Abstract
DNA double-strand breaks can be repaired by illegitimate recombination without extended sequence homology. A distinct mechanism namely microhomology-mediated recombination occurs between a few basepairs of homology that is associated with deletions. Ionizing radiation and restriction enzymes have been shown to increase the frequency of nonhomologous integration in yeast. However, the mechanism of such enhanced recombination events is not known. Here, we report that both ionizing radiation and restriction enzymes increase the frequency of microhomology-mediated integration. Irradiated yeast cells displayed 77% microhomology-mediated integration, compared to 27% in unirradiated cells. Radiation-induced integration exhibited lack of deletions at genomic insertion sites, implying that such events are likely to occur at undamaged sites. Restriction enzymes also enhanced integration events at random non-restriction sites via microhomology-mediated recombination. Furthermore, generation of a site-specific I-SceI-mediated double-strand break induces microhomology-mediated integration randomly throughout the genome. Taken together, these results suggest that double-strand breaks induce a genome-wide microhomology-mediated illegitimate recombination pathway that facilitates integration probably in trans at non-targeted sites and might be involved in generation of large deletions and other genomic rearrangements.
INTRODUCTION
Double-strand breaks (DSBs) can be produced exogenously by ionizing radiation and chemical DNA damaging agents or by endogenous free radicals generated during cellular metabolism. Eukaryotic cells have evolved two main pathways to repair DSBs: homologous recombination relies on extensive sequence homology between the damaged DNA and donor template, whereas illegitimate recombination, or nonhomologous end joining (NHEJ), involves end joining in the absence of DNA sequence homology (1). Some illegitimate recombination events are characterized by a few basepairs (bp) of homology shared at the ends of the two recombination junctions, so called microhomology-mediated recombination (MHMR) (2). This class of events is independent of the end-binding Yku70/Yku80 heterodimer, and thus is distinct from the classical NHEJ pathway (3–7). As well, MHMR is a mutagenic pathway that is often associated with deletion of the sequences spanning one of the microhomology regions and the intervening sequence.
MHMR has been implicated in generation of large deletions and other genomic rearrangements in mammalian cells. Microhomology is often found at the recombination junctions of radiation-induced genomic rearrangements (8,9), implying that radiation-induced DSBs are repaired preferentially by MHMR. Furthermore, 50% of large deletions mediated by MHMR are associated with human disorders (10–14).
In yeast, DSBs are mainly repaired by homologous recombination, but different mechanisms of illegitimate recombination have been evident in the absence of a homologous donor or an essential gene in the homologous recombination pathway (2,7,15). By transformation of a URA3-containing fragment into haploid yeast cells lacking the URA3 gene and subsequent selection for Ura+ transformants (2,3), three classes of spontaneous illegitimate recombination events were identified. The first class is due to MHMR, characterized by 2–4 bp of microhomologies at the junctions (2). The second class is mediated by topoisomerase I where the transforming DNA integrates next to the preferred cleavage sites of topoisomerase I, CTT or GTT (16). The final class involves in vivo ligation of transforming DNA to mitochondrial DNA fragments that seem to be capable of autonomous replication (2).
Co-transformation of a recombination substrate with restriction enzymes BamHI, BglII and KpnI increases the efficiency of DNA integration by several fold and the majority of events integrated into the restriction sites introduced by the co-transformed restriction enzyme (3,17), this process being termed restriction enzyme-mediated integration (REMI). The simple interpretation is that the co-transformed restriction enzymes create DSBs at the genomic restriction sites that attract the transforming DNA ends. Restriction enzymes were subsequently used to facilitate integration in Dictyostelium discoideum for random mutagenesis and simultaneous tagging of the mutant gene (18).
Ionizing radiation enhances the frequency of nonhomologous integration (NHI) in yeast (19). In addition, other DNA damaging agents including UV, bleomycin, camptothecin, VP-16 and hydrogen peroxide also enhance gene integration in mammalian cells (20–25). These DNA damaging agents may eventually lead to the formation of DSBs and suggest that DSBs could be the inducing agent for the enhancement of integration.
The DNA sequences of recombining junctions have been determined for very few DNA damage-enhanced illegitimate recombination events and the possibility that DNA damage induces illegitimate recombination events at undamaged sites has not previously been investigated. In this manuscript, we investigate the effects of ionizing radiation and restriction enzymes on NHI by sequence analysis of the integration target sites. We report here that both ionizing radiation and restriction enzymes increase the frequency of microhomology-mediated integration. In addition, a site-specific I-SceI-mediated DSB induces microhomology-mediated integration randomly throughout the genome, suggesting that DSBs induce MHMR probably in trans at non-targeted sites.
MATERIALS AND METHODS
Strains and Media
Experiments were performed in the haploid Saccharomyces cerevisiae strain RSY12 (MATa leu2-3,112 his3-11,15 ura3▵::HIS3), in which the entire URA3 open reading frame and promoter sequence was replaced by the HIS3 gene (17). Strain YWY200 was constructed in the RSY12 background by integration of plasmid pWY203 containing I-SceI gene under the GAL1 promoter into LYS2 locus (Yap,W. and Schiestl, R. H., unpublished results). The construct containing the 24-bp I-SceI recognition site and a LEU2 marker flanked by 50 bp of upstream and downstream sequence of TRP1 was generated by PCR and transformed into YWY200 to generate strain YWY200-I-SceI. Escherichia coli strain DH5α was used for the maintenance and amplification of plasmid DNA.
Plasmids
YEplac195 contains the URA3 marker for selection and the two-micron origin of replication and was used to control for transformation efficiency. Plasmids pM150 and pM151 were described previously (17) and contain a 1.1 kb URA3 HindIII fragment.
Transformation
For yeast transformation the ‘Lithium Acetate/Single-Stranded DNA/PEG’ transformation method was used (26). In previous studies, ∼10% of spontaneous NHI events were ligated to mitochondrial DNA fragments (2). The occurrence of these events is probably related to the fact that some mitochondrial DNA fragments can function as an origin of replication in the nucleus (27). We excluded these mitochondrial events in the current study by checking the frequency of loss of the Ura3 phenotype on medium containing 5-fluoro-orotic acid (FOA) because these events are not due to integrations into genomic DNA.
Molecular techniques
Standard methods were followed except as noted below. pM151 was digested with BglII and pM150 with KpnI and prepared as described (17). The spontaneous and radiation-induced integration target sites were determined by direct genomic sequencing. Yeast genomic DNA was purified using the genomic purification tips (100/T) (Qiagen, Valencia, CA, USA). Fifteen microgram of genomic DNA was used for a double big dye sequencing reaction (28) with the primer pM151-3559 5′-GTGCACCATATG CGGTGTGAAATACC-3 to sequence the forward direction and the primer pM151-71 5′-AATCTAAGTCTGTGCTCCTTCCTTCG-3′ to sequence the reverse direction. Sequencing products were purified by Centri-Sep columns (Princeton Separations, Adelphia, NJ, USA) and loaded on an ABI 371 sequencer.
Restriction enzyme-induced integration
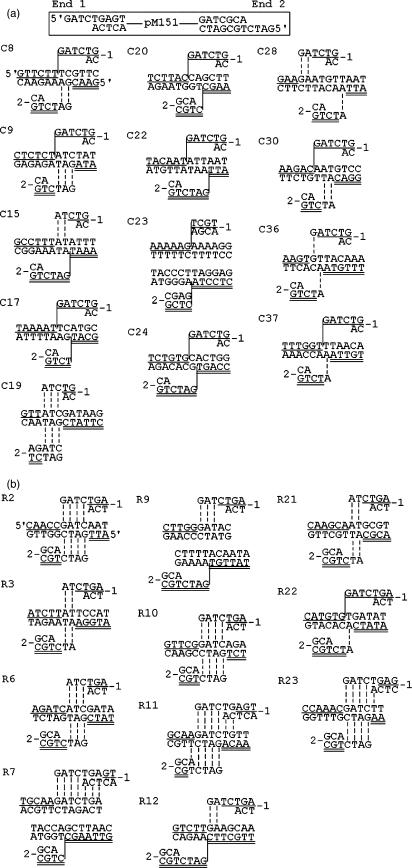
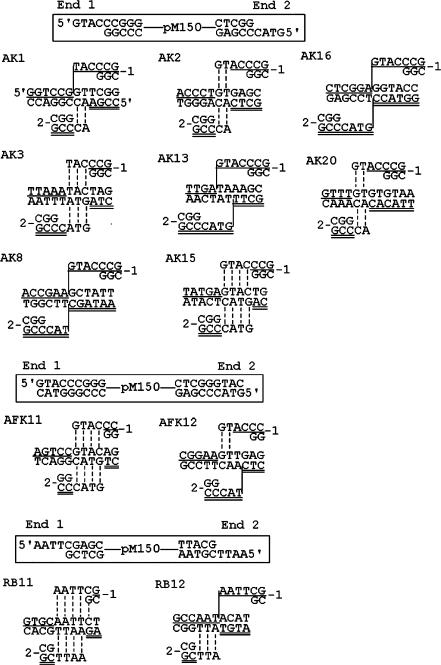
Plasmid DNA was digested with restriction enzyme, precipitated with ethanol and resuspended in 200 µl of 0.01 M Tris-HCl (pH 7.5), 0.05 M EDTA, 1% sodium dodecyl sulfate and 100 µg of proteinase K/ml. After 30 min incubation at 37°C, the sample was extracted with phenol–chloroform–isoamyl alcohol, precipitated with ethanol, washed with 70% ethanol and vacuum dried. The pellet was dissolved in sterile water, and the yeast cells were then transformed with this solution. Also, 200U of restriction enzyme and 1/10 volume of the restriction enzyme buffer were added to the transformation mixtures. The restriction enzyme-induced integration target sites were cloned by plasmid rescue as described previously (17). Asp718 was purchased from Roche (Basel, Switzerland). All other enzymes were purchased from New England Biolabs (Ipswich, MA, USA). Thirty-eight junctions from 19 integration events were sequenced in which both junctions resulted from NHI (Figures 4 and 5B). Also included in our analysis are three junctions from NHI events that were previously published [eventBH10 in Figure 2A, events SB3 and SB13 in Figure 2B in ref. (17)].
Figure 4.
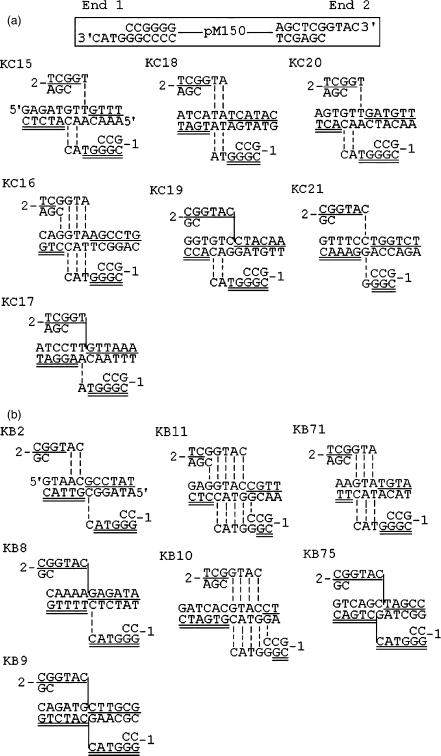
Target sites of NHI events of KpnI-linearized pM150 (3′PSS) in wild-type strain RSY12. (A) Spontaneous events. (B) After co-transformation with BglII. The genomic location, locus and gene of the target sites are as follows: KC15, Chr. III, YCL067C, HMLALPHA2; KC16, Chr. IX, YIL137C, RBF108; KC17, Chr. XI, YKL197C, PEX1; KC18, Chr. II, 21590, intergenic region; KC19, Chr. XIV, YNL298W, CLA4; KC20, Chr. II, YBL104C, hypo. ORF; KC21, Chr. VII, 144125, intergenic region; KB2, Chr. VII, YGL062W, PYC1; KB8, Chr. IV, YDL042C, SIR2; KB9, Chr. III, YCR061W, hypo. ORF; KB11, Chr. IV, 11262, intergenic region; KB10, Chr. XIII, YML043C, RRN11; KB71, Chr. III, YCL057W, PRD1; KB75, Chr. XIII, YML086C, ALO1.
Figure 5.
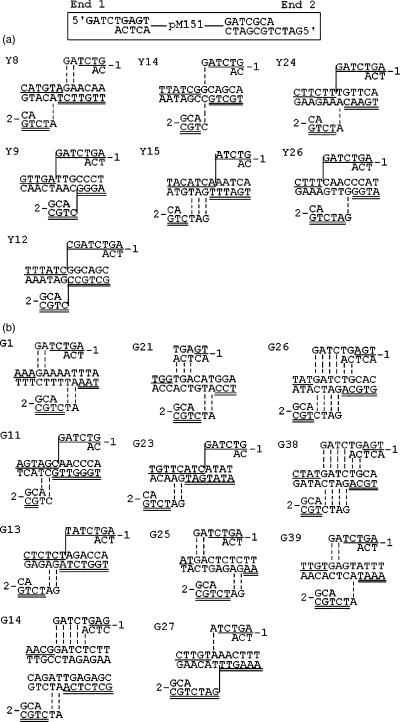
Target sites of NHI events of BglII-linearized pM151 in (A) YWY200 strain in the absence of the I-SceI site in galactose-growing medium. (B) YWY200-SceI strain in the presence of the I-SceI site at Chr. IV upon galactose induction of I-SceI endonuclease. The genomic location, locus and gene of the target sites are as follows: Y8, Chr. V, ARS521; Y12, Chr. XII, YLR306W, UBC12; Y15, Chr. XI, YKL044W, hypo. ORF; Y24, Chr. VII, YGR148C, RPL24B; Y26, Chr. XII, NTS1-2; G11, Chr. XII, NTS1-2; G13, Chr. VII, YGR142W, BTN2; G14, Chr. XII, RDN37-2, a 939 bp deletion; G21, Chr. IV, YDR030C, RAD28; G23, Chr. VI, YFL065C, hypo, ORF; G25, Chr. XII, RDN25-2; G26, Chr. XIII, YML103C, NUP188; G27, Chr. VII, YGL131C, SNT2; G38, Chr. XVIII, YML103C, NUP188; G39, Chr. II, YBR140C, IRA1.
Figure 2.
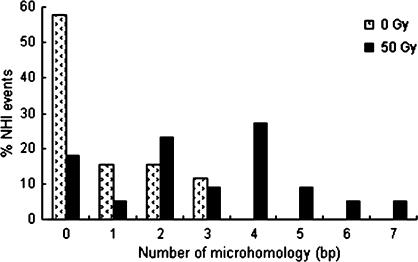
The distribution of microhomology usage in spontaneous and radiation-induced NHI events in strain RSY12.
γ-Irradiation
After transformation of plasmid DNA, yeast cells were plated onto synthetic complete medium lacking uracil and exposed to 50 Gy of γ-ray using a Mark-I irradiator (Shepherd and Associates, San Fernando, CA, USA), with a Cs-137 source and a dose rate of 529 cGy/min.
Statistical analysis
The frequencies of microhomology-mediated recombination with and without exposure to γ-ray, restriction enzymes and I-SceI endonuclease were analyzed by the χ2 test.
RESULTS
To examine the effect of γ-irradiation on NHI, a BglII-linearized plasmid pM151 containing the URA3 gene was transformed into yeast strain RSY12, which lacks any homology to the URA3 gene. Cells were then exposed to 50 Gy of γ-rays which causes an average of two DSBs per cell (29). Cells expressing the Ura+ phenotype are the results of the URA3 containing plasmid pM151 that has either integrated into the genome or has captured an origin of replication. In parallel, a self-replicating plasmid YEplac195 that contains a two-micron origin and the URA3 gene was used to transform RSY12 to normalize changes in the transformation efficiency caused by radiation. The frequency of spontaneous NHI events was about 0.75 per μg DNA per 104 YEplac195 transformants. Exposure of yeast cells to γ-irradiation in three different experiments caused an average of 4.7-fold increase in NHI frequency, which is similar to previous results (19). In addition, γ-irradiation caused an average of 6.3-fold increase in NHI frequency using a SalI-linearized plasmid pM151 as integrating DNA, indicating that radiation-induced integration is not specific to the BglII-linearized substrate (data not shown).
To investigate the mechanism of radiation-enhanced integration, we isolated genomic DNA from yeast colonies that underwent NHI events and sequenced the regions spanning the junctions between genomic DNA and the integrated plasmid. Obtained sequences were aligned with yeast chromosomal DNA sequences in the Saccharomyces Genome Database (SGD) to identify integration target sites. We randomly selected and analyzed 13 stable transformants of spontaneous integration events and 11 transformants derived after irradiation. Among the spontaneous events, 7 out of 26 junctions (27%) contained 2–3 bp of microhomology between the protruding single-stranded (PSS) ends of the integrating DNA and the genomic target sites (Figure 1A). Remarkably, among integration events in irradiated cells, 17 out of 22 junctions (77%) displayed 2–7 bp of microhomology (Figure 1B). This 2.9-fold increase in the fraction of MHMR events is significantly different (P < 0.001) in comparison to spontaneous events. Forty-five percent of events in irradiated yeast cells utilized 4 or more bp of microhomology (27% with 4 bp, 9% with 5 bp, 4.5% with both 6 and 7 bp) while no events with 4 or more bp were observed in unirradiated cells (Figure 2) indicating that microhomology length is increased in irradiated cells in comparison to spontaneous events (P < 0.0005).
Figure 1.
Target sequences of NHI events of BgIII-linearized pM151 in wild-type strain RSY12. (A) Spontaneous events. (B) After exposure to 50Gy of γ-irradiation. Shown are the plasmid DNA substrate, the target sequences and the genomic locations into which the plasmid integrated. The ends of the integrating DNA are shown above and below the genomic target sequences. The numbers 1 and 2 correspond to the sequence of the 5′ (1) and 3′ (2) PSS ends of the integrating DNA. The region of microhomology between the ends of integrating DNA and genomic target DNA is visualized by vertical dashed lines, with the crossover occurring at either one of the dashed lines. If there is no overlapping microhomology sequence between the recombining DNAs, the site of crossover is represented by vertical solid line. End 1 of the integrating DNA sequence is shown as single underlined sequence and after the crossover continuing at the target. End 2 is double underlined continuing after the crossover. The genomic location, locus and gene of the target sites are as follows: C8, Chr. (Chromosome) I, promoter of YAL022C, FUN26; C9, Chr. VI, promoter of tA(AGC), tRNA; C15, Chr. XVI, YPL070W, MUK1; C17, CHR. VIII, YIL071C, PCI8; C19, Chr. XII, RDN37-1, 35SrRNA; C20, Chr. XII, RDN5-1, 5SrRNA; C22, Chr. II, promoter of YBL079W, NUP170; C23, Chr. VI, hypo. (hypothetical) ORF; C24, Chr. XVI, YPR124W, CTR1; C28, Chr. XI 151654, intergenic region; C30, Chr. XIV, YNL012W, SPO1; C36, Chr. XVI 630665, intergenic region; C37, Chr. XIV 669282, intergenic region; R2, Chr. IV, YDR131C, hypo. ORF; R3, Chr. XIII, promoter of YMR251W, GTO3; Chr. XV, ROL089C, HAL9; R7, Chr. V, YER135C hypo. ORF/Chr. XII, RDN5-1, 5SrRNA; R9, Chr. XV, YOR313C, SPS4/Chr. XIII, YMR310C, hypo. ORF; R10, Chr. II, promoter of YBR001C, NTH2; R11, Chr. V, YER135C, hypo. ORF; R12, Chr. IV, YKR017C, hypo. ORF; R21, Chr. II, YBL060W, hypo. ORF; R22, Chr. II, YBL060W, hypo. ORF; R23, Chr. IV, YDR258C, HSP78.
As radiation-induced DSBs are modified and required to be processed prior to religation (30), deletions are often found at the breakpoints (31,32). However, the majority of spontaneous and radiation-induced integration events had the genomic sequences flanking the integrating substrate maintained without any deletion, implying that spontaneous and radiation-induced integrations are likely to occur at sites other than the sites of DSBs. All spontaneous and radiation-induced illegitimate integration events were randomly distributed throughout the yeast genome without bias for any particular chromosome position, expressed gene or ORF.
Previous experiments have shown co-transformation with restriction enzymes increase the efficiency of DNA integration into genomic sites of the restriction enzymes (3,17). When Asp718-linearized plasmid was transformed into yeast cells in the presence of KpnI, the integration efficiency increased 2- to 3-fold yet less than 1/3 integrated at the KpnI sites (17). Furthermore, when BglII was co-transformed with a KpnI-linearized plasmid, no integrations were observed at the BglII sites (17). These results suggest that co-transformation of linearized DNA substrate with restriction enzymes may lead to increased integration efficiency into genomic sites other than the ones recognized by the restriction enzymes. In this work, we continued our previous studies and analyzed the events obtained after co-transformation with restriction enzymes by sequence analysis. We examined the following enzyme combinations: BamHI-BamHI, SalI-BglII, EcoRI-BglII, KpnI-BglII, Asp718-KpnI and Asp718 (filled PSS ends)-KpnI, in which the former enzyme was used to linearize the integrating plasmid pM150 and the latter enzyme was added during the transformation process (Table 1). We classified all events after restriction enzyme addition into different integration classes (restriction enzyme-mediated integration (REMI) and NHI) and compared this distribution with the control experiments lacking co-transformation with restriction enzymes.
Table 1.
Classification of integration junctions in the presence and absence of restriction enzymes
| Linearizing enzyme | Co-transformation enzymeb | Relative frequency of integrationc | Fold increase in integration | Junctions | Length of microhomology (bp)d | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | REMI mediatede | NHI | 5 | 4 | 3 | 2 | 0–1 | ||||
| BamHI | – | 2.1 ± 0.1 | – | 14 | 0 | 14 | 0 | 0 | 7 | 1 | 6 |
| EcoRI | – | 1.1 ± 0.1 | – | 8 | 0 | 8 | 0 | 0 | 3 | 1 | 4 |
| KpnI | – | 1.8 ± 0.4 | – | 14 | 0 | 14 | 0 | 1 | 1 | 5 | 7 |
| Controla | 36 | 0 | 36 | 0 | 1 | 11 | 7 | 17 | |||
| BamHI | BamHI | 11.5 ± 0.7 | 5.5 ± 1.2** | 8 | 7 | 1 | 0 | 1 | 0 | 0 | 0 |
| SalI | BglII | 10.8 ± 2.1 | 8.3 ± 1.2** | 18 | 16 | 2 | 0 | 1 | 0 | 1 | 0 |
| EcoRI | BglII′ | 3.2 ± 0.3 | 2.9 ± 0.3** | 8 | 4 | 4 | 1 | 1 | 1 | 0 | 1 |
| KpnI | BglII | 4.3 ± 0.8 | 2.4 ± 0.8** | 14 | 0 | 14 | 3 | 1 | 2 | 1 | 7 |
| Asp718 | KpnI | 5.8 ± 0.8 | 2.3 ± 0.3** | 22 | 6 | 16 | 0 | 2 | 2 | 5 | 7 |
| Asp718 filled | KpnI | 3.26 ± 0.56 | 1.4** | 8 | 4 | 4 | 0 | 2 | 0 | 1 | 1 |
| Total | 78 | 37 | 41 | 4 | 8 | 5 | 8 | 16 | |||
aIn the control, the BamHI or EcoRI-linearized YIplac211 or the KpnI-linearized pM150 plasmid was transformed into RSY12 without addition of restriction enzyme. The junction sequences which resulted are shown in Figures 1 and 4 in ref. (3) (BamHI and EcoRI-linearized YIplac211) and Figure 4A (KpnI-linearized pM150).
bFor example, in the combination of BamHI and BamHI, BamHI-linearized pM150 plasmid was transformed into RSY12 in the presence of BamHI. In the combination of SalI and BglII, SalI-linearized pM150 plasmid was transformed into RSY12 in the presence of BglII. The KpnI-BglII junction sequences are shown in Figure 4B and the junction sequences of other restriction enzymes-induced integration events are shown in Figure 3.
cData are taken from ref. (1). Number of integration events per microgram of DNA per 104 2 μm transformants. This number represents the average of five of more experiments ± the standard error. **, t-test indicates a statistically significant difference (P < 0.05) from control values.
dNumber of bases at the ends of integrating plasmid DNA which is homologous to the genomic target sequences.
eREMI stands for restriction enzyme-mediated integration.
Among events from co-transformation with restriction enzymes, 12 out of 41 junctions (29%) displayed 4 or more bp of microhomologies between the PSS ends of the integrating DNA and the genomic sequences whereas only 1 out of 36 junctions (2.7%) was observed in the control (P < 0.005) (Table 1, Figures 3 and 4B). We compared 14 junctions from spontaneous integrations of a 3′ PSS substrate linearized by KpnI (Figure 4A) with 14 junctions obtained from co-transformation with BglII, producing a 5′PSS (Figure 4B). The distances to the closest BglII sites for each of the 14 integration junctions were in agreement with a random distribution in relation to those BglII sites in the genome (data not shown). This suggests that a specific pathway utilizing microhomology was induced by restriction enzymes increasing integration efficiency in trans.
Figure 3.
Target sequences of restriction enzyme-induced NHI events. In Asp718-KpnI (AK) events, KpnI was added to Asp718-linearized plasmid pM150 during transformation. In EcoRI-BglII (RB) events, BglII was added to EcoRI-linearized plasmid pM150. In Asp718 (filled)-KpnI (AFK) events, the ends of Asp718-linearized plasmid pM150 were filled before transformation. KpnI was added to the transformation mixture. The genomic location, locus and gene of the target sites are as follows: AK1, Chr. VII, YGL086W, MAD1; AK3, Intergenic region; AK8, Chr. XII, YLR207W, HRD3; AK2, intergenic region; AK13, Chr. XV, YOR269W, PAC1; AK15, Chr. II, YBR176W, ECM31; AK16, YDR205C; AK20, Chr. VII, YGL081W, hypo. ORF; RB11, intergenic region; RB12, Chr. X, YJR036C, HUL4; AFK11, Chr. IX, YIL085C, KTR7; AFK12, Chr. IV, YDL197C, ASF2.
Restriction enzymes sometimes show ‘star activity’ in the presence of detergents in the buffer or with inappropriate buffers (33) (www.neb.com). It is possible that the enzymes show such star activity under the conditions that occur in the nuclei of cells. The most common types of star activity are single base-pair substitutions or truncations of outer bases (www.neb.com). However, none of the integration sites could be accounted for by the restriction enzyme cutting at such common star activity sites.
To determine if integration occurs close to the site of breakage or at random locations throughout the genome, we overexpressed the I-SceI endonuclease to generate a single DSB at a defined site in the genome. I-SceI is a meganuclease that cleaves at a 24-bp recognition site not present in the yeast genome (34). Cells overexpressing the endonuclease, and having a recombinant I-SceI site, undergo competition between cleavage and religation resulting in a constitutive DSB. Strains YWY200 and YWY200-I-SceI express the I-SceI enzyme under the GAL1 promoter, inserted at the LYS2 locus, while the YWY200-I-SceI strain in addition contains an I-SceI recognition site at the TRP1 locus. Upon 6 h of galactose induction in the YWY200-I-SceI strain, cleavage of the recognition site in the genome by the overexpressed I-SceI was verified by PCR analysis (data not shown). Strains YWY200 and YWY200-I-SceI were grown in complete medium with 2% glucose or 2% galactose for 6 h and transformed with BglII-linearized pM151 to determine the NHI frequency. After induction on galactose-containing medium, the relative frequency of NHI (transformants per μg DNA per 104 YEplac195 transformants) in the strain containing the I-SceI site increased by 4.8-fold, from 0.11 ± 0.05 to 0.50 ± 0.14 whereas in control cells on galactose, not containing the I-SceI site, there was no increase in the NHI frequency; 0.11 ± 0.01 and 0.13 ± 0.04, respectively.
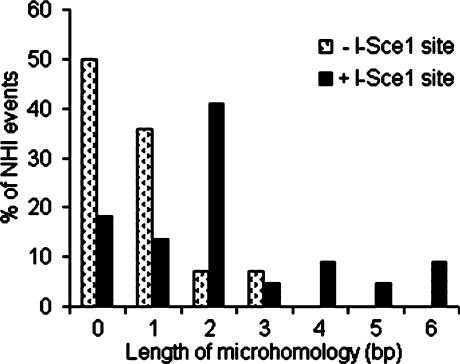
After induction in galactose media, we sequenced 7 and 11 transformants derived from YWY200 and YWY200-I-SceI cells respectively (Figure 5). In the YWY200 cells, 2 out of 14 (14%) junctions showed 2–3 bp of microhomologies whereas 15 out of 22 (68%) junctions from the YWY200-I-SceI cells displayed 2–6 bp of microhomologies, resulting in a significant difference in the occurrence of microhomology usage (P < 0.005) (Figure 6). In seven integration events from YWY200 cells combined with 13 spontaneous events from the isogenic wild-type RSY12 strain (Figure 1A), 9 out of 40 (23%) junctions showed 2–3 bp of microhomologies versus 16 out of 22 (73%) junctions from the YWY200-I-SceI cells utilized microhomologies from 2 to 6 bp, which is also significantly different (P < 0.0005). Strikingly, the integrations from the YWY200-I-SceI cells were distributed randomly over the genome, as opposed to being targeted in proximity to the I-SceI-mediated DSB (Figure 5), indicating that DSBs induce MHMR that is probably in trans at random, non-targeted sites.
Figure 6.
The distribution of microhomology usage in I-SceI-induced NHI events in the absence and presence of the I-SceI site.
DISCUSSION
Our results show that a MHMR pathway is induced after exposure to ionizing radiation, restriction enzymes and I-SceI endonuclease. Radiation caused a 3-fold increase in microhomology-mediated integration. Radiation-induced integration showed lack of deletions at genomic insertion sites, implying that such events are likely to occur at sites that are not damaged. Similarly, restriction enzymes and I-SceI endonuclease increase the frequency of microhomology-mediated integration. Restriction enzymes induce microhomology-mediated integration at non-restriction sites that are undamaged. Furthermore, the I-SceI induced microhomology-mediated integration events were distributed randomly throughout the genome, as opposed to being targeted in proximity to the I-SceI-mediated DSB, suggesting that radiation-induced MHMR events may also occur at undamaged sites. Taken together, these results demonstrate that DSBs induce MHMR that is probably in trans at non-targeted sites.
Ionizing radiation and other DNA damaging agents have previously been shown to increase the frequency of NHI in mammalian cells (20–25), and there is evidence for DNA damage or a DSB-induced pathway of MHMR (8,9). Moreover, the recombined products of MHMR show deletions from the site of breakage to the closest region of microhomology (32). However, in these studies only the rejoining of the DSB at the target site was examined, and illegitimate recombination events at non-targeted sites were not detectable because of their design. Consequently, the author's interpretation in these studies was that during the repair of a DSB, degradation of DNA ends ensues until microhomology is encountered. Few studies have reported ionizing radiation-induced rearrangements not targeted at the DSB. These include radiation-enhanced formation of the λbio transducing phage during prophage induction (35,36). Noteworthy, radiation-enhanced excision by recombination was not observed between random sequences but between hotspot sites of microhomology (36). The author's interpretation was that it is unlikely that these events were DSB mediated and therefore another radiation-induced DNA damage at these sites of microhomology might have enhanced the efficiency (36). This is an unlikely but theoretically possible explanation of our radiation-induced integration events into the genome. However, this possibility does not account for restriction enzyme and I-SceI-induced integration events that are most likely in trans to the DNA damage.
We utilized a system that overexpresses I-SceI endonuclease to constitutively generate a DSB at a specific locus, which enables us to examine whether the enhanced integration events were targeted to the site of DSB or occurred at non-targeted sites throughout the genome. Similar to ionizing radiation and restriction enzymes, I-SceI endonuclease induces microhomology-mediated recombination. Strikingly, all of the I-SceI-mediated DSB-induced integration events were distributed randomly at non-targeted sites throughout the genome, as opposed to being targeted in proximity to the I-SceI-mediated DSB. These results demonstrate that DSBs induce a genome-wide MHMR pathway that facilitates integration probably in trans at non-targeted sites.
Ionizing radiation generates single-stranded breaks, DSBs, base damage and DNA crosslinks. It is possible that radiation-induced integration events occur at these damaged sites. Based on the observations that deletions are frequently observed at the sites of DSBs in yeast (4,15,37) and mammalian cells (9,38); if the integrating substrate was targeted to the damaged site, deletions would have been expected to occur at the site of damage until microhomology was encountered, similar to microhomology-driven deletions in plasmid end-joining assays (39,40). However, this possibility is excluded by our results, which show lack of deletions at the genomic insertion sites in 9 out of 11 independent radiation-induced MHMR events. Furthermore, the probability of radiation-induced damage coinciding with the sites of microhomologies is extremely low. It has been shown that radiation-induced damage occur rather randomly (29). The expected random occurrence for 4, 5, 6 and 7 nt of microhomology is ∼1, 0.3, 0.09 and 0.03%, respectively (5), whereas our frequencies of usage of these microhomologies after irradiation were 27, 9, 4.5 and 4.5%, respectively (P < 0.005). In addition, it is very unlikely that radiation-induced damage occurs at the specific sequences GATC or TCGA that is homologous to the ends of the integrating substrate linearized by BglII or SalI. Even though unlikely it is still possible that radiation-induced integration occurs at the subset of damaged sites that coincide with the microhomology for MHMR.
Among the radiation-induced integration events, the total loss of nucleotides at the PSS ends is 9 out of 88 compared to 23 out of 104 in spontaneous events (P < 0.05) indicating that radiation protected the ends of the integrating DNA substrate. Of the restriction enzyme induced NHI events, 14 out of 56 nt were lost from spontaneous integrations of a KpnI-linearized substrate compared to only 3 out of 56 after co-transformation with BglII (P < 0.005). Thus co-transformation with restriction enzymes, like radiation, seems to protect DNA ends from degradation. This may reflect a faster or more efficient microhomology search after introduction of DSBs somewhere else in the genome. Alternatively, a hypothetical activity that preserves the PSS ends induced by DSBs might have a similar effect and increase the availability of plasmid PSS ends for microhomology search.
A time course study showed that exposure of yeast cells to radiation 6 h before transformation of the integrating substrate increases the frequency of NHI by 2-fold (data not shown), indicating that an increase in microhomology usage after irradiation is associated with an inducible process since the majority of radiation-induced damage is repaired within 2 h in yeast (41). Based on our data, it is likely that a factor is induced that facilitates microhomology search and mediates MHMR.
MHMR has shown to be independent of KU that is distinct from the prototypical NHEJ pathway (3–7). The possibility that induced MHMR could be mediated by a type of homologous recombination with low fidelity is much less likely since homologous integration requires at least 30 bp of homology on each end of the DNA fragment (42,43), as opposed to only a few base pairs of homology in the induced MHMR events. Alternatively, the induced MHMR may occur via a pathway similar to single-strand annealing that takes place between two direct repeats; however, this is also not likely since homologous sequences as small as 29 bp was observed in only 0.2% of the time in single-strand annealing (44).
Under environmental stress, a process called adaptive or stationary-phase mutations arise, which are different from spontaneous growth-dependent mutations in E. coli (45,46). Harris et al. showed that the adaptive mutation is dependent on the RecA-RecBCD homologous recombination pathway (47). The adaptive mutations occur at unselected genes throughout the bacterial chromosome (48,49). The transient global mutagenesis increases the rate of evolution enormously and produces a small population of cells with a hypermutable state that can adapt to a new environment, which is advantageous when genetic variation is a limiting factor for evolution (50). Similarly, cancer cells assume a mutator phenotype to produce high levels of genomic instability during cancer development. Similar mechanisms may be involved in evolution and carcinogenesis. According to our data, it is plausible that a DSB-induced genome-wide MHMR pathway could lead to large-scale genomic rearrangements after a single DSB end invades another genomic location. Such a phenomenon may provide benefits to evolve genetic variants that have growth advantages under genotoxic stress. We propose that this inducible MHMR pathway could be a potential mechanism of adaptive evolution and carcinogenesis in eukaryotes.
ACKNOWLEDGEMENTS
This work was supported by grant No. 1 U19 A1 67769-01 National Institute of Allergy and Infectious Diseases, N.I.H. to William McBride, Project 1 to R.H.S. and a fellowship of the UCLA Chemistry-Biology Interface Training Program with USPHS National Research Service Award GM08496 and a research fellowship of the UC Toxic Substances Research and Teaching Program, both to C.Y.C. Funding to pay the Open Access publication charges for this article was provided by the N.I.H.
Conflict of interest statement. None declared.
REFERENCES
- 1.Derbyshire MK, Epstein LH, Young CS, Munz PL, Fishel R. Nonhomologous recombination in human cells. Mol. Cell. Biol. 1994;14:156–169. doi: 10.1128/mcb.14.1.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schiestl RH, Dominska M, Petes TD. Transformation of Saccharomyces cerevisiae with nonhomologous DNA: illegitimate integration of transforming DNA into yeast chromosomes and in vivo ligation of transforming DNA to mitochondrial DNA sequences. Mol. Cell. Biol. 1993;13:2697–2705. doi: 10.1128/mcb.13.5.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schiestl RH, Petes TD. Integration of DNA fragments by illegitimate recombination in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1991;88:7585–7589. doi: 10.1073/pnas.88.17.7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu X, Gabriel A. Ku-dependent and Ku-independent end-joining pathways lead to chromosomal rearrangements during double-strand break repair in Saccharomyces cerevisiae. Genetics. 2003;163:843–856. doi: 10.1093/genetics/163.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roth DB, Porter TN, Wilson JH. Mechanisms of nonhomologous recombination in mammalian cells. Mol. Cell. Biol. 1985;5:2599–2607. doi: 10.1128/mcb.5.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feldmann E, Schmiemann V, Goedecke W, Reichenberger S, Pfeiffer P. DNA double-strand break repair in cell-free extracts from Ku80-deficient cells: implications for Ku serving as an alignment factor in non-homologous DNA end joining. Nucleic Acids Res. 2000;28:2585–2596. doi: 10.1093/nar/28.13.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boulton SJ, Jackson SP. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 1996;15:5093–5103. [PMC free article] [PubMed] [Google Scholar]
- 8.Nohmi T, Suzuki M, Masumura K, Yamada M, Matsui K, Ueda O, Suzuki H, Katoh M, Ikeda H, et al. Spi(-) selection: an efficient method to detect gamma-ray-induced deletions in transgenic mice. Environ. Mol. Mutagen. 1999;34:9–15. doi: 10.1002/(sici)1098-2280(1999)34:1<9::aid-em2>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 9.Morris T, Thacker J. Formation of large deletions by illegitimate recombination in the HPRT gene of primary human fibroblasts. Proc. Natl. Acad. Sci. USA. 1993;90:1392–1396. doi: 10.1073/pnas.90.4.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woods-Samuels P, Kazazian HH, Jr, Antonarakis SE. Nonhomologous recombination in the human genome: deletions in the human factor VIII gene. Genomics. 1991;10:94–101. doi: 10.1016/0888-7543(91)90489-2. [DOI] [PubMed] [Google Scholar]
- 11.Love DR, England SB, Speer A, Marsden RF, Bloomfield JF, Roche AL, Cross GS, Mountford RC, Smith TJ, et al. Sequences of junction fragments in the deletion-prone region of the dystrophin gene. Genomics. 1991;10:57–67. doi: 10.1016/0888-7543(91)90484-v. [DOI] [PubMed] [Google Scholar]
- 12.Kornreich R, Bishop DF, Desnick RJ. Alpha-galactosidase A gene rearrangements causing Fabry disease. Identification of short direct repeats at breakpoints in an Alu-rich gene. J. Biol. Chem. 1990;265:9319–9326. [PubMed] [Google Scholar]
- 13.Henthorn PS, Smithies O, Mager DL. Molecular analysis of deletions in the human beta-globin gene cluster: deletion junctions and locations of breakpoints. Genomics. 1990;6:226–237. doi: 10.1016/0888-7543(90)90561-8. [DOI] [PubMed] [Google Scholar]
- 14.Krawczak M, Cooper DN. Gene deletions causing human genetic disease: mechanisms of mutagenesis and the role of the local DNA sequence environment. Hum. Genet. 1991;86:425–441. doi: 10.1007/BF00194629. [DOI] [PubMed] [Google Scholar]
- 15.Kramer KM, Brock JA, Bloom K, Moore JK, Haber JE. Two different types of double-strand breaks in Saccharomyces cerevisiae are repaired by similar RAD52-independent, nonhomologous recombination events. Mol. Cell. Biol. 1994;14:1293–1301. doi: 10.1128/mcb.14.2.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu J, Schiestl RH. Topoisomerase I involvement in illegitimate recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:1805–1812. doi: 10.1128/mcb.16.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manivasakam P, Schiestl RH. Nonhomologous end joining during restriction enzyme-mediated DNA integration in Saccharomyces cerevisiae. Mol. Cell. Biol. 1998;18:1736–1745. doi: 10.1128/mcb.18.3.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adachi H, Hasebe T, Yoshinaga K, Ohta T, Sutoh K. Isolation of Dictyostelium discoideum cytokinesis mutants by restriction enzyme-mediated integration of the blasticidin S resistance marker. Biochem. Biophys. Res. Commun. 1994;205:1808–1814. doi: 10.1006/bbrc.1994.2880. [DOI] [PubMed] [Google Scholar]
- 19.Kiechle M, Manivasakam P, Eckardt-Schupp F, Schiestl RH, Friedl AA. Promoter-trapping in Saccharomyces cerevisiae by radiation-assisted fragment insertion. Nucleic Acids Res. 2002;30:e136. doi: 10.1093/nar/gnf136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stevens CW, Zeng M, Cerniglia GJ. Ionizing radiation greatly improves gene transfer efficiency in mammalian cells. Hum. Gene. Ther. 1996;7:1727–1734. doi: 10.1089/hum.1996.7.14-1727. [DOI] [PubMed] [Google Scholar]
- 21.Nakayama C, Adachi N, Koyama H. Bleomycin enhances random integration of transfected DNA into a human genome. Mutat. Res. 1998;409:1–10. doi: 10.1016/s0921-8777(98)00036-6. [DOI] [PubMed] [Google Scholar]
- 22.Shcherbakova OG, Filatov MV. Camptothecin enhances random integration of transfected DNA into the genome of mammalian cells. Biochim. Biophys. Acta. 2000;1495:1–3. doi: 10.1016/s0167-4889(99)00151-2. [DOI] [PubMed] [Google Scholar]
- 23.Stevens CW, Cerniglia GJ, Giandomenico AR, Koch CJ. DNA damaging agents improve stable gene transfer efficiency in mammalian cells. Radiat. Oncol. Investig. 1998;6:1–9. doi: 10.1002/(sici)1520-6823(1998)6:1<1::aid-roi1>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 24.Perez CF, Botchan MR, Tobias CA. DNA-mediated gene transfer efficiency is enhanced by ionizing and ultraviolet irradiation of rodent cells in vitro. I. Kinetics of enhancement. Radiat. Res. 1985;104:200–213. [PubMed] [Google Scholar]
- 25.Fujimaki K, Aratani Y, Fujisawa S, Motomura S, Okubo T, Koyama H. DNA topoisomerase II inhibitors enhance random integration of transfected vectors into human chromosomes. Somat. Cell Mol. Genet. 1996;22:279–290. doi: 10.1007/BF02369567. [DOI] [PubMed] [Google Scholar]
- 26.Gietz D, St. Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blanc H. Two modules from the hypersuppressive rho- mitochondrial DNA are required for plasmid replication in yeast. Gene. 1984;30:47–61. doi: 10.1016/0378-1119(84)90104-5. [DOI] [PubMed] [Google Scholar]
- 28.Horecka J, Jigami Y. Identifying tagged transposon insertion sites in yeast by direct genomic sequencing. Yeast. 2000;16:967–970. doi: 10.1002/1097-0061(200007)16:10<967::AID-YEA597>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 29.Kraxenberger A, Friedl AA, Kellerer AM. Computer simulation of pulsed field gel runs allows the quantitation of radiation-induced double-strand breaks in yeast. Electrophoresis. 1994;15:128–136. doi: 10.1002/elps.1150150122. [DOI] [PubMed] [Google Scholar]
- 30.Henner WD, Grunberg SM, Haseltine WA. Enzyme action at 3′ termini of ionizing radiation-induced DNA strand breaks. J. Biol. Chem. 1983;258:15198–15205. [PubMed] [Google Scholar]
- 31.Odersky A, Panyutin IV, Panyutin IG, Schunck C, Feldmann E, Goedecke W, Neumann RD, Obe G, Pfeiffer P. Repair of sequence-specific 125I-induced double-strand breaks by nonhomologous DNA end joining in mammalian cell-free extracts. J. Biol. Chem. 2002;277:11756–11764. doi: 10.1074/jbc.M111304200. [DOI] [PubMed] [Google Scholar]
- 32.Lutze LH, Cleaver J, Morgan WF, Winegar RA. Mechanisms involved in rejoining DNA double-strand breaks induced by ionizing radiation and restriction enzymes. Mutat. Res. 1993;299:225–232. doi: 10.1016/0165-1218(93)90099-y. [DOI] [PubMed] [Google Scholar]
- 33.Nasri M, Thomas D. Relaxation of recognition sequence of specific endonuclease HindIII. Nucleic Acids Res. 1986;14:811–821. doi: 10.1093/nar/14.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plessis A, Perrin A, Haber JE, Dujon B. Site-specific recombination determined by I-SceI, a mitochondrial group I intron-encoded endonuclease expressed in the yeast nucleus. Genetics. 1992;130:451–460. doi: 10.1093/genetics/130.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Onda M, Yamaguchi J, Hanada K, Asami Y, Ikeda H. Role of DNA ligase in the illegitimate recombination that generates lambdabio-transducing phages in Escherichia coli. Genetics. 2001;158:29–39. doi: 10.1093/genetics/158.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shanado Y, Hanada K, Ikeda H. Suppression of gamma ray-induced illegitimate recombination in Escherichia coli by the DNA-binding protein H-NS. Mol. Genet. Genomics. 2001;265:242–248. doi: 10.1007/s004380000399. [DOI] [PubMed] [Google Scholar]
- 37.Ma JL, Kim EM, Haber JE, Lee SE. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol. Cell. Biol. 2003;23:8820–8828. doi: 10.1128/MCB.23.23.8820-8828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bao CY, Ma AH, Evans HH, Horng MF, Mencl J, Hui TE, Sedwick WD. Molecular analysis of hypoxanthine phosphoribosyltransferase gene deletions induced by alpha- and X-radiation in human lymphoblastoid cells. Mutat. Res. 1995;326:1–15. doi: 10.1016/0027-5107(94)00152-u. [DOI] [PubMed] [Google Scholar]
- 39.Roth DB, Wilson JH. Nonhomologous recombination in mammalian cells: role for short sequence homologies in the joining reaction. Mol. Cell. Biol. 1986;6:4295–4304. doi: 10.1128/mcb.6.12.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Conley EC, Saunders VA, Jackson V, Saunders JR. Mechanism of intramolecular recyclization and deletion formation following transformation of Escherichia coli with linearized plasmid DNA. Nucleic Acids Res. 1986;14:8919–8932. doi: 10.1093/nar/14.22.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frankenberg-Schwager M. Review of repair kinetics for DNA damage induced in eukaryotic cells in vitro by ionizing radiation. Radiother. Oncol. 1989;14:307–320. doi: 10.1016/0167-8140(89)90143-6. [DOI] [PubMed] [Google Scholar]
- 42.Hua SB, Qiu M, Chan E, Zhu L, Luo Y. Minimum length of sequence homology required for in vivo cloning by homologous recombination in yeast. Plasmid. 1997;38:91–96. doi: 10.1006/plas.1997.1305. [DOI] [PubMed] [Google Scholar]
- 43.Manivasakam P, Weber SC, McElver J, Schiestl RH. Micro-homology mediated PCR targeting in Saccharomyces cerevisiae. Nucleic Acids Res. 1995;23:2799–2800. doi: 10.1093/nar/23.14.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sugawara N, Ira G, Haber JE. DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Mol. Cell. Biol. 2000;20:5300–5309. doi: 10.1128/mcb.20.14.5300-5309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosenberg SM. In pursuit of a molecular mechanism for adaptive mutation. Genome. 1994;37:893–899. doi: 10.1139/g94-127. [DOI] [PubMed] [Google Scholar]
- 46.Foster PL. Adaptive mutation: the uses of adversity. Annu. Rev. Microbiol. 1993;47:467–504. doi: 10.1146/annurev.mi.47.100193.002343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harris RS, Longerich S, Rosenberg SM. Recombination in adaptive mutation. Science. 1994;264:258–260. doi: 10.1126/science.8146657. [DOI] [PubMed] [Google Scholar]
- 48.Torkelson J, Harris RS, Lombardo MJ, Nagendran J, Thulin C, Rosenberg SM. Genome-wide hypermutation in a subpopulation of stationary-phase cells underlies recombination-dependent adaptive mutation. EMBO J. 1997;16:3303–3311. doi: 10.1093/emboj/16.11.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Foster PL. Nonadaptive mutations occur on the F′ episome during adaptive mutation conditions in Escherichia coli. J. Bacteriol. 1997;179:1550–1554. doi: 10.1128/jb.179.5.1550-1554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosenberg SM, Hastings PJ. Microbiology and evolution. Modulating mutation rates in the wild. Science. 2003;300:1382–1383. doi: 10.1126/science.1085691. [DOI] [PubMed] [Google Scholar]