExo− variant displayed a wild-type-like phenotype (data not shown). The smaller size of serine could allow one of its rotameric conformations to locate its hydroxyl group in a position suitable to contact the O8 group of 8oxodG(syn). Additionally, the absence of the bulky aromatic ring in mutant PolY390SExo− could favour a better accommodation of the nitrogen base of 8oxodG(syn) that would permit the establishment of the above-mentioned interaction. Thus, allowing this mutant to discriminate against 8oxodG:dC elongation at a wild-type level. In this extension step, the aromatic group of Tyr would be dispensable, their role being restricted to confer specificity during the insertion step.
Figure 5.
Modelling of 8oxodG(anti):dC and 8oxodG(syn):dA base pairs poised for extension in ϕ29 DNA polymerase active site. Modelling an 8oxodG(syn):dA base pair shows the potential contact through a H-bond (green dotted line) between the O8 atom of 8oxodG and the OH group of Tyr390, in accordance to (20). van der Waals surfaces are shown for base pairs (light brown) and for Tyr390 side chain (coloured according to its electrostaticity). 8oxodG(anti):dC [coordinates obtained from PDB1 Q9Y (20)], and 8oxodG(syn):dA [PDB1 TK8 (19)] base pairs were first modelled into RB69 DNA polymerase active site (PDB1 IG9) by a structural fitting of this pair with the dG:dC (template/primer) terminus from the RB69 DNA polymerase ternary complex. Superposition of RB69 and ϕ29 (PDB1 XHX) DNA polymerase active sites allowed us to model both base pairs at the active site of the latter.
Our biochemical results, together with the structural models, let to predict the dual role of the conserved Tyr residue of motif B of family B DNA polymerases during the bypass of the 8oxodG lesions of the DNA by means of its aromatic and hydroxyl groups. The former will dictate the preferential 8oxodG(anti) conformation during the nucleotide insertion step, and as a consequence will favour the non-mutagenic dC incorporation, while the latter will do it during further base-pair extension, stabilizing the 8oxodG(syn) form promoting extension of the mispair 8oxodG:dA. Although the role of this Tyr as a key residue for controlling nucleotide misincorporation on natural DNA has been extensively described in family B DNA polymerases (55–57), this is the first time that the specific contribution of its chemical substituents during the two steps of 8oxodG replication is biochemically demonstrated. On the other hand, contrarily to thermophilic A-family DNA polymerases, currently used for PCR, as Taq polymerase from Thermus aquaticus that incorporates indistinguishably dC and dA opposite 8oxodG (58), preferential incorporation of dC opposite 8oxodG makes ϕ29 DNA polymerase one of the most suitable enzymes to amplify faithfully DNA substrates that, as ancient DNAs, could be widely oxidized.
8oxodGTP incorporation and extension by ϕ29 DNA polymerase
In addition to promote lesions in the DNA, the presence inside cells of ROS also generates oxidatively altered purines and pyrimidines, the most abundant being 8oxodGTP (14). Although organisms synthesize 8oxodGTPases to sanitize the dNTP pool, the incorporation of such modified nucleotide could have adverse consequences as pairing opposite adenine of the template strand would result in A:T to C:G transversions (59).
Primer extension analyses similar to those described above were performed to analyse the templating nucleotide preference (dA versus dC) during 8oxodGMP insertion by ϕ29 DNA polymerase, using the 15/21-mer hybrid molecules sp1/sp1cC+5 and sp1/sp1cA+5 described in the Materials and Methods section (see also Supplementary Figure S3). As it can be seen in Table 3, ϕ29 DNA polymerase inserts 8oxodG preferentially opposite dC, although 2000-fold less efficiently than insertion of the unmodified dGMP. Additionally, qualitative analysis of the extension of both dA:8oxodG and dC:8oxodG base pairs obtained after insertion of 8oxodG in these molecules (see Materials and Methods section), show the preferential extension of the correct dC:8oxodG base pair (Figure 6), with an efficiency similar to that of the normal dC:dG base pair (the appearance of the 18- and 20-mer bands is due to the pairing of 8oxodG with the templating dA and dC at these positions, respectively). These are very unusual results as most of DNA polymerases assayed displayed a favoured 8oxodG insertion opposite dA, as T7 and γ DNA polymerases from family A (22,60), β and λ DNA polymerases from family X (39,45,47) and Dpo4 and Dbh DNA polymerases belonging to family Y (61). Solely HIV-RT, Pol Ik (family A), E. coli DNA pol II (family B) and DNA polymerase from African Swine Fever Virus (family X) showed an 8oxodG insertion pattern similar to that described for ϕ29 DNA polymerase (22,45). Conversely, PolIk and ϕ29 DNA polymerases are the only enzymes studied so far exhibiting a prone extension of the correct dC:8oxodG base pair (22) (this study).
Table 3.
Insertion of 8oxodG opposite dA and dC by PolwtExo− mutant
| Km (μM) | Kcat (s−1) | (Kcat/Km)(μM−1s−1) | |
|---|---|---|---|
| dG insertion opposite dC | 1.71 × 10−3 ± 0.16 × 10−3 | 3.68 | 2.15 × 103 |
| 8oxodG insertion opposite dC | 3.27 ± 0.58 | 3.48 | 1.07 |
| 8oxodG insertion opposite dA | 9.32 ± 1.25 | 3.3 | 0.35 |
(Kcat/Km) stands for the catalytic efficiency displayed by mutant polymerase for the formation of the indicated base pair.
Figure 6.
Extension of dC:8oxodG and dA:8oxodG base pairs. As substrate, 1.2 nM of the 5′-labelled hybrid molecule depicted at the top of the figure was incubated at 25°C, in the presence of either 100 nM dGTP or 64 µM 8oxodGTP to obtain the dC:dG, dC:8oxodG and dA:8oxodG base pairs poised for extension after addition of the indicated dATP concentrations. Samples were incubated at 25°C for 15 s and quenched by adding 3 µl of gel loading buffer. Reactions were analysed by electrophoresis in 8 M urea–20% polyacrylamide gels.
At present, DNA polymerase ternary complexes with 8oxodGTP as incoming nucleotide have not been solved. This fact precludes the analysis of the structural rationale for 8oxodG insertion preferences. In Figure 7, we have modelled dC:8oxodGTP(anti) and dA:8oxodGTP(syn) base pairs into the RB69 DNA polymerase active site. As it happens with a non-modified incoming nucleotide, Lys560 residue, placed at the fingers subdomain, will make a hydrogen bond to one of the two negatively charged equatorial oxygens of the α phosphate of the incoming 8oxodGTP(anti), enhancing catalysis by allowing the proper stabilization of the catalytic state (8). Nevertheless, protrusion into the major groove of N1, N2 and O6 atoms of the incoming 8oxodGTP(syn) could led to a sterical and/or electrostatic clash between the N2 atom and the edge of the side chain of Lys560 (red patch in Figure 7), affecting the proper interaction between this residue and the α phosphate of the 8oxodGTP(syn) and, as a consequence, its insertion. Structural alignments of both RB69 and ϕ29 DNA polymerases have allowed us to identify ϕ29 DNA polymerase Lys383 as the homologous lysine residue (62). By the contrary, we have not found a structural rationale for the preferential extension of the dC:8oxodG base pair, as once inserted, the 8oxodGMP(syn) should be translocated backwards to the primer terminus site. At this location, the only contacts between the protein and the 3′-end nucleotide are through the phosphodiester bond. The impaired elongation of the incorrect base pair could suggest that the syn conformation of the 3′ terminus either promotes a subtle distortion in the proper orientation of the attacking 3′-OH group to form the phosphodiester bond with the next incoming nucleotide, or a slightly variation in the angle of the glycosidic bond that could make difficult the establishment of the proper stacking interactions between the nitrogen bases of the incoming nucleotide and primer terminus. Difficulties in the translocation step due to the presence of 8oxodGMP(syn) at the 3′ primer terminus cannot be ruled out.
Figure 7.
Modelling of nascent dC:8oxodGTP(anti) and dA:8oxodGTP(syn) base pairs in the RB69 DNA polymerase active site. Modelling a dA:8oxodGTP(syn) base pair shows the potential steric hindrance between the N2 atom of 8oxodGTP and the edge of the side chain of Lys560 (red patch). van der Waals surfaces are shown for base pairs (light brown) and for Lys560 side chain (coloured according to its electrostaticity). Green dotted line represents the hydrogen bond between Lys560 and the α phosphate of the incoming 8oxodG(anti). Modelling was performed essentially as described above.
As mentioned above, several DNA polymerases from eukaryotes and prokaryotes can use 8oxodGTP as incoming nucleotide, leading to transversions. Surveillance mechanisms have evolved to maintain a low frequency of these types of mutations, owing to the action of enzymes able to degrade such mutagenic substrates (14). Thus, E. coli synthesizes MutT protein that hydrolyses 8oxodGTP to 8oxodGMP (59), this latter form being unable to be rephosphorylated, preventing insertion of 8oxodGTP during DNA synthesis. Analogously, mammalian cells code for MTH1, a protein showing a similar enzymatic activity (63–65). However, proteins as E. coli MutY, which excises a dA mispaired with 8-oxodG as part of the process to restore the original G:C base (66) could have a deleterious effect if 8oxodG had been the erroneously incorporated nucleotide. Bacillus subtilis, the host of bacteriophage ϕ29, encodes two potential 8oxodGTPases that could prevent the mutagenic effects of 8oxodGTP, as the lack of both proteins sensitized growing cells to oxidizing agents (67). Thus, if bacteriophage ϕ29 had to develop under conditions that could increase the pool of 8oxodGTP inside the cell, as severe oxidative stress or dysfunctions in the activity of any 8oxodGTPase, the discrimination against 8oxodGMP insertion as well as the great fidelity exhibited by ϕ29 DNA polymerase during the unlikely insertion and further extension of 8oxodGMP would guarantee the maintenance of the genetic information of the bacteriophage.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We are grateful to Dr Javier Saturno for the preparation of the ϕ29 DNA polymerase mutants PolY390FExo− and PolY390SExo−, to José M. Lázaro for the purification of the proteins and to Dr Luis Blanco for critical reading of the manuscript and for helpful discussions. Spanish Ministry of Education and Science (BFU 2005-00733 to M.S.); Institutional grant from Fundación Ramón Areces to the Centro de Biología Molecular “Severo Ochoa”. Funding to pay the Open Access publication charges for this article was provided by Research grant BFU 2005-00733 from the Spanish Ministry of Education and Science.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kunkel TA. DNA replication fidelity. J. Biol. Chem. 2004;279:16895–16898. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- 2.Kool ET. Replication of non-hydrogen bonded bases by DNA polymerases: a mechanism for steric matching. Biopolymers. 1998;48:3–17. doi: 10.1002/(SICI)1097-0282(1998)48:1<3::AID-BIP2>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 3.Kunkel TA, Bebenek K. DNA replication fidelity. Annu. Rev. Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 4.Zhang X, Lee I, Berdis AJ. The use of nonnatural nucleotides to probe the contributions of shape complementarity and pi-electron surface area during DNA polymerization. Biochemistry. 2005;44:13101–13110. doi: 10.1021/bi050585f. [DOI] [PubMed] [Google Scholar]
- 5.Doubliè S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 6.Li Y, Korolev S, Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steitz TA. DNA polymerases: structural diversity and common mechanisms. J. Biol. Chem. 1999;274:17395–17398. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 8.Franklin MC, Wang J, Steitz TA. Structure of the replicating complex of a pol alpha family DNA polymerase. Cell. 2001;105:657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 9.Johnson SJ, Beese LS. Structures of mismatch replication errors observed in a DNA polymerase. Cell. 2004;116:803–816. doi: 10.1016/s0092-8674(04)00252-1. [DOI] [PubMed] [Google Scholar]
- 10.Meyer AS, Blandino M, Spratt TE. Escherichia coli DNA polymerase I (Klenow fragment) uses a hydrogen-bonding fork from Arg668 to the primer terminus and incoming deoxynucleotide triphosphate to catalyze DNA replication. J. Biol. Chem. 2004;279:33043–33046. doi: 10.1074/jbc.C400232200. [DOI] [PubMed] [Google Scholar]
- 11.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 12.Marnett LJ. Oxyradicals and DNA damage. Carcinogenesis. 2000;21:361–370. doi: 10.1093/carcin/21.3.361. [DOI] [PubMed] [Google Scholar]
- 13.Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 14.Sekiguchi M, Tsuzuki T. Oxidative nucleotide damage: consequences and prevention. Oncogene. 2002;21:8895–8904. doi: 10.1038/sj.onc.1206023. [DOI] [PubMed] [Google Scholar]
- 15.Kouchakdjian M, Bodepudi V, Shibutani S, Eisenberg M, Johnson F, Grollman AP, Patel DJ. NMR structural studies of the ionizing radiation adduct 7-hydro-8-oxodeoxyguanosine (8-oxo-7H-dG) opposite deoxyadenosine in a DNA duplex. 8-Oxo-7H-dG(syn).dA(anti) alignment at lesion site. Biochemistry. 1991;30:1403–1412. doi: 10.1021/bi00219a034. [DOI] [PubMed] [Google Scholar]
- 16.Oda Y, Uesugi S, Ikehara M, Nishimura S, Kawase Y, Ishikawa H, Inoue H, Ohtsuka E. NMR studies of a DNA containing 8-hydroxydeoxyguanosine. Nucleic Acids Res. 1991;19:1407–1412. doi: 10.1093/nar/19.7.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McAuley-Hecht KE, Leonard GA, Gibson NJ, Thomson JB, Watson WP, Hunter WN, Brown T. Crystal structure of a DNA duplex containing 8-hydroxydeoxyguanine-adenine base pairs. Biochemistry. 1994;33:10266–10270. doi: 10.1021/bi00200a006. [DOI] [PubMed] [Google Scholar]
- 18.Lipscomb LA, Peek ME, Morningstar ML, Verghis SM, Miller EM, Rich A, Essigmann JM, Williams LD. X-ray structure of a DNA decamer containing 7,8-dihydro-8-oxoguanine. Proc. Natl. Acad. Sci USA. 1995;92:719–723. doi: 10.1073/pnas.92.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brieba LG, Eichman BF, Kokoska RJ, Doublie S, Kunkel TA, Ellenberger T. Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase. EMBO J. 2004;23:3452–3461. doi: 10.1038/sj.emboj.7600354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freisinger E, Grollman AP, Miller H, Kisker C. Lesion (in)tolerance reveals insights into DNA replication fidelity. EMBO J. 2004;23:1494–1505. doi: 10.1038/sj.emboj.7600158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furge LL, Guengerich FP. Analysis of nucleotide insertion and extension at 8-oxo-7,8-dihydroguanine by replicative T7 polymerase exo- and human immunodeficiency virus-1 reverse transcriptase using steady-state and pre-steady-state kinetics. Biochemistry. 1997;36:6475–6487. doi: 10.1021/bi9627267. [DOI] [PubMed] [Google Scholar]
- 22.Einolf HJ, Schnetz-Boutaud N, Guengerich FP. Steady-state and pre-steady-state kinetic analysis of 8-oxo-7,8-dihydroguanosine triphosphate incorporation and extension by replicative and repair DNA polymerases. Biochemistry. 1998;37:13300–13312. doi: 10.1021/bi981346d. [DOI] [PubMed] [Google Scholar]
- 23.Einolf HJ, Guengerich FP. Fidelity of nucleotide insertion at 8-oxo-7,8-dihydroguanine by mammalian DNA polymerase delta. Steady-state and pre-steady-state kinetic analysis. J. Biol. Chem. 2001;276:3764–3771. doi: 10.1074/jbc.M006696200. [DOI] [PubMed] [Google Scholar]
- 24.Brieba LG, Kokoska RJ, Bebenek K, Kunkel TA, Ellenberger T. A lysine residue in the fingers subdomain of T7 DNA polymerase modulates the miscoding potential of 8-oxo-7,8-dihydroguanosine. Structure (Camb.) 2005;13:1653–1659. doi: 10.1016/j.str.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 25.Esteban JA, Salas M, Blanco L. Fidelity of ϕ29 DNA polymerase. Comparison between protein-primed initiation and DNA polymerization. J. Biol. Chem. 1993;268:2719–2726. [PubMed] [Google Scholar]
- 26.Garmendia C, Bernad A, Esteban JA, Blanco L, Salas M. The bacteriophage ϕ29 DNA polymerase, a proofreading enzyme. J. Biol. Chem. 1992;267:2594–2599. [PubMed] [Google Scholar]
- 27.Blanco L, Bernad A, Lázaro JM, Martín G, Garmendia C, Salas M. Highly efficient DNA synthesis by the phage ϕ29 DNA polymerase. Symmetrical mode of DNA replication. J. Biol. Chem. 1989;264:8935–8940. [PubMed] [Google Scholar]
- 28.Huber HE, Tabor S, Richardson CC. Escherichia coli thioredoxin stabilizes complexes of bacteriophage T7 DNA polymerase and primed templates. J. Biol. Chem. 1987;262:16224–16232. [PubMed] [Google Scholar]
- 29.Jonsson ZO, Hubscher U. Proliferating cell nuclear antigen: more than a clamp for DNA polymerases. Bioessays. 1997;19:967–975. doi: 10.1002/bies.950191106. [DOI] [PubMed] [Google Scholar]
- 30.Kong XP, Onrust R, O'Donnell M, Kuriyan J. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992;69:425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- 31.Tabor S, Huber HE, Richardson CC. Escherichia coli thioredoxin confers processivity on the DNA polymerase activity of the gene 5 protein of bacteriophage T7. J. Biol. Chem. 1987;262:16212–16223. [PubMed] [Google Scholar]
- 32.Dean FB, Hosono S, Fang L, Wu X, Faruqi AF, Bray-Ward P, Sun Z, Zong Q, Du Y, et al. Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl Acad. Sci. USA. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dean FB, Nelson JR, Giesler TL, Lasken RS. Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 2001;11:1095–1099. doi: 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blanco L, Bernad A, Blasco MA, Salas M. A general structure for DNA-dependent DNA polymerases. Gene. 1991;100:27–38. doi: 10.1016/0378-1119(91)90346-d. [DOI] [PubMed] [Google Scholar]
- 35.Kamtekar S, Berman AJ, Wang J, Lázaro JM, de Vega M, Blanco L, Salas M, Steitz TA. Insights into strand displacement and processivity from the crystal structure of the protein-primed DNA polymerase of bacteriophage. Mol. Cell. 2004;16:609–618. doi: 10.1016/j.molcel.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 36.Lázaro JM, Blanco L, Salas M. Purification of bacteriophage ϕ29 DNA polymerase. Methods Enzymol. 1995;262:42–49. doi: 10.1016/0076-6879(95)62007-9. [DOI] [PubMed] [Google Scholar]
- 37.Bloom LB, Otto MR, Beechem JM, Goodman MF. Influence of 5′-nearest neighbors on the insertion kinetics of the fluorescent nucleotide analog 2-aminopurine by Klenow fragment. Biochemistry. 1993;32:11247–11258. doi: 10.1021/bi00092a039. [DOI] [PubMed] [Google Scholar]
- 38.Creighton S, Bloom LB, Goodman MF. Gel fidelity assay measuring nucleotide misinsertion, exonucleolytic proofreading, and lesion bypass efficiencies. Methods Enzymol. 1995;262:232–256. doi: 10.1016/0076-6879(95)62021-4. [DOI] [PubMed] [Google Scholar]
- 39.Brown JA, Duym WW, Fowler JD, Suo Z. Single-turnover kinetic analysis of the mutagenic potential of 8-oxo-7,8-dihydro-2′-deoxyguanosine during gap-filling synthesis catalyzed by human DNA polymerases lambda and beta. J. Mol. Biol. 2007;367:1258–1269. doi: 10.1016/j.jmb.2007.01.069. [DOI] [PubMed] [Google Scholar]
- 40.Saturno J, Lázaro JM, Esteban FJ, Blanco L, Salas M. ϕ29 DNA polymerase residue Lys383, invariant at motif B of DNA-dependent polymerases, is involved in dNTP binding. J. Mol. Biol. 1997;269:313–325. doi: 10.1006/jmbi.1997.1053. [DOI] [PubMed] [Google Scholar]
- 41.Carlson KD, Washington MT. Mechanism of efficient and accurate nucleotide incorporation opposite 7,8-dihydro-8-oxoguanine by Saccharomyces cerevisiae DNA polymerase eta. Mol. Cell. Biol. 2005;25:2169–2176. doi: 10.1128/MCB.25.6.2169-2176.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haracska L, Prakash S, Prakash L. Yeast DNA polymerase zeta is an efficient extender of primer ends opposite from 7,8-dihydro-8-oxoguanine and O6-methylguanine. Mol. Cell. Biol. 2003;23:1453–1459. doi: 10.1128/MCB.23.4.1453-1459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haracska L, Yu SL, Johnson RE, Prakash L, Prakash S. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase eta. Nat. Genet. 2000;25:458–461. doi: 10.1038/78169. [DOI] [PubMed] [Google Scholar]
- 44.Hsu GW, Ober M, Carell T, Beese LS. Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase. Nature. 2004;431:217–221. doi: 10.1038/nature02908. [DOI] [PubMed] [Google Scholar]
- 45.Kumar S, Lamarche BJ, Tsai MD. Use of damaged DNA and dNTP substrates by the error-prone DNA polymerase X from African Swine fever virus. Biochemistry. 2007;46:3814–3825. doi: 10.1021/bi061501l. [DOI] [PubMed] [Google Scholar]
- 46.Lowe LG, Guengerich FP. Steady-state and pre-steady-state kinetic analysis of dNTP insertion opposite 8-oxo-7,8-dihydroguanine by Escherichia coli polymerases I exo- and II exo. Biochemistry. 1996;35:9840–9849. doi: 10.1021/bi960485x. [DOI] [PubMed] [Google Scholar]
- 47.Miller H, Prasad R, Wilson SH, Johnson F, Grollman AP. 8-oxodGTP incorporation by DNA polymerase beta is modified by active-site residue Asn279. Biochemistry. 2000;39:1029–1033. doi: 10.1021/bi991789x. [DOI] [PubMed] [Google Scholar]
- 48.Neeley WL, Delaney S, Alekseyev YO, Jarosz DF, Delaney JC, Walker GC, Essigmann JM. DNA polymerase V allows bypass of toxic guanine oxidation products in vivo. J. Biol. Chem. 2007;282:12741–12748. doi: 10.1074/jbc.M700575200. [DOI] [PubMed] [Google Scholar]
- 49.Rechkoblit O, Malinina L, Cheng Y, Kuryavyi V, Broyde S, Geacintov NE, Patel DJ. Stepwise translocation of Dpo4 polymerase during error-free bypass of an oxoG lesion. PLoS Biol. 2006;4:e11. doi: 10.1371/journal.pbio.0040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vaisman A, Woodgate R. Unique misinsertion specificity of poliota may decrease the mutagenic potential of deaminated cytosines. EMBO J. 2001;20:6520–6529. doi: 10.1093/emboj/20.22.6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zang H, Irimia A, Choi JY, Angel KC, Loukachevitch LV, Egli M, Guengerich FP. Efficient and high fidelity incorporation of dCTP opposite 7,8-dihydro-8-oxodeoxyguanosine by Sulfolobus solfataricus DNA polymerase Dpo4. J. Biol. Chem. 2006;281:2358–2372. doi: 10.1074/jbc.M510889200. [DOI] [PubMed] [Google Scholar]
- 52.Saturno J, Blanco L, Salas M, Esteban JA. A novel kinetic analysis to calculate nucleotide affinity of proofreading DNA polymerases. Application to ϕ29 DNA polymerase fidelity mutants. J. Biol. Chem. 1995;270:31235–31243. doi: 10.1074/jbc.270.52.31235. [DOI] [PubMed] [Google Scholar]
- 53.Blasco MA, Lázaro JM, Bernad A, Blanco L, Salas M. ϕ29 DNA polymerase active site. Mutants in conserved residues Tyr254 and Tyr390 are affected in dNTP binding. J. Biol. Chem. 1992;267:19427–19434. [PubMed] [Google Scholar]
- 54.Lone S, Romano LJ. The role of specific amino acid residues in the active site of Escherichia coli DNA polymerase I on translesion DNA synthesis across from and past an N-2-aminofluorene adduct. Biochemistry. 2007;46:2599–2607. doi: 10.1021/bi061324o. [DOI] [PubMed] [Google Scholar]
- 55.Blasco MA, Lázaro JM, Bernad A, Blanco L, Salas M. ϕ29 DNA polymerase active site. Mutants in conserved residues Tyr254 and Tyr390 are affected in dNTP binding. J. Biol. Chem. 1992;267:19427–19434. [PubMed] [Google Scholar]
- 56.Saturno J, Blanco L, Salas M, Esteban JA. A novel kinetic analysis to calculate nucleotide affinity of proofreading DNA polymerases. Application to ϕ29 DNA polymerase fidelity mutants. J. Biol. Chem. 1995;270:31235–31243. doi: 10.1074/jbc.270.52.31235. [DOI] [PubMed] [Google Scholar]
- 57.Yang G, Wang J, Konigsberg W. Base selectivity is impaired by mutants that perturb hydrogen bonding networks in the RB69 DNA polymerase active site. Biochemistry. 2005;44:3338–3346. doi: 10.1021/bi047921x. [DOI] [PubMed] [Google Scholar]
- 58.Belousova EA, Rechkunova NI, Lavrik OI. Thermostable DNA polymerases can perform translesion synthesis using 8-oxoguanine and tetrahydrofuran-containing DNA templates. Biochim. Biophys. Acta. 2006;1764:97–104. doi: 10.1016/j.bbapap.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 59.Maki H, Sekiguchi M. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature. 1992;355:273–275. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- 60.Hanes JW, Thal DM, Johnson KA. Incorporation and replication of 8-oxo-deoxyguanosine by the human mitochondrial DNA polymerase. J. Biol. Chem. 2006;281:36241–36248. doi: 10.1074/jbc.M607965200. [DOI] [PubMed] [Google Scholar]
- 61.Shimizu M, Gruz P, Kamiya H, Kim SR, Pisani FM, Masutani C, Kanke Y, Harashima H, Hanaoka F, et al. Erroneous incorporation of oxidized DNA precursors by Y-family DNA polymerases. EMBO Rep. 2003;4:269–273. doi: 10.1038/sj.embor.embor765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Salas M, de Vega M. Bacteriophage protein-primed DNA replication. KL, Ithaca: Hefferon; 2006. [Google Scholar]
- 63.Mo JY, Maki H, Sekiguchi M. Hydrolytic elimination of a mutagenic nucleotide, 8-oxodGTP, by human 18-kilodalton protein: sanitization of nucleotide pool. Proc. Natl Acad. Sci. USA. 1992;89:11021–11025. doi: 10.1073/pnas.89.22.11021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sakumi K, Furuichi M, Tsuzuki T, Kakuma T, Kawabata S, Maki H, Sekiguchi M. Cloning and expression of cDNA for a human enzyme that hydrolyzes 8-oxo-dGTP, a mutagenic substrate for DNA synthesis. J. Biol. Chem. 1993;268:23524–23530. [PubMed] [Google Scholar]
- 65.Furuichi M, Yoshida MC, Oda H, Tajiri T, Nakabeppu Y, Tsuzuki T, Sekiguchi M. Genomic structure and chromosome location of the human mutT homologue gene MTH1 encoding 8-oxo-dGTPase for prevention of A:T to C:G transversion. Genomics. 1994;24:485–490. doi: 10.1006/geno.1994.1657. [DOI] [PubMed] [Google Scholar]
- 66.Delaney S, Neeley WL, Delaney JC, Essigmann JM. The substrate specificity of MutY for hyperoxidized guanine lesions in vivo. Biochemistry. 2007;46:1448–1455. doi: 10.1021/bi061174h. [DOI] [PubMed] [Google Scholar]
- 67.Castellanos-Juárez FX, Álvarez-Álvarez C, Yasbin RE, Setlow B, Setlow P, Pedraza-Reyes M. YtkD and MutT protect vegetative cells but not spores of Bacillus subtilis from oxidative stress. J. Bacteriol. 2006;188:2285–2289. doi: 10.1128/JB.188.6.2285-2289.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.