Abstract
Selenocysteine (Sec) and pyrrolysine (Pyl) are rare amino acids that are cotranslationally inserted into proteins and known as the 21st and 22nd amino acids in the genetic code. Sec and Pyl are encoded by UGA and UAG codons, respectively, which normally serve as stop signals. Herein, we report on unusually large selenoproteomes and pyrroproteomes in a symbiont metagenomic dataset of a marine gutless worm, Olavius algarvensis. We identified 99 selenoprotein genes that clustered into 30 families, including 17 new selenoprotein genes that belong to six families. In addition, several Pyl-containing proteins were identified in this dataset. Most selenoproteins and Pyl-containing proteins were present in a single deltaproteobacterium, δ1 symbiont, which contained the largest number of both selenoproteins and Pyl-containing proteins of any organism reported to date. Our data contrast with the previous observations that symbionts and host-associated bacteria either lose Sec utilization or possess a limited number of selenoproteins, and suggest that the environment in the gutless worm promotes Sec and Pyl utilization. Anaerobic conditions and consistent selenium supply might be the factors that support the use of amino acids that extend the genetic code.
INTRODUCTION
Selenium (Se) is an essential micronutrient due to its requirement for biosynthesis and function of the 21st amino acid, selenocysteine (Sec). This amino acid is typically found in the active sites of a small number of selenoproteins in all three domains of life: archaea, bacteria and eukaryotes (1–4). Biosynthesis of Sec and its cotranslational insertion into polypeptides require a complex molecular machinery that recodes in-frame UGA codons, which normally function as stop signals, to serve as Sec codons (5–9). Although the occurrence of selenoprotein genes is limited, the Sec UGA codon has become the first addition to the universal genetic code since the code was deciphered 40 years ago (10).
The mechanism of Sec insertion differs in the three domains of life. In bacteria, this process has been most thoroughly elucidated in Escherichia coli (1,2,6). Translation of bacterial selenoprotein mRNA requires both a selenocysteine insertion sequence (SECIS) element, which is a stem-loop structure immediately downstream of Sec-encoding UGA codon (5,11,12), and trans-acting factors dedicated to Sec incorporation (8). In archaea and eukaryotes, SECIS elements are located in 3′-UTRs and some factors involved in Sec biosynthesis and insertion are different. Recent identification of Sec synthase, SecS, in eukaryotes, which is different from the bacterial Sec synthase, SelA, provided important insights into Sec biosynthesis in these organisms (13).
Recently, an additional rare amino acid pyrrolysine (Pyl), was identified, which expanded the canonical genetic code to 22 amino acids (14,15). Pyl is inserted in response to UAG codon in several methanogenic archaea (14). Although the mechanism of Pyl biosynthesis and incorporation into protein is not fully understood, the presence of a tRNApyl gene (pylT) with the CUA anticodon and of class II aminoacyl-tRNA synthetase gene (pylS) argued for cotranslational incorporation of Pyl (15). In Desulfitobacterium hafniense, a single bacterium, in which a Pyl-containing protein was found, PylS consists of two proteins: PylSn and PylSc (15).
In recent years, large-scale genome sequencing projects, including both organism-specific and environmental metagenomic projects, provided a large volume of gene and protein sequence information. However, selenoprotein genes are almost universally misannotated in these datasets because UGA has the dual function of encoding Sec and terminating translation, and only the latter function is recognized by current annotation programs. Several bioinformatics tools have been developed to address this problem and can be used to identify selenoprotein genes (16–22). These programs have successfully identified many new selenoproteins in both prokaryotic and eukaryotic genomes, as well as in the Sargasso Sea environmental samples (23).
Complex symbiotic relationships between bacteria and multicellular eukaryotes have evolved in several environments, but science has traditionally focused on interactions that are pathogenic (24). Recently, there has been increased recognition of symbiotic interactions that benefit both the microorganism and the host (25). A recent metagenomic analysis of the symbiotic microbial consortium of the marine oligochaete Olavius algarvensis, a worm lacking a mouth, gut and nephridia, revealed four major co-occurring symbionts, which belong to Deltaproteobacteria (δ1 and δ4) and Gammaproteobacteria (γ1 and γ3), as well as one minor Spirochaete species. Since some Deltaproteobacteria are selenoprotein-rich organisms (27), we analyzed the selenoproteomes of these symbionts to examine a possible relationship between selenium and symbiosis.
To characterize selenoproteome in these symbionts, we adopted a Sec/cysteine(Cys) homology-based search approach, which has been successfully used to characterize the selenoproteomes of both prokaryotes (22) and one of the largest prokaryotic sequencing projects, the Sargasso Sea microbial sequencing project (23). We detected known selenoproteins present in this metagenomic dataset and identified several novel selenoproteins. Interestingly, one deltaproteobacterium, δ1 symbiont, contains at least 57 selenoproteins, which is the largest number of selenoproteins reported to date in any organism. In addition, several Pyl-containing proteins were identified and most were also found in the same δ1 symbiont. Our results provide new insights into understanding evolution and function of these rare amino acids.
MATERIALS AND METHODS
Databases and resources
Assembled sequences of the Olavius symbionts’ metagenome were obtained from NCBI with the project accession number AASZ00000000 (ftp://ftp.ncbi.nih.gov/genbank/wgs/wgs.AASZ.1.gbff.gz). The database contained 5597 genomic sequences, which corresponded to a total of 23.7 million nucleotides. Non-redundant (NR) protein database was downloaded from NCBI ftp server. This dataset contained a total of 4 644 764 protein sequences (1 603 127 260 amino acids). BLAST (28) was also obtained from NCBI.
Identification of Cys/TGA pairs in homologous sequences and minimal ORFs
Each Cys-containing protein sequence in the NR database was initially searched against the Olavius symbionts’ metagenomic database for possible TGA/TAG/TAA-containing homologs using TBLASTN with default parameters. Only local alignments, in which Cys in the query protein was aligned with TGA codon in the nucleotide sequence from the Olavius symbionts’ metagenomic database, were selected for further analysis. For each TGA-containing nucleotide sequence identified in the metagenomic database, regions upstream and downstream of the putative in-frame TGA codon were analyzed to identify a minimal ORF. If a stop codon was found between the in-frame TGA codon and an initiation codon (ATG or GTG), such a TGA-containing sequence was discarded.
Analyses of TGA-flanking regions and sequence clustering
We analyzed the conservation of TGA-flanking regions in all six reading frames using BLASTX. If the best hit, which covered the TGA codon with at least a 10-nt overlap, was in a different reading frame than the TGA codon, the corresponding sequence was filtered out. RPS-BLAST was then used to search against conserved domains database (CDD). If the best hit which covered the TGA codon with at least a five-residue overlap was in a different reading frame or additional stop codons appeared within the conserved domain in the same frame, the sequence was removed.
We used BL2SEQ to cluster remaining protein sequences into different groups. If a local alignment of two proteins had an E-value below 10−4 and was at least 20 amino acid long, as well as the predicted Sec residues were located at the same position or very close (no more than three residues apart) in the alignment, the two proteins were assigned to the same cluster.
Cysteine conservation and selenoprotein classification
All clusters were automatically searched against NCBI NR and microbial databases using BLASTX and TBLASTX. Each predicted ORF containing an in-frame TGA was considered further only if at least two corresponding Cys-containing homologs were detected and the proportion of TGA/Cys pairs in the set of homologs was >50%.
The remaining clusters were analyzed for occurrence of bacterial SECIS elements, located immediately downstream of the in-frame TGA codons, using bSECISearch program (19). The final clusters were manually analyzed and divided into three groups: known selenoproteins, new selenoproteins (clusters containing at least two different sequences with conserved in-frame TGA codons) and selenoprotein candidates (clusters containing only one sequence). It should be noted that sequencing errors that generate in-frame UGA codons could not be excluded for selenoprotein candidates.
Identification of Pyl operon proteins and known Pyl-containing proteins
PylT and PylS sequences from Methanosarcina barkeri (accession number AY064401) were used to search for possible homologs in the metagenomic dataset. Candidate tRNAPyl was further analyzed to identify structural features associated with known tRNAPyl, including a six base-pair acceptor stem and a base between the D and acceptor stems (15). Other genes in the Pyl operon (pylB, pylC, pylD) were also analyzed by comparative sequence analyses.
The TBLASTN program with default parameters was used to search for known Pyl-containing methylamine methyltransferases. Open reading frames (ORFs) and conservation of UAG-flanking regions were examined manually. Multiple alignments were generated with ClustalW (29).
RESULTS
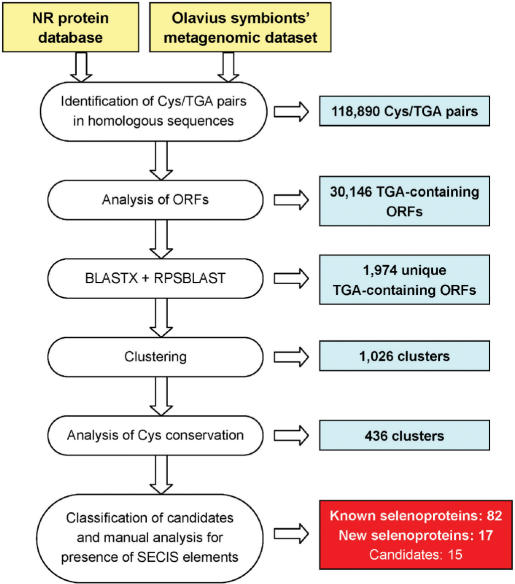
To identify selenoprotein genes in the Olavius symbiont metagenomic dataset, we employed an algorithm that we previously used to identify selenoproteins in the Sargasso Sea microbial dataset (23). This technique takes advantage of the fact that almost all selenoproteins have Cys-containing homologs in different organisms. Intermediate results for each step in the search process are shown in Figure 1. In addition, an independent BLAST homology search for Sec-containing homologs of all known selenoprotein families was performed.
Figure 1.
A schematic diagram of the search algorithm. Details of the search process are provided in Materials and methods section.
Identification of known selenoproteins in the Olavius symbionts’ metagenome
A total of 82 selenoprotein genes, which belong to 24 previously described selenoprotein families, were identified (Table 1). Considering that only four major symbionts were identified in the Olavius symbionts’ metagenomic dataset, each selenoprotein could be mapped into the exact organism, from which the sequence was derived. Essentially all selenoproteins were found to map to symbionts δ1 and δ4. The former organism contained 44 homologs of known selenoproteins, already the largest number of selenoproteins reported to date in any organism [a previous record holder is also a deltaproteobacterium, Syntrophobacter fumaroxidans, which has 31 selenoprotein genes, see (27)]. In addition, several selenoproteins were found in sequences not mapped to any of the four symbionts (designated as unassigned sequences). In contract, no selenoprotein genes could be identified in symbionts γ1 and γ3. All identified selenoprotein genes were misannotated in the original dataset. Several selenoprotein families detected in the dataset were represented by 2–12 selenoprotein genes, whereas six families, DsbG-like, peroxiredoxin (Prx), thioredoxin (Trx), glutaredoxin (Grx), NADH oxidase and UGSC-containing protein [unpublished data; this is a selenoprotein of unknown function that also occurs in Hyphomonas neptunium (30) and detected in the environmental sequencing project of the microbial communities in the North Pacific Subtropical Gyre (31)], were represented by single sequences. Sequencing errors that generate in-frame TGA codons in these sequences cannot be excluded; however, the fact that they correspond to known selenoproteins and possess strong predicted SECIS elements argue that they are true selenoproteins. Many of the detected selenoprotein families also had Cys-containing homologs in the metagenomic database (Table 1).
Table 1.
Known selenoprotein families identified in the Olavius algarvensis symbionts
| Protein family | Total selenoproteins | Olavius symbionts | Number of Cys homolog | ||||
|---|---|---|---|---|---|---|---|
| δ1 | δ4 | γ1 | γ3 | Unassigned | |||
| Detected selenoproteins (24 families) | |||||||
| F420-reducing hydrogenase, delta subunit (FrhD) | 12 | 5 | 2 | 0 | 0 | 5 | 6 |
| Heterodisulfide reductase, subunit A (HdrA) | 10 | 4 | 4 | 0 | 0 | 2 | 3 |
| Rhodanese-related sulfurtransferase | 8 | 4 | 2 | 0 | 0 | 2 | 0 |
| AhpD-like* | 7 | 5 | 1 | 0 | 0 | 1 | 4 |
| Prx-like thiol:disulfide oxidoreductase* | 6 | 2 | 3 | 0 | 0 | 1 | 4 |
| Proline reductase (PR)* | 5 | 5 | 0 | 0 | 0 | 0 | 0 |
| Formate dehydrogenase alpha subunit (FdhA) | 4 | 2 | 1 | 0 | 0 | 1 | >10 |
| Sulfurtransferase COG2897 | 3 | 1 | 2 | 0 | 0 | 0 | 4 |
| DsrE-like* | 3 | 2 | 1 | 0 | 0 | 0 | 0 |
| DsbA-like* | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| F420-reducing hydrogenase, alpha subunit (FrhA) | 2 | 1 | 1 | 0 | 0 | 0 | 3 |
| Selenophosphate synthetase (SelD) | 2 | 1 | 1 | 0 | 0 | 0 | 1 |
| HesB-like | 2 | 1 | 0 | 0 | 0 | 1 | 0 |
| Fe-S oxidoreductase (GlpC) | 2 | 1 | 1 | 0 | 0 | 0 | 10 |
| Distant AhpD homolog* | 2 | 2 | 0 | 0 | 0 | 0 | 2 |
| Sulfurtransferase COG0607 | 2 | 1 | 0 | 0 | 0 | 1 | >10 |
| Methione sulfoxide reductase A (MsrA)* | 2 | 1 | 1 | 0 | 0 | 0 | 6 |
| Methylated-DNA-protein-cysteine methyltransferase | 2 | 0 | 0 | 0 | 0 | 2 | 8 |
| DsbG-like* | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| Peroxiredoxin (Prx)* | 1 | 1 | 0 | 0 | 0 | 0 | 4 |
| Thioredoxin (Trx)* | 1 | 1 | 0 | 0 | 0 | 0 | >10 |
| NADH oxidase | 1 | 1 | 0 | 0 | 0 | 0 | 1 |
| Glutaredoxin* | 1 | 0 | 0 | 0 | 0 | 1 | 2 |
| UGSC-containing protein* | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| Known selenoprotein families not detected (17 families) | |||||||
| SelW-like* | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Glutathione peroxidase (GPx)* | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Homolog of AhpF N-terminal domain* | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| Thiol:disulfide interchange protein* | 0 | 0 | 0 | 0 | 0 | 0 | 8 |
| Glycine reductase selenoprotein A (GrdA) | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Glycine reductase selenoprotein B (GrdB) | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Arsenate reductase* | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Molybdopterin biosynthesis MoeB protein | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| Glutathione S-transferase (GST)* | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Deiodinase-like* | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Thiol-disulfide isomerase-like protein* | 0 | 0 | 0 | 0 | 0 | 0 | 5 |
| Hypothetical protein 1* | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OsmC-like protein* | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| NADH:ubiquinone oxidoreductase | 0 | 0 | 0 | 0 | 0 | 0 | 9 |
| Radical SAM domain protein | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Putative mercuric transport protein | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Cation-transporting ATPase, E1-E2 family | 0 | 0 | 0 | 0 | 0 | 0 | 7 |
| Total | 82 | 44 | 20 | 0 | 0 | 18 | |
*Homologs of known thiol-based oxidoreductases or thioredoxin-like fold proteins.
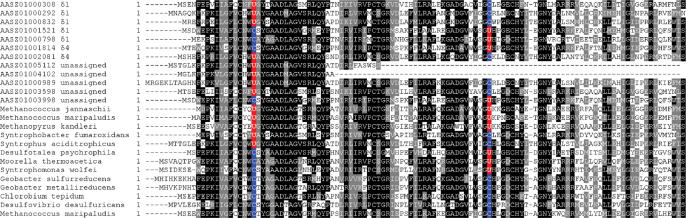
Several selenoprotein families had a particularly high representation in the Olavius symbionts dataset. The most abundant family was F420-reducing hydrogenase delta subunit (FrhD), which included 12 selenoprotein genes. Figure 2 shows a multiple alignment of this family. This selenoprotein family was previously found in both methanogenic archaea and bacteria. In archaea, its Sec-containing forms contain two Sec residues. In contrast, only one of the two Sec residues was found in different Sec-containing homologs in bacteria, including all metagenomic sequences in the current study. Such flexibility in replacing functionally important Cys with Sec has not been described previously.
Figure 2.
Multiple sequence alignment of FrhD family. Conserved residues are highlighted. Sec (U) and the corresponding Cys (C) residues are shown in red and blue, respectively.
Heterodisulfide reductase subunit A (HdrA) was the second most abundant selenoprotein family, which was represented by 10 selenoprotein genes. It is interesting that most of the HdrA sequences were found to cluster with FrhD sequences. This finding is consistent with our previous hypothesis that the hdrA-frhD-frhG-frhA cluster could be laterally transferred between Sec-decoding archaea and Deltaproteobacteria (27). A rhodanese-related sulfurtransferase [8 genes, (19)], AhpD-like (7 genes), Prx-like thiol:disulfide oxidoreductase (6 genes) and proline reductase (PR, 5 genes) were the next most abundant selenoprotein families. These six families accounted for 58.5% of known selenoprotein sequences, suggesting importance of their functions in the symbiosis involving Deltaproteobacteria and the host worm. Other detected selenoprotein families included formate dehydrogenase alpha subunit (FdhA), F420-reducing hydrogenase alpha subunit (FrhA), selenophosphate synthetase (SelD), HesB-like, Fe-S oxidoreductase (GlpC), methionine sulfoxide reductase A (MsrA) and several other selenoprotein families.
Most of these selenoproteins were redox proteins, which used Sec either to coordinate redox-active metals or for thiol/disulfide-based redox catalysis. Moreover, among 24 selenoprotein families detected in the symbionts’ metagenomic dataset, at least 17 (67 sequences, 81.7%) were homologs of known thiol oxidoreductases or possessed Trx-like fold (Table 1). Many of these selenoproteins contained a conserved UxxC/UxxS/CxxU/TxxU redox motif.
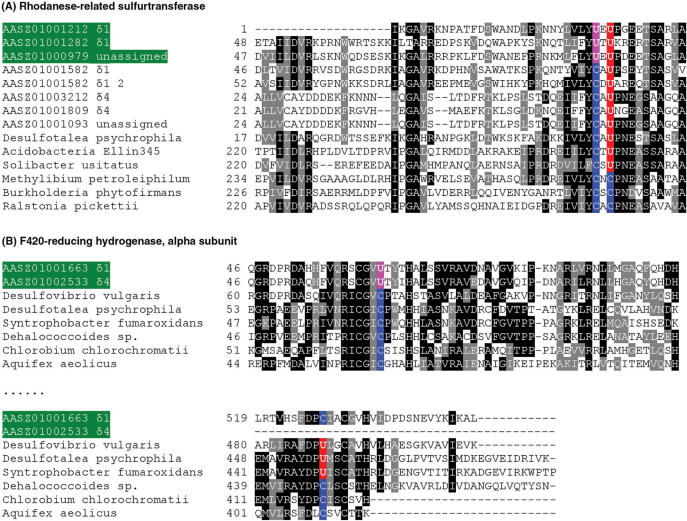
In two known selenoprotein genes, new Sec positions were identified. Interestingly, in a rhodanese-related sulfurtransferase family, a new protein form was detected wherein a second Sec evolved in the protein, thus resulting in a UxU motif (Figure 3A). In addition, a new Sec was observed in FrhA, which resulted in a CxxU motif compared to the previously known UxxC motif (Figure 3B).
Figure 3.
Multiple sequence alignment of several known selenoprotein families containing new features. New Sec positions are shown in pink. Contigs containing these new features are also highlighted in green background. (A) Rhodanese-related sulfurtransferase; (B) F420-reducing hydrogenase, alpha subunit.
New selenoproteins identified in the Olavius symbionts’ metagenome
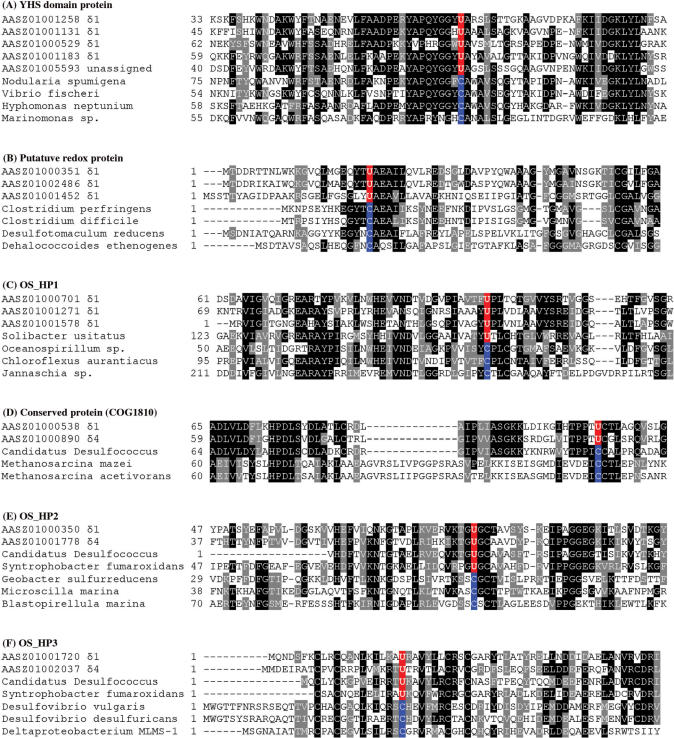
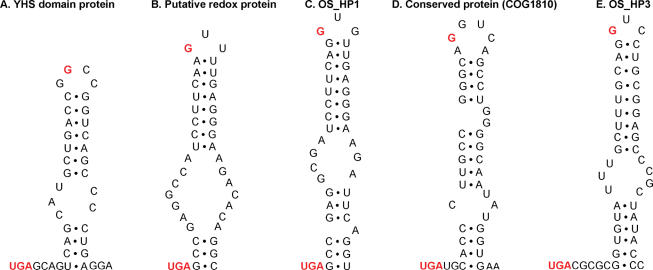
In addition to homologs of previously described selenoproteins, we identified six new selenoprotein families, which were represented by at least two individual TGA-containing ORFs (total of 17 genes, Table 2). Most of these new families did not correspond to domains of known function and were not homologous to protein families with known functions. Multiple alignments of these new selenoproteins and their Cys-containing homologs (Figure 4) highlight sequence conservation of Sec/Cys pairs and their flanking regions. All new selenoproteins contained stable stem-loop structures downstream of Sec-encoding TGA codons that resembled bacterial SECIS elements. Representative predicted SECIS elements found in these new selenoprotein families are shown in Figure 5.
Table 2.
Novel selenoproteins identified in the Olavius algarvensis symbionts
| Protein family | Total selenoproteins | Olavius symbionts | Number of Cys homolog | ||||
|---|---|---|---|---|---|---|---|
| δ1 | δ4 | γ1 | γ3 | Unassigned | |||
| YHS domain protein | 5 | 4 | 0 | 0 | 0 | 1 | 2 |
| Putative redox protein | 3 | 3 | 0 | 0 | 0 | 0 | 2 |
| OS_HP1* | 3 | 3 | 0 | 0 | 0 | 0 | 2 |
| Conserved protein COG1810 | 2 | 1 | 1 | 0 | 0 | 0 | 0 |
| OS_HP2 | 2 | 1 | 1 | 0 | 0 | 0 | 0 |
| OS_HP3 | 2 | 1 | 1 | 0 | 0 | 0 | 0 |
| Total | 17 | 13 | 3 | 0 | 0 | 1 | 6 |
*OS_HP, Olavius symbiont's hypothetical protein.
Figure 4.
Multiple sequence alignments of new selenoproteins and their Cys homologs. The alignments show Sec-flanking regions in detected proteins. Both selenoprotein sequences detected in the Olavius symbionts’ metagenome dataset and their Sec/Cys-containing homologs present in indicated organisms are shown. Conserved residues are highlighted. Predicted Sec (U) and the corresponding Cys (C) residues in other homologs are shown in red and blue, respectively.
Figure 5.
Predicted bacterial SECIS elements in representative sequences of new selenoprotein families. Only sequences downstream of in-frame UGA codons are shown. In-frame UGA codons and conserved guanosines in the apical loop are shown in red. (A) YHS domain protein, AASZ01000529; (B) Putative redox protein, AASZ01002486; (C) OS_HP1, AASZ01000351; (D) Conserved protein (COG1810), AASZ01000538; (E) OS_HP3, AASZ01001720.
We also detected at least 15 additional TGA-containing sequences, which showed similarity neither to known and new selenoproteins nor to each other. No definitive conclusion can be made regarding these sequences because of the possibility of sequencing errors. However, some of them contained candidate SECIS elements. Moreover, a small number of TGA-containing homologs of candidate selenoproteins, which have no conserved Cys homologs, but were previously predicted in sequenced bacterial genomes using bSECISearch (19), were identified. Future experimental verification is needed for these selenoprotein candidates.
Pyl-containing proteins detected in the Olavius symbionts’ dataset
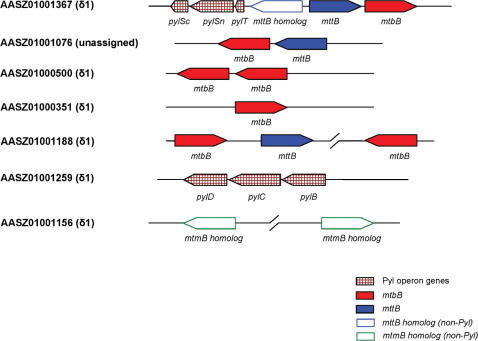
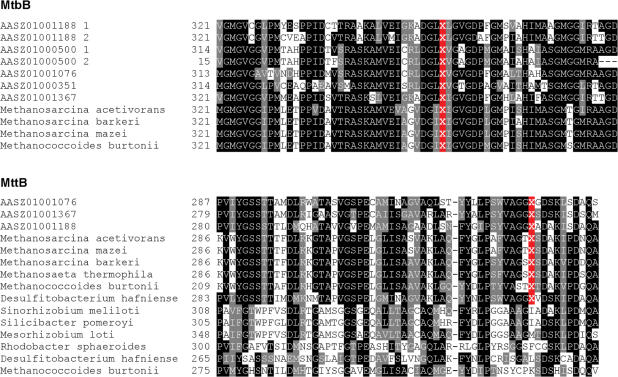
Pyl has been identified in the active sites of several methylamine methyltransferase families, including monomethylamine methyltransferase (MtmB), dimethylamine methyltransferase (MtbB) and trimethylamine methyltransferase (MttB), in several methanogenic archaea (14,15). However, only one gram-positive bacterium, D. hafniense, has been found that possesses a single Pyl-containing MttB homolog. Recently, a transposase family was identified as a new Pyl-containing protein family (32). Besides pylT and pylS, a pylB-pylC-pylD gene operon (especially pylD) was proposed to be specific for Pyl utilization (32). We examined the occurrence of both Pyl-containing proteins and Pyl operon genes. To our surprise, a total of 10 Pyl-containing methylamine methyltransferase sequences (belonging to MtbB and MttB families) were identified and eight were found in the δ1 endosymbiont which also had pylT, pylSn, pylSc and pylB-pylC-pylD genes (Table 3). Several genes were clustered or were present in the same operon (Figure 6). An alignment of these sequences and their homologs is shown in Figure 7.
Table 3.
Known Pyl-containing proteins and Pyl operon proteins identified in the Olavius algarvensis symbionts
| Protein family | Total sequences | The Olavius symbionts | Other homologs | ||||
|---|---|---|---|---|---|---|---|
| δ1 | δ4 | γ1 | γ3 | Unassigned | |||
| Known Pyl-containing proteins | |||||||
| MtmB | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| MtbB | 7 | 6 | 0 | 0 | 0 | 1 | 0 |
| MttB | 3 | 2 | 0 | 0 | 0 | 1 | >10 |
| Pyl biosynthesis and insertion components | |||||||
| PylSn | 1 | 1 | 0 | 0 | 0 | 0 | |
| PylSc | 1 | 1 | 0 | 0 | 0 | 0 | |
| PylB | 1 | 1 | 0 | 0 | 0 | 0 | |
| PylC | 1 | 1 | 0 | 0 | 0 | 0 | |
| PylD | 1 | 1 | 0 | 0 | 0 | 0 | |
| PylT | 1 | 1 | 0 | 0 | 0 | 0 | |
Figure 6.
Occurrence of genes for Pyl-containing proteins and Pyl operon proteins in Olavius symbionts’ metagenomic sequences. The mtbB and mttB genes and other Pyl operon genes are shown by the indicated color scheme in contigs containing these genes.
Figure 7.
Multiple alignments of Pyl-flanking regions in methylamine methyltransferase families (MtbB and MttB). Pyl is shown by X and its location in the alignment is highlighted in red.
It was proposed that Pyl is inserted by UAG codons with the help of a putative pyrrolysine insertion sequence (PYLIS) element, which was predicted to be located downstream of the Pyl-encoding UAG codon in Pyl-containing protein mRNAs (33). Although the presence of such element in archaea is questionable, it is reasonable that there should be a certain cis-element to distinguish the Pyl-encoding UAG codon from stop codon in bacteria (32). To search for candidate PYLIS elements in bacteria, sequences downstream of in-frame UAG codons and in putative 5′- and 3′-UTRs of methylamine methyltransferase mRNAs in both D. hafniense and the δ1 symbiont were analyzed manually for possible conserved structures and sequence features within these structures. Our analyses revealed no obvious common structure shared by all members of these methylamine methyltransferase families.
Relationship between different symbiotic conditions and Sec utilization
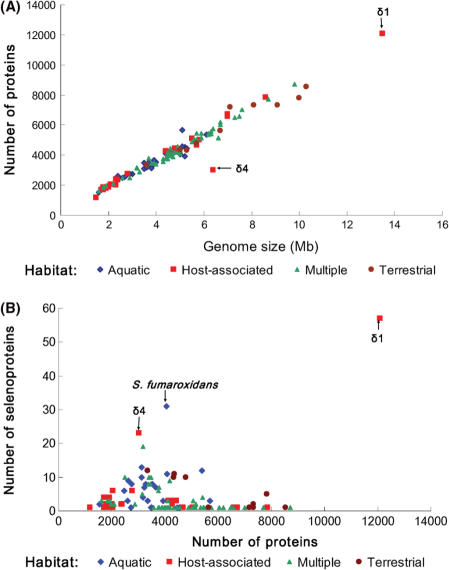
Although δ1 and δ4 endosymbionts belong to the selenoprotein-rich phylum Deltaproteobacteria, they are host-associated organisms. In contrast, most selenoprotein-rich organisms identified previously are free-living organisms (27). To investigate the relationships between habitats, genome/proteome size and Sec utilization in bacteria, we carried out an exhaustive homology search of all known selenoprotein families against 450 sequenced bacterial genomes. A total of 116 Sec-utilizing organisms were found. Characteristics of selenoproteomes, genome size, proteome size and habitats for these organisms are shown in Table S1, and Figure 8 illustrates correlations among these properties. For Sec-containing organisms, regardless of habitat, the proteome size was proportional to the genome size (Figure 8A). No obvious correlation was observed between the size of selenoproteome and the size of proteome. However, a trend could be seen wherein host-associated organisms possess the smallest selenoproteomes compared to free-living organisms (Figure 8B).
Figure 8.
Relationship among habitats, genome size, proteome size and selenoproteomes. Sec-containing organisms were classified into four groups based on different habitats: aquatic, host-associated, multiple and terrestrial. (A) Correlation between genome size and proteome size. (B) Correlation between proteome size and selenoproteomes. δ1 and δ4 symbionts are indicated in the figure.
We found that δ1 and δ4 symbionts were outliers with respect to selenoproteome size, especially when compared with other host-associated bacteria (Figure 8B). Table 4 shows a comprehensive list of sequenced host-associated selenoprotein-containing bacteria and their living conditions. In contrast to selenoprotein-rich δ1 and δ4 symbionts, most of these organisms had FdhA and/or SelD as their only selenoproteins. One possibility is that δ1 and δ4 symbionts are located below the worm cuticle, where essentially no oxygen is present, whereas other parasites, most of which are facultative anaerobic, microaerobic and aerobic, are located in mouth, respiratory tract or gastrointestinal tract, which are exposed to at least some oxygen (34). We previously found that decrease in oxygen concentration correlates with increase in Sec utilization (27). Olavius algarvensis is the first marine host identified to date which lives in obligate and species-specific associations with Sec-containing bacterial symbionts. Presumably, these deltaproteobacterial symbionts take advantage of a relatively constant supply of selenium in sea water and have increased their demand for this trace element.
Table 4.
Selenoproteins in sequenced symbiotic/host-associated bacteria
| Phylum | Organism | Total number of proteins | Number of selenoproteins | Habitat | Oxygen requirement |
|---|---|---|---|---|---|
| Actinobacteria | Collinsella aerofaciens | 2367 | 2 | Human gut | Anaerobic |
| Mycobacterium smegmatis | 6716 | 1 | Human smegma | Aerobic | |
| Mycobacterium avium | 5120 | 1 | Lung | Aerobic | |
| Betaproteobacteria/ Burkholderiaceae | Burkholderia mallei | 5025 | 1 | Mammals | Aerobic |
| Burkholderia multivorans | 6604 | 1 | Human lung | Aerobic | |
| Burkholderia phymatum | 7845 | 1 | Root nodules of tropical legumes | Aerobic | |
| Deltaproteobacteriadelta | Lawsonia intracellularis | 1185 | 1 | Mucosa of the lower intestinal tract in animals | Facultative |
| δ1 symbiont | 12084 | 57 | Below the worm cuticle of Olavius algarvensis | Anaerobic | |
| δ4 symbiont | 3012 | 23 | Below the worm cuticle of Olavius algarvensis | Anaerobic | |
| Epsilonproteobacteria | Campylobacter concisus | 2039 | 6 | Human oral cavity and gastrointestinal tract | Microaerophilic |
| Campylobacter curvus | 1921 | 4 | Human oral cavity and gastrointestinal tract | Microaerophilic | |
| Campylobacter fetus | 1719 | 4 | Human blood | Microaerophilic | |
| Helicobacter hepaticus | 1875 | 1 | Mucosal layer of the gastrointestinal tract | Microaerophilic | |
| Wolinella succinogenes | 2043 | 1 | Gastrointestinal tract | Microaerophilic | |
| Gammaproteobacteria/ Enterobacteriales | Escherichia coli | 4243 | 3 | Lower intestine | Facultative |
| Photorhabdus luminescens | 4683 | 1 | The gut of an entomopathogenic nematode | Facultative | |
| Salmonella enterica | 4427 | 3 | Gastrointestinal tract in animals | Facultative | |
| Salmonella typhimurium | 4425 | 3 | Gastrointestinal tract in animals | Facultative | |
| Shigella boydii | 4136 | 3 | Gastrointestinal tract in animals | Facultative | |
| Shigella dysenteriae | 4274 | 2 | Gastrointestinal tract in animals | Facultative | |
| Shigella flexneri 2a | 4182 | 3 | Gastrointestinal tract in animals | Facultative | |
| Shigella sonnei | 4223 | 3 | Gastrointestinal tract in animals | Facultative | |
| Gammaproteobacteria/ Pasteurellaceae | Actinobacillus pleuropneumoniae | 2012 | 2 | Lower respiratory tract of pigs | Facultative |
| Actinobacillus succinogenes | 1883 | 2 | Bovine rumen | Anaerobic | |
| Haemophilus ducreyi | 1717 | 1 | Animal mucous membranes | Anaerobic | |
| Haemophilus influenzae | 1791 | 2 | Animal mucous membranes | Facultative | |
| Mannheimia succiniciproducens | 2380 | 2 | Bovine rumen | Anaerobic | |
| Pasteurella multocida | 2015 | 1 | Mucous membranes of the intestinal, genital and respiratory tissues | Facultative | |
| Spirochaetales | Treponema denticola | 2767 | 6 | Oral cavity | Anaerobic |
DISCUSSION
Whole-genome shotgun and metagenomic sequencing projects have provided a new and powerful tool in the study of community organization and metabolism in natural microbial communities (35–37). Recently, such methods have been extended to analyze symbiotic relationships. One project involved an analysis of microbes from a marine oligochaete O. algarvensis, which lacks a mouth, gut, anus or nephridial excretory system, and contains several bacterial endosymbionts that are located just below the worm cuticle (26). These endosymbionts include two sulfur-oxidizing gammaproteobacteria (γ1 and γ3) and two sulfate-reducing deltaproteobacteria (δ1 and δ4). Identification of selenoprotein genes in such an unusual symbiotic system may help understand the role of selenium and other micronutrients in the intricate interactions that form such a complex, adaptive consortium.
In the present study, we employed a procedure that analyzes Sec/Cys pairs in homologous sequences to characterize the selenoproteomes of symbiotic microorganisms in the gutless worm. A total of 82 genes that belonged to 24 previously described prokaryotic selenoprotein families and 17 sequences that belonged to six new selenoprotein families were identified. Most selenoproteins were found to occur in δ1 symbiont, which contained 44 known selenoproteins (21 families) and 13 new selenoproteins (6 families). Although the genome size of δ1 symbiont is ∼13.5 Mb, which is larger than most other deltaproteobacteria, its reconstruction revealed a single species (26). If this is the case, then our study identified an organism, which has the largest selenoproteome reported to date (57 selenoproteins) of any organism, including eukaryotes and archaea.
Most detected selenoproteins were homologs of thiol-based redox enzymes and contained conserved redox motifs. In contrast, such known redox motifs were largely absent in new selenoproteins identified in the metagenomic dataset. In addition, analysis of secondary structures revealed that these new selenoproteins did not contain thioredoxin-like fold, which is a dominant fold in selenoproteins identified in several marine environmental sequencing projects (23,38). Perhaps, additional redox reactions that are carried out by new selenoproteins occur in these symbionts.
Besides the unusually high number of selenoproteins, 10 Pyl-containing proteins were identified in the metagenomic dataset. δ1 contained eight of these sequences that belonged to MtbB and MttB families. Thus, the δ1 symbiont is also the organism, which has the largest number of Pyl-containing proteins in bacteria. Previously, only one bacterial protein, from D. hafniense, was known to possess Pyl. Therefore, identifying so many pyrroproteins in the same bacterium is truly remarkable.
We previously proposed that UAG may be an ambiguous codon in some archaea, wherein it could serve as either Pyl codon or a stop signal. However, in D. hafniense, UAG is frequently used as a stop signal, suggesting an unknown mechanism that allows ribosomes to recognize function of specific UAG codons. By analogy to Sec, which is inserted with the help of SECIS elements, PYLIS elements may be present in bacterial pyrroprotein genes. However, our analysis of genes coding for Pyl-containing proteins revealed no common RNA structures. Additional RNA structure searches should be carried out in the future. The current set of Pyl-containing proteins provides an excellent dataset for further interrogation.
Given that most symbiotic and host-associated bacteria have lost the ability to utilize Sec or only possess a limited number of selenoproteins, the dramatic abundance of selenoproteins in the two endosymbiotic deltaproteobacteria, especially δ1 that also contains many Pyl-containing proteins, is remarkable, raising a series of questions regarding evolution and function of these proteins, as well as their roles in symbiosis. It has been suggested that most selenoproteins evolved from their Cys-containing homologs and anaerobic environments could support the use of Sec (27). Compared to most other symbionts and host-associated organisms, which seem to live under aerobic or microaerobic conditions, the obligate anaerobic environment of the two symbionts may be one reason for evolution of new selenoproteins. In addition, compared to the environments where other hosts live, seawater could provide a constant supply of selenium for Sec biosynthesis in these symbionts. An alternative hypothesis is that the host worm needs more efficient metabolism and waste management, which are provided by its symbionts because of the lack of digestive and excretory systems. These special needs might have led to selective advantage of harboring multiple symbionts that utilize amino acids that provide catalytic advantages to various metabolic systems, such as Sec in many redox proteins and Pyl in methylamine methyltransferases.
Symbiotic deltaproteobacteria in the gutless worm evolved as organisms that support the broadest use of the genetic code, utilizing 63 of 64 codons to code for 22 amino acids. It would be interesting to examine if this and other symbiotic systems provide selective advantage to further expand the genetic code, either utilizing a third stop signal, UAA, or using some codons to insert multiple non-canonical or common amino acids.
CONCLUSIONS
In this study, we report a comprehensive analysis of Sec and Pyl utilization in the Olavius symbiont metagenomic database by identifying selenoproteins and Pyl-containing proteins. An organism, δ1 symbiont, which contains the largest number of both selenoproteins and pyrroproteins in any organism was identified. This dataset provides opportunities for addressing critical questions regarding evolutionary factors that influence utilization of Sec and Pyl, further extension of the genetic code and understanding of molecular mechanisms of recoding.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
ACKNOWLEDGEMENTS
Supported by NIH GM061603 to V.N.G. We thank the Research Computing Facility of the University of Nebraska – Lincoln for the use of Prairiefire supercomputer, and Drs Dmitri Fomenko and Alexey Lobanov for helpful comments. Funding to pay the Open Access publication charges for the article was provided by NIH GM061603.
Conflict of interest statement. None declared.
REFERENCES
- 1.Böck A, Forchhammer K, Heider J, Leinfelder W, Sawers G, Veprek B, Zinoni F. Selenocysteine: the 21st amino acid. Mol. Microbiol. 1991;5:515–520. doi: 10.1111/j.1365-2958.1991.tb00722.x. [DOI] [PubMed] [Google Scholar]
- 2.Stadtman TC. Selenocysteine. Annu. Rev. Biochem. 1996;65:83–100. doi: 10.1146/annurev.bi.65.070196.000503. [DOI] [PubMed] [Google Scholar]
- 3.Gladyshev VN, Hatfield DL. Selenocysteine-containing proteins in mammals. J. Biomed. Sci. 1999;6:151–160. doi: 10.1007/BF02255899. [DOI] [PubMed] [Google Scholar]
- 4.Hatfield DL, Gladyshev VN. How selenium has altered our understanding of the genetic code. Mol. Cell. Biol. 2002;22:3565–3576. doi: 10.1128/MCB.22.11.3565-3576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Low S, Berry MJ. Knowing when not to stop: selenocysteine incorporation in eukaryotes. Trends Biochem. Sci. 1996;21:203–208. [PubMed] [Google Scholar]
- 6.Böck A. Biosynthesis of selenoproteins—an overview. Biofactors. 2000;11:77–78. doi: 10.1002/biof.5520110122. [DOI] [PubMed] [Google Scholar]
- 7.Rother M, Resch A, Wilting R, Böck A. Selenoprotein synthesis in archaea. Biofactors. 2001;14:75–83. doi: 10.1002/biof.5520140111. [DOI] [PubMed] [Google Scholar]
- 8.Driscoll DM, Copeland PR. Mechanism and regulation of selenoprotein synthesis. Annu. Rev. Nutr. 2003;23:17–40. doi: 10.1146/annurev.nutr.23.011702.073318. [DOI] [PubMed] [Google Scholar]
- 9.Copeland PR, Stepanik VA, Driscoll DM. Insight into mammalian selenocysteine insertion: domain structure and ribosome binding properties of Sec insertion sequence binding protein 2. Mol. Cell. Biol. 2001;21:1491–1498. doi: 10.1128/MCB.21.5.1491-1498.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nirenberg M, Caskey T, Marshall R, Brimacombe R, Kellog D, Doctor BP, Hatfield D, Levin J, Rothman F, et al. The RNA code in protein synthesis. Cold Spring Harb. Symp. Quant. Biol. 1966;31:11–24. doi: 10.1101/sqb.1966.031.01.008. [DOI] [PubMed] [Google Scholar]
- 11.Thanbichler M, Böck A. The function of SECIS RNA in translational control of gene expression in Escherichia coli. EMBO J. 2002;21:6925–6934. doi: 10.1093/emboj/cdf673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Z, Reches M, Groisman I, Engelberg-Kulka H. The nature of the minimal ‘selenocysteine insertion sequence’ (SECIS) in Escherichia coli. Nucleic Acids Res. 1998;26:896–902. doi: 10.1093/nar/26.4.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu XM, Carlson BA, Mix H, Zhang Y, Saira K, Glass RS, Berry MJ, Gladyshev VN, Hatfield DL. Biosynthesis of selenocysteine on its tRNA in eukaryotes. PLoS Biol. 2006;5:e4. doi: 10.1371/journal.pbio.0050004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hao B, Gong W, Ferguson TK, James CM, Krzycki JA, Chan MK. A new UAG-encoded residue in the structure of a methanogen methyltransferase. Science. 2002;296:1462–1466. doi: 10.1126/science.1069556. [DOI] [PubMed] [Google Scholar]
- 15.Srinivasan G, James CM, Krzycki JA. Pyrrolysine encoded by UAG in Archaea: charging of a UAG-decoding specialized tRNA. Science. 2002;296:1459–1462. doi: 10.1126/science.1069588. [DOI] [PubMed] [Google Scholar]
- 16.Kryukov GV, Kryukov VM, Gladyshev VN. New mammalian selenocysteine-containing proteins identified with an algorithm that searches for selenocysteine insertion sequence elements. J. Biol. Chem. 1999;274:33888–33897. doi: 10.1074/jbc.274.48.33888. [DOI] [PubMed] [Google Scholar]
- 17.Lescure A, Gautheret D, Carbon P, Krol A. Novel selenoproteins identified in silico and in vivo by using a conserved RNA structural motif. J. Biol. Chem. 1999;274:38147–38154. doi: 10.1074/jbc.274.53.38147. [DOI] [PubMed] [Google Scholar]
- 18.Castellano S, Morozova N, Morey M, Berry MJ, Serras F, Corominas M, Guigo R. In silico identification of novel selenoproteins in the Drosophila melanogaster genome. EMBO Rep. 2001;2:697–702. doi: 10.1093/embo-reports/kve151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Gladyshev VN. An algorithm for identification of bacterial selenocysteine insertion sequence elements and selenoprotein genes. Bioinformatics. 2005;21:2580–2589. doi: 10.1093/bioinformatics/bti400. [DOI] [PubMed] [Google Scholar]
- 20.Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, Gladyshev VN. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 21.Castellano S, Novoselov SV, Kryukov GV, Lescure A, Blanco E, Krol A, Gladyshev VN, Guigo R. Reconsidering the evolution of eukaryotic selenoproteins: a novel nonmammalian family with scattered phylogenetic distribution. EMBO Rep. 2004;5:71–77. doi: 10.1038/sj.embor.7400036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kryukov GV, Gladyshev VN. The prokaryotic selenoproteome. EMBO Rep. 2004;5:538–543. doi: 10.1038/sj.embor.7400126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Fomenko DE, Gladyshev VN. The microbial selenoproteome of the Sargasso Sea. Genome Biol. 2005;6:R37. doi: 10.1186/gb-2005-6-4-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walker A, Crossman LC. This place is big enough for both of us. Nat. Rev. Microbiol. 2007;5:90–92. doi: 10.1038/nrmicro1601. [DOI] [PubMed] [Google Scholar]
- 25.Ruby EG, Henderson B, McFall-Ngai M. We get by with a little help from our (little) friends. Science. 2004;303:1305–1307. doi: 10.1126/science.1094662. [DOI] [PubMed] [Google Scholar]
- 26.Woyke T, Teeling H, Ivanova NN, Huntemann M, Richter M, Gloeckner FO, Boffelli D, Anderson IJ, Barry KW, et al. Symbiosis insights through metagenomic analysis of a microbial consortium. Nature. 2006;443:950–955. doi: 10.1038/nature05192. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Romero H, Salinas G, Gladyshev VN. Dynamic evolution of selenocysteine utilization in bacteria: a balance between selenoprotein loss and evolution of selenocysteine from redox active cysteine residues. Genome Biol. 2006;7:R94. doi: 10.1186/gb-2006-7-10-r94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 29.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Badger JH, Hoover TR, Brun YV, Weiner RM, Laub MT, Alexandre G, Mrazek J, Ren Q, Paulsen IT, et al. Comparative genomic evidence for a close relationship between the dimorphic prosthecate bacteria Hyphomonas neptunium and Caulobacter crescentus. J. Bacteriol. 2006;188:6841–6850. doi: 10.1128/JB.00111-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard NU, Martinez A, Sullivan MB, Edwards R, et al. Community genomics among stratified microbial assemblages in the ocean's interior. Science. 2006;311:496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Baranov PV, Atkins JF, Gladyshev VN. Pyrrolysine and selenocysteine use dissimilar decoding strategies. J. Biol. Chem. 2005;280:20740–20751. doi: 10.1074/jbc.M501458200. [DOI] [PubMed] [Google Scholar]
- 33.Longstaff DG, Blight SK, Zhang L, Green-Church KB, Krzycki JA. In vivo contextual requirements for UAG translation as pyrrolysine. Mol. Microbiol. 2007;63:229–241. doi: 10.1111/j.1365-2958.2006.05500.x. [DOI] [PubMed] [Google Scholar]
- 34.Dhebri AR, Afify SE. Free gas in the peritoneal cavity: the final hazard of diathermy. Postgrad. Med. J. 2002;78:496–497. doi: 10.1136/pmj.78.922.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hallam SJ, Putnam N, Preston CM, Detter JC, Rokhsar D, Richardson PM, DeLong EF. Reverse methanogenesis: testing the hypothesis with environmental genomics. Science. 2004;305:1457–1462. doi: 10.1126/science.1100025. [DOI] [PubMed] [Google Scholar]
- 36.Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW, Podar M, Short JM, Mathur EJ, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 37.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fomenko DE, Xing W, Adair BM, Thomas DJ, Gladyshev VN. High-throughput identification of catalytic redox-active cysteine residues. Science. 2007;315:387–389. doi: 10.1126/science.1133114. [DOI] [PubMed] [Google Scholar]