Abstract
Genomic RNA circularization has been proposed for several RNA viruses. In this study, we examined if the 5′ and 3′ ends of the 9-kb HIV-1 RNA genome can interact. In vitro assays demonstrated a specific interaction between transcripts encompassing the 5′ and 3′ terminal 1 kb, suggesting that the HIV-1 RNA genome can circularize. Truncation of the transcripts indicated that the 5′–3′ interaction is formed by 600–700 nt in the gag open reading frame and the terminal 123 nt of the genomic RNA. Detailed RNA structure probing indicates that sequences flanking the 3′ TAR hairpin interact with complementary sequences in the gag gene. Phylogenetic analysis indicates that all HIV-1 subtypes can form the 5′/3′ interaction despite considerable sequence divergence, suggesting an important role of RNA circularization in the HIV-1 replication cycle.
INTRODUCTION
RNA structures play important roles in the HIV-1 life cycle and several structured motifs are clustered in the untranslated regions (UTR) that are present at both ends of the viral RNA. The 5′UTR contains the repeat region (R), U5 region (unique at the 5′ terminus) and the leader sequence. The R region folds the TAR and the polyA hairpins (Figure 1), and TAR binds the viral Tat protein to activate transcription (1–3). Among many other RNA signals, the dimer initiation site (DIS) is located in the HIV-1 leader RNA and forms a stem-loop structure with a palindromic sequence. Direct base pairing between the loop-exposed palindromes of two DIS hairpins results in RNA dimerization. The 3′UTR is subdivided into the U3 (unique at the 3′ terminus) and the R region. The 3′R also folds the TAR hairpin, but the polyA hairpin is shortened due to polyadenylation (Figure 1).
Figure 1.
Organization of the HIV-1 genomic RNA. The HIV-1 genomic RNA, the untranslated R, U5, U3 regions and all open reading frames are indicated. The secondary structures of the 5′ and 3′R domains are shown on top. To map the sequences involved in the 5′/3′ interaction we made transcripts that are truncated from the 3′ terminus by 100 nt steps. Their ability to form the 5′/3′ interaction is indicated by +/−.
HIV-1 full-length RNA serves both as mRNA for the Gag-Pol proteins and as genomic RNA that is packaged in virion particles. It therefore contains characteristics for both functions. HIV-1 genomic RNA is 5′-capped and 3′-polyadenylated like cellular mRNAs, but it performs some unique virus-specific functions, such as dimerization, packaging in virion particles and reverse transcription. Reverse transcription is initiated within the leader RNA and the initial copy DNA of the 5′R is transferred to the 3′ to allow copying of the full-length genome (4,5). It has been suggested that the 5′/3′ ends should be close together in virions to facilitate this obligatory strand transfer step (4).
Cellular mRNAs and the RNA genomes of several viruses were demonstrated to have a 5′/3′ interaction (6–12). Specifically, genome circularization plays a role in RNA viruses that lack the cap structure (the Picornaviridae family), the polyA tail (the Reoviridae family) or both elements (the Flaviridae family). In some cases, a protein bridge is involved in RNA circularization, e.g. for members of the Picornaviridae and the Reoviridae family (8,10,13,14). Sequences in the 5′ and 3′ end of the RNA genome base pair in members of the Flaviridae family and the LTR-retrotransposon Ty1 (6,7,9,11,12). The retrotransposon Ty1 shares several features with HIV-1, including its genome organization and the process of reverse transcription. Ty1 RNA circularization was shown to be involved in reverse transcription initiation (7). It has been suggested that RNA circularization is a common feature of all RNA viruses (10). We therefore set out to test this for HIV-1 RNA using in vitro assays.
MATERIALS AND METHODS
DNA templates for in vitro transcription
The full-length HIV-1 molecular clone pLAI was used as a template for PCR amplification of certain genome regions (15). Mutant GC1 contains a GCGC deletion in the palindromic sequence of the DIS hairpin (16). The DNA templates for in vitro transcription were obtained by PCR amplification with sense primers with 5′-flanking T7 RNA polymerase promoter sequence and antisense primers (Table 1). The PCR fragments were ethanol-precipitated and dissolved in water.
Table 1.
PCR primers used in this study
| Name | Positiona | Primer sequence (5′–3′)b |
|---|---|---|
| T7-2 | 1–20 | TAATACGACTCACTATAGGG |
| TCTCTCTGGTTAGACCAG | ||
| T7-iscan | 8206–8226 | CTAATACGACTCACTATAGGG |
| AATCTCCTACAGTATTGG | ||
| T7scanG | 601–620 | CTAATACGACTCACTATAGGG |
| ATCAAAGGATAGAGATAA | ||
| T7-1 | 9100–9181 | CTAATACGACTCACTATAGGG |
| AGTGGCGAGCCCTCAGATG | ||
| cir1 | 81–100 | ACTTGAAGCACTCAAGGCAA |
| cir2 | 181–200 | AGTCCCTGTTCGGGCGCCAC |
| cir3 | 281–300 | GGCGTACTCACCAGTCGCCT |
| cir4 | 381–400 | CTTAACCGAATTTTTTCCCA |
| cir5 | 481–500 | TTCTGATGTTTCTAACAGGC |
| cir6 | 581–600 | GCACACAATAGAGGGTTGCT |
| cir7 | 681–700 | GTGTCAGCTGCTGCTTGCTG |
| cir8 | 781/800 | CCATCGATTTAAAGTTCTAG |
| cir9 | 881/900 | TTAGCATGGTGTTTAAATCT |
| scanA | 1016–1036 | ATGAGAGAACCAAGGGGAAG |
| cira | 8306–8326 | ATAACCCTATCTCTCCCCTC |
| cirb | 8406–8426 | GACCACTTGCCACCCATCTT |
| circ | 8506–8526 | TTTTTCCAGGTCTCGAGATG |
| cird | 8606–8626 | GAGGTGTGACTGGAAAACCC |
| cire | 8706–8726 | CTTCGTTGGGAGTGAATTAG |
| cirf | 8806–8826 | AAAGGTCAGTGGATATCTAG |
| cirg | 8906–8926 | TTCCATGCAGGCTCACAGGG |
| cirh | 9006–9026 | CAGTTCTTGAAGTACTCCGG |
| ciri | 9106–9126 | TATGCAGCATCTGAGGGCTC |
| scani | 9209–9229 | AGCTTGCCTTGAGTGCTTCA |
adenotes the coresponding nucleotide position of the HIV-1 LAI genome. bT7-promotor is underlined; the transcription site is indicated in italic.
In vitro transcription
In vitro transcription was performed with the Ambion MegaShortscript T7 transcription kit according to the manufacturer's instructions. The transcripts were separated on a 6% denaturing polyacrylamide gel containing urea, subsequently excised after visualization by UV shadowing and eluted from the gel fragments by overnight incubation at 30°C in 1× TBE. The RNA was subsequently ethanol-precipitated and dissolved in water. The transcripts were purified by Nucaway columns (Ambion). The quality of the unlabeled RNA was analyzed on a 2% agarose gel containing ethidium bromide, and the concentration of the RNA was determined by UV spectrometry (Smartspec™ 3000, Bio-Rad).
End-labeled transcripts
For 5′ end-labeling, the transcripts were dephosphorylated and end-labeled with [γ-P32]-ATP (Amersham) according to the KinaseMax (Ambion) protocol. Transcript 3′ end-labeling was performed with T4 RNA ligase (Ambion) and [32P]pCp (Amersham) according to the 3′ end-labeling protocol available at the Ambion website. End-labeled transcripts were purified on NucAway Spin Columns (Ambion).
Electrophoretic mobility shift assay
Electrophoretic mobility shift assays were performed in 20 μl reactions, containing a physiological buffer (final concentration: 125 mM K acetate, 2.5 mM Mg acetate, 25 mM HEPES, pH 7.0), ∼200 counts/s of 32P-labeled transcript and different concentrations of unlabeled transcripts (end concentrations of 0, 5, 20, 80 and 320 nM). This mixture was heated at 85°C for 2 min to denature the transcripts and slowly cooled to room temperature for refolding. After addition of 4 μl non-denaturing loading buffer (25% glycerol with bromophenol blue dye), 10 μl was loaded on a 4% 0.25× TBM non-denaturing polyacrylamide gel (Boehringer Mannheim). Electrophoresis was performed at 150 V for at least 3 h for short transcripts or 100 V overnight for large transcripts. Subsequently, gels were dried and exposed overnight to a PhosphorImager screen. Quantification was performed with the ImageQuant software.
RNA stability measurements
RNA complexes were formed in 200 μl physiological buffer (final concentration: 125 mM K acetate, 2.5 mM Mg acetate, 25 mM HEPES, pH 7.0), ∼2000 counts/s of 32P-labeled 3′ transcript with unlabeled transcript (final concentration 320 nM). This mixture was heated at 85°C for 2 min to denature the transcripts and slowly cooled to room temperature for refolding. The mixture was split in 20 μl samples and kept on ice. Non-radiolabeled 3′ transcript was added (320 nM) to prevent regeneration of the labeled gag–U3R interaction upon melting. To determine the melting temperature, the samples were incubated at different temperatures for 5 min and subsequently frozen in dry ice. After addition of 4 μl loading buffer (25% glycerol with bromophenol blue dye), 10 μl was loaded on a 4% 0.25× TBM non-denaturing polyacrylamide gel (Boehringer Mannheim). Electrophoresis was performed at 150 V for 3 h. Subsequently, gels were dried and exposed overnight to a PhosphorImager screen. Quantification was performed with the ImageQuant software.
RNA structure probing
Radiolabeled 5′ or 3′ transcripts (1000 count/s) were either incubated alone or mixed with non-radiolabeled 5′ or 3′ transcript (320 nM) in 90 μl physiological salt buffer (final concentration: 125 mM K acetate, 2.5 mM Mg acetate, 25 mM HEPES, pH 7.0) at 85°C for 2 min and slowly cooled to room temperature. The sample was split into 4 × 20 μl and kept at room temperature. Lead acetate (5 mM final concentration) was added for 2.5, 5 and 10 min. Cleavage by lead ions was stopped by addition of EDTA (100 mM final concentration) and one volume of formamide containing loading buffer. A hydrolysis ladder was made by incubating the radiolabeled transcripts in hydrolysis buffer (50 mM Na2CO3/NaHCO3, pH 9.2) at 95°C for 5, 10 and 15 min, followed by addition of loading buffer. All samples were heated at 95°C for 2 min and separated on a 6% denaturing acrylamide gel at 50 W for 3 h. The gel was dried and exposed overnight to a PhosphorImager screens. Gels were analyzed by ImageQuant.
RESULTS
5′/3′ interaction in HIV-1 RNA
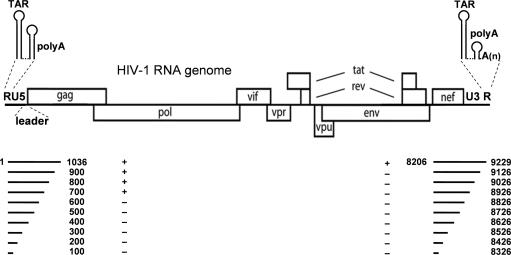
To determine if RNA circularization occurs in the HIV-1 genome, we screened for an interaction between two transcripts encompassing the 5′- and the 3′-terminal 1 kb of the HIV-1 genome, fragments 1–1036 and 8206–9229 (Figure 1). To prevent homodimerization of the 5′ transcript, we used the GC1 mutant with a deletion in the loop of the DIS hairpin. This mutation has previously been demonstrated to effectively block in vitro HIV-1 RNA dimerization (17). We first checked for the lack of homodimerization. Incubation of the radiolabeled 5′ transcript with increasing amounts of non-radiolabeled 5′ transcript in physiological salt buffer and analysis on a non-denaturing gel did not show homodimers (Figure 2, first 5 lanes), demonstrating the effectiveness of the DIS mutation. The 3′ transcript is not expected to form homodimers, although a small amount of slow-migrating complexes are observed (20%) at the highest RNA concentration (290 nM, lane 10). We next mixed the 5′ and 3′ transcripts. The mixture was heated in physiological salt buffer to 75°C and slowly cooled to room temperature and analyzed on gel. The radiolabeled 5′ transcript was incubated with increasing amounts of non-labeled 3′ transcript, and the radiolabeled 3′ transcript was incubated with an increasing amount of non-labeled 5′ transcript. Both experiments show an apparent band shift signal (Figure 2, marked by arrow). The radiolabeled RNA was efficiently shifted in this complex at relatively low concentration of the non-labeled RNA. The formation of homodimers and heterodimers was quantified as shown in Figure 2B. These results indicate that the 5′ and 3′ ends of HIV-1 genomic RNA can interact.
Figure 2.
The 5′/3′ interaction within HIV-1 RNA. Electrophoretic mobility shift assay of 5′ and 3′ 1-kb HIV-1 transcripts. (A) Radiolabeled 5′ and 3′ 1-kb transcripts were incubated with increasing amounts of the equivalent non-radiolabeled transcripts to check for homodimerization (first 10 lanes). To analyze whether the 5′ and the 3′ transcripts interact, the radiolabeled 5′ 1-kb transcripts were incubated with increasing amounts of non-radiolabeled 3′ 1-kb transcripts and the radiolabeled 3′ 1-kb transcripts were incubated with increasing amounts of non-radiolabeled 5′ 1-kb transcripts (last 10 lanes). (B) The efficiency of dimer formation was calculated by dividing the amount of shifted transcripts by the total amount of transcripts. Three independent experiments were quantified.
Mapping the 5′/3′ interaction domains
To map the sequences involved in the 5′/3′ interaction, we made a 3′-truncated set of both transcripts with steps of 100 nt (Figure 1). The radiolabeled 5′ set is shown upon gel analysis in the left panel of Figure 3A. These transcripts were subsequently incubated with non-labeled 3′ 1-kb transcript (320 nM) (Figure 3A, right panel). Only the four largest 5′ transcripts are able to interact with the 3′ transcript, yielding a prominent band shift (marked with arrow). Transcripts of 600 nt or shorter do not interact, indicating that the interaction domain requires sequences between position 600 and 700 of the gag coding sequence. The 3′-truncated set of 3′ transcripts is shown in the left panel of Figure 3B. Upon incubation with non-labeled 5′ RNA (320 nM), only the largest transcript that includes the 3′ terminal 123 nt of HIV-1 genomic RNA is able to shift (Figure 3B, right panel). These interaction results are summarized in Figure 1 with +/− signs. The analysis implicates gag sequences (position 600–700) and 3′UTR sequences (position 9127–9229) in the 5′/3′ interaction. This 3′ domain encodes the U3 and R sequences, the latter includes the 3′ TAR and polyA hairpins (Figure 1).
Figure 3.
Mapping the minimal sequences that form the 5′/3′ interaction. (A) The 5′ 1-kb transcript was used to generate a set of radiolabeled transcripts with 3′ 100 nt truncations. These transcripts were either incubated alone (left panel) or with non-radiolabeled 3′ 1-kb transcripts (right panel). The arrow indicates the 5′/3′ complex (B). The 3′ 1-kb transcript was used to generate a set of radiolabeled transcripts with 3′ 100 nt truncations. These transcripts were either incubated alone (left panel) or with non-radiolabeled 5′ 1-kb transcripts (right panel). The results are summarized in Figure 1 with +/− signs.
Gag sequences interact with the extreme 3′ end of HIV-1 RNA
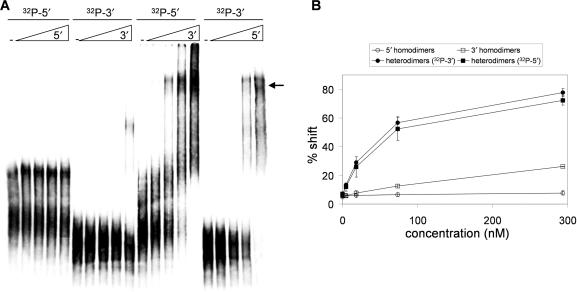
To confirm the involvement of nt 600–700 and 9127–9229 in heterodimer formation, we performed band shift assays with short transcripts encompassing these gag and U3R sequences. These radiolabeled transcripts do not form significant homodimer levels (Figure 4A), although the U3R transcript forms ∼10% slowly migrating complexes at the highest RNA concentration. This result is consistent with that of the band shift assay performed with the 1-kb transcripts. Most importantly, a prominent heterodimer complex is formed upon mixing of the transcripts (Figure 4A, marked with arrow). The signals were quantified and the percentage bandshift was calculated (Figure 4B). When the radiolabeled gag RNA is incubated with increasing amounts of non-radiolabeled 3′U3R RNA or vice versa, the transcripts shift efficiently (80 and 100%, respectively). These results demonstrate the minimal sequence requirements for the 5′/3′ interaction in gag (600–700) and U3R (9127–9229).
Figure 4.
Gag sequences interact with the extreme 3′ end of HIV-1 RNA. (A) Radiolabeled gag (600–700) and U3R (9127–9229) transcripts were incubated with increasing concentrations of the same non-radiolabeled transcripts to check for homodimerization (first 10 lanes). To test the 5′/3′ interaction, the radiolabeled gag transcript was incubated with increasing amounts of non-radiolabeled U3R transcript and vice versa (last 10 lanes). (B) The percentage gel shift was calculated by dividing the shifted transcript (see arrow) by the total amount of transcripts. The results of three independent experiments were quantified.
Melting temperature of the 5′/3′ complex
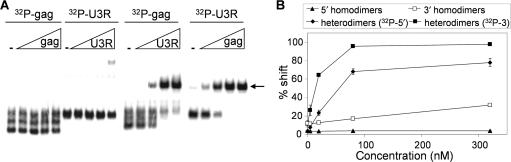
To further analyze whether the correct 5′/3′ interaction partners were mapped in the truncation series, we compared the melting temperature of the complexes formed with the 1-kb 5′/3′ transcripts versus the minimal gag/U3R domains. A large batch of the respective complexes was made in physiological salt buffer by heating to 75°C and slowly cooling to room temperature and samples were aliquoted. The samples were incubated at different temperatures for 5 min, snap-cooled and analyzed on a native acrylamide gel, of which the appropriate sections are shown (Figure 5). To prevent renewed complex formation after heat-induced dissociation, a large amount of non-radiolabeled RNA corresponding to the radiolabeled transcript was present. The Tm is ∼57°C for the complex with 1-kb transcripts and 55°C for the gag–U3R transcripts, indicating that essentially the same base-pairing interaction is formed. This result confirms that the minimal gag and U3R domains are sufficient for the 5′/3′ interaction.
Figure 5.
Melting temperature of the gag–U3R interaction. (A) Complexes were made with 5′ and 3′ 1-kb transcripts (left), but also with the minimal gag and U3R transcripts (right). Samples were incubated at the indicated temperatures for 5 min and quickly frozen in dry ice before gel analysis. (B) The fraction-shifted complex (see arrow) was calculated by dividing the shifted transcripts by the total transcript. The results of three independent experiments were quantified and the ratio of the samples incubated at 25°C was set at one.
RNA structure probing of the gag–U3R interaction
We next performed RNA structure probing of the minimal gag and U3R transcripts before and after complex formation. We used lead (Pb2+) acetate to specifically probe the single-stranded, unpaired nucleotides. The samples were subsequently denatured and separated on a sequencing gel. The Pb2+-induced cleavage pattern of the individual gag transcript shows reactivity along the entire transcript, indicating the absence of prominent secondary structure (Figure 6A). In the presence of unlabeled U3R transcript, a profound footprint is induced in the gag RNA. This footprint domain shows reduced Pb2+ reactivity in two regions (marked 1 and 3 in Figure 6A), with a hyperreactive region in between (marked 2).
Figure 6.
RNA structure probing of the gag–U3R interaction. (A) 5′ radiolabeled gag and U3R transcripts were either incubated alone or with the other non-radiolabeled U3R transcript as indicated (left panel). The same experiments were performed with 3′ radiolabeled transcripts (right panel). Subsequently, the transcripts were treated with lead acetate (5 mM) for different time periods (0, 2.5, 5 and 10 min). The lane denoted OH− indicates the hydrolysis ladder upon alkali treatment. The polyA, TAR and U3 sequences are indicated by black lines. The numbered stretches are discussed in the text and correlate with specific domains in panel B. (B) RNA base-pairing model of the gag–U3R interaction based on RNA structure probing. Gag 619–696 nt and U3R 9109–9229 nt (LAI coordinates) are shown. Blue arrows indicate nucleotides that are cleaved by lead ions and thus likely to be single stranded. Red circles indicate nucleotides that are cleaved by lead ions in monomeric RNA, but become protected upon gag–U3R complex formation. Blue circles indicate nucleotides that become more exposed upon gag–U3R interaction.
The Pb2+-induced cleavage pattern of the individual U3R transcript shows an extended domain that lacks sensitivity with a hyperreactive segment in the middle. This pattern correlates with the stable secondary structure of the 3′ TAR hairpin (1). The hyperreactivity of the TAR UUU-bulge is caused by the magnesium-binding pocket that strongly induces cleavage in the presence of Pb2+ ions (18). Upon mixing with the gag transcript, the TAR footprint remains unaltered, but several flanking sequences become protected (marked 4 and 6).
To analyze the Pb2+-induced cleavage pattern within the downstream half of the transcripts, we repeated the experiment with 3′ labeled transcripts (Figure 6A, right panel). This experiment revealed clear signals in the gag transcript that become protected upon complex formation (marked 1, 2, 7 and 9). Sequences that become protected were also observed in the U3R transcript (marked 4, 6 and 10).
Analysis of the cleavage patterns, combined with Mfold analysis (19), resulted in a prediction of the gag–U3R interaction (Figure 6B). The base-pairing model shows that the two segments flanking the 3′ TAR hairpin in U3R interact with partially complementary sequences in gag. The TAR hairpin itself remains unaffected by formation of the 5′/3′ interaction. The gag–U3R interaction protects certain nucleotides against Pb2+-induced cleavage (indicated by red circles). The nucleotides of the gag–U3R heterodimer that are cleaved by Pb2+ ions are mainly located in internal loops (indicated by blue arrows).
The gag–U3R interaction is conserved among the HIV-1 subtypes
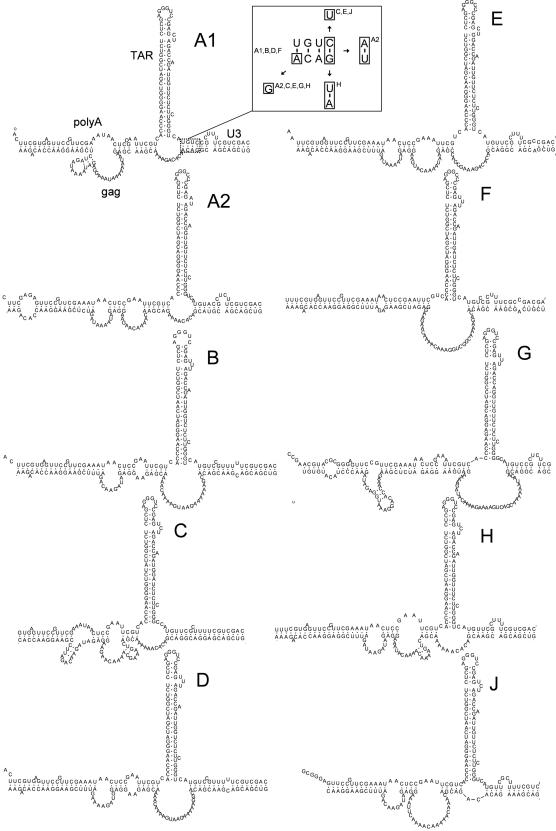
To provide phylogenetic support for the proposed 5′/3′ interaction, we analyzed the RNA genome of different HIV-1 subtypes. A representative sequence was selected for subtype A1, A2, B, C, D, AE, F, G, H and J. We fused fragments 600–700 nt and 9079–9229 nt (subtype B LAI coordinates) in silico and analyzed the RNA secondary structure with Mfold (19). The proposed 5′/3′ interaction can be formed by all HIV-1 subtypes, despite significant sequence variation (Figure 7). All subtypes form a similar long distance base-pairing interaction between gag and U3. Within the gag–U3 interaction, a typical 4-nucleotide pyrimidine bulge that is flanked by two stretches of interacting nucleotides is apparent in several subtypes. One of these stretches, which is boxed in subtype A1, is analyzed in more detail in the insert of Figure 7. This stretch can be formed by all subtypes and is supported by some interesting base-pairing co-variations. In particular, the marked C-G base pair of subtype A1, B, D and F (insert of Figure 7) is changed to U-G in subtype C, E and J, and true co-variations are also seen: A-U for subtype A2 and U-A for subtype H. The observation that the 5′/3′ interaction is conserved among the HIV-1 subtypes strongly suggests a role for this interaction in HIV-1 replication.
Figure 7.
Phylogenetic analysis of the gag–U3R interaction. Representative genomic sequences of HIV-1 subtypes A through J were analyzed with Mfold and edited by RnaViz. The sequences used in our analysis; A1 AF004885, A2 AF286237, B K02030, C U46016, D K03454, E U54771, F AF005494, G AF061641, H AF005496, J AF082395. A small segment formed by the interaction between gag and U3 (boxed in subtype A1) is analyzed in the boxed insert for co-variations. The boxed nucleotides show the nucleotides that are different among HIV-1 subtypes.
DISCUSSION
We identified an RNA–RNA interaction between the 5′ and the 3′ ends of the HIV-1 RNA genome. Mapping of the minimal domains indicated that the interaction is formed by a 5′ sequence in the gag open reading frame and the 3′-terminal U3R region. Transcripts comprising these short sequences form a complex with a similar Tm as the complex formed by the 1-kb transcripts, confirming that we identified the minimal sequences that constitute the 5′/3′ interaction. Detailed RNA structure probing indicates that sequences flanking the 3′ TAR hairpin base pair with gag sequences. This gag–U3R interaction can be formed by all HIV-1 subtypes despite considerable sequence variation, thus providing phylogenetic support for the proposed interaction. Further experimentation is needed to elucidate the exact role of RNA circularization in the HIV-1 life cycle.
The proposed gag–U3R interaction is supported by the result of two previous studies. Kanevsky et al. (20) analyzed the role of secondary structure in the 5′ and 3′R regions on the strand transfer process of reverse transcription. This study used RNA structure probing to show that the TAR hairpin is present at both ends, but the polyA hairpin is presented differently, either in extended form (5′) or shortened form (3′) due to 3′-polyadenylation (Figure 1). Interestingly, this study also placed the R region within the gag coding sequence. In this context, the shorter 3′ polyA hairpin is no longer observed, possibly due to pairing with gag sequences. Because the RNA structure probing data reveal a surprisingly strong correlation with our probing data, the same interaction between gag and polyA sequences may be formed. In another study, Gee et al. (21) used computational folding predictions of the 5′ and the 3′ polyA hairpin on transcripts encompassing the 5′-terminal 662 nt fused to the 3′ region of subtypes AE, AG, B and D. The 5′ polyA sequence predominantly folds the polyA hairpin, but the 3′ polyA sequence preferentially interacts with a sequence in gag. This structure, which was predicted for all four subtypes, is the same as the gag–U3R interaction described in this study. Since only the 5′ and 3′ 1000 nt were examined in this study, we cannot exclude the possibility that other sequences might influence the gag–U3R interaction. Computational RNA structure predictions of complete genomic sequences revealed that such interactions are not expected (21).
Circularization of HIV-1 RNA seems exclusive for unspliced genomic RNA because all spliced subgenomic mRNAs lack the gag interaction domain. The gag–U3R interaction may distinguish genomic RNA from spliced transcripts and could thus function in genomic RNA-specific processes, e.g. packaging and reverse transcription. Especially for the latter process, the gag–U3R distance interaction may bring both ends of the genomic RNA close together. During reverse transcription, the (–) strong-stop DNA is transferred to the 3′R sequence to allow reverse transcription to proceed. Bringing the 5′ and the 3′ ends of the genomic RNA together could facilitate this strand transfer step (4,5). Furthermore, a long distance RNA interaction between the 5′ polyA region and a sequence in gag was described recently (22). Surprisingly, this gag sequence is located only 170 nt upstream of the gag domain that interacts with U3R. Combining the two long-distance RNA interactions would bring the 5′R and 3′R sequences in very close proximity, which may facilitate strand transfer. The gag–U3R interaction may also positively influence polyadenylation at the 3′ polyA site, which is inhibited by the polyA hairpin as present in 5′R (21,23).
RNA circularization has been described for several positive-stranded RNA viruses (12). Circularization plays an important role in essential steps of the viral replication cycle such as translation, transcription and reverse transcription (6–12). The 5′/3′ interaction in HIV-1 is similar to the long-distance interaction described for the retrotransposon Ty1. For Ty1, gag sequences base pair with the U3 domain, and circularization of the retrotransposon RNA genome is required for efficient reverse transcription initiation (7). For HIV-1 RNA, there are multiple sequence motifs near the 5′ end that regulate initiation of reverse transcription (24–26), but there is no evidence that the 3′ end is involved. Further research will be needed to address this issue.
The HIV-1 genome contains many inhibitory sequences (INS) that block RNA export from the nucleus to the cytoplasm (27,28). One INS element is located within the gag sequence that is involved in the gag–U3R interaction (29). The gag sequence has been reported to bind polyA-binding protein (PABP1), which usually interacts with the 3′-polyA tail of mRNAs (30). Gag mutations that prevent binding of PABP1 increase gag translation significantly (29). It has been speculated that PABP1 binds to the 3′ polyA tail at low concentrations, but also to the INS in gag at higher concentrations. Since PABP1 can multimerize, the proteins bound to gag and the 3′ polyA tail could also interact, thus forming a 5′/3′-protein bridge. The possibility that PABP1 modulates the gag–U3R interaction is an interesting option.
ACKNOWLEDGEMENTS
RNA research in the Berkhout lab is sponsored by ZonMw (Vici grant) and NWO-CW (Top grant). Funding to pay the Open Access publication charges for this article was provided by the TOP grant.
Conflict of interest statement. None declared.
REFERENCES
- 1.Baudin F, Marquet R, Isel C, Darlix JL, Ehresmann B, Ehresmann C. Functional sites in the 5′ region of human immunodeficiency virus type 1 RNA form defined structural domains. J. Mol. Biol. 1993;229:382–397. doi: 10.1006/jmbi.1993.1041. [DOI] [PubMed] [Google Scholar]
- 2.Berkhout B, Silverman RH, Jeang KT. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell. 1989;59:273–282. doi: 10.1016/0092-8674(89)90289-4. [DOI] [PubMed] [Google Scholar]
- 3.Marciniak RA, Sharp PA. HIV-1 Tat protein promotes formation of more-processive elongation complexes. EMBO J. 1991;10:4189–4196. doi: 10.1002/j.1460-2075.1991.tb04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkhout B, van Wamel J, Klaver B. Requirements for DNA strand transfer during reverse transcription in mutant HIV-1 virions. J. Mol. Biol. 1995;252:59–69. doi: 10.1006/jmbi.1994.0475. [DOI] [PubMed] [Google Scholar]
- 5.Berkhout B, Vastenhouw NL, Klasens BI, Huthoff H. Structural features in the HIV-1 repeat region facilitate strand transfer during reverse transcription. RNA. 2001;7:1097–1114. doi: 10.1017/s1355838201002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alvarez DE, Lodeiro MF, Luduena SJ, Pietrasanta LI, Gamarnik AV. Long-range RNA-RNA interactions circularize the dengue virus genome. J. Virol. 2005;79:6631–6643. doi: 10.1128/JVI.79.11.6631-6643.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cristofari G, Bampi C, Wilhelm M, Wilhelm FX, Darlix JL. A 5′-3′ long-range interaction in Ty1 RNA controls its reverse transcription and retrotransposition. EMBO J. 2002;21:4368–4379. doi: 10.1093/emboj/cdf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Groft CM, Burley SK. Recognition of eIF4G by rotavirus NSP3 reveals a basis for mRNA circularization. Mol. Cell. 2002;9:1273–1283. doi: 10.1016/s1097-2765(02)00555-5. [DOI] [PubMed] [Google Scholar]
- 9.Guo L, Allen EM, Miller WA. Base-pairing between untranslated regions facilitates translation of uncapped, nonpolyadenylated viral RNA. Mol. Cell. 2001;7:1103–1109. doi: 10.1016/s1097-2765(01)00252-0. [DOI] [PubMed] [Google Scholar]
- 10.Herold J, Andino R. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell. 2001;7:581–591. doi: 10.1016/S1097-2765(01)00205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khromykh AA, Meka H, Guyatt KJ, Westaway EG. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 2001;75:6719–6728. doi: 10.1128/JVI.75.14.6719-6728.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.You S, Falgout B, Markoff L, Padmanabhan R. In vitro RNA synthesis from exogenous dengue viral RNA templates requires long range interactions between 5′- and 3′-terminal regions that influence RNA structure. J. Biol. Chem. 2001;276:15581–15591. doi: 10.1074/jbc.M010923200. [DOI] [PubMed] [Google Scholar]
- 13.Piron M, Delaunay T, Grosclaude J, Poncet D. Identification of the RNA-binding, dimerization, and eIF4GI-binding domains of rotavirus nonstructural protein NSP3. J. Virol. 1999;73:5411–5421. doi: 10.1128/jvi.73.7.5411-5421.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Padilla-Noriega L, Paniagua O, Guzman-Leon S. Rotavirus protein NSP3 shuts off host cell protein synthesis. Virology. 2002;298:1–7. doi: 10.1006/viro.2002.1477. [DOI] [PubMed] [Google Scholar]
- 15.Peden K, Emerman M, Montagnier L. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology. 1991;185:661–672. doi: 10.1016/0042-6822(91)90537-l. [DOI] [PubMed] [Google Scholar]
- 16.Berkhout B, van Wamel JLB. Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J. Virol. 1996;70:6723–6732. doi: 10.1128/jvi.70.10.6723-6732.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersen ES, Jeeninga RE, Damgaard CK, Berkhout B, Kjems J. Dimerization and template switching in the 5′ untranslated region between various subtypes of Human Immunodeficiency Virus type 1. J. Virol. 2003;77:3020–3030. doi: 10.1128/JVI.77.5.3020-3030.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olejniczak M, Gdaniec Z, Fischer A, Grabarkiewicz T, Bielecki L, Adamiak RW. The bulge region of HIV-1 TAR RNA binds metal ions in solution. Nucleic Acids Res. 2002;30:4241–4249. doi: 10.1093/nar/gkf541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanevsky I, Chaminade F, Ficheux D, Moumen A, Gorelick R, Negroni M, Darlix JL, Fosse P. Specific interactions between HIV-1 nucleocapsid protein and the TAR element. J. Mol. Biol. 2005;348:1059–1077. doi: 10.1016/j.jmb.2005.03.046. [DOI] [PubMed] [Google Scholar]
- 21.Gee AH, Kasprzak W, Shapiro BA. Structural differentiation of the HIV-1 polyA signals. J. Biomol. Struct. Dyn. 2006;23:417–428. doi: 10.1080/07391102.2006.10531236. [DOI] [PubMed] [Google Scholar]
- 22.Paillart JC, Skripkin E, Ehresmann B, Ehresmann C, Marquet R. In vitro evidence for a long range pseudoknot in the 5′-untranslated and matrix coding regions of HIV-1 genomic RNA. J. Biol. Chem. 2002;277:5995–6004. doi: 10.1074/jbc.M108972200. [DOI] [PubMed] [Google Scholar]
- 23.Das AT, Klaver B, Berkhout B. A hairpin structure in the R region of the Human Immunodeficiency Virus type 1 RNA genome is instrumental in polyadenylation site selection. J. Virol. 1999;73:81–91. doi: 10.1128/jvi.73.1.81-91.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Isel C, Marquet R, Keith G, Ehresmann C, Ehresmann B. Modified nucleotides of tRNA(3Lys) modulate primer/template loop-loop interaction in the initiation complex of HIV-1 reverse transcription. J. Biol. Chem. 1993;268:25269–25272. [PubMed] [Google Scholar]
- 25.Beerens N, Groot F, Berkhout B. Initiation of HIV-1 reverse transcription is regulated by a primer activation signal. J. Biol. Chem. 2001;276:31247–31256. doi: 10.1074/jbc.M102441200. [DOI] [PubMed] [Google Scholar]
- 26.Iwatani Y, Rosen AE, Guo J, Musier-Forsyth K, Levin JG. Efficient initiation of HIV-1 reverse transcription in vitro. Requirement for RNA sequences downstream of the primer binding site abrogated by nucleocapsid protein-dependent primer-template interactions. J. Biol. Chem. 2003;278:14185–14195. doi: 10.1074/jbc.M211618200. [DOI] [PubMed] [Google Scholar]
- 27.Schwartz S, Campbell M, Nasioulas G, Harrison J, Felber BK, Pavlakis GN. Mutational inactivation of an inhibitory sequence in human immunodeficiency virus type 1 results in Rev-independent gag expression. J. Virol. 1992;66:7176–7182. doi: 10.1128/jvi.66.12.7176-7182.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwartz S, Felber BK, Pavlakis GN. Distinct RNA sequences in the gag region of human immunodeficiency virus type 1 decrease RNA stability and inhibit expression in the absence of Rev protein. J. Virol. 1992;66:150–159. doi: 10.1128/jvi.66.1.150-159.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Afonina E, Neumann M, Pavlakis GN. Preferential binding of poly(A)-binding protein 1 to an inhibitory RNA element in the human immunodeficiency virus type 1 gag mRNA. J. Biol. Chem. 1997;272:2307–2311. doi: 10.1074/jbc.272.4.2307. [DOI] [PubMed] [Google Scholar]
- 30.Burd CG, Matunis EL, Dreyfuss G. The multiple RNA-binding domains of the mRNA poly(A)-binding protein have different RNA-binding activities. Mol. Cell. Biol. 1991;11:3419–3424. doi: 10.1128/mcb.11.7.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]