Abstract
5S rRNA is an integral component of the large ribosomal subunit in virtually all living organisms. Polyamine binding to 5S rRNA was investigated by cross-linking of N1-azidobenzamidino (ABA)-spermine to naked 5S rRNA or 50S ribosomal subunits and whole ribosomes from Escherichia coli cells. ABA-spermine cross-linking sites were kinetically measured and their positions in 5S rRNA were localized by primer extension analysis. Helices III and V, and loops A, C, D and E in naked 5S rRNA were found to be preferred polyamine binding sites. When 50S ribosomal subunits or poly(U)-programmed 70S ribosomes bearing tRNAPhe at the E-site and AcPhe-tRNA at the P-site were targeted, the susceptibility of 5S rRNA to ABA-spermine was greatly reduced. Regardless of 5S rRNA assembly status, binding of spermine induced significant changes in the 5S rRNA conformation; loop A adopted an apparent ‘loosening’ of its structure, while loops C, D, E and helices III and V achieved a more compact folding. Poly(U)-programmed 70S ribosomes possessing 5S rRNA cross-linked with spermine were more efficient than control ribosomes in tRNA binding, peptidyl transferase activity and translocation. Our results support the notion that 5S rRNA serves as a signal transducer between regions of 23S rRNA responsible for principal ribosomal functions.
INTRODUCTION
The large subunit of Escherichia coli ribosomes comprises two RNA species, 23S and 5S rRNA, and 33 proteins (1). 5S rRNA is the smallest RNA component of the ribosome, 120 nucleotides long. Its secondary structure has been extensively investigated using phylogenetic data and a suite of biophysical and biochemical techniques, including NMR spectroscopy and microarrays (2,3). It consists of five helices (I–V), two hairpin loops (C and D), two internal loops (B and E) and a hinge loop (A) organized in a three-helix junction.
The tertiary structure of E. coli 5S rRNA in solution is not known yet. The most detailed picture of the tertiary structure of E. coli 5S rRNA has emerged from models reconstructed on the basis of crystallographic and cryo-EM analysis of the large ribosomal subunit or the whole 70S ribosome from bacteria and archaea (4–11). Beyond this contribution, these studies revealed that 5S rRNA, in conjunction with proteins L5, L18 and L25, forms the central protuberance of the large ribosomal subunit. Extending from there it connects the upper part of the large subunit with the peptidyl transferase (PTase) center and the binding site of elongation factor EF-G (Figure 1). Consistent with this 5S rRNA topography, cross-linking and foot printing analysis (12–16), site-directed mutagenesis studies (17) and in vitro screening of RNA motifs interacting with 5S rRNA (18) have indicated that 5S rRNA interacts with multiple functional regions of 23S rRNA in E. coli ribosomes, including helices 38 and 39, the terminal loop of helix 89, and sites between nucleosides 2272–2345 in domain V. In parallel, mutagenesis studies in 5S rRNA from Saccharomyces cerevisiae (19,20) and functional substitution of 5S rRNA within Thermus aquaticus ribosomes by antibiotics interacting simultaneously with domains II and V of 23S rRNA (21) demonstrated that 5S rRNA is actively involved in different ribosomal functions. Combined with early studies on the reconstitution of active 50S subunits (22, 23), this extensive investigation on the structural and functional role of 5S rRNA led to the hypothesis that 5S rRNA assists in stabilizing the PTase center and in facilitating communication between different functional centers of the ribosome. Nevertheless, despite the tendency in this series of experiments to attribute all of the observed effects to 5S rRNA, it is in certain cases difficult to distinguish the role of 5S rRNA per se from that of the associated ribosomal proteins. An alternative experimental approach to investigate the functional role of 5S rRNA is to specifically alter its folding and to correlate the resulting conformational changes with alterations in the function of the 50S ribosomal subunit.
Figure 1.
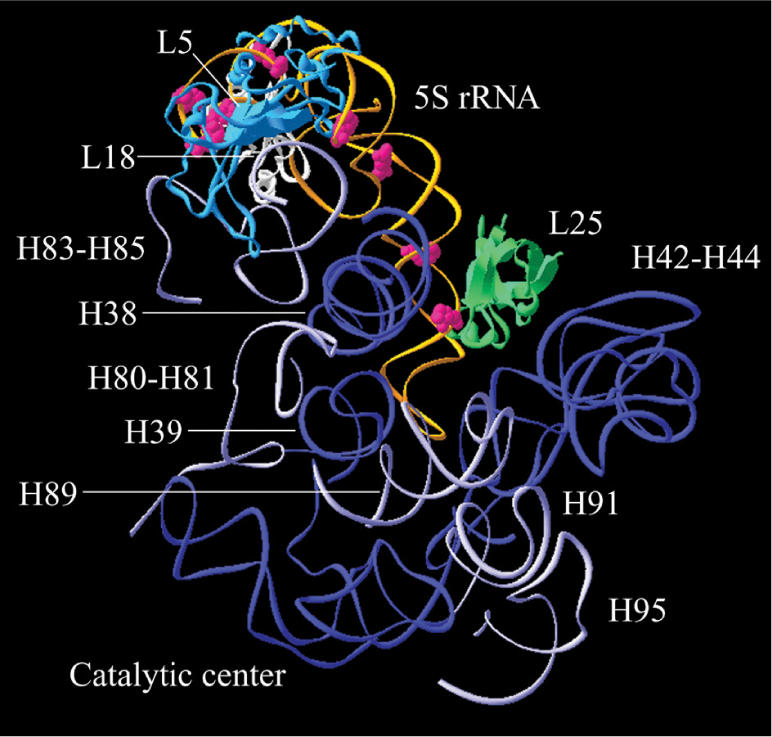
Three-dimensional representation showing the ABA-spermine cross-linking sites in 5S rRNA. Cross-links are superimposed on a tertiary structure of 5S rRNA (in orange) reconstructed on the basis of crystallographic data obtained with E. coli ribosomes (8) using the Swiss-PdbViewer. The labeled nucleosides with ABA-spermine are shown in red. Ribosomal proteins (L5, L18 and L25) and 23S rRNA regions complexed with 5S rRNA are also presented.
Polyamines, like other monovalent and divalent ions, cause on 5S rRNA a conformational switch to a more compact form (24). For instance, spermine has been found to stabilize the secondary and tertiary structure of 5S rRNA quite strongly, more so and in a different way than Mg2+ (25). This was attributed to the hydrogen bonds afforded by all the primary and secondary amines of spermine. Obviously, to unveil the molecular basis of spermine effect on the structure and function of 5S rRNA, the localization of polyamine binding sites on 5S rRNA is a prerequisite.
In the present study, mapping of spermine binding sites in 5S rRNA was achieved by photo-affinity labeling of 5S rRNA, either free in solution or as part of 50S subunits or whole ribosomes. In each case, the cross-linking sites were identified by primer extension analysis. As expected, binding of spermine to 5S rRNA caused changes in the conformation of 5S rRNA. The influence of these changes on several ribosomal properties was investigated.
MATERIALS AND METHODS
Materials
[γ-32P] ATP and [α-32P] GTP were obtained from Izotop (Budapest, Hungary). L-[2, 6 3H] Phenylalanine was from Moravek Biochemicals Inc. (Brea, CA, USA). [14C] Spermine tetrahydrochloride was from Amerscham Biosciences Inc. (Piscataway, NJ, USA). RNase H was purchased from Promega (Madison, WI, USA), and AMV reverse transcriptase from Roche Diagnostics (Mannheim, Germany). The dNTPs and ddNTPs were from Boehringer (Mannheim, Germany). Spermine tetrahydrochloride, dimethyl sulfate (DMS), DMS stop solution, puromycin dihydrochloride, thiostrepton and heterogenous tRNA from E. coli were from Sigma (St Louis, MO, USA). Cellulose nitrate filters (type HA; 0.45 μM pore size) were from Millipore Corporation (Bedford, MA, USA), while thin plastic sheet of PEI-cellulose F were from Merck KGaA (Darmstadt, Germany). 17-deoxynucleotides, used as primers in the primer extension analysis, were synthesized by Invitrogen (Paisley, UK). Elongation factor G (EF-G) from E. coli was kindly provided by Prof. K. H. Nierhaus (Max-Planck Institute, Berlin). ABA-spermine was synthesized from methyl-4-azidobenzoimidate and spermine, and purified according to Clark et al. (26).
Biochemical preparations
Polyamine-depleted 70S ribosomes, native 50S and 30S subunits, and partially purified translation factors were prepared from E. coli K12 cells, as reported previously (27). 23S rRNA, 5S rRNA and total proteins (TP50) were isolated from 50S ribosomal subunits and activated prior to their use by incubation for 20 min at 42°C in a buffer containing 40 mM HEPES-KOH, pH 7.2, 20 mM magnesium acetate and 100 mM NH4Cl (28). Ac[3H]Phe-tRNA was prepared from heterogeneous E. coli tRNA (Sigma), charged to ∼80%, 100% being 28 pmol of [3H]Phe per A260 unit (28). Post-translocation complex of poly(U)-programmed 70S ribosomes (POST-complex) bearing tRNAPhe at the E-site and Ac[3H]Phe-tRNA at the P-site, pre-translocation complex (PRE-complex) containing tRNAPhe at the P-site and Ac[3H]Phe-tRNA at the A-site and a ribosomal complex (P-complex) carrying a single tRNAPhe in the P-site were prepared as described by Dinos et al. (29). The state of tRNA binding (hybrid versus classical) in the prepared ribosomal complexes was established using radioactivity and puromycin-reactivity measurements, as well as chemical protection assays (30). POST-complex prepared according to this experimental protocol was reactive against puromycin to ∼90%. Reconstituted 50S ribosomal subunits from TP50, 23S and 5S rRNA were prepared by a two-step incubation procedure (23). When required, 5S rRNA labeled at the 3′-end with [5′-32P] pCp and T4 RNA ligase (31) or photolabeled by ABA-[14C]spermine was used in the reconstitution experiments. After reconstitution, each one of the samples was loaded on a 10–30% linear sucrose gradient in buffer A containing 100 mM Tris–HCl, pH 7.2, 6 mM MgCl2, 100 mM NH4Cl and 6 mM 2-mercaptoethanol. The gradients were centrifuged for 6 h at 85 000g, at 4°C. The percentage of total reconstitution was then estimated by processing the gradients for optical scanning at 260 nm and radioactivity counting.
Photoaffinity labeling and mapping of ABA-spermine cross-linking sites in 5S rRNA
Naked 5S rRNA, 50S ribosomal subunits or POST-complex were photolabeled with ABA-spermine and purified as shown previously (32). The ABA-spermine cross-linking sites in 5S rRNA were determined by primer-extension analysis (33), making use of the fact that by linking the photoprobe to a nucleoside, it acts as a barrier for reverse transcriptase. The DNA primer used was complementary to the 5S rRNA region encompassing nucleosides 104–120. The entire 5S rRNA, except its extreme 3′-end (nucleosides 95–120), was analyzed in this way. To check the extreme 3′-end of 5S rRNA, modified 5S rRNA with ABA-SPM was radioactively labeled at its 3′-end with [5′-32P] pCp and T4 RNA ligase and then extended by a 38-oligonucleotide [5′-(A)21(G)10(A)7-3′]. The product was purified by gel electrophoresis on an 8% (w/v) polyacrylamide/7M urea gel, excised and ethanol precipitated. Such extended 5S rRNA was analyzed by primer extension using a deoxy-nucleotide complementary to the extended arm, 5′-(T)7(C)10-3′, as primer. The stops of reverse transcriptase reaction were visualized on a gel autoradiogram (33). Controls with untreated 5S rRNA or samples photolabeled in the simultaneous presence of a 250-fold excess of spermine were run in parallel in order to distinguish nicks in the RNA, non-specific photoincorporation or autonomous pauses of reverse transcriptase. Only bands reproduced at least three times were taken into account. To confirm the results of primer extension analysis and to search for possible photoincorporation into positions non-recognizable by reverse transcriptase, samples were first hybridized with selected pairs of 11-deoxynucleotides complementary to 5S rRNA at positions 40 nts apart, digested with RNase H, and applied to a 8% (w/v) polyacrylamide/7M urea gel for analysis (32). 5S rRNA, untreated or photolabeled, was modified with DMS (32), and then subjected to primer extension analysis.
Kinetics of ABA-spermine cross-linking
The number of cross-linking sites, the dissociation constant (Kd) of the reversible complex between 5S rRNA (R) and the photoprobe (L) and the Hill coefficient were estimated as shown previously (34). We assumed that photoincorporation of ABA-spermine into 5S rRNA proceeds as an irreversible pseudo-first-order reaction,
where RLn is the encounter complex between 5S rRNA and the photoprobe (non-covalently associated) and RLn* represents the irreversible complex formed after irradiation of the sample.
Biochemical assays
To evaluate the ability of the reconstituted 50S subunits to associate with native 30S subunits, 5 A260 units of each subunit were incubated in buffer A for 30 min at 37°C. The mixture was then applied to a linear 10–30% sucrose gradient in buffer A, centrifuged for 6 h at 85 000g and then analyzed by optical scanning at 260 nm.
Binding of Ac[3H]Phe-tRNA to the ribosomal P- and A-sites was performed in buffer B (40 mM HEPES-KOH, pH 7.2, 6 mM magnesium acetate, 100 mM NH4Cl and 6 mM 2-mercaptoethanol) containing or not containing 50 μM spermine, by incubating Ac[3H]Phe-tRNA with poly(U)-programmed 70S ribosomes pre-filled (A-site binding) or not pre-filled (total binding) at their P-sites by tRNAPhe (32). The Ac[3H]Phe-tRNA binding was measured by nitrocellulose filtration. The P-site bound Ac[3H]Phe-tRNA was estimated from the total binding by titrating the resultant Ac[3H]Phe-tRNA·poly(U)·70S ribosome complex with puromycin (2 mM, 2 min at 25°C).
PTase activity of the POST-complex was assessed by the puromycin reaction carried out at 25°C in buffer B. When required, 50 μM spermine was also included in the incubation mixture. The catalytic rate constant (kcat) of PTase and the affinity constant (Ks) of puromycin were estimated as described previously (28).
For translocation assays, aliquots of PRE-complex (1.6 pmol) were added in 12 μl of buffer B containing 0.015 μM EF-G and 0.12 mM GTP. When desired, spermine at 50 μM was also included in buffer B. The mixtures were incubated at 25°C for specified time intervals. Translocation was monitored by titrating the resultant POST-complex with a solution of puromycin and thiostrepton (28). Spontaneous translocation was measured in the absence of EF-G. In another series of experiments, increasing concentrations of EF-G were added in buffer B containing 0.12 mM GTP and PRE-complex at a fixed concentration. The reaction was carried out for 5 min at 25°C, after which translocation was again estimated by reaction with puromycin. Possible interference in the PRE-complex samples by POST-complex was measured at the start of the translocation time-course and subtracted.
To estimate the capacity of ribosomes for EF-G binding, PRE-complex was prepared either from native subunits (control samples) or from native 30S subunits and reconstituted large ribosomal subunits (29). Aliquots of these complexes (1.6 pmol) were incubated at 25°C for 30 min in 12 μl of buffer B containing 1 μM EF-G, 10 μM GTP, 5 μCi of [α-32P]GTP (400 Ci/mmol), and 0.5 mM fusidic acid. When desired, spermine at 50 μM was also included in the incubation mixture. Six microliters of each incubation mixture were filtered through nitro-cellulose filters, washed twice with 2 ml of ice-cold buffer B, and the radioactivity retained on the filters was determined by scintillation counting. Control experiments were performed in which ribosomes were incubated in the absence of EF-G, and the radioactivity measured was subtracted. P-complex and empty ribosomes are also capable of binding EF-G and triggering the GTPase activity of EF-G (35,36). Therefore, the participation of such complexes in our experimental system was quantified and used to correct the raw data, applying Equation (1),
| 1 |
where Bi represents the EF-G·GTP binding per i-type ribosomal complex, αi is the percentage of each i-type ribosomal complex in the mixed ribosomal population and Bt is the total EF-G·GTP binding.
Ribosome-dependent GTP hydrolysis catalyzed by EF-G was determined in 15 μl of buffer B containing 1.6 pmol of PRE-complex, 8.8 pmol of EF-G, 50 μM GTP and 5 μCi of [α-32P]GTP (400 Ci/mmol). The mixture was incubated in the presence or absence of 50 μM spermine at 4°C for specified time intervals. An aliquot (3 μl) was withdrawn at each time-point, and the reaction in this aliquot stopped by adding 1 μl of 11 M formic acid. The samples were kept on ice for 5 min, centrifuged at 10 000g for 10 min, and then analyzed by thin-layer chromatography on PEI-cellulose F in 1 M formic acid and 1 M lithium chloride. The extent of hydrolysis was quantified using a FUJIFILM phosphoimager. [α-32P]GTP hydrolysis by ribosomes solely were measured separately and subtracted. In addition, the capacity of P-complex and empty ribosomes to trigger the GTPase activity of EF-G was separately measured and taken into account in data processing.
Statistics
One-way ANOVA was used to estimate the mean values, data variability and significant differences between means.
RESULTS
Localization of ABA-spermine cross-linking sites in 5S rRNA
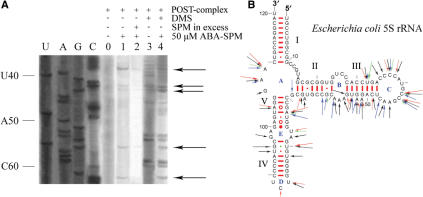
The arylazido group, ABA, is positioned 9 Å from the N1-amino group of spermine. In the dark, the photoprobe interacts in a reversible fashion with 5S rRNA. By irradiation at 300 nm, however, the arylazido group is converted to nitrene, which reacts rapidly and covalently with a variety of adjacent groups of the target molecule. Precise identification of the cross-linking sites was achieved by primer extension analysis, taking advantage of the fact that reverse transcriptase pauses or stops one position before an ABA-spermine-modified nucleoside. The modified nucleosides were then determined by electrophoretic and autoradiographic analysis of the reverse transciptase products. To make the above analysis possible in the extreme 3′-end region of 5S rRNA, the target molecule was extended after its labeling with ABA-spermine by ligating the 5′-(A)21(G10)(A)7-3′ oligonucleotide at its 3′-end and using a primer complementary to this extension. A representative auto-radiogram obtained by primer-extension analysis is shown in Figure 2A. The cross-linking sites of ABA-spermine in 5S rRNA, either free in solution or assembled into 50S subunit and POST-complex, are summarized in a secondary structure model of 5S rRNA (Figure 2B). Detailed presentation of the 5S rRNA nucleosides labeled under various conditions is given in Supplementary Table 1. The authentic character of the cross-linking sites was confirmed by RNase H cleavage of 5S rRNA modified with ABA-[14C] spermine, in the presence of selected pairs of 11-deoxynucleotide complementary to sequences located 40 nts apart in the primary structure of 5S rRNA (Supplementary Table 2).
Figure 2.
Primer extension analysis of ABA-spermine cross-linking in 5S rRNA. (A) 5S rRNA isolated from POST-complex photolabeled with ABA-spermine was monitored by primer extension analysis. The primer for the reverse transcriptase was complementary to 5S rRNA positions 104–120. U, A, G and C are dideoxy sequencing lanes. Lane 0, control (non-photolabeled sample); lane 1, sample photolabeled with 50 μM ABA-spermine; lane 2, sample like that used in lane 1, but photolabeled in the simultaneous presence of spermine in excess; lane 3, sample modified by DMS; lane 4, sample treated like that in lane 1, and then modified by DMS. The stops of reverse transcriptase reaction due to ABA-spermine cross-linking are shown by arrows. (B) Summary of ABA-spermine cross-linking sites, superimposed on a secondary structure diagram of E. coli 5S rRNA (cited at http://www.rna.icmb.utexas.edu). Cross-links are marked with red (cross-linking in naked 5S rRNA), blue (cross-linking in 50S subunit) and green arrows (cross-linking in POST-complex). Long arrows, strong cross-links; medium arrows, intermediate-strength cross-links; short arrows, weak cross-links. For comparison, cross-links obtained by labeling naked 5S rRNA with 300 μM ABA-spermine are also shown in black.
Except for helix I, ABA-spermine cross-linking was observed throughout the 5S rRNA molecule in both helices and loops of the secondary structure, but it was detected more frequently in helix III and loop C (Figure 2B). The primarily labeled nucleosides in single-stranded regions were A and C (84%), while in double-stranded regions the label was more equally distributed among the four nucleosides. In most cases, reverse transcriptase stopped at a single nucleoside. Only three exceptions were recorded, in which two adjacent nucleosides were modified; the one closer to the 3′-terminus corresponded to a band with stronger intensity. Such a profile is probably due to a ‘stuttering’ of reverse transcriptase, observed also previously (28,32). Evidence for site-specific photo-incorporation was sought by running in parallel control experiments, in which spermine was added in excess in the incubation mixture during photolabeling. As shown in Figure 2A, the authentic stops of reverse transciptase are essentially abolished under such conditions. As detected by additional experiments, spermine exhibited the best competition potency, compared with spermidine and putrescine. Monovalent ions, such as Na+ or NH4+ did not compete, while Mg2+ decreased the labeling of 5S rRNA in a specific and dose-dependent manner. At 10 mM Mg2+, a 10% reduction in photolabeling was recorded; nucleosides U32, A73, A52, A53, C71, A73 and U80 were the cross-linking residues most affected by Mg2+. In contrast, the cross-linking pattern for the remaining positions was not significantly altered.
An important feature of Figure 2B is that the cross-linking pattern depends on the photoprobe concentration and the assembly status of the target molecule. With regards to the former factor, an increase of ABA-spermine concentration from 50 to 300 μM enhanced the susceptibility of A52, A53 and G56 to the photoprobe, while it reduced the cross-linking at nucleosides G67 and A73. In addition, the increase of photoprobe concentration resulted in enrichment of cross-linking by additional sites dispersed throughout the 5S rRNA molecule. Assembly of 50S subunit resulted in protection of several positions from cross-linking by ABA-spermine, especially of nucleosides belonging to helices III and V and to loops C, D and E. In contrast, it favored cross-linking to helix II and loop B. Finally, when 50S subunit was incorporated into the ribosomal POST-complex, a conspicuous change in the susceptibility of several nucleosides against ABA-spermine was recorded. Namely, nucleosides A46, C47, A52, A59, C62, G69 and A73 were freshly protected, while C42 and A108 exhibited increased protection.
Chemical probing experiments using DMS indicated that the protection pattern of 5S rRNA changes, when it is incorporated into 50 subunits. In agreement with previous observations (37), nucleosides C11, A15, C35, A57, A58, A59, A66, A73 and A99 became protected, while nucleoside A78 became more accessible. Formation of the ribosomal POST-complex reduced the susceptibility of nucleosides C38 and A45 against DMS, but increased the reactivity of A15 and A52. On the other hand, cross-linking of ABA-spermine induced further alterations in the reactivity of 5S rRNA towards DMS by increasing the accessibility of some positions and decreasing the accessibility of others. The results of the latter experiments are summarized in Table 1.
Table 1.
Reactivity against DMS of 5S rRNA naked or incorporated into the ribosomal 50S subunit and POST-complexa
| Regions of 5S rRNA | Target molecule: free 5S rRNA Experimental conditions:b | 50S ribosomal subunit | POST-complex | |||
|---|---|---|---|---|---|---|
| A | B | A | B | A | B | |
| Helix I | G10, C114 | |||||
| Helix II | C63, A66, G67 | C63, A66, G67 | G61, C62, A66 | G61, C62, A66 | G61, C62, A66 | G61, A66 |
| Helix III | U32, A34, C49, A52, A53, U55, G56 | U32, C49, A52, A53, U55, G56 | C30, U32, C49, A52, A53, U55 | U32, A52, A53, U55 | U32, A52, A53 | U32, (A52), (A53) |
| Helix IV | U80, U82, C90, U95, G96 | U80 | C90 | C90 | ||
| Helix V | A73, A104 | A73, A104 | C71, A73, U74, A104 | A73, (A104) | C71,A73, U74, G76, A104 | A73, (A104) |
| Loop A | C11, C12, A15, G107, A108, A109 | C11, C12, A15, A108, A109 | C11, C12, A15, A108, A109 | C11, C12, A15, G69, A108, A109 | C11, C12, A15, A108, A109 | C11, C12, A15, A108,A109 |
| Loop B | C26, C27, A57, A58, A59 | A57, A58, A59 | A57, A58, A59 | (A58), A59 | (A58), A59 | A58, A59 |
| Loop C | C35, C36, C37, C38, A39, C42, C43, A45, A46 | C35, C36, C37, C38, (A39), C42, C43, (A45) | C35, C36, C37, C38, A39, C42, C43, A45, A46 | (C35), (C36), C37, (C38), (A39), C42, C43, (A45),A46, C47 | A35, C36, C37, C38, A39, C42, C43, A45 | (C35), (C36), C37, (C38), (A39), C42, C43, (A45) |
| Loop D | U87, C88 | U87, C88 | U87, (C88) | (C88) | (C88) | (C88) |
| Loop E | A78, G98, A99 | U77, (A78), (A99) | U77, (A78) | (A78) | U77, (A78) | U77, A78 |
aNucleosides exhibiting increased reactivity compared with that of control samples (non-labeled) are indicated in bold, while nucleosides with decreased reactivity are shown in parentheses. bSamples were photolabeled with 300 μM (condition A) or with 50 μM ABA-spermine (condition B).
Kinetics of ABA-spermine cross-linking to 5S rRNA
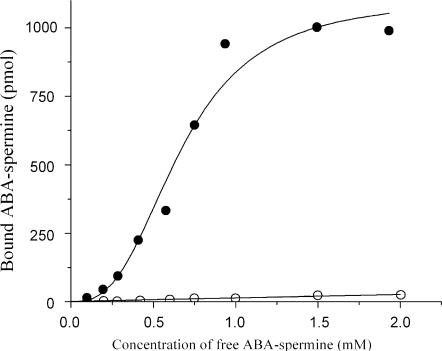
Photolabeling data at various concentrations of ABA-spermine established a sigmoidal hyperbola (Figure 3). Therefore, they were fitted to Equation (2),
| 2 |
using the Hill hyperbola function of the Microcal Origin 6.00 program. In Equation (2), [L] is the concentration of the free photoprobe, B is the amount of the covalently bound photoprobe to naked 5S rRNA, Bmax is the value of B at saturation conditions, n is the Hill coefficient, and Kd is the overall dissociation constant of the encounter complex between the photoprobe and 5S rRNA. From the value of Bmax and the amount of 5S rRNA used in the assay, we estimated a value for the number of cross-linking sites equal to 6.35.
Figure 3.
ABA-spermine photoincorporation into naked 5S rRNA at various concentrations of the photoprobe. Photoaffinity labeling was carried out in 15 μl of buffer 40 mM HEPES-KOH, pH 7.2, 6 mM magnesium acetate and 100 mM NH4Cl, also containing 174 pmol of 5S rRNA, and ABA-spermine at the indicated concentrations; (filled circles), specific binding; (open circles), non-specific binding estimated by performing photolabeling in the simultaneous presence of spermine in excess.
Effect of 5S rRNA photolabeling on its ability to assemble into 50S ribosomal particles and 70S ribosomes
The crude product derived from reconstitution experiments using photolabeled 5S rRNA, TP-50 and wild-type 23S rRNA was found to exhibit decreased capability to form 70S ribosomes upon association with native 30S ribosomal subunits, compared with that obtained by utilizing wild-type 5S rRNA. Two factors may cause such an effect. First, photolabeling of 5S rRNA molecule may directly influence the ability of the reconstituted particles to associate with 30S subunits. Second, reduced ability could be the result of an impaired reconstitution, and thus of a decrease in the net fraction of the 50S subunits. In order to discriminate between these two possibilities, the crude product was subjected to sucrose gradient centrifugation and the yield of reconstitution was estimated. As indicated in Table 2, ribosomal particles deprived of 5S rRNA sedimented at about 47S. Such particles, upon association with native 30S subunits, led to the formation of 62S ribosomal complexes. When 5S rRNA photolabeled with 50 μM ABA-spermine was used as a component of the reconstitution mixture, both 50S and 47S particles were obtained, thus resulting in a decreased yield of 50S subunits. However, isolated 50S subunits from this mixture were fully active at associating with native 30S subunits and forming 70S ribosomes. In contrast, photolabeling of 5S rRNA with 300 μM ABA-spermine prior to its use in the reconstitution experiments, disturbed more dramatically both the yield of 50S subunits and their capability to associate with native 30S subunits.
Table 2.
Effect of 5S rRNA photolabeling on 50S ribosomal subunit assembly and the ability of the assembled particles to associate with native 30S ribosomal subunits
| 5S rRNA speciesa | % Reconstitutionb | % Association of ribosomal subunitsc |
|---|---|---|
| Wild-type | 100 | 70.0 ± 2.0 |
| None | n.dd | 65.7 ± 5.0 |
| Labeled with 50 μM ABA-spermine | 52.5 ± 7.0 | 73.5 ± 5.2 |
| Labeled with 300 μM ABA-spermine | 15.8 ± 3.0 | 22.5 ± 3.7 |
a5S rRNA samples were 3′-labeled with [32P] before reconstitution into 50S particles. bThe values of % reconstitution have been normalized to the yield of reconstitution achieved with wild-type 5S rRNA. cNative 30S subunits were added in a two-molar excess over 50S particles in order to yield the maximum level of association. The yield of association is expressed as % of the 50S particle input. dNo material with 50S sedimentation coefficient was detected in these experiments. Instead, 47S particles were obtained and a heavy shoulder of about 62S was seen upon their association with 30S subunits.
Effect of 5S rRNA photolabeling on the binding of AcPhe-tRNA to poly(U)-programmed ribosomes and the PTase activity
When 70S ribosomes obtained from reconstituted 50S subunits and native 30S subunits were poly(U)-programmed and incubated with equimolar amount of AcPhe-tRNA, their capacity for binding to the P- and A-site was found 0.171 and 0.092, respectively. AcPhe-tRNA binding to both sites was remarkably improved in the presence of 50 μM spermine (Table 3). At higher molar ratio of AcPhe-tRNA to ribosomes (2:1) or at higher concentrations of spermine (300 μM), the binding was further improved. In this series of experiments, wild-type 5S rRNA was used in the reconstitution of 50S subunits. Next, 70S ribosomes containing 5S rRNA photolabeled with 50 μM ABA-spermine were prepared and tested under identical conditions, but in the absence of free spermine. As shown in Table 3, cross-linking of ABA-spermine to 5S rRNA caused a slight, but statistically significant improvement in the binding properties of the modified ribosomes. Moreover, cross-linking of ABA-spermine to 5S rRNA stimulated by 30% the catalytic rate constant of PTase, without affecting essentially the affinity of ribosomes towards puromycin. In agreement with previous studies (22,23), 70S ribosomes deprived of 5S rRNA displayed extremely low capability to bind AcPhe-tRNA, particularly at the A-site. These ribosomes also exhibited impaired PTase activity (Table 3).
Table 3.
Effect of ABA-spermine cross-linking in 5S rRNA of poly(U)-programmed ribosomes on their capacity for AcPhe-tRNA binding and PTase activity
| Ribosomal speciesa | P-site bound AcPhe-tRNA per ribosomeb | A-site bound AcPhe-tRNA per ribosomeb | PTase activityc (mM−1 min−1) |
|---|---|---|---|
| Unlabeled (-spermine) | 0.171 ± 0.008 | 0.092 ± 0.006 | 3.16 ± 0.18 |
| Unlabeled (+50 μM spermine) | 0.420 ± 0.028 | 0.190 ± 0.014 | 5.12 ± 0.32 |
| Labeled in 5S rRNA | 0.218 ± 0.023 | 0.100 ± 0.010 | 4.07 ± 0.20 |
| Labeled in 5S rRNA (+50 μM spermine) | 0.407 ± 0.015 | 0.185 ± 0.014 | 5.01 ± 0.25 |
| Deprived of 5S rRNA (-spermine) | 0.051 ± 0.005 | 0.004 ± 0.003 | 0.30 ± 0.02 |
| Deprived of 5S rRNA (+50 μM spermine) | 0.071 ± 0.005 | 0.006 ± 0.003 | 0.35 ± 0.02 |
a70S ribosomes were prepared from reconstituted 50S ribosomal subunits and native 30S subunits. For the preparation of 5S rRNA-deprived ribosomes, 47S ribosomal particles instead of 50S subunit were associated with native 30S subunits. bBinding was monitored in 200 μl of buffer B containing 0.4 mM GTP, 83.2 pmol Ac[3H]Phe-tRNA and 83.2 pmol 70S or 62S poly(U)-programmed ribosomes pre-filled (A-site binding) or not pre-filled (total binding) at their P-site by tRNAPhe. The Ac[3H]Phe-tRNA binding at 25°C was measured by nitrocellulose filtration. The P-site bound AcPhe-tRNA was estimated from the total binding by titration with puromycin. cEach species of POST-complex reacted with puromycin in buffer B. When desired, 50 μM spermine was also included in the reaction mixture. The PTase activity was estimated by the ratio kcat/Ks.
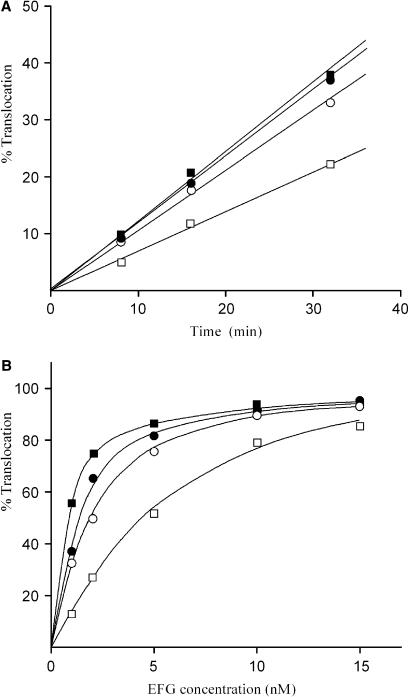
Effect of 5S rRNA photolabeling on translocation
Two species of PRE-complex were tested for their efficiency to translocate AcPhe-tRNA from the A- to the P-site: one reconstituted from wild-type 5S rRNA, and the other reconstituted from 5S rRNA photolabeled with 50 μM ABA-spermine. For the construction of PRE-complexes, poly(U)-programmed 70S ribosomes were incubated for 20 min at 37°C with tRNAPhe (molar ratio to ribosomes 2 : 1) in order to pre-fill the P-site. Chemical protection assays verified that tRNAPhe sampled the classical P/P binding state. Using [32P]tRNAPhe, we found that occupation of the P-site in this complex (P-complex) ranged from 40% to 75%, depending on the 5S rRNA species (photolabeled or wild-type) and the ionic conditions used. Subsequently, Ac[3H]Phe-tRNA was added (molar ratio 2:1) and incubated for an additional 20 min at 37°C to allow non-enzymatic A-site binding (PRE-complex). Ribosomal complexes prepared in this way were reactive against puromycin to ∼20%. In addition, chemical protection assays indicated very low, <10%, but measurable protection of nucleoside 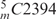 in 23S rRNA (E-site) against DMS, while radioactivity measurements detected no release of the bound [32P] isotope from the ribosome. This means that more than 80% of the bound AcPhe-tRNA sampled the classical A/A binding state. Our results are in agreement with previous observations that N-protected aminoacyl-tRNAs bound in a pre-translocation state on the ribosome generally occupy the classical A/A site (38). The small amount of AcPhe-tRNA which was reactive toward puromycin, probably translocating spontaneously from the A/A to the P/P site during preparation of the PRE-complexes, was measured in each case and taken into account in data processing. Translocation was studied in buffer B containing 0.12 mM GTP, in the absence or in the presence of 50 μM spermine. Spermine 50 μM was chosen on the basis of previous findings that such a concentration is optimum for translocation (28). EF-G at 15 nM promoted relatively fast translocation that was almost complete within 2 min, independently of whether free spermine was present or absent. In the absence of EF-G, however, translocation was much slower and proceeded in a linear fashion at least up to 30 min. Spermine either free or covalently bound to 5S rRNA improved spontaneous translocation (Figure 4A). To examine the effect of spermine on EF-G requirements for efficient translocation, increasing amounts of EF-G were added in buffer B containing PRE-complex at fixed concentration, and translocation was allowed to proceed for 5 min at 25°C. As shown in Figure 4B, the extent of translocation in each case followed a hyperbolic curve reaching the same plateau at high concentrations of EF-G. Nevertheless, the concentration of EF-G at which 50% translocation was achieved, differed. Namely, PRE-complexes exposed to free spermine or possessing covalently bound spermine at their 5S rRNA required lower concentrations of EF-G for efficient translocation.
in 23S rRNA (E-site) against DMS, while radioactivity measurements detected no release of the bound [32P] isotope from the ribosome. This means that more than 80% of the bound AcPhe-tRNA sampled the classical A/A binding state. Our results are in agreement with previous observations that N-protected aminoacyl-tRNAs bound in a pre-translocation state on the ribosome generally occupy the classical A/A site (38). The small amount of AcPhe-tRNA which was reactive toward puromycin, probably translocating spontaneously from the A/A to the P/P site during preparation of the PRE-complexes, was measured in each case and taken into account in data processing. Translocation was studied in buffer B containing 0.12 mM GTP, in the absence or in the presence of 50 μM spermine. Spermine 50 μM was chosen on the basis of previous findings that such a concentration is optimum for translocation (28). EF-G at 15 nM promoted relatively fast translocation that was almost complete within 2 min, independently of whether free spermine was present or absent. In the absence of EF-G, however, translocation was much slower and proceeded in a linear fashion at least up to 30 min. Spermine either free or covalently bound to 5S rRNA improved spontaneous translocation (Figure 4A). To examine the effect of spermine on EF-G requirements for efficient translocation, increasing amounts of EF-G were added in buffer B containing PRE-complex at fixed concentration, and translocation was allowed to proceed for 5 min at 25°C. As shown in Figure 4B, the extent of translocation in each case followed a hyperbolic curve reaching the same plateau at high concentrations of EF-G. Nevertheless, the concentration of EF-G at which 50% translocation was achieved, differed. Namely, PRE-complexes exposed to free spermine or possessing covalently bound spermine at their 5S rRNA required lower concentrations of EF-G for efficient translocation.
Figure 4.
Effect of ABA-spermine cross-linking to 5S rRNA on translocation. (A) Time course of spontaneous translocation. Aliquots of PRE-complex, i.e. poly(U)-programmed 70S ribosomes occupied at their P- and A-sites with tRNAPhe and Ac[3H]Phe-tRNA, respectively, possessing (squares) wild-type or (circles) photolabeled 5S rRNA by 50 μM ABA-spermine, were incubated at 25°C for the indicated time intervals, in buffer B containing 0.12 mM GTP, (filled symbols) in the presence or (open symbols) in the absence of 50 μM free spermine. (B) Dependence of translocation on EF-G concentration. PRE-complex was incubated with different amounts of EF-G for 5 min at 25°C. The symbols are the same as those used in (A). In each plot, translocation of 100% represents conversion of all PRE-complex into POST-complex.
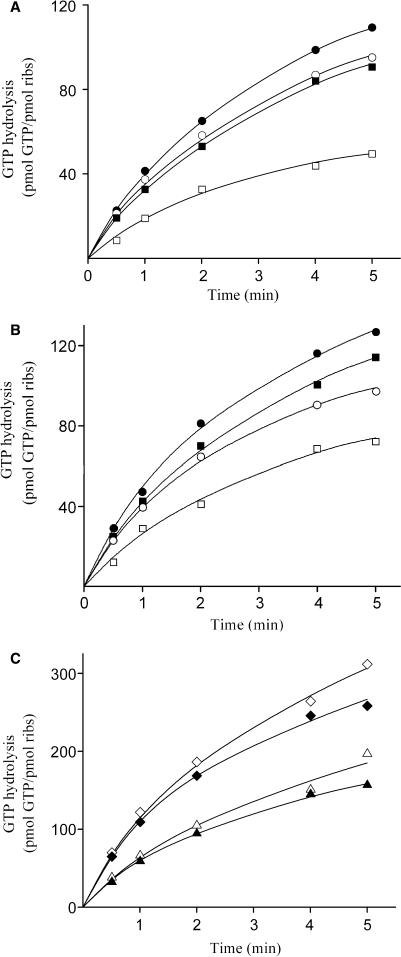
The sparing effect of spermine on EF-G-requirements could be explained either by a beneficial effect of spermine on the binding of EF-G to ribosomes, and/or by an influence of spermine on the ribosome-dependent GTP hydrolysis catalyzed by EF-G. To clear this point, we applied the following two approaches: First, we used fusidic acid, an antibiotic that binds to the EF-G·GDP-ribosome complex and prevents dissociation of EF-G·GDP from the ribosome. Under these conditions, the EF-G·GDP-ribosome complex is stalled in a post-translocation state (39). Employment of [α-32P] GTP in the assay allows the indirect estimation of EF-G binding, by measuring the trapped radioactivity on ribosomes relative to controls incubated in the absence of EF-G. The results from these experiments are summarized in Table 4. It is evident that exposure of whole PRE-complexes to free spermine or covalent attachment of spermine to their 5S rRNA promotes EF-G binding. Similar treatment of P-complex or empty ribosomes results in analogous improvement of EF-G binding. Nevertheless, it is apparent from the data shown in Table 4 that the interaction of EF-G with the ribosome is generally stimulated when deacylated tRNA occupies the P-site. In agreement with previous observations (22), ribosomes deprived of 5S rRNA were almost inactive to bind EF-G. Second, the capacity of PRE-complex to activate hydrolysis of GTP by EF-G was determined. As shown in Figure 5, the initial velocity of the reaction was found to be almost identical for either ribosomes reconstituted with wild-type or ribosomes containing photolabeled 5S rRNA, and similar to that possessed by native ribosomes, regardless of whether free spermine was present or not in the reaction mixture [compare panels (A) and (B) in Figure 5]. Similar behavior was exhibited by P-complex and empty ribosomes (Figure 5C). In agreement with previous studies (36,40), empty ribosomes or PRE-complex were less active in stimulating the GTPase activity of EF-G, compared with P-complex. Interestingly, ribosomal complexes deprived of 5S rRNA exhibited considerably reduced efficiency to activate the GTP hydrolysis by EF-G, much less than that reported by Dohme and Nierhaus (23).
Table 4.
Binding of EF-G·GTP complex to native and reconstituted ribosomes
| Ribosomal speciesa | EF-G·GTP bound per ribosomeb | |||||
|---|---|---|---|---|---|---|
| -spermine | +spermine | |||||
| PRE- | P- | empty | PRE- | P- | empty | |
| Native | 0.48 | 0.79 | 0.52 | 0.93 | 0.98 | 0.94 |
| 5S rRNA-deprived | 0.02 | – | – | 0.09 | – | – |
| Reconstituted with wild-type 5S-rRNA | 0.17 | 0.25 | 0.19 | 0.44 | 0.57 | 0.48 |
| Reconstituted with photolabeled 5S-rRNA | 0.37 | 0.54 | 0.40 | 0.42 | 0.52 | 0.45 |
aRibosomal complexes were prepared from native subunits, or from native 30S subunits and reconstituted 50S subunits (the latter with wild-type or modified 5S rRNA by 50 μM ABA-spermine). For the preparation of poly(U)-programmed 62S ribosomes, 5S rRNA was omitted during the reconstitution of the large ribosomal subunit. bThe binding mixture (12 μl) contained 40 mM HEPES-KOH, pH 7.2, 6 mM magnesium acetate, 100 mM NH4Cl, 6 mM 2-mercaptoethanol, 1.6 pmol ribosomal complex, 1 μM EF-G, 10 μM GTP, 5 μCi of [α-32P]GTP, 0.5 mM fusidic acid and, when desired, 50 μM spermine. The mixture was incubated at 25°C for 30 min. The binding was measured by nitrocellulose filtration. Controls run in parallel without EF-G, were subtracted. The values corresponding to each binding state (PRE-complex, P-complex, empty ribosomes) have been corrected for the interference by coexisting ribosomal complexes.
Figure 5.
Effect of ABA-spermine cross-linking to 5S rRNA on the capacity of ribosomes to activate EF-G-catalyzed GTP hydrolysis. (A) Time course of GTP hydrolysis in the presence of 50 μM spermine. The reaction was carried out at 4°C in buffer B (15 μl) containing 1.6 pmol of PRE-complex formed either from (filled circles) native 30S and 50S subunits, or from native 30S subunits and 50S subunits reconstituted from (filled squares) wild-type 5S rRNA, (open circles) photolabeled 5S rRNA by 50 μM ABA-spermine, or (open squares) deprived of 5S rRNA; 8.8 pmol of EF-G, 50 μM GTP, 5 μCi of [α-32P]GTP (400 Ci/mmol), and 50 μM spermine. An aliquot (3 μl) was withdrawn at each time-point and the reaction stopped by adding 1 μl of 11 M formic acid. The reaction products were analyzed by thinlayer chromatography on PEI-cellulose F, as described in Materials and Methods section. (B) Time course of GTP hydrolysis in the absence of spermine. The reactions were performed and analyzed as in (A), but in the absence of spermine. (C) Time course of GTP hydrolysis, using (diamonds) P-complex or (triangles) empty 70S ribosomes. The ribosomal complexes were formed from native 30S subunits and 50S subunits reconstituted from (open symbols) wild-type 5S rRNA or (filled symbols) 5S rRNA photolabeled by 50 μM ABA-spermine. The reactions were performed and analyzed as in (B). The data presented in panels (A), (B) and (C) have been corrected for the interference by other species coexisting with the ribosomal complex under investigation.
DISCUSSION
In the present study, mapping of spermine binding sites in 5S rRNA was achieved by a photoaffinity labeling approach combined with primer extension analysis. The rationale is that attachment of ABA-spermine to a nucleoside acts as a barrier for reverse transcriptase. This technique bypasses several disadvantages associated with other cross-linking methods utilizing homobifunctional reagents (41,42). It has been successfully applied so far for mapping spermine binding sites in AcPhe-tRNA free or bound to the P-site of E. coli ribosomes (34), as well as in ribosomal proteins (27) and 16S and 23S rRNA naked or incorporated into ribosomes (28,32). It should be mentioned that, apart from monovalent and divalent cations, polyamines too are important components of the ionic environment of ribosomes, playing an essential role in the structural integrity of ribosomes and hence, in translational accuracy and efficiency (43). Although the existence of spermine in E. coli cells is questionable (44,45), accumulated evidence supports the notion that almost all of the cellular functions of the naturally occurring polyamines can be fulfilled by spermine (43). On the other hand, due to its four positive charges and the hydrogen bonds afforded by its primary and secondary amines, spermine is the most effective polyamine in stabilizing the RNA folding. Therefore, it can be experimentally used in micromolar concentrations, a very advantageous fact in bypassing artifacts associated with non-specific binding, frequently encountered in the application of photolabeling techniques.
The number of ABA-spermine cross-linking sites in 5S rRNA, determined in the present study by primer extension analysis, is higher than that calculated by Hill-plot analysis. This contradiction can be explained by the fact that apparently different cross-links localized to adjacent positions in the structure of 5S rRNA may represent the same binding site. To some degree, non-specific binding may also account for it. In fact, as revealed by competition experiments using natural polyamines (Figure 3) or Mg2+ ions as competitors, most of the cross-linking sites appear to be specific. The remaining sites are placed on the cytosolic surface of the large ribosomal subunit (C71), at the top edge of the central protuberance (U32, A52, and A53) or are located in defined electro-negative pockets (A73, U80). Nevertheless, independent of the specificity, polyamine binding to certain regions may be beneficial for the stabilization of the 5S rRNA tertiary structure and its communication with functional regions of 23S rRNA. Noteworthy is the binding of spermine within and around loop E, a region implicated in the binding of ribosomal protein L25. Crystal structure analysis of 5S rRNA fragments encompassing the loop E sequence, alone (46) or in complex with protein L25 (47), has demonstrated that loop E contains seven non-Watson–Crick base-pairs stabilized by several Mg2+ ions. Interestingly, molecular dynamics simulations revealed extensive binding of monovalent ions in the 5S rRNA loop E motif, even in the presence of 4–5 mM Mg2+ (48,49). Most of the monovalent ion binding sites coincide (G98, U80) or are in close proximity (G105, G72, U77) to cross-linking sites of ABA-spermine. Another issue of importance is related to the occupancy of distant sites from the symmetry center (the G75-A101 pair) by freely diffusing ligands, like charged chemical groups belonging to proteins or drugs. Indeed, in a complex of L25 with a 5S rRNA fragment studied by Lu and Steitz (47), an amino group of L25 binds close to U80. Therefore, polyamine binding to this site may modulate directly the interaction of 5S rRNA with protein L25. In contrast, sites located close to the symmetry center display high electronegative potential. This implies that ions associated with these regions may be more difficult to displace. In line, we observed that binding of ABA-spermine to A73 is highly resistant to competition by monovalent cations. Other examples of cross-linking, which may interfere in protein binding, are related to helices II and III, and loops B and C. Crystallographic studies (6–8) as well as foot printing and cross-linking data [reviewed in (9)] have indicated that these regions constitute the primary binding sites for proteins L5 and L18. Detailed analysis of Thermus thermophilus ribosomal protein L5 in complex with a 34-nt fragment comprising helix III and loop C of E. coli 5S rRNA revealed that nucleosides C42 and C43 are involved in both hydrophobic interactions and hydrogen bonding with no polar and side chain atoms of L5, respectively (50). It is noteworthy that these residues of loop C are susceptible to ABA-spermine cross-linking, regardless of the 5S rRNA assembly status or the photoprobe concentration.
Photolabeling of 5S rRNA with 50 μM ABA-spermine causes profound changes in the susceptibility of several nucleosides in 5S rRNA against DMS (Table 1). Therefore, it is tempting to suggest that binding of spermine to 5S rRNA promotes changes in its folding. Namely, loop A adopts an apparent ‘loosening’ of its structure, while loops C, D, E and helix III achieve a more tight structure. These findings are consistent with previous observations obtained by means of calorimetric studies (24,25). Photolabeling of 5S rRNA with 300 μM ABA-spermine results to a conformer, which does not meet the operational definition of the ‘A’ form (51). 50S subunits reconstituted from 5S rRNA modified in this way associate with native small ribosomal subunits at low yield (Table 2) and are functionally inactive.
The yield of the 50S rRNA subunit reconstitution obtained by 5S rRNA photolabeled with 50 μM ABA-spermine was found to be approximately half that achieved with wild-type 5S rRNA. Kakegawa et al. (52) indicated that the in vitro reconstitution of 50S particles from 23S rRNA, 5S rRNA and TP50 is not influenced greatly by polyamines. On the other hand, there is evidence that 5S rRNA undergoes conformational changes during its assembly into the central protuberance of the 50S subunit (37). In agreement, we observed that some of the ABA-spermine crosslinks in 5S rRNA get more pronounced in the 50S subunit than in the isolated state, clearly indicating that 5S rRNA undergoes conformational changes upon incorporation into ribosomes. Therefore, it is tempting to suggest that ABA-spermine cross-linking, although not directly affecting the reconstitution process, may hinder the required conformational changes by stabilizing a rigid conformation in 5S rRNA. In support of this hypothesis, labeling of 5S rRNA with 300 μM ABA-spermine reduces further its incorporation into 50S subunits. In contrast to 50S subunits containing 5S rRNA photolabeled with ABA-spermine at 300 μM, those assembled from 5S rRNA photolabeled with ABA-spermine at 50 μM were fully active in associating with native 30S subunits.
The loop B →loop C arm in 5S rRNA is the most affected region by ABA-spermine photolabeling. A great body of experimental data has previously demonstrated that this region participates in several signal transmission chains connecting 5S rRNA with P-site-bound tRNA. For instance, this region binds protein L5 which interacts with the T-loop of P-site bound tRNA (5–8,19,50). Our data show that photolabeling of 5S rRNA with 50 μM ABA-spermine results in a subtle but visible stimulation of the AcPhe-tRNA binding to the P-site. Binding to the A-site is not affected, albeit loop B →loop C arm is near the tip of H38 of 23S rRNA (15) which communicates via protein S13 of the small subunit (bridge B1a) with the anticodon region of A-site-bound tRNA. In fact, several studies revealed that loss of contacts between protein S13 and H38 has little consequences on tRNA binding (53–56). Whole ribosomes containing either wild-type or photolabeled 5S rRNA, exhibit greater capacity for AcPhe-tRNA binding to the A-site in the presence of free spermine than in its absence. In this case, however, 30S subunit and AcPhe-tRNA also interact with spermine, a fact that favors tRNA binding (32). In agreement with previous findings (23), we observed that omission of 5S rRNA from the large subunit suppresses AcPhe-tRNA binding, in particular to the A-site. Such ribosomes, even though provided with free spermine, cannot recover from the lack of 5S rRNA.
Ribosomes containing photolabeled 5S rRNA exhibit 30% higher PTase activity than those containing wild-type 5S rRNA. Catalysis of peptide bond formation is mediated almost entirely through precise alignment of the A- and P-substrates within the active center, coupled to substrate-assisted catalysis (57). Therefore, it is tempting to suggest that besides the effect of the 5S rRNA photolabeling on the affinity of P-site, the signals transmitted from 5S rRNA via protein L5 to the P-site may also affect the functional positioning of AcPhe-tRNA within the catalytic center. Since the affinity of puromycin for the A-site is not altered upon 5S rRNA photolabeleling, analogous effects on the A-site cannot be postulated. Ribosomes deprived of 5S rRNA retain 10% of their PTase activity, in agreement with previous observations according to which 5S rRNA enhances ribosomal activity (22,23) but is not absolutely essential for it (58).
Spontaneous translocation is an inherent property of the ribosome itself, even though extremely slow (59,60). This is verified by the present work, also showing that reversible interaction of spermine with whole ribosomes or covalent binding of ABA-spermine to 5S rRNA stimulate EF-G-independent translocation. This prompted us to suggest that certain changes in the folding of ribosomal RNA may be beneficial to the translocation of tRNAs. Supporting evidence is provided by DMS-protection results, indicating that the region around the G1338-U1341 ridge in the small-subunit head and A790 terminal loop in the platform achieves a more ‘open'structure upon ABA-spermine photo-incorporation. Crystallographic studies in E. coli ribosomes have postulated that the gap left between the G1338-U1341 ridge and A790 operates as a ‘lock’, controlling the passage of the P-site tRNA anticodon stem-loop (8,61). Steady-state data derived in the present work show that translocation in the presence of high concentrations of EF-G and GTP is fast, even in the absence of polyamines. Therefore, monitoring the ABA-spermine cross-linking effect on EF-G-dependent translocation is impossible, without using rapid kinetic techniques. Following an alternative approach, we found that interaction of spermine with the whole ribosome or with its 5S rRNA has a sparing effect on EF-G requirements. Further experiments revealed that photoincorporation of ABA-spermine into 5S rRNA does not affect the initial rate of EF-G catalyzed GTP hydrolysis by 70S ribosomes, but enhances the binding of EF-G to ribosomes. In agreement with previous observations (22,23), we found that ribosomes deprived of 5S rRNA retain some EF-G-dependent GTPase activity but are unable to bind efficiently EF-G, even in the presence of spermine. The EF-G binding site in ribosome consists of the GTPase-associated center (GAC) located in domain II of 23S rRNA (helices H42-H44) and the sarcin–ricin loop (SRL) located in domain VI of 23S rRNA (helix H95) (1). Direct contacts of 5S rRNA with GAC, SRL or EF-G have not been identified so far in E. coli ribosomes. Nevertheless, accumulated evidence suggests that several allosteric signal transmission pathways set 5S rRNA off coordinating the PTase reaction with subsequent EF-G·GTP binding and GTP hydrolysis (12–18,62).
In conclusion, a comprehensive view of the 5S rRNA nucleotide residues involved in spermine binding is obtained by the present study. The results suggest that binding of spermine to 5S rRNA causes conformational changes in certain regions of the molecule. Consequently, these changes have beneficial effects on the ribosome functioning. The general improvement observed in various functional tests is an important argument that the identified sites in 5S rRNA may have an important functional relevance to the overall active structure of the ribosome.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Dennis Synetos for critical reading of the manuscript and Panagiotis Karahalios for assistance with 5S rRNA preparation. We are also grateful to K. H. Nierhaus for providing us with EF-G, and Spyridoula Agelakopoulou for technical advice. This work was supported by a grant from European Social Fund, Operational Program for Educational and Vocational Training II (program IRAKLEITOS). Funding to pay the Open Access publication charges for this article was provided by the University of Patras.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wilson DN, Blaha G, Connell SR, Ivanov PV, Jenke H, Stelzl U, Teraoka Y, Nierhaus KH. Protein synthesis at atomic resolution: mechanistics of translation in the light of highly resolved structures for the ribosome. Curr. Protein Pept. Sci. 2003;3:1–53. doi: 10.2174/1389203023380846. [DOI] [PubMed] [Google Scholar]
- 2.Szymanski M, Barciszewska MZ, Erdmanm VA, Barciszewski J. 5S rRNA: structure and interactions. Biochem. J. 2003;371:641–651. doi: 10.1042/BJ20020872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kierzek E, Kierzek R, Turner DH, Catrina IE. Facilitating RNA structure prediction with microarrays. Biochemistry. 2006;45:581–593. doi: 10.1021/bi051409+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matadeen R, Patwardhan A, Gowen B, Orlova EV, Pape T, Cuff M, Mueller F, Brimacombe R, van Heel M. The Escherichia coli large ribosomal subunit at 7.5 Å resolution. Structure. 1999;7:1575–1583. doi: 10.1016/s0969-2126(00)88348-3. [DOI] [PubMed] [Google Scholar]
- 5.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 6.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JHD, Noller HF. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 7.Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, Bartels H, Franceschi F, Yonath A. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001;107:679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- 8.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JHD. Structure of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 9.Mueller F, Sommer I, Baranov P, Matadeen R, Stoldt M, Wöhnert J, Görlach M, van Heel M, Brimacombe R. The 3D arrangement of the 23S and 5S rRNA in the Escherichia coli 50S ribosomal subunit based on a cryo-electron microscopic reconstruction at 7.5 Å resolution. J. Mol. Biol. 2000;298:35–59. doi: 10.1006/jmbi.2000.3635. [DOI] [PubMed] [Google Scholar]
- 10.Gao H, Sengupta J, Valle M, Korostelev A, Eswar N, Stagg SM, van Roey P, Agrawal RK, Harvey SC, et al. Study of the structural dynamics of the E. coli 70S ribosome using real-space refinement. Cell. 2003;113:789–801. doi: 10.1016/s0092-8674(03)00427-6. [DOI] [PubMed] [Google Scholar]
- 11.Skibinska L, Banachowicz E, Gapiński J, Patkowski A, Barciszewski J. Structural similarity of E. coli 5S rRNA in solution and within the ribosome. Biopolymers. 2004;73:316–325. doi: 10.1002/bip.10598. [DOI] [PubMed] [Google Scholar]
- 12.Dontsova O, Tishkov V, Dokudovskaya S, Bogdanov A, Dőring T, Rinke-Appel J, Thamm S, Greuer B, Brimacombe R. Stem-loop IV of 5S rRNA lies close to the peptidyltransferase center. Proc. Natl Acad. Sci., USA. 1994;91:4125–4129. doi: 10.1073/pnas.91.10.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dokudovskaya S, Dontsova O, Shpanchenko O, Bogdanov A, Brimacombe R. Loop IV of 5S ribosomal RNA has contacts both to domain II and to domain V of the 23S rRNA. RNA. 1996;2:146–152. [PMC free article] [PubMed] [Google Scholar]
- 14.Sergiev P, Dokudovskaya S, Romanova E, Topin A, Bogdanov A, Brimacombe R, Dontsova O. The environment of 5S rRNA in the ribosome: cross-links to the GTPase associated area of 23S rRNA. Nucleic Acids Res. 1998;26:2519–2525. doi: 10.1093/nar/26.11.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osswald M, Brimacombe R. The environment of 5S rRNA in the ribosome: cross-links to 23S rRNA from sites within helices II and III of the 5S molecule. Nucleic Acids Res. 1999;27:2283–2290. doi: 10.1093/nar/27.11.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Østergaard P, Phan H, Johansen LB, Egebjerg J, Østergaard L, Porse BT, Garrett RA. Assembly of proteins and 5S rRNA to transcripts of the major structural domains of 23S rRNA. J. Mol. Biol. 1998;284:227–240. doi: 10.1006/jmbi.1998.2185. [DOI] [PubMed] [Google Scholar]
- 17.Sergier PV, Bogdanov AA, Dahlberg AE, Dontsova O. Mutations at position A960 of E. coli 23S ribosomal RNA influence the structure of 5S ribosomal RNA and the peptidyltrasferase region of 23S ribosomal RNA. J. Mol. Biol. 2000;299:379–389. doi: 10.1006/jmbi.2000.3739. [DOI] [PubMed] [Google Scholar]
- 18.Ko J, Lee Y, Park I, Cho B. Identification of a structural motif of 23S rRNA interacting with 5S rRNA. FEBS Lett. 2001;508:300–304. doi: 10.1016/s0014-5793(01)03068-x. [DOI] [PubMed] [Google Scholar]
- 19.Smith MW, Meskauskas A, Wang,F. Sergiev PV, Dinman JD. Saturation mutagenesis of 5S rRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 2001;21:8264–8275. doi: 10.1128/MCB.21.24.8264-8275.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiparisov S, Petrov A, Meskauskas A, Sergiev P, Dontsova OA, Dinman JD. Structural and functional analysis of 5S rRNA in Saccharomyces cerevisiae. Mol. Genet. Genomics. 2005;274:235–247. doi: 10.1007/s00438-005-0020-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khaitovich P, Mankin AS. Effect of antibiotics on large ribosomal subunit assembly reveals possible function of 5S rRNA. J. Mol. Biol. 1999;291:1025–1034. doi: 10.1006/jmbi.1999.3030. [DOI] [PubMed] [Google Scholar]
- 22.Erdmann VA, Fahnestock S, Higo K, Nomura M. Role of 5S rRNA in the functions of 50S ribosomal subunits. Proc. Natl Acad. Sci. USA. 1971;68:2932–2936. doi: 10.1073/pnas.68.12.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dohme F, Nierhaus KH. Role of 5S rRNA in assembly and function of the 50S subunit from Escherichia coli. Proc. Natl Acad. Sci. USA. 1976;73:2221–2225. doi: 10.1073/pnas.73.7.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kao TH, Crothers DM. A proton-coupled conformational switch of Escherichia coli 5S ribosomal RNA. Proc. Natl Acad. Sci. USA. 1980;77:3360–3364. doi: 10.1073/pnas.77.6.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barciszewski J, Bratek-Wiewiórowska MD, Górnicki P, Nasket-Barciszewska M, Wiewiórowski M, Zielenkiewicz A, Zielenkiewicz W. Comparative calorimetric studies on the dynamic conformation of plant 5S rRNA. I. Thermal unfolding pattern of lupin seeks and wheat germ 5S rRNAs, also in the presence of magnesium and sperminium cations. Nucleic Acids Res. 1988;16:685–701. doi: 10.1093/nar/16.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clark E, Swank RA, Morgan JE, Basu H, Matthews HR. Two new photoaffinity polyamines appear to alter the helical twist of DNA in nucleosome core particles. Biochemistry. 1991;30:4009–4020. doi: 10.1021/bi00230a028. [DOI] [PubMed] [Google Scholar]
- 27.Amarantos I, Xaplanteri MA, Choli-Papadopoulou T, Kalpaxis DL. Effects of two photoreactive spermine analogues on peptide bond formation and their application for labeling proteins in Escherichia coli functional ribosomal complexes. Biochemistry. 2001;40:7641–7650. doi: 10.1021/bi010010s. [DOI] [PubMed] [Google Scholar]
- 28.Xaplanteri MA, Petropoulos AD, Dinos GP, Kalpaxis DL. Localization of spermine binding sites in 23S rRNA by photoaffinity labeling: parsing the spermine contribution to ribosomal 50S subunit functions. Nucleic Acid Res. 2005;33:2792–2805. doi: 10.1093/nar/gki557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dinos G, Wilson DN, Teraoka Y, Szaflarski W, Fucini P, Kalpaxis DL, Nierhaus KH. Dissecting the ribosomal inhibition mechanisms of edeine and pactamycin: the universally conserved residues G693 and C795 regulate P-site tRNA binding. Mol. Cell. 2004;13:113–124. doi: 10.1016/s1097-2765(04)00002-4. [DOI] [PubMed] [Google Scholar]
- 30.Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 31.Bruce AG, Uhlenbeck OC. Reactions at the termini of tRNA with T4 RNA ligase. Nucleic Acids Res. 1978;5:3665–3677. doi: 10.1093/nar/5.10.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amarantos I, Zarkadis IK, Kalpaxis DL. The identification of spermine binding sites in 16S rRNA allows interpretation of the spermine effect on ribosomal 30S subunit functions. Nucleic Acids Res. 2002;30:2832–2843. doi: 10.1093/nar/gkf404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stern S, Moazed D, Noller HF. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1998;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- 34.Amarantos I, Kalpaxis DL. Photoaffinity polyamines: interactions with AcPhe-tRNA free in solution or bound at the P-site of Escherichia coli ribosomes. Nucleic Acids Res. 2000;28:3733–3742. doi: 10.1093/nar/28.19.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zavialov AV, Ehrenberg M. Peptidyl-tRNA regulates the GTPase activity of translation factors. Cell. 2003;114:113–122. doi: 10.1016/s0092-8674(03)00478-1. [DOI] [PubMed] [Google Scholar]
- 36.Sergiev PV, Lesnyak DV, Burakovsky DE, Kiparisov SV, Leonov AA, Bogdanov AA, Brimacombe R, Dontsova OA. Alteration in location of a conserved GTPase-associated center of the ribosome induced by mutagenesis influences the structure of peptidyltransferase center and activity of elongation factor G. J. Biol. Chem. 2005;280:31882–31889. doi: 10.1074/jbc.M505670200. [DOI] [PubMed] [Google Scholar]
- 37.Shpanchenko OV, Dontsova OA, Bogdanov AA, Nierhaus KH. Structure of 5S rRNA within the Escherichia coli ribosome: iodine-induced cleavage patterns of phosphorothioate derivatives. RNA. 1998;4:1154–1164. doi: 10.1017/s1355838298980359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharma D, Southworth DR, Green R. EF-G-independent reactivity of a pre-translocation-state ribosome complex with the aminoacyl tRNA substrate puromycin supports an intermediate (hybrid) state of tRNA binding. RNA. 2004;10:102–113. doi: 10.1261/rna.5148704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bowen WS, Van Dyke N, Murgola EJ, Lodmell JS, Hill WE. Interaction of thiostrepton and elongation factor-G with the ribosomal protein L11-binding domain. J. Biol. Chem. 2005;280:2934–2943. doi: 10.1074/jbc.M407008200. [DOI] [PubMed] [Google Scholar]
- 40.Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature. 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- 41.Bernabeu C, Vazquez D, Ballesta JPG. Proteins associated with rRNA in the Escherichia coli ribosomes. Biochim. Biophys. Acta. 1978;518:290–297. doi: 10.1016/0005-2787(78)90185-5. [DOI] [PubMed] [Google Scholar]
- 42.Kakegawa T, Sato E, Hirose S, Igarashi K. Polyamine binding sites on Escherichia coli ribosomes. Arch. Biochem. Biophys. 1986;251:413–420. doi: 10.1016/0003-9861(86)90347-4. [DOI] [PubMed] [Google Scholar]
- 43.Cohen SS. A Guide to Polyamines. New York, NY: Oxford University Press; 1998. [Google Scholar]
- 44.Tabor CW, Tabor H. 1,4-diaminobutane (putrescine), spermidine and spermine. Annu. Rev. Biochem. 1976;45:285–306. doi: 10.1146/annurev.bi.45.070176.001441. [DOI] [PubMed] [Google Scholar]
- 45.Kamekura M, Hamana K, Matsuzaki S. Polyamine contents and amino acid decarboxylation activities of extremely halophilic archaebacteria and some eubacteria. FEMS Microbiol. Lett. 1987;43:301–305. [Google Scholar]
- 46.Correll CC, Freeborn B, Moore PB, Steitz TA. Metals, motifs, and recognition in the crystal structure of a 5S rRNA domain. Cell. 1997;91:705–712. doi: 10.1016/s0092-8674(00)80457-2. [DOI] [PubMed] [Google Scholar]
- 47.Lu M, Steitz TA. structure of Escherichia coli ribosomal protein L25 complexed with 5S rRNA fragment at 1.8-Å resolution. Proc. Natl Acad. Sci. USA. 2000;97:2023–2028. doi: 10.1073/pnas.97.5.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Réblová K, Špačková N, Štefl R, Csaszar K, Koča J, Leontis NB, Šponer J. Non-Watson-Crick basepairing and hydration in RNA motifs: molecular dynamics of 5S rRNA loop E. Biophys. J. 2003;84:3564–3582. doi: 10.1016/S0006-3495(03)75089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Auffinger P, Bielecki L, Westhof E. Symmetric K+ and Mg2+ ion-binding sites in the 5S rRNA loop E inferred from molecular dynamics simulations. J. Mol. Biol. 2004;335:555–571. doi: 10.1016/j.jmb.2003.10.057. [DOI] [PubMed] [Google Scholar]
- 50.Perederina A, Nevskaya N, Nikonov O, Nikulin A, Dumas P, Yao M, Tanaka I, Garber M, Gongadze G, et al. Detailed analysis of RNA-protein interactions within the bacterial ribosomal protein L5/5S rRNA complex. RNA. 2002;8:1548–1557. [PMC free article] [PubMed] [Google Scholar]
- 51.Göringer HU, Szymkowiak C, Wagner R. Escherichia coli 5S RNA A characterization by enzymatic and chemical methods and B conformers. Eur. J. Biochem. 1984;144:25–34. doi: 10.1111/j.1432-1033.1984.tb08426.x. [DOI] [PubMed] [Google Scholar]
- 52.Kakegawa T, Hirose S, Kashiwagi K, Igarashi K. Effect of polyamines on in vitro reconstitution of ribosomal subunits. Eur. J. Biochem. 1986;158:265–269. doi: 10.1111/j.1432-1033.1986.tb09746.x. [DOI] [PubMed] [Google Scholar]
- 53.Cukras AR, Green R. Multiple effects of S13 in modulating the strength of intersubunit interactions in the ribosome during translation. J. Mol. Biol. 2005;349:47–59. doi: 10.1016/j.jmb.2005.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liiv A, O′Connor M. Mutations in the intersubunit bridge regions of 23S rRNA. J. Biol. Chem. 2006;281:29850–29862. doi: 10.1074/jbc.M603013200. [DOI] [PubMed] [Google Scholar]
- 55.Sergiev PV, Kiparisov SV, Burakovsky DE, Lesnyak DV, Leonov AA, Bogdanov AA, Dontsova OA. The conserved A-site finger of the 23S rRNA: just one of the inter subunit bridges or a part of the allosteric communication pathway? J. Mol. Biol. 2005;353:116–123. doi: 10.1016/j.jmb.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 56.Komoda T, Sato NS, Phelps SS, Namda N, Joseph S, Suzuki T. The A-site finger in 23S rRNA acts as a functional attenuator for translocation. J. Biol. Chem. 2006;281:32303–32309. doi: 10.1074/jbc.M607058200. [DOI] [PubMed] [Google Scholar]
- 57.Rodnina MV, Beringer M, Wintermeyer W. How ribosomes make peptide bonds. Trends Biochem. Sci. 2007;32:20–26. doi: 10.1016/j.tibs.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 58.Schulze H, Nierhaus KH. Minimal set of ribosomal components for reconstitution of the peptidyltransferase activity. EMBO J. 1982;1:609–613. doi: 10.1002/j.1460-2075.1982.tb01216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bergemann K, Nierhaus KH. Spontaneous, elongation factor G independent translocation in Escherichia coli ribosomes. J. Biol. Chem. 1983;258:15105–15113. [PubMed] [Google Scholar]
- 60.Southworth DR, Brunelle JL, Green R. EFG-independent translocation of the mRNA: tRNA complex is promoted by modification of the ribosome with thiol-specific reagents. J. Mol. Biol. 2002;324:611–623. doi: 10.1016/s0022-2836(02)01196-8. [DOI] [PubMed] [Google Scholar]
- 61.Berk V, Zhang W, Pai RD, Cate JHD. Structural basis for mRNA and tRNA positioning on the ribosome. Proc. Natl Acad. Sci. USA. 2006;103:15830–15834. doi: 10.1073/pnas.0607541103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chan Y-L, Dresios J, Wool IG. A pathway for the transmission of allosteric signals in the ribosome through a network of RNA tertiary interactions. J. Mol. Biol. 2006;355:1014–1025. doi: 10.1016/j.jmb.2005.11.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.