Abstract
The combinatorial control of gene expression by the association of members of different families of transcription factors is a common theme in eukaryotic transcriptional control. The MADS-box transcription factors SRF and Mcm1 represent paradigms for such regulation through their interaction with numerous partner proteins. For example, in Saccharomyces cerevisiae, Mcm1 interacts with the forkhead transcription factor Fkh2. Here, we identify a novel interaction between SRF and the Forkhead transcription factor FOXK1 in human cells. The importance of this interaction is shown for the regulation of the SRF target genes SM α-actin and PPGB. The binding of FOXK1 to the SM α-actin and PPGB promoters requires the presence of SRF on the promoter. FOXK1 acts as a transcriptional repressor and it represses SM α-actin and PPGB expression. Thus FOXK1 represents an additional member of the growing repertoire of transcription factors that can interact with SRF and modulate the transcriptional output from SRF-regulated promoters.
INTRODUCTION
The regulation of eukaryotic promoters is complex and involves the combinatorial action of transcription factors (1). These transcription factors often form distinct modules which coregulate the expression of groups of genes with related function. One example of such a module, is the complex formed by the association of the Forkhead transcription factor Fkh2p and the MADS-box transcription factor Mcm1p in Saccharomyces cerevisiae. This complex controls the cyclical activation of a cohort of genes expressed during the late G2 and M phases of the cell cycle (2–5). Mcm1p itself functions in a number of alternative complexes with other coregulators to target genes involved in different biological processes (6,7). Indeed, combinatorial interactions with other transcription factors are typical of members of the MADS-box transcription factor family, including the mammalian protein SRF. Moreover, some of these combinatorial interactions appear to be evolutionarily conserved as, for example, both SRF and Mcm1p show interactions with homeodomain transcription factors (8–11).
The yeast forkhead protein Fkh2p contains two defined domains, the Forkhead DNA-binding domain and the phospho-peptide binding FHA domain. In Fkh2p, the FHA domain acts as a transcriptional activation domain that functions through phosphorylation-dependent recruitment of the coactivator Ndd1p (12,13). There are over 40 human Forkhead transcription factors (14). However, only two of these are known to possess FHA domains in addition to their Forkhead DNA-binding domain, FOXK1 and FOXK2. The mouse version of FOXK1, Foxk1/MNF exists as two isoforms, MNFα and MNFβ which differ through alternative splicing leading to the production of the C-terminally truncated MNFβ isoform (15). Foxk1/MNF has been implicated in the correct functioning of myogenic stem cells (16), which is in part due to proliferative defects caused by Foxk1/MNF loss (17). Molecularly, MNFβ has been shown to act as a transcriptional repressor protein, but the role of MNFα is unclear (15). Other forkhead proteins such as FOXM1 have been linked to controlling cell cycle-dependent expression of genes expressed at the G2-M boundary during the cell cycle (18), suggesting that Forkhead proteins play a similar role in cell cycle control in yeast and mammalian systems.
Mcm1p and SRF are highly related proteins, especially within their DNA-binding domains, and this is reflected by their ability to bind similar DNA motifs known as CArG boxes (7). Moreover, many of the protein–protein interfaces are conserved, exemplified by the observation that the yeast Forkhead transcription factor Fkh2p can form ternary DNA bound complexes with both Mcm1p and SRF (19). Thus, we hypothesized that in an analogous manner to the yeast Mcm1p-Fkh2p complex, human SRF might also interact with a Forkhead transcription factor to control gene expression. Here we demonstrate that SRF interacts physically and functionally with FOXK1, demonstrating the evolutionary conservation of this transcription factor module.
MATERIALS AND METHODS
Plasmid constructs
The following plasmids were used in mammalian cell transfections. The L8G5E1a-Luc and LexA-VP16 constructs were provided by C. Lemercier (20). pCH110 (Pharmacia), pEF1/myc-His/LacZ (Invitrogen), p6xFOX-Luc (kindly provided by R. Costa; 21), p5xCArG-Luc (Stratagene), pGL3-CDC25C-luc (pAS2608) (kindly provided by K. Engeland), pGL3-SM α-actin-Luc/pAS2268 (containing the rat SM α-actin promoter sequences −713 to +51 bp; kindly provided by S. Pham; 22), pAS2511, pAS2512 and pAS2513 (containing the rat SM α-actin promoter sequences −713 to +51 bp and mutations in either CArG box A, CArG box B or both CArG boxes A and B, respectively), were made by Quikchange mutagenesis using the primer-template combinations ADS1617/ADS1618-pAS2268, ADS1619/ADS1620-pAS2268, ADS1619/ADS1620-pAS2511, respectively. pCGNSRF (encoding HA-tagged full-length SRF, kindly provided by R. Prywes) was described previously.
pAS2256 [encoding CMV-driven full-length His-Flag tagged FOXK1(1–733)] was constructed by a two-step procedure. First, the HindIII/XbaI fragment from pAS1169 was cloned into the same sites in pCMV5 (encoding amino acids 97–733) to create pAS1173. Next, a HindIII/AscI-cleaved PCR fragment (primer pair ADS1315/ADS1316 and Image clone 30345138/pAS1186 as a template) was ligated into the same sites in pAS1173 to create pAS2256. pAS2265 [encoding CMV-driven full-length His-Flag tagged FOXK1(1–733)(H355A)] was constructed by ligating a KpnI/XbaI fragment from pAS2259 into the same sites in pCMV5. pAS1175 [encoding CMV-driven full-length His-Flag tagged FOXM1b(1–763)] was constructed by ligating the HindIII/XbaI fragment from pAS1171 into the same sites in pCMV5. pAS2257 [encoding CMV-driven FOXK1(1–262) fused to the GAL4 DNA-binding domain] was constructed by ligating a BamHI/XbaI-cleaved PCR fragment (primer pair ADS1315/ADS1297 and pAS2256 as a template) was ligated into the same sites in pAS2063 (23).
For bacterial expression, pGEX-KG and pAS58 (encoding GST-coreSRF[amino acids 132–222]; 24) have been described previously.
For in vitro transcription/translation, pAS1242 [encoding full-length Fkh2p(1–862)] has been described previously (19). pAS2255 [encoding T3-driven full-length His-Flag tagged FOXK1(1–733)] was constructed by a two-step procedure. First, HindIII/XhoI-cleaved PCR fragment (encoding amino acids 97–733; primer pair ADS1168/ADS1169 and Image clone 5168241/pAS1184 as a template) was cloned into the same sites in pAS728 (25) to create pAS1169. Next, a HindIII/AscI-cleaved PCR fragment (primer pair ADS1315/ADS1316 and Image clone 30345138/pAS1186 as a template) was ligated into the same sites in pAS1169 to create pAS2255. pAS1168 [encoding T3-driven full-length His-Flag tagged FOXK1(216–418)] was constructed by ligating a NcoI/XhoI-cleaved PCR fragment (primer pair ADS1166/ADS1167 and Image clone 5168241/pAS1184 as a template) into the same sites in pAS728. pAS2259 and pAS2258 [encoding T3-driven full-length His-Flag tagged FOXK1(1–733)(H355A) and FOXK1(216–488)(H355A), respectively] were created by QuikChange mutagenesis using the primer pair ADS1342/ADS1343 on the templates pAS2255 and pAS1168, respectively. pAS1171 [encoding T3-driven full-length His-Flag-tagged FOXM1b(1–748)] was constructed by ligating a HindIII/XhoI-cleaved PCR fragment (primer pair ADS1177/ADS1178 and Image clone 3834244/pAS1181 as a template) into the same sites in pAS728.
Tissue culture, cell transfection, reporter gene assays, RT-PCR and RNA interference
A total of 293 cells and muscle-derived RD18 rhabdomyosarcoma cells were grown in DMEM supplemented with 10% foetal bovine serum. Transfections were performed with Polyfect (Qiagen) for 293 cells or Lipofectamine 2000 (Invitrogen) for RD18 cells according to the manufacturer's instructions.
For reporter gene assays, typically 0.25 μg of reporter plasmid and 50 ng of pEF1/myc-His/LacZ or pCH110 were co-transfected with 0.005–2 μg of expression plasmids. Cell extracts were prepared and equal amounts of protein were used in luciferase and β-galactosidase assays as described previously (26).
Real time RT-PCR was carried out as described previously (27). The following primer-pairs were used for RT-PCR experiments. FOXK1: ADS1372 (5′-CGAGTTCGAGTTCCTCATGC-3′) and ADS1373 (5′-GGGAGATCTGGGGGTACAGT-3′), SM α-actin: ADS1358 (5′-GCGTGGCTATTCCTTCGTTA-3′) and ADS1359 (5′-ATGAAGGATGGCTGGAACAG-3′), PPGB, ADS1348 (5′-AGCTGCTTCCACCTACCTCA-3′) and ADS1349 (5′-CTTCTGGTTGAGGGAATCCA-3′), SRF ADS1027 and ADS1028 (28), GAPDH, ADS4007 (5′-ACAGTCAGCCGCATCTTCTT-3′) and ADS4008 (5′-TTGATTTTGGAGGGATCTCG-3′) and 18S internal control, ADS4005 (5′-TCAAGAACGAAAGTCGGAGGTT-3′) and ADS4006, (5′-GGACATCTAAGGGCATCACAG-3′).
siRNA against FOXK1, and matched GAPDH control, were constructed by the Silencer™ siRNA construction kit (Ambion). Human FOXK1 target sequences were: FOXK1-1 5′-TTGTGATAGAGCGACGTGACCTGTCTC-3′ (ADS1403/4) and FOXK1-2 5′-TTCTGCACAAAGCGGTAACCCTGTCTC-3′ (ADS1405/6) (italicized residues represent sequences used in the in vitro transcription process in producing the constructs). The siRNAs against SRF and matched control siRNA (Santa Cruz) were made synthetically. To carry out RNA interference (RNAi), a two-step transfection protocol was carried out in 12-well plates as described previously (27).
Western blot, co-immunoprecipitation and GST pulldown analysis
Western blotting was carried out with the primary antibodies; Flag (Sigma), GAPDH (Abcam), Erk2 (sc-154; Santa Cruz) and SRF (Santa Cruz) essentially as described previously (27).
Coimmunoprecipitation analysis of overexpressed proteins was performed using protein A sepharose beads (Sigma) as described previously (29). GST pulldown analysis was performed essentially as described previously (24) with purified bacterially expressed recombinant GST-coreSRF and in vitro translated FOXK1 derivatives.
Gel retardation and in vitro biotinylated DNA-binding assays
Gel retardation assays were performed as described previously using a fragment from the SWI5 promoter (4), and coreSRF purified from bacteria (24) and in vitro translated FOXK1 and Fkh2p derivatives.
The in vitro biotinylated DNA-binding assays were performed essentially as described previously (30). Total 2 × 106 HEK293 cells were cotransfected with 8 μg Flag tagged FOXK1 plasmids and 200 pmol control siRNA or siSRF using lipofectamine 2000. Twenty-four hours after transfection, the whole-cell extracts were incubated with 1 μg biotinylated Rat SM α-actin promoter DNA or 500 pmol of CArG-A element, which were immobilized on streptavidin–agarose beads (Dynabead M-280 streptavidin), in binding buffer [50 mM HEPES·KOH, pH 7.9/150 mM NaCl/0.5% Triton X-100/2 mM EDTA/20 mM NaF/1 mM Na3VO4/20 μg/ml poly(dI-dC) and protease inhibitors] at 4°C for 60 min. The beads were washed four times with the binding buffer, and precipitated proteins were analysed by western blotting. The biotin-linked Rat α-actin promoter was generated by PCR using the template pAS2268 (for the wild-type promoter) or pAS2513 (for the mutant promoter with both CArG boxes disrupted and the primers ADS1692 (5′-AAGGGTCAGCGATAAACCAA-3′) and ADS1693 (Biotin-CTTACCCTGATGGCGACTG-3′). The biotin-linked site containing CArG element A was created by annealing the oligonucleotides ADS1694 (biotin-CCTGTCTTTGCTCCTTGTTTGGGAAGCGAGTGGG) and ADS1695 (CCCACTCGCTTCCCAAACAAGGAGCAAAGACAGG).
ChIP assays
Chromatin immunoprecipitation (ChIP) assays using control IgG (Upstate) or antisera specific to SRF (Santa Cruz), E2A (Santa Cruz: E2A, E12 (V-18) SC-349) and FOXK1 (Abcam) were performed as described previously (31) except that cross-linking was performed for 10 min. Bound promoters were detected by PCR using primers for the human SM α-actin promoter (ADS1650, 5′-CTCTGGGCATTTCTGCAGTT-3′ and ADS1651, 5′-TTCTGCTCTCCTCCCACTTG-3′), the human PPGB promoter (ADS1696, 5′-ACTTAGCCGTCCACAACAGG-3′ and ADS1697 5′-GGGGACTGGAAGTCATGTGT-3′) or for the SRF promoter or intron 3 as described previously (28).
RESULTS
FOXK1 interacts with SRF in vivo and in vitro
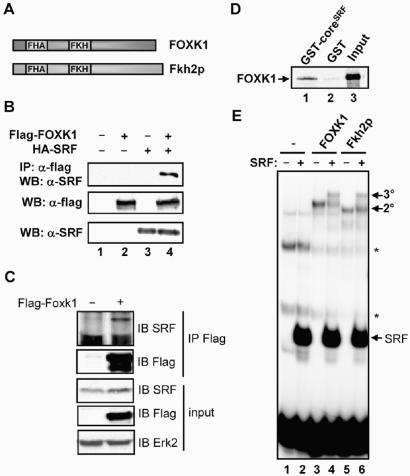
The Forkhead transcription factor Fkh2p interacts with the MADS-box transcription factor Mcm1p in S. cerevisiae. To establish whether a similar interaction could be detected between the closest human homologue of Mcm1p, SRF and a forkhead transcription factor, we first cloned the full-length human homologue of Foxk1/MNF, by combining the sequences from two IMAGE clones. The sequence we cloned was identical to that recently identified by in silico approaches (32). Human FOXK1 shares a conserved domain structure with mouse Foxk1/MNF and yeast Fkh2p, with an FHA domain preceding the Forkhead DNA-binding domain (Figure 1A). Overall, there is 90% sequence identity between human and mouse FOXK1/Foxk1. The sequence conservation between human FOXK1 and yeast Fkh2p is substantially lower (20% identity) and is concentrated mainly in the FHA and Forkhead domains.
Figure 1.
FOXK1 and SRF interact in vivo and in vitro. (A) Schematic of the domain structure of Foxk1 and Fkh2p. (B) Co-immunoprecipitation of FOXK1 with SRF from 293 cells transfected with constructs encoding Flag-FOXK1 and/or HA-SRF where indicated. FOXK1 proteins were immunoprecipitated by Flag antibody, and precipitated proteins detected by immunoblotting (IB) using SRF or Flag (for FOXK1) antibodies. (C) Co-immunoprecipitation of FOXK1 with endogenous SRF from 293 cells transfected with a construct encoding Flag-FOXK1. FOXK1 proteins were immunoprecipitated by Flag antibody, and precipitated proteins detected by immunoblotting (IB) using SRF or Flag (for FOXK1) antibodies. (D) GST pull-down analysis of FOXK1 interaction with SRF. GST-coreSRF or GST alone was used in a GST pull-down assay with full-length in vitro translated FOXK1. (E) Gel retardation analysis of the binding of coreSRF, FOXK1 and Fkh2p (where indicated) to a fragment of the SWI5 promoter. The locations of complexes formed with coreSRF alone and binary FOXK1-SWI5 (2°) or ternary FOXK1-coreSRF-SWI5 (3°) are indicated by arrows. Asterisks represents non-specific bands arising from the reticulocyte lysate.
Next, we co-expressed epitope-tagged versions of FOXK1 and SRF in human 293 cells and carried out co-immunoprecipitation analysis. Co-precipitation of SRF and FOXK1 was observed which was dependent on the expression of both transcription factors (Figure 1B). To establish whether endogenous SRF could interact with FOXK1, we expressed Flag epitope-tagged FOXK1 and attempted to co-precipitate SRF. Endogenous SRF was co-precipitated with FOXK1 (Figure 1C). We were however unable to detect interactions between SRF and FOXK1 expressed at endogenous levels, most likely due to the low levels of expression of these proteins coupled with the likely substoichiometric association of these transcription factors (data not shown). Interactions between yeast Fkh2p and Mcm1p occur with the DNA-binding domain of Mcm1p. We therefore tested whether we could detect analogous interactions between FOXK1 and SRF in vitro. Indeed, the DNA-binding domain of SRF (coreSRF) was sufficient for interaction with FOXK1 in a GST pulldown assay (Figure 1D). This interaction was specific as different forkhead proteins FOXM1b and FOXN3 were unable to form specific complexes with coreSRF (data not shown). Moreover, both FOXK1 and Fkh2p could form complexes with SRF on a fragment derived from the SWI5 promoter (Figure 1E), a known binding site for the Fkh2p-Mcm1p complex, which contains a composite binding motif for both SRF and the forkhead transcription factor (2–4). In contrast, FOXM1b was unable to form a similar complex (data not shown).
Collectively, these data demonstrate that in common with their yeast counterparts the human Forkhead FOXK1 and MADS-box SRF transcription factors can interact in vivo and in vitro.
FOXK1 is a repressor and inhibits SRF-dependent promoter activity
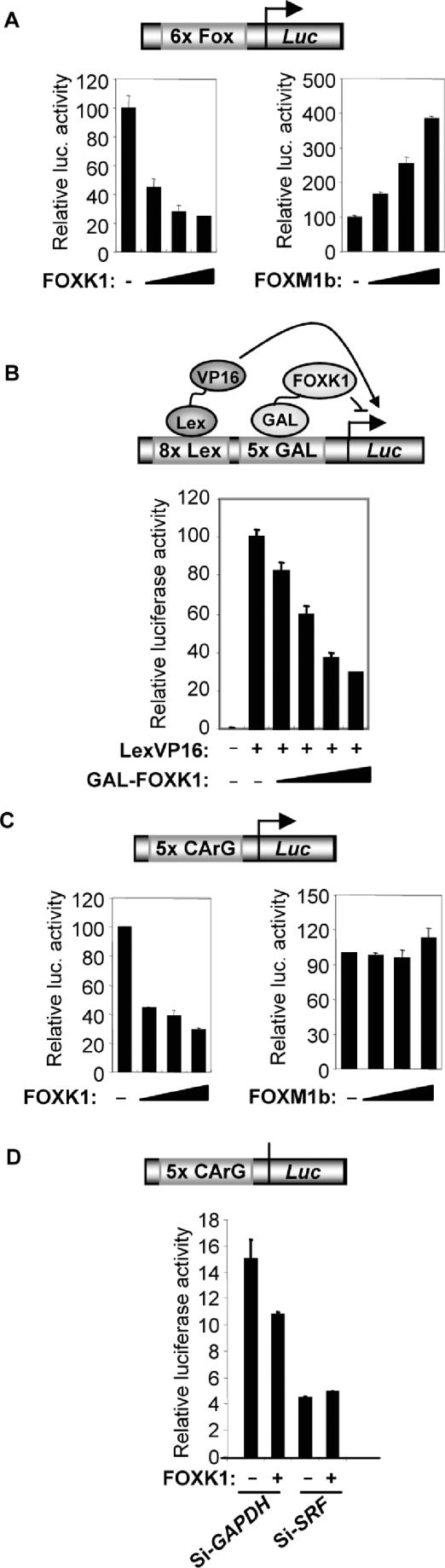
A short splice form of the mouse homologue of FOXK1, MNFβ has previously been shown to act as a transcriptional repressor (15). Full-length FOXK1 acts as a potent repressor of a reporter gene controlled by multimerized forkhead binding sites (Figure 2A). In contrast, FOXM1b acts as a transcriptional activator in this assay (Figure 2A). The N-terminal part of FOXK1 which includes the FHA domain is sufficient to repress transcription, as revealed by the ability of a GAL-FOXK1(1–262) fusion protein to repress transcription (Figure 2B). This effect is specific, as the GAL4 DNA-binding domain alone or other control GAL4 fusions do not cause such dose-dependent decreases in reporter activity (data not shown).
Figure 2.
FOXK1 represses SRF-dependent promoter–reporter activity. (A–D) Luciferase reporter assays with the indicated promoter–reporter plasmids (shown schematically at top of each graph). (A) A 6xFox-Luc reporter construct (250 ng) was transfected in the presence of increasing amounts of Flag-tagged FOXK1 (0, 100, 200 and 400 ng) or FOXM1b (0, 200, 400 and 800 ng) expression constructs in 293 cells. (B) A Lex-GAL-Luc reporter construct (250 ng) was transfected in the presence and absence of Lex-VP16 (100 ng) and increasing amounts of GAL-FOXK1(1–262) (0, 5, 10, 50 and 100 ng) constructs in 293 cells. (C) A 5xCArG-Luc reporter construct (250 ng) was transfected in the presence of increasing amounts of Flag-tagged FOXK1 (0, 100, 200 and 400 ng) or FOXM1b (0, 200, 400 and 800 ng) expression constructs in 293 cells. Data in A–C are presented relative to the activity of the reporter in the absence of FOXK1/FOXM1b (taken as 100%) and are the average of triplicate samples. (D) A 5xCArG-Luc reporter construct (250 ng) was transfected in the presence and absence of a Flag-tagged FOXK1 expression construct (100 ng) in 293 cells. siRNA duplexes against GAPDH or SRF were cotransfected where indicated. Data are the average of triplicate samples.
To establish potential functional interactions with SRF, we examined the activity of FOXK1 on a reporter driven by multimerized SRF binding sites (CArG-luc). FOXK1 functioned as repressor of this reporter gene while the alternative forkhead transcription factor FOXM1b had little effect on the activity of this reporter (Figure 2C). Importantly, upon depletion of SRF by siRNA, the repressive effect of FOXK1 on the CArG-luc reporter was lost (Figure 2D). Thus, FOXK1 can repress SRF-dependent promoter activation.
FOXK1 and SRF coregulate the expression of SM α-actin
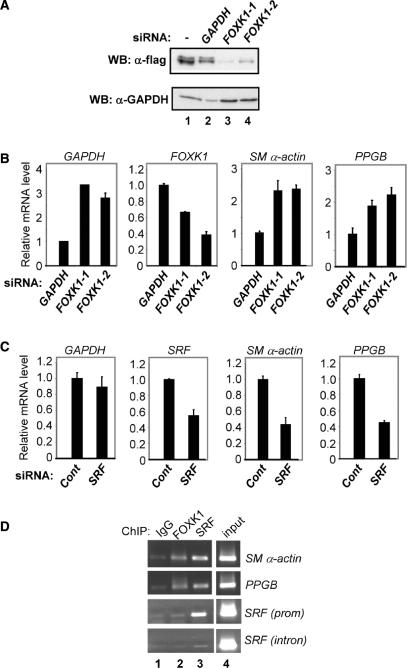
A previous microarray study has identified a number of potential FOXK1 target genes through the analysis of muscle side population cells derived from foxk1/mnf knockout mice (33). Similarly, a number of SRF target genes were identified through microarray analysis following the expression of SRF-VP16 fusion proteins in ES cells derived from srf knockout mice (34). Two of the deregulated genes in both cases were smooth muscle (SM) α-actin and PPGB (encoding a lysosomal protective protein) which were induced by either loss of Foxk1/MNF or expression of SRF-VP16, making this a likely candidate for detecting functional interactions between these two transcription factors.
First, we confirmed the involvement of human FOXK1 and SRF in controlling SM α-actin and PPGB expression. Two different siRNAs were derived which caused the depletion of FOXK1 expression at both the protein and RNA levels (Figure 3A and B). Upon depletion of FOXK1 in 293 cells, we observed a large increase in SM α-actin and PPGB levels with both siRNA constructs, consistent with a role for FOXK1 in repressing the activity of these genes (Figure 3B). Similar results were obtained in muscle-derived RD18 rhabdomyosarcoma cells (data not shown). Conversely, the depletion of SRF caused a substantial decrease in the expression of SM α-actin and PPGB expression (Figure 3C), confirming the ability of SRF to activate the expression of these genes. Indeed, other studies have previously also implicated SRF in controlling the activity of SM α-actin (35). Importantly, ChIP analysis demonstrated that both FOXK1 and SRF could be found associated with the endogenous SM α-actin and PPGB genes in 293 (Figure 3E) and RD18 cells (data not shown). In contrast, only SRF was found to associate with the srf promoter and neither protein associated with an intron found in the srf gene (Figure 3E, bottom panels). Thus, both FOXK1 and SRF can directly associate with the promoter regions and control the expression of SM α-actin and PPGB.
Figure 3.
FOXK1 and SRF control SM α-actin expression. (A) Western analysis of the expression of Flag-tagged FOXK1 expression in 293 cells cotransfected with siRNA duplexes against GAPDH or FOXK1 where indicated. (B–C) Real time RT-PCR analysis of the expression of the indicated genes in 293 cells transfected with, (B) RNAi duplexes against GAPDH or FOXK1, or (C) and RNAi duplex against SRF or a control duplex. Data are representative of 2–3 independent experiments and the average of triplicate samples. (D) ChIP analysis using the indicated antibodies in 293 cells. Coprecipitating SM α-actin promoter, PPGB promoter, SRF promoter or SRF intron 3 was detected by PCR.
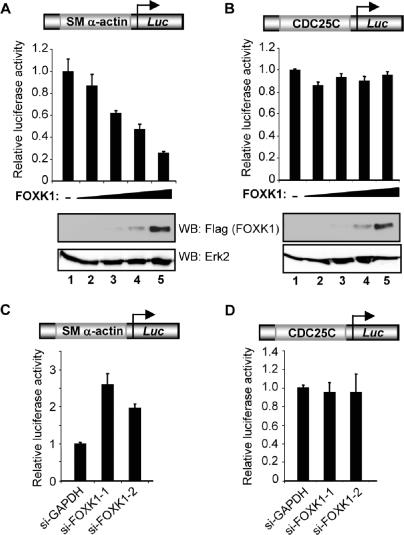
To establish whether FOXK1 could affect the activity of the SM α-actin promoter, we carried out reporter gene assays. Overexpression of FOXK1 caused the dose-dependent repression of a SM α-actin promoter–reporter construct (Figure 4A). In contrast, at the same levels of expression, little effect was observed on the activity of a CDC25C promoter–reporter construct (Figure 4B). Conversely, knockdown of FOXK1 expression caused an enhancement in the activity of the SM α-actin promoter (Figure 4C). This effect was specific, as no effect was seen on the CDC25C promoter (Figure 4D). Thus, FOXK1 acts to repress the activity of the SM α-actin promoter.
Figure 4.
FOXK1 represses SM α-actin promoter activity. (A and B) Luciferase reporter assays with the SM α-actin (A) or CDC25C (B) promoter–reporter plasmid (250 ng) (shown schematically at top of each graph). RD18 cells were cotransfected with increasing amounts of FOXK1 expression construct (0, 50, 100, 200 and 400 ng). Western blots of FOXK1 and Erk2 (control) expression are shown below each graph. (C and D) Luciferase reporter assays with the SM α-actin (C) or CDC25C (D) promoter–reporter plasmid (250 ng) (shown schematically at top of each graph). Total 293 cells were cotransfected with increasing amounts of FOXK1 expression construct (0, 50, 100, 200 and 400 ng). Data are presented relative to reporter alone (taken as 1) and are the average of triplicate samples.
FOXK1 regulates SM α-actin expression in a DNA-binding independent manner
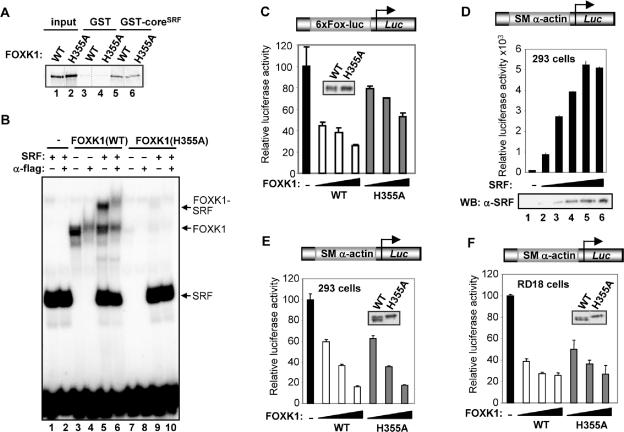
FOXK1 might regulate the SM α-actin promoter through either direct promoter binding or indirectly through association with SRF. We therefore created a mutant form of FOXK1 containing a point mutation in a region that is predicted to disrupt its DNA-binding surface [FOXK1(H355A)]. This point mutation did not affect interactions between FOXK1 and SRF (Figure 5A), but severely compromised the ability of FOXK1 to bind to DNA (Figure 5B, lanes 7–9). Indeed, FOXK1(H355A) exhibited much reduced repressive ability on a reporter driven by six forkhead binding sites (6xFox-luc; Figure 5C).
Figure 5.
FOXK1 represses SM α-actin expression in a DNA-binding independent manner. (A) GST pull-down analysis of GST-coreSRF or GST with full-length in vitro translated wild-type (WT) FOXK1 or FOXK1(H355A). (B) Gel retardation analysis of the binding of either coreSRF, or truncated versions (amino acids 216–488) of FOXK1(WT) and FOXK1(H355A) (where indicated) to a fragment of the SWI5 promoter. The locations of complexes formed with coreSRF alone and binary FOXK1-SWI5 (2°) or ternary FOXK1-coreSRF-SWI5 (3°) are indicated by arrows. Anti-Flag antibody is added where indicated to demonstrate the presence of FOXK1 in the complexes. (C–F) Luciferase reporter assays with the 6xFox site (C) or SM α-actin promoter–reporter plasmid (D–F) (250 ng) (shown schematically at top of each graph). (C) 293 cells were transfected increasing amounts (0, 100, 200 and 400 ng) of wild-type (WT) or H355A mutant FOXK1 expression constructs. (D) 293 cells were transfected with increasing amounts of SRF expression construct (0, 1, 5, 10, 20 and 50 ng). A western blot of SRF expression is shown below. Data are presented relative to reporter alone and are the average of triplicate samples. (E and F) 293 (E) or RD18 (F) cells were transfected increasing amounts (0, 100, 200 and 400 ng) of wild-type (WT) or H355A mutant FOXK1 expression constructs. Cells were also cotransfected with an SRF expression construct (20 ng) in (E). Western blots of FOXK1 expression in cells transfected with 200 ng expression construct are shown as insets. Data are presented relative to the activity of the reporter in the absence of FOXK1 (taken as 100%) and are the average of triplicate samples.
Next, we demonstrated that increasing amounts of SRF were able to activate the SM α-actin promoter in 293 cells (Figure 5D). Conversely, when we co-expressed increasing amounts of wild-type FOXK1 with SRF, we were able to demonstrate that FOXK1 was able to repress the activity of this SRF-activated reporter in this cell type (Figure 5E). Moreover, the DNA-binding defective FOXK1(H355A) also repressed the activity of the SM α-actin promoter to a similar extent as the wild-type protein (Figure 5E) suggesting that FOXK1 does not need to bind to DNA directly. Indeed, in agreement with this, we were unable to detect significant binding of wild-type FOXK1 to fragments of the SM α-actin promoter in vitro (data not shown). Similarly, both wild-type FOXK1 and FOXK1(H355A) were able to repress the SM α-actin promoter in the presence of endogenous levels of SRF in RD18 cells (Figure 5F).
Together these data demonstrate that FOXK1 can regulate the SM α-actin promoter in a DNA-binding independent manner.
FOXK1 regulates the expression of SM α-actin through SRF
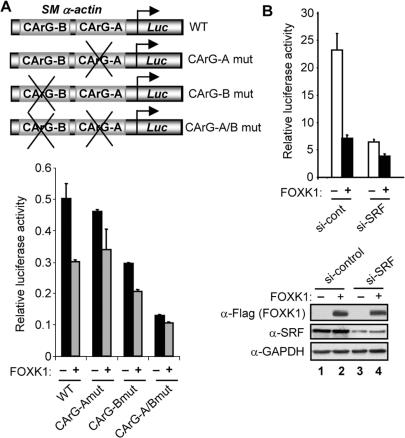
A likely mechanism through which FOXK1 regulates the SM α-actin promoter is via direct interactions with SRF. To establish whether this is the case, we first carried out reporter assays using a series of SM α-actin promoter–reporter constructs in which either one or both of the two SRF binding sites were mutated (Figure 6A). FOXK1 was able to repress the activity of the wild-type promoter, but this activity was reduced upon mutation of either of the two SRF binding sites. Moreover, the repressive activity of FOXK1 was virtually abolished upon mutation of both SRF binding sites (CArG-A/Bmut; Figure 6A). Next, we depleted endogenous SRF levels by siRNA treatment (Figure 6B). Upon depletion of SRF, the ability of FOXK1 to repress the SM α-actin promoter was blunted.
Figure 6.
FOXK1 represses SM α-actin expression in a SRF-dependent manner. (A) Luciferase reporter assay with the indicated wild-type (WT) and mutant SM α-actin promoter–reporter plasmids (250 ng) (shown schematically at top of figure). Total 293 cells were transfected with each reporter in the absence and presence of a FOXK1 expression construct (200 ng). (B) Luciferase reporter assay in 293 cells transfected with the wild-type (WT) SM α-actin promoter–reporter plasmid (250 ng) in the absence and presence of a FOXK1 expression construct (200 ng) and either control siRNA duplexes or siRNA duplexes against SRF. A western blot showing the expression of FOXK1, SRF and GAPDH expression are shown below the graph. Data in A and B are presented relative to the activity of cotransfected pCH110 and are the average of triplicate samples.
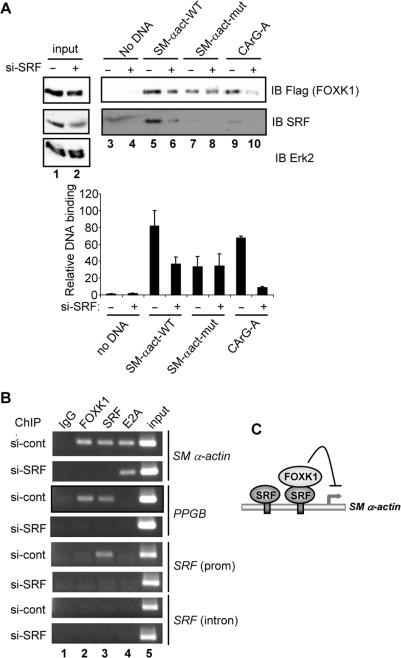
Finally, we asked whether SRF was required for FOXK1 recruitment to the SM α-actin promoter. We were unable to detect FOXK1 binding to the SM α-actin promoter by gel retardation analysis (data not shown), therefore, we used an immobilized biotinylated-DNA-binding assay to detect FOXK1-DNA interactions in vitro. Either the wild type or a mutant version (with both CArG boxes disrupted) of the SM α-actin promoter were coupled to streptavidin-linked beads. As a further control, we created duplexes which spanned just the proximal CArG (CArG-A) from this promoter. Extracts from 293 cells were used in the binding assays from cells that had been transfected with Flag-tagged FOXK1 in the presence of control siRNA or siRNA constructs against SRF. Binding of FOXK1 to the wild-type SM α-actin promoter could be detected and this binding was reduced upon depletion of SRF (Figure 7A, lanes 5 and 6). Similar effects were seen on the isolated CArG-A element (Figure 7A, lanes 9 and 10). These reductions in binding mirrored the reductions in SRF binding. In comparison to the wild-type promoter, the mutant SM α-actin promoter showed reduced levels of both SRF and FOXK1 binding (Figure 7A, lanes 7 and 8). Thus, SRF is required for efficient recruitment of FOXK1 to the SM α-actin promoter.
Figure 7.
FOXK1 binds to the SM α-actin expression in a SRF-dependent manner. (A) In vitro biotinylated DNA-binding assay. Binding to the indicated DNA fragments of proteins from extracts of 293 cells transfected with a FOXK1 expression vector and siRNAs against SRF where indicated. Input proteins and proteins remaining bound to the DNA after washing were detected by immunoblotting (IB) with the indicated antibodies. The SM-αact fragments contain 743 bp of the SM α-actin promoter surrounding either the intact WT or mutated (mut) CArG boxes. The CArG-A site contains only CArG box A. Quantification of FOXK1 binding in two independent experiments is shown graphically below the figure. (B) ChIP analysis using an IgG control or antibodies against FOXK1, SRF or E2A antibody in 293 cells. Cells were transfected with either a control siRNA duplex (cont) or a siRNA against SRF. Precipitated SM α-actin promoter was detected by PCR. (C) Model for how FOXK1 controls the activity of SRF target genes through SRF-dependent recruitment, and subsequent transcriptional repression.
To establish whether the same mechanism is operative in vivo, ChIP analysis was used to monitor the occupancy of the SM α-actin promoter upon SRF depletion. The reduction of SRF levels caused the expected reduction of SRF binding to the SM α-actin promoter. However, the binding of FOXK1 was also compromised (Figure 7B, top two panels). This reduction was not due to a general effect on SM α-actin promoter occupancy as the binding of an alternative transcription factor E2A (36) was maintained upon SRF depletion. Similar results were obtained on the PPGB promoter, where SRF depletion led to reductions in both SRF and FOXK1 binding (Figure 7B, panels 3 and 4).
Together these results therefore demonstrate that SRF is required for the FOXK1 to regulate and be recruited to the SM α-actin promoter. Thus, FOXK1 and SRF interact both physically and functionally to control the activity of target genes such as SM α-actin and PPGB.
DISCUSSION
Complexes between Forkhead and MADS-box transcription factors have previously been shown to be an important common combination involved in controlling the cyclical expression of cell cycle genes in S. cerevisiae. Here, we demonstrate that this combination of transcription factors is also functionally important in human cells, adding to the repertoire of transcription factor modules that function in metazoan systems. Specifically, we demonstrate that the human Forkhead transcription factor FOXK1 functionally interacts with the MADS-box protein SRF.
FOXK1 can form complexes with SRF in the absence of DNA through binding to the minimal core DNA-binding domain of SRF which includes the MADS-box (Figure 1). Indeed, a mutant form of FOXK1 that cannot bind to DNA efficiently can still control SRF-dependent promoter activity in vivo (Figure 5). However, SRF is required for the efficient recruitment of FOXK1 to target promoters in vitro and in vivo (Figures 2 and 7). This suggests a model whereby SRF acts as a platform to recruit FOXK1 (Figure 7C). FOXK1 can then repress promoter activity. These observations are fully consistent with the known roles of SRF and other MADS-box proteins in acting as a platform for the assembly of many different types of transcriptional regulatory complexes, some of which like MRTFs make minimal DNA interactions (7,37).
The paradigm for interactions between Forkhead and MADS-box transcription factors is the yeast Fkh2p-Mcm1p complex (2–5). However, while there are important overall similarities between the Fkh2p-Mcm1p and human FOXK1-SRF complexes, their modes of interaction and regulation are not identical. Both Fkh2p and FOXK1 share a similar domain structure, with both possessing an N-terminally located FHA domain in addition to the Forkhead DNA-binding domain. FOXK1 is a transcriptional repressor protein, and Fkh2p can also repress transcription of its target genes during the early part of the cell cycle (3–5). The mouse homologue of FOXK1, Foxk1/MNFβ represses transcription through the recruitment of the Sin3 corepressor (38) and Fkh2p can also bind to Sin3 in vitro (our unpublished data). In contrast, to date, no transcriptional activation capacity has been identified for mammalian FOXK1 proteins, and the region encompassing the FHA domain has repressive activity rather than the transactivation ability exhibited by the same region in Fkh2p (Figure 2; 12,13,38). Moreover, to date, we have been unable to establish a role for the FOXK1-SRF complex in cell cycle control (our unpublished data), and instead, an alternative forkhead protein FOXM1 appears to perform the major role in controlling G2-M phase transcription in mammalian cells (18). Our data indicate that FOXM1 does not function through binding and changing the activity of SRF (Figure 2, data not shown).
There also seem to be important differences between human and mouse FOXK1 proteins. In mice, there are two isoforms of Foxk1/MNF, MNFα and the shorter splice form MNFβ. However, only the latter apparently shows DNA-binding and transcriptional regulatory capacity (15). In contrast, full-length human FOXK1 (equivalent to MNFα) can bind to DNA and regulate transcription (Figures 1 and 2). Secondly, mutations designed to disrupt the phosphopeptide binding activity of the FHA domain of MNFβ partially diminished the repressive activity of MNFβ (15) but were without effect in FOXK1 (data not shown). It is currently unclear why these proteins apparently function differently but these observations might reflect important evolutionary differences.
To establish the importance of the FOXK1-SRF interaction, we demonstrated that this complex functions on the SM α-actin and PPGB genes, and that FOXK1 has a repressive role in the complex. However, FOXK1 does not seem to be an obligate partner for SRF as we could not detect FOXK1 binding to the different target gene, SRF (Figure 3). Thus, FOXK1 is likely to be a substoichimetric partner for SRF, as suggested by our inability to co-immunoprecipitate endogenous FOXK1 and SRF. Biologically, FOXK1 is likely to restrict the expression of SM α-actin expression in non-smooth muscle cell types such as the stem cell-like myogenic side population cells (33). Recently, another forkhead transcription factor Foxo4 was shown to repress SM α-actin expression in proliferating smooth muscle cells (39). In common with FOXK1, Foxo4 repressed transcription in a DNA-binding independent manner and achieved this through interacting with and inhibiting the SRF-myocardin activator complex. Direct interactions between Foxo4 with SRF were not however shown. A different SRF partner protein Elk-1 was also shown to inhibit the expression of a number of smooth muscle-specific genes (40). However, Elk-1 is ineffective against SM α-actin. Thus, several different ways might have been devised to reduce the expression of different smooth muscle genes in different cell types through impacting on the activity of SRF.
In summary, we have identified a novel combination of functionally interacting human transcription factors, the Forkhead protein FOXK1 and MADS-box protein, SRF. Future studies will focus on how common this mode of SRF target gene regulation is in human cells.
ACKNOWLEDGEMENTS
We thank Anne Clancy for excellent technical assistance; Paul Shore and Stefan Roberts and members of our laboratory for comments on the manuscript and stimulating discussions; Robert Costa, Ron Prywes, Kurt Engeland, Saadi Kochbin, Pier-Luigi Lollini, Gary Owens and Sem Phan for reagents. This work was supported by grants from the Wellcome Trust and by the IZKF at the University of Leipzig (D02). Funding to pay this Open Access publication charges for this article was provided by the Wellcome Trust.
Conflict of interest statement. None declared.
REFERENCES
- 1.Remenyi A, Scholer HR, Wilmanns M. Combinatorial control of gene expression. Nat. Struct. Mol. Biol. 2004;11:812–815. doi: 10.1038/nsmb820. [DOI] [PubMed] [Google Scholar]
- 2.Koranda M, Schleiffer A, Endler L, Ammerer G. Forkhead-like transcription factors recruit Ndd1 to the chromatin of G2/M-specific promoters. Nature. 2000;406:94–98. doi: 10.1038/35017589. [DOI] [PubMed] [Google Scholar]
- 3.Kumar R, Reynolds DM, Shevchenko A, Shevchenko A, Goldstone SD, Dalton S. Forkhead transcription factors, Fkh1p and Fkh2p, collaborate with Mcm1p to control transcription required for M-phase. Curr. Biol. 2000;10:896–906. doi: 10.1016/s0960-9822(00)00618-7. [DOI] [PubMed] [Google Scholar]
- 4.Pic A, Lim FL, Ross SJ, Veal EA, Johnson AL, Sultan MR, West AG, Johnston LH, Sharrocks AD, et al. The forkhead protein Fkh2 is a component of the yeast cell cycle transcription factor SFF. EMBO J. 2000;19:3750–3761. doi: 10.1093/emboj/19.14.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu G, Spellman PT, Volpe T, Brown PO, Botstein D, Davis TN, Futcher B. Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature. 2000;406:90–94. doi: 10.1038/35017581. [DOI] [PubMed] [Google Scholar]
- 6.Shore P, Sharrocks AD. The MADS-box family of transcription factors. Eur. J. Biochem. 1995;229:1–13. doi: 10.1111/j.1432-1033.1995.tb20430.x. [DOI] [PubMed] [Google Scholar]
- 7.Messenguy F, Dubois E. Role of MADS box proteins and their cofactors in combinatorial control of gene expression and cell development. Gene. 2003;316:1–21. doi: 10.1016/s0378-1119(03)00747-9. [DOI] [PubMed] [Google Scholar]
- 8.Chen CY, Schwartz RJ. Recruitment of the tinman homolog Nkx-2.5 by serum response factor activates cardiac alpha-actin gene transcription. Mol. Cell. Biol. 1996;16:6372–6384. doi: 10.1128/mcb.16.11.6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grueneberg DA, Simon KJ, Brennan K, Gilman M. Sequence-specific targeting of nuclear signal transduction pathways by homeodomain proteins. Mol. Cell. Biol. 1995;15:3318–3326. doi: 10.1128/mcb.15.6.3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pramila T, Miles S, GuhaThakurta D, Jemiolo D, Breeden LL. Conserved homeodomain proteins interact with MADS box protein Mcm1 to restrict ECB-dependent transcription to the M/G1 phase of the cell cycle. Genes Dev. 2002;16:3034–3045. doi: 10.1101/gad.1034302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayashi K, Nakamura S, Nishida W, Sobue K. Bone morphogenetic protein-induced MSX1 and MSX2 inhibit myocardin-dependent smooth muscle gene transcription. Mol. Cell. Biol. 2006;26:9456–9470. doi: 10.1128/MCB.00759-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darieva Z, Pic-Taylor A, Boros J, Spanos A, Geymonat M, Reece RJ, Sedgwick SG, Sharrocks AD, Morgan BA. Cell cycle-regulated transcription through the FHA domain of Fkh2p and the coactivator Ndd1p. Curr. Biol. 2003;13:1740–1745. doi: 10.1016/j.cub.2003.08.053. [DOI] [PubMed] [Google Scholar]
- 13.Reynolds D, Shi BJ, McLean C, Katsis F, Kemp B, Dalton S. Recruitment of Thr 319-phosphorylated Ndd1p to the FHA domain of Fkh2p requires Clb kinase activity: a mechanism for CLB cluster gene activation. Genes Dev. 2003;17:1789–1802. doi: 10.1101/gad.1074103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaestner KH, Knochel W, Martinez DE. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 2000;14:142–146. [PubMed] [Google Scholar]
- 15.Yang Q, Bassel-Duby R, Williams RS. Transient expression of a winged-helix protein, MNF-beta, during myogenesis. Mol. Cell. Biol. 1997;17:5236–5243. doi: 10.1128/mcb.17.9.5236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garry DJ, Meeson A, Elterman J, Zhao Y, Yang P, Bassel-Duby R, Williams RS. Myogenic stem cell function is impaired in mice lacking the forkhead/winged helix protein MNF. Proc. Natl Acad. Sci. USA. 2000;97:5416–5421. doi: 10.1073/pnas.100501197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hawke TJ, Jiang N, Garry DJ. Absence of p21CIP rescues myogenic progenitor cell proliferative and regenerative capacity in Foxk1 null mice. J. Biol. Chem. 2003;278:4015–4020. doi: 10.1074/jbc.M209200200. [DOI] [PubMed] [Google Scholar]
- 18.Laoukili J, Stahl M, Medema RH. FoxM1: at the crossroads of ageing and cancer. Biochim. Biophys. Acta. 2007;1775:92–102. doi: 10.1016/j.bbcan.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 19.Boros J, Lim FL, Darieva Z, Pic-Taylor A, Harman R, Morgan BA, Sharrocks AD. Molecular determinants of the cell-cycle regulated Mcm1p-Fkh2p transcription factor complex. Nucleic Acids Res. 2003;31:2279–2288. doi: 10.1093/nar/gkg347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemercier C, Verdel A, Galloo B, Curtet S, Brocard MP, Khochbin S. mHDA1/HDAC5 histone deacetylase interacts with and represses MEF2A transcriptional activity. J. Biol. Chem. 2000;275:15594–15599. doi: 10.1074/jbc.M908437199. [DOI] [PubMed] [Google Scholar]
- 21.Major ML, Lepe R, Costa RH. Forkhead box M1B transcriptional activity requires binding of Cdk-cyclin complexes for phosphorylation-dependent recruitment of p300/CBP coactivators. Mol. Cell. Biol. 2004;24:2649–2661. doi: 10.1128/MCB.24.7.2649-2661.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu B, Wu Z, Phan SH. Smad3 mediates transforming growth factor-beta-induced alpha-smooth muscle actin expression. Am. J. Respir. Cell Mol. Biol. 2003;29:397–404. doi: 10.1165/rcmb.2003-0063OC. [DOI] [PubMed] [Google Scholar]
- 23.Yang SH, Jaffray E, Hay RT, Sharrocks AD. Dynamic interplay of the SUMO and ERK pathways in regulating Elk-1 transcriptional activity. Mol. Cell. 2003;12:63–74. doi: 10.1016/s1097-2765(03)00265-x. [DOI] [PubMed] [Google Scholar]
- 24.Shore P, Sharrocks AD. The transcription factors Elk-1 and serum response factor interact by direct protein-protein contacts mediated by a short region of Elk-1. Mol. Cell. Biol. 1994;14:3283–3291. doi: 10.1128/mcb.14.5.3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ling Y, Lakey J, Roberts EC, Sharrocks AD. Molecular characterisation of the B-box protein-protein interaction motif of the ETS-domain transcription factors Elk-1. EMBO J. 1997;16:2431–2440. doi: 10.1093/emboj/16.9.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang SH, Yates PR, Whitmarsh AJ, Davis RJ, Sharrocks AD. The Elk-1 ETS-domain transcription factor contains a MAP kinase targeting motif. Mol. Cell. Biol. 1998;18:710–720. doi: 10.1128/mcb.18.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang SH, Sharrocks AD. PIASx acts as an Elk-1 coactivator by facilitating derepression. EMBO J. 2005;24:2161–2171. doi: 10.1038/sj.emboj.7600690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kasza A, O’donnell A, Gascoigne K, Zeef LA, Hayes A, Sharrocks AD. The ETS domain transcription factor Elk-1 regulates the expression of its partner protein, SRF. J. Biol. Chem. 2005;280:1149–1155. doi: 10.1074/jbc.M411161200. [DOI] [PubMed] [Google Scholar]
- 29.Yang SH, Vickers E, Brehm A, Kouzarides T, Sharrocks AD. Temporal recruitment of the mSin3A-histone deacetylase corepressor complex to the ETS domain transcription factor Elk-1. Mol. Cell. Biol. 2001;21:2802–2814. doi: 10.1128/MCB.21.8.2802-2814.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuzaki H, Daitoku H, Hatta M, Aoyama H, Yoshimochi K, Fukamizu A. Acetylation of Foxo1 alters its DNA-binding ability and sensitivity to phosphorylation. Proc. Natl Acad. Sci. USA. 2005;102:11278–11283. doi: 10.1073/pnas.0502738102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang SH, Sharrocks AD. PIASxalpha differentially regulates the amplitudes of transcriptional responses following activation of the ERK and p38 MAPK pathways. Mol. Cell. 2006;22:477–487. doi: 10.1016/j.molcel.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 32.Katoh M, Katoh M. Identification and characterization of human FOXK1 gene in silico. Int. J. Mol. Med. 2004;14:127–132. [PubMed] [Google Scholar]
- 33.Meeson AP, Hawke TJ, Graham S, Jiang N, Elterman J, Hutcheson K, Dimaio JM, Gallardo TD, Garry DJ. Cellular and molecular regulation of skeletal muscle side population cells. Stem Cells. 2004;22:1305–1320. doi: 10.1634/stemcells.2004-0077. [DOI] [PubMed] [Google Scholar]
- 34.Philippar U, Schratt G, Dieterich C, Muller JM, Galgoczy P, Engel FB, Keating MT, Gertler F, Schule R, et al. The SRF target gene Fhl2 antagonizes RhoA/MAL-dependent activation of SRF. Mol. Cell. 2004;16:867–880. doi: 10.1016/j.molcel.2004.11.039. [DOI] [PubMed] [Google Scholar]
- 35.Shimizu RT, Blank RS, Jervis R, Lawrenz-Smith SC, Owens GK. The smooth muscle alpha-actin gene promoter is differentially regulated in smooth muscle versus non-smooth muscle cells. J. Biol. Chem. 1995;270:7631–7643. doi: 10.1074/jbc.270.13.7631. [DOI] [PubMed] [Google Scholar]
- 36.Kumar MS, Hendrix JA, Johnson AD, Owens GK. Smooth muscle alpha-actin gene requires two E-boxes for proper expression in vivo and is a target of class I basic helix-loop-helix proteins. Circ. Res. 2003;92:840–847. doi: 10.1161/01.RES.0000069031.55281.7C. [DOI] [PubMed] [Google Scholar]
- 37.Posern G, Treisman R. Actin’ together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 2006;16:588–596. doi: 10.1016/j.tcb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 38.Yang Q, Kong Y, Rothermel B, Garry DJ, Bassel-Duby R, Williams RS. The winged-helix/forkhead protein myocyte nuclear factor beta (MNF-beta) forms a co-repressor complex with mammalian sin3B. Biochem. J. 2000;345:335–343. [PMC free article] [PubMed] [Google Scholar]
- 39.Liu ZP, Wang Z, Yanagisawa H, Olson EN. Phenotypic modulation of smooth muscle cells through interaction of Foxo4 and myocardin. Dev. Cell. 2005;9:261–270. doi: 10.1016/j.devcel.2005.05.017. [DOI] [PubMed] [Google Scholar]
- 40.Zhou J, Hu G, Herring BP. Smooth muscle-specific genes are differentially sensitive to inhibition by Elk-1. Mol. Cell. Biol. 2005;25:9874–9885. doi: 10.1128/MCB.25.22.9874-9885.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]