Abstract
Eukaryotic genomes contain many endogenous retroviral sequences (ERVs). ERVs are often severely mutated, therefore difficult to detect. A platform independent (Java) program package, RetroTector© (ReTe), was constructed. It has three basic modules: (i) detection of candidate long terminal repeats (LTRs), (ii) detection of chains of conserved retroviral motifs fulfilling distance constraints and (iii) attempted reconstruction of original retroviral protein sequences, combining alignment, codon statistics and properties of protein ends. Other features are prediction of additional open reading frames, automated database collection, graphical presentation and automatic classification. ReTe favors elements >1000-bp long due to its dependence on order of and distances between retroviral fragments. It detects single or low-copy-number elements. ReTe assigned a ‘retroviral’ score of 890–2827 to 10 exogenous retroviruses from seven genera, and accurately predicted their genes. In a simulated model, ReTe was robust against mutational decay. The human genome was analyzed in 1–2 days on a LINUX cluster. Retroviral sequences were detected in divergent vertebrate genomes. Most ReTe detected chains were coincident with Repeatmasker output and the HERVd database. ReTe did not report most of the evolutionary old HERV-L related and MalR sequences, and is not yet tailored for single LTR detection. Nevertheless, ReTe rationally detects and annotates many retroviral sequences.
INTRODUCTION
Retroviruses occasionally integrate into the germ line and may then be transmitted vertically to new generations as ‘endogenous’ retroviral sequences (ERVs) (1). A substantial part of extant eukaryotic genomes consists of ERVs (2–5). ERVs are one of many kinds of transposable genetic elements (1). Transposons far outnumber conventional genes in higher eukaryotic genomes (6–8). ERVs are often severely mutated, which makes them difficult to recognize. Detection, classification and pathophysiological studies of ERVs are accelerating.
ERV detection has mostly been conducted by BLAST algorithms using the non-redundant (nr) sequence database at NCBI (http://www.ncbi.nlm.nih.gov/), or using the BLAT search at the UCSC genome browser interface (http://genome.ucsc.edu/). This requires a preconceived notion of the query sequence and, although computer-aided, is largely a time-consuming manual process. A further difficulty is that current ERV classification is nonsystematic, which complicates evaluation of recognition techniques. The primary classification principle for human ERVs (HERVs) has been tRNA complementary sequences in the primer binding site (PBS) (9). The RepBase nomenclature (6) is based on nucleotide identity to machine-generated consensus sequences (10) of repetitive elements. Although efficient and pervasive, this approach does not in itself identify the repetitive element as retroviral. This is completed by manual inspection, a slow and sometimes error-prone process. RepeatMasker (7,11), is a system for genome wide screening for repetitive sequences, based on RepBase. It gradually developed from simple detection to a degree of characterization of the repeats. The characterization is however still limited. HERVd (12,13), in its turn, is a derivative of RepeatMasker. These sequence collections are the main references. They are further described below.
A number of algorithms have been developed for sequence searching, see e.g. (14,15). In general, they are not suitable for the task of large-scale identification of ERVs in genomic material. This is because the conserved features of ERVs are short and sparse, while the intervening sequences are highly variable, even before degradation by mutations sets in. Published methods for retrieval of retroviral sequences from genomic databases either center on detection of long terminal repeat (LTR) pairs, specific conserved sequences, or general repeat detection. To our knowledge, there is however no comprehensive attempt to both detect ERVs and to characterize their internal structure.
More and more vertebrate genomes have been sequenced. A generic tool for detection of a broad range of retroviral sequences, which is not limited to primate genomes, is needed. We have therefore developed a procedure, which concentrates on the conserved features (motifs). The intervening regions come in only as rough measures of distances between motifs, though they are the subject of follow-up analysis in regions focussed by the primary search. The search procedure for each individual motif may be chosen according to its characteristics, though so far straightforward codon-by-codon comparison with a consensus amino acid sequence dominates. Modules for search, analysis and result presentation have been united as a package, RetroTector©, ReTe, with large scope for modification to meet particular needs, even possibly for other tasks than ERV searching.
ReTe is an expert system, which strives to embody and generalize present knowledge of retroviral genomic structures. It uses a combination of several novel heuristic algorithms. The primary algorithm is based on the principle of ‘fragment threading’. It first detects candidates for the LTRs, then different conserved retroviral motifs. Having reduced the search space, more time-consuming and exhaustive algorithms come into play. The LTRs and motifs are then connected into chains, indicating more or less complete ERVs. Finally, it attempts to reconstruct the four major retroviral proteins Gag, Pro, Pol and Env. The findings are collected into a database for convenient retrieval. Data are presented in an interactive graphical format, akin to the format used in textbooks (1).
MATERIALS AND METHODS
Data set
Reference retroviral sequences were collected from GenBank. Whole genomic sequences (human genome versions hg15, hg16, hg17 and hg18, chimpanzee genome versions panTro1 and panTro2, wild red jungle fowl genome versions galGal1 and galGal3, dog genome canFam2, Rhesus macaque rheMac2, and Mouse genome mm8) were downloaded via the UCSC Genome Browser (http://genome.ucsc.edu/). Results from the analyses of the various assemblies will be published separately. Most of the results in this methodological paper are based on early versions (hg15 and hg16) of the human genome assembly. However, more recent data, from hg18, panTro2, rheMac2, canFam2, musMus8 and galGal3, and the corresponding RepeatMasker output files, are also included. Reference retroviral sequences RSV (J20342 and NC_001407), ALV (NC_001408), MMTV (NC_001503), MPMV (NC_001550), JSRV (NC_001494), FLV (NC_001940), MLV (J02255 and NC_001501), HTLV1 (NC_001436), HTLV2 (M10060 and NC_001488), WDSV (NC_001867), Snakehead retrovirus (NC_001724), Xen1 (AJ506107), HIV (K03455 and NC_001802) and HFV (NC_001736), were analyzed with ReTe. The errantiviruses ZAM (AJ00387) and CER1 (U15406) were also analyzed.
Hardware
The genomic analyses were performed on (i) seven Dell Optiplex 260 office computers with 2–2.5 GHz Pentium processors, and 40 GB hard disks, and (ii) at the Uppmax (www.uppmax.uu.se) computer cluster of AMD Opteron 250, 850 and 875 CPUs running Scientific Linux 4.2. The latter configuration yielded 2–5 times shorter execution times.
Algorithms
Fragment threading
We coined this term to describe the central procedure in ReTe. It depends on a database of conserved motifs, constraints on the distances between motif ‘hits’ and a matrix of similarities between amino acids. From a programmer's point of view, ‘motifs’ are procedures for detection of conserved ERV traits in the face of mutations. The bulk of the motifs operate through simple comparison (using the acid similarity matrix) against a conserved amino acid sequence, but there are several other types (Table 1, which also includes motifs for other purposes). Each motif is connected to one or more retrovirus genera (at present alpha-, beta-, gamma-, delta-, epsilon-, spuma- and lentiretroviruses, and the related viruses Gypsy and Copia). The constraints on the distances between motif ‘hits’ are based on the position distances in known retroviruses, extended with a ‘safety margin’. At present the motifs and constraints are adapted primarily to vertebrate, especially primate, sequences, but are also flexible to change in order to accommodate other sequence analyses.
Table 1.
Motifs utilized by ReTe at present. Some of the Motifs are used in the ‘fragment threading’, others in LTR search or in putein construction
| Name | Used in | Characteristics |
|---|---|---|
| AcidMotif | RetroVID | Compares to conserved amino acid sequence |
| AcidNNMotif | RetroVID | Uses neural network trained on known peptides |
| SplitAcidMotif | RetroVID | Like several AcidMotifs at prescribed distances |
| BaseMotif | RetroVID, LTRID | Compares to conserved nucleotide sequence |
| HyPhobMotif | RetroVID | Searches for hydrophobic region in Gag |
| LTRMotif | RetroVID | Encapsulates LTR candidate found by LTRID |
| PPTMotif | RetroVID | Special algorithm for polypurine tract |
| SpliceAcceptorMotif | ORFID | Searches for splice acceptor consensus |
| SpliceDonorMotif | ORFID | Searches for splice donor consensus |
| SlipperyMotif | ORFID | Searches for XXXYYYZ |
| ProteaseCleavageMotif | ORFID | Searches for wPf, fPv and yPi |
| PseudoKnotMotif | ORFID | Searches for pseudoknot-like structures |
| FrameShifterMotif | ORFID | Combination of SlipperyMotif and PseudoKnotMotif |
| PuteinStartMotif | ORFID | Compares to leading amino acid sequences in known proteins |
| PuteinStartMotif | ORFID | Compares to trailing amino acid sequences in known proteins |
| SiSeqMotif | ORFID | Scores with weight matrix for signal sequence of von Heijne |
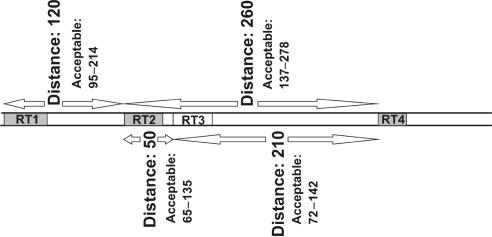
The principle of ‘fragment threading’ is illustrated in Figure 1. The most likely of the often many possible combinations of motif hits is chosen according to a heuristic procedure. Motif hits are combined into ‘chains’ satisfying distance constraints, corresponding to potential ERVs, though ‘broken’ chains violating one or two constraints are also possible, to account for ERVs containing insertions or deletions (indels). To evaluate the chain, it is assigned a score and a retroviral genus (or more than one in ambiguous cases) through a vector procedure: Each motif hit is assigned a vector. Its direction is dependent on its genus and its length depends on a weight factor for the motif (see Supplementary Data S3), and how well the hit fits the motif. The motif-hit vectors are summed (with some modifications) into a vector for the whole chain. The length of this vector determines the chain score and its direction determines the retroviral genus assigned.
Figure 1.
The principle of ‘fragment threading’. Three motif hits (RT1, RT2 and RT4) are within accepted distances from each other, whereas one (RT3) is not. Motifs RT1, RT2 and RT4 can therefore be utilized to build a proviral chain.
‘Fragment threading’ is simple in principle, but in order not to miss mutilated or previously unknown ERVs, the motif hit and distance constraints must be so lax that an exhaustive search of all possible combinations is not practical. This ‘combinatorial explosion’ has been countered in several ways: (i) The search is hierarchical. The motifs are grouped into 14 ‘subgenes’ (5′LTR, PBS, MA, CA, NC, DU, PR, RT (incl. RNH), DL, IN, SU, TM, PPT and 3′LTR) according to established retrovirus terminology (DL is here used to denote a dUTPase sequence integrated in the integrase region). Exhaustive ‘fragment threading’ of motif hits is applied within each subgene (except the first two and last two, which contain only one motif each) to generate subgene hits. The subgene hits are then threaded to form chains, with a limit on the number of hits tried for each subgene. (ii) Another limit is set on the length of gaps in the subgene sequence. (iii) A subset of the motifs (notably the PBS and PPT subgenes) is normally not used in the primary search, but only in refining already found chains. (iv) Long sequences are split into chunks (typically 115-kb long with 15-kb overlap) before processing. The 15-kb overlap is sufficient to minimize loss of ERVs, normally up to 10-kb long (1), in the sequence chunk border region.
Sequence statistics
The algorithms below utilize results from two unpublished studies by Blomberg:
A search for oligomers which differed in frequency between ORFs and the two alternative reading frames was made in the collection of reference retroviral sequences. A systematic evaluation of gag, pro, pol and env sequences showed that hexamers yielded higher ORF selectivities than tri-, tetra- and pentamers. For example, four especially selective hexamers were GATACG, CGCAGG, CTAGAA and GAAGAT, which were 4.1–6 times more frequent in retroviral ORFs relative to the two alternative reading frames. They encode the dipeptides DT, RR, LE and ED, respectively. It seems that these combinations are less likely to occur by chance in overlapping non-coding retroviral reading frames. A list of 31 hexamers with 3–6 times selectivity for an ORF in that reading frame, relative to the other two frames, is included in ReTe. Its negative control was provided by a list of 700 non-ORF hexamers. These had no increased frequency in ORFs relative to non-ORFs.
A set of LTR selective split octamers was collected after a systematic evaluation of split octamer motifs in the database of reference retroviral genomes. All combinations of two tetramers occurring within a distance range of 0–60 nt positions from each other were tested for LTR selectivity versus (i) retroviral non-LTR sequences, and (ii) a random sequence of 100 Mb. This resulted in a list of 1256 binary tetramer combinations, each with a range for the distance between them. The LTR/non-LTR selectivities were 10–165, whereas LTR/random selectivities for the same set were 0.91–44.54. The two selectivity criteria varied rather independently, indicating a considerably non-random distribution of split octamers in retroviral non-LTR sequences. For example, four especially selective combinations were TCTG<8–12>CCCC, CCCC<6–8>CACC, GACA<10–14>CTGT and GTGC<6–8>AACA, which all were 12–165 times LTR/non-LTR selective and 3–45 times LTR/random selective. The functional basis behind this selectivity is obscure. A similar approach was used before (16–18).
Alignment through dynamic programming
Several variants of this standard procedure are employed and will be referred to by these abbreviations:
A1: Pairwise nucleotide alignment, essentially according to Huang (19).
A2: Nucleotide alignment which is aborted if it does not seem promising; thus non-exhaustive but fast.
A3: Pairwise amino acid alignment, essentially according to Huang (19), using the amino acid similarity matrix.
A4: Alignment of a nucleotide sequence to a set of known, aligned peptides, between two predetermined endpoints. Path elements are scored either, depending on which is greater, (a) by the similarity score between the codon in the sequence and the best available amino acid in the alignment or (b) by an estimate of the general suitability of the reading frame of the codon, based on stop codon density, glycosylation site density (for Env), reading frame of nearby motif hits and presence of nucleotide hexamers known to be frequent in or outside retroviral ORFs, respectively. This was progressively evaluated over a window of 200 nt.
Neural networks
Standard multilayered perceptron networks trained by back projection, see e.g. (20).
Implementation
Design
ReTe is written in Java and should run on any computer with Java runtime 1.4.3 or later. For full functionality an SQL database manager, preferably MySQL, should be available. ReTe has been extensively used and tested under the Windows (with Sun Java runtime), MacOS X 10.4 (i.e. UNIX), LINUX (Red Hat 9, Shrike) and Scientific Linux operating systems. In the programming, the shareware source http://www.jibble.org/epsgraphics/ (.eps file module) has been utilized.
ReTe is designed for searching entire genomes, i.e. the algorithms were chosen for speed rather than refinement. Also, information outside the ERV proper, such as integration repeats, also referred to as ‘target site duplications’, is utilized by ReTe. The design is flexible, with numerous variable parameters and facilities for plugin extensions (in the form of Java classes). Further information and documentation about ReTe is available at the URL: http://www.kvir.uu.se/RetroTector/RetroTectorProject.html.
The variable parameters allow the user to adjust the unavoidable tradeoff between speed, sensitivity and selectivity. The standard settings (see the URL above) provide for the processing of a genome in about one month processor time (2 GHz Pentium) with sensitivity prioritized over selectivity. The processing time can be drastically reduced by running the program on a cluster of faster processors. The selectivity may be increased in retrospect by disregarding low-scoring results.
ReTe contains modules for various operations that can be executed from a menu. However, for many of them execution is normally initiated by a script file, generated by another module, containing necessary information. The script files thus serve to connect the different modules. One of the modules (SweepScripts) handles automatic execution of scripts, so that an entire chromosome can be processed automatically.
Sequence of operations
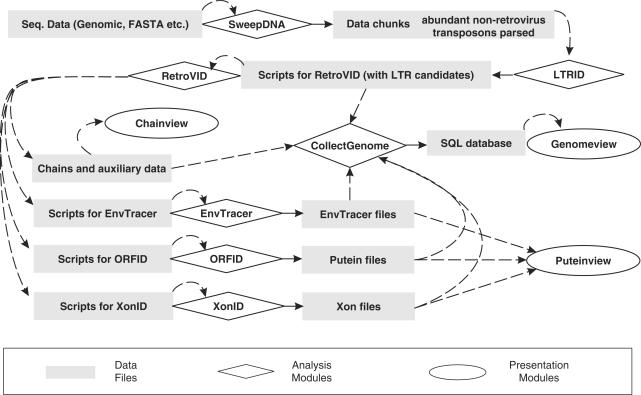
Typically, in analyzing a chromosome, the following procedure is performed automatically (Figure 2): (i) the module SweepDNA cuts the DNA sequence into chunks as mentioned above. It also tries to identify ALUs and LINE L1 fragments and possibly other frequent nonretroviral species-specific transposons, using algorithm A2. These are excluded from the further analyses. (ii) The LTRID module identifies possible LTRs, paired and unpaired. (iii) The RetroVID module identifies possible ERVs through ‘fragment threading’, also utilizing the LTR candidates found by LTRID as motif hits belonging to the 5′LTR and 3′LTR subgenes. For chains exceeding a score threshold, it also generates scripts for the ORFID and XonID modules. If a chain has no motif hits in Env, but there are hits in IN and 3′LTR separated by a motif-empty stretch, suggesting the presence of at least a fragment of env, RetroVID generates a script for EnvTracer. (iv) The ORFID module generates putative proteins, ‘puteins’, in an attempt to reconstruct the original retroviral Gag, Pol, Pro and Env. (v) The XonID module gives hints about possible exons not found by ORFID. (vi) EnvTracer attempts to find a likely Env, employing similar principles as XonID. (vii) The CollectGenome module collects the selected output data into an SQL database.
Figure 2.
Flow of events during a RetroTector© analysis.
Core modules of ReTe
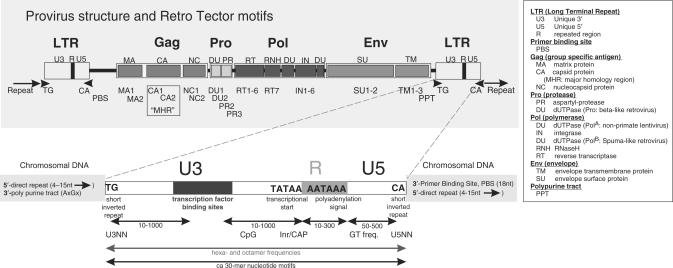
‘LTRID’ is a module that identifies potential LTRs (Figure 3). LTRID first aims to find the polyadenylation signal, always present in an LTR, either as the characteristic sequence (AATAAA, ATTAAA or AGTAAA) or as a high score by a neural network trained for this purpose. It detects the R-U5 portion of LTRs of many Alpha-, Beta- and Gammaretroviruslike sequences. After detection of the polyadenylation signal, LTRID calculates a score for the LTR candidate by searching for other LTR characteristics (GT accumulation, a further neural network, TATA box, characteristic nucleotide sequences, binding sites of selected transcription factors, CpG-rich regions) within realistic distances from the polyadenylation signal. All of these LTR-specific features were found using the database of annotated reference retroviral genomes (Blomberg, J., unpublished data).
Figure 3.
LTR features utilized by RetroTector©, in the proviral model context. A combination of obligate and alternative landmarks is used to select LTR candidates, which are further selected by pairing and proviral chain distance criteria. Upper panel: ERV structure overview, with standard terms and ReTe motif group names below. Lower panel: LTR features utilized by ReTe. Constraints between them are also shown. Motifs and their abbreviations are explained in Supplementary Data S3.
If the LTRID score exceeds a specified threshold, the single LTR candidate is accepted and included in the script for RetroVID (see below). So also are pairs of similar (by algorithm A2) LTR candidates separated by a realistic distance, irrespective of LTR scores. In both cases, start and end points of the LTRs are suggested based on similarities to the characteristic direct and short inverted repeats formed during the retroviral integration and flanking the provirus (1).
LTRID recognizes LTR pairs adequately, though with many false positives. Pairs of ALUs, LINEs or other transposons not found and masked by SweepDNA may be reported as LTR pairs. Identification of solitary LTRs is at present not satisfactory. However, inclusion of HMMs, see e.g. (21), will probably improve this.
‘RetroVID’ is normally initiated by a script generated by LTRID and containing its findings of LTR candidates. These are included as motif hits in the subsequent procedure. The other motifs are given a score threshold by sampling each of them in 1000 positions along the target sequence, the score threshold for motif hits then being determined by the statistics of these scores, typically at mean +5.5 SD, thus adjusting score thresholds to local genetic noise. A subset of the motifs is then scored in all positions and hits recorded where the score threshold is exceeded. The hits are combined into chains through ‘fragment threading’, and a subset of non-overlapping, high-scoring chains is subsequently selected. The selected chains are further refined, mainly by including the full set of motifs and by making a renewed attempt to include LTRs, by searching for pairs with algorithm A1. The resulting chains are output to one file, and if appropriate, scripts for ORFID, EnvTracer and XonID are also generated.
‘ORFID’ is normally started by a script generated by RetroVID, containing information about detected motif hits and ranges for the start and end of the protein. There is one script for each gene in which motif hits were found. Moreover, if the genus of the chain was ambiguous, a set of ORFID scripts for each genus are generated. ORFID then constructs a putative protein, or ‘putein’, passing through most of the motif hits. Very weak or otherwise doubtful motif hits are ignored. The frequent post-integrational mutations in ERVs makes it especially important to have multiple criteria for continuously selecting the most likely reading frame. Essentially, ORFID strives for an optimal pattern of frame shifts using algorithm A4.
All codons between frame shifts are included in the putein, with no attempt to identify indels. A4 is applied between each pair of consecutive motif hits, where it is in general reasonably stable. It is also applied to the end portions, but is then combined with a procedure that evaluates the fitness of each position within the range as starting- (or end-) point for a protein. The details of this procedure are different for each gene and retroviral genus and built on similarity to known protein ends, the relations of known viral proteins to stop codons, Kozak start consensus (22), protease cleavage sites, slippery sequences, pseudoknots, splice sites and von Heijne signal sequence (23). If puteins are made for adjacent genes, they are also adjusted to each other. This selection of putein ends is not always satisfactory and will be improved. ORFID also identifies the longest ORF coinciding with the putein, as a possible present-day coding sequence.
‘XonID’ identifies other possible exons than the four fundamental retroviral (gag, pro, pol and env) genes, since ORFID only constructs rather obvious puteins. XonID may be applied to search for more vague traces of exons, combining criterion (ii) in algorithm A4 with data about canonical splice sites and start/stop codons.
‘EnvTracer’ is based on similar principles as XonID, but is specialized to finding likely env reading frames. It is focussed on long open reading frames not recognized by ORFID, rich in predicted N-glycosylation sites, which occur between the predicted end of pol and a 3′LTR.
Other modules
Within ReTe, there are about 20 other command modules, some for visualization and storage of results, others mainly for maintenance and debugging. Of particular interest are the modules:
‘PseuGID’ is applied if a chain has none or very short LTRs, suggesting that the ERV may be a processed pseudogene (24). RetroVID generates a script for this module, which searches for the structures characteristic of processed pseudogenes. This function has not yet been fully tested.
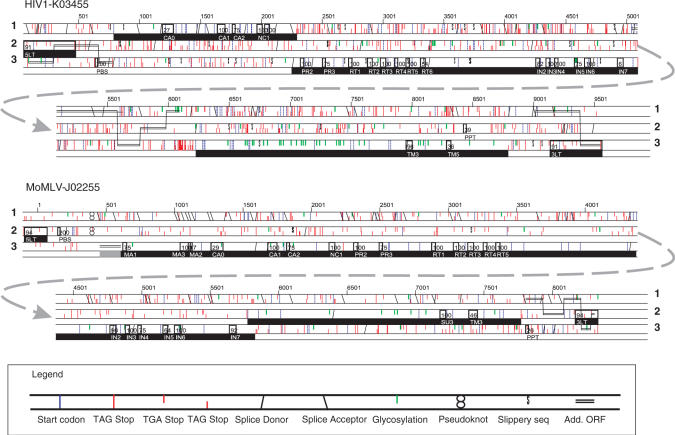
‘Chainview’ displays one chain at a time in detailed graphics (Figure 4; Supplementary Data S2) and in detailed text. Apart from the motif hits and the course of the predicted chain, it may also display an analysis of LTR structure, puteins and EnvTracer and XonID output related to the chain, start and stop codons, splice donors/acceptors and several other features.
Figure 4.
Chainview picture of HIV and MLV. Symbols are explained below the proviral renditions.
‘Puteinview’ shows details, such as the full amino acid sequence of a putein or an exon suggested by XonID or EnvTracer.
‘CollectGenome’: The mass of text files containing all the results from a ReTe analysis may be forbidding. This module extracts selected results and collects them into an SQL database, with separate tables for LTR candidates, chains and puteins. Thereby it also compares each chain to a set of RepBase consensus sequences and a set of known annotated retrovirus sequences, using algorithm A1, and Pol puteins to a set of known Pol proteins (using algorithm A3) for classification purposes. Judgment of the content compared to nonretroviral repetitive sequences is conducted as an internal control.
‘Genomeview’ may be used to inspect the database generated by CollectGenome. It shows the distribution of LTR candidates and chains within the chromosomes. Chains and their relation to the RepBase reference sequences and known retroviruses may also be viewed graphically (with less detail than Chainview).
‘RetroTectorShell’: This is a Windows-specific program separate from ReTe, written in Visual FoxPro (VFP). It was developed in parallel with the Java ReTe kernel for user interaction and data handling. It performs similar functions as CollectGenome and Genomeview, but has a somewhat different profile. Features so far found only in RetroTectorShell are: (i) A mechanism for collecting ReTe output into a VFP table. Each genome's retroviral content is thus contained in a single table. It disregards single LTRs and alternative ORFs. (ii) A BLAST-like algorithm for searching chains in this table according to protein or nucleic acid similarity.
RESULTS
Evaluation of ReTe using an artificial data set
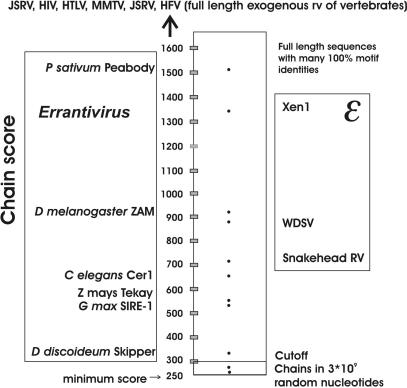
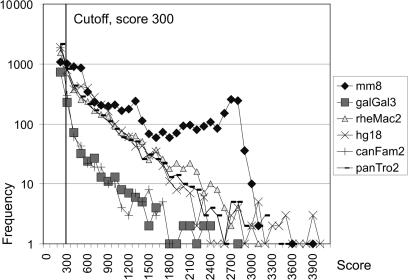
A 3 × 109 nt stretch of random nucleotide sequence with equal A, T, G and C frequencies was run. Two chains with a score above the minimum (250), 258 and 273, resulted. Thus, none were above 300. The experience from many genomic analyses, primarily from the human genome versions hg15–hg18, has shown that a score cutoff of 300 almost totally eliminates spurious chains results from motif-hit combinations occurring by chance (Figure 7; ROC curves in Supplementary Data S10–S15).
Figure 7.
Chain scores with retroviral, retroviruslike and random sequences. Retroviruslike (errantiviral; gypsy elements) sequences of slime molds, insects and plants are shown to the left. Epsilonretroviral sequences of amphibians and fish are shown to the right. Scores of chains detected in a 108 random sequence are shown below the cutoff. The chains from 108 random nucleotides were obtained before the changed settings described in the text.
Evaluation of ReTe with simulated mutated retroviral sequences
In order to test the detection limits of ReTe with regard to degraded retroviral sequences (i.e. very old insertions), a test set with artificially degraded sequences was created. These sequences were based on the complete genome of HIV-1, isolate MNCG (Genbank accession no M17449), which was degraded according to four different mutational models, with decay ranging from 1 to 60% mutation (see Supplemental Data S1 for further details). The resulting test set was analyzed with ReTe and also with BLAST (see below), for comparison. Four different mutational models were selected: (i) Random substitutions with equal probabilities (Jukes–Cantor); (ii) Higher probability of transition over transversion (Kimura 2-parameter model); (iii) The Kimura 2-parameter model with insertions and deletions, with frequencies of indels applying to human pseudogenes (24) (‘indel model’); (iv) Simulation of an active retrovirus, where both endogenous and exogenous phases are subjected to purifying selection during each round of infection and replication (‘Exogenous model’). Mutations harming features important for the retroviral function will not persist in the viral population. HIV is a highly replicative retrovirus. The Los Alamos National Laboratory (http://hiv-web.lanl.gov/content/index) presents a large database with sequence data for many subtypes, including a set of full-length HIV genomes aligned with respect to nucleotide codon triplets. The ‘exogenous’ model uses this aligned data set to test which mutations are allowed.
The ReTe analysis utilized default settings. BLAST (version 2.2.6, obtained from NCBI, http://www.ncbi.nlm.nih.gov/), run locally under Linux Red Hat 9, with default settings except that word length was 7. The sequences were matched against the reference sequence HIVMNCG, as well as against the entire non-redundant nucleotide database (see Supplementary Data S1 for further details).
Sensitivity and specificity
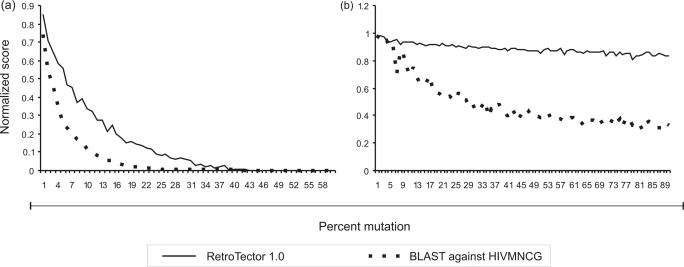
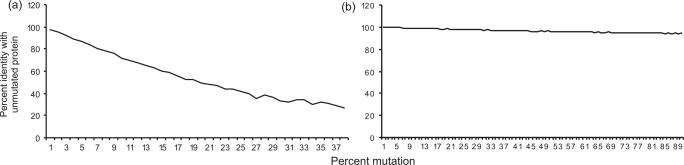
In the simulated evolution model based on HIV, ReTe chain detection was more resistant to mutational decay than BLAST sequence detection. ReTe also attempts to reconstruct the retroviral proteins. Sequence similarity in the form of percent identity between the Pol puteins and the original HIVMNCG Pol protein, shows that ReTe can detect even extensively mutated and evolutionarily distant Pol sequences (Figures 5, 6 and 7; Supplementary Data S1).
Figure 5.
Simulation of mutation of an endogenous and an exogenous retrovirus. Average scores for 20 sequences at each level of mutation, divided by maximum score for unmutated HIVMNCG, when analyzed with ReTe and BLAST, are shown. (a) Normalized score for sequences in the endogenous (indel) model. ReTe is more tolerant to mutation than BLAST, when scoring a sequence as retroviral. (b) Normalized score for sequences in the exogenous model. These sequences receive high scores throughout the analysis when analyzed with ReTe. BLAST, on the other hand, does not as readily recognize the sequences as descendant from HIVMNCG. Further information is given in the Supplementary Data, S1.
Figure 6.
Sequence similarity (percent identity, gap positions excluded) for the Pol puteins compared to the unmutated HIVMNCG Pol protein. Average for 20 puteins at each level of mutation. (a) Endogenous (indel) model puteins: sequence identity. (b) Exogenous model puteins: sequence identity. Further information is given in the Supplementary Data, S1.
Evaluation of ReTe with whole genome sequences from a variety of species
As seen in Figures 7 and 8, chain scores in the human genome version hg18 ranged from the cutoff of 300–4400. High scoring (>2000) chains were from exogenous retroviruses, and from structurally intact endogenous proviruses like the betaretroviruslike HERV-K(HML2) (25). Incomplete proviruses, with LTR-gag-env-LTR etc. scored 250–400 points. Errantivirus sequences like gypsy scored 250–950. Pseudovirus sequences like copia scored 200–350, or not at all. Epsilonretrovirus chains scored 350–1050.
Figure 8.
Frequencies of scores of the chains reported by ReTe version 1.0 from the human (hg18), chimpanzee (panTro2), rhesus (rheMac2), dog (canFam2), mouse (mm8) and chicken (galGal3) genome assemblies.
As shown in Tables 2 and 3, and Figure 7, ReTe can detect retroviral sequences in a wide variety of genomes, also from less complete assemblies (e.g. panTro1). The four major genes were detected in many of the chains. However, env sequences were less common than the other three. Detection of the env gene, the least conserved of the four major retroviral genes, poses special difficulties. There are few conserved motifs in its SU portion, and the conserved motifs in the transmembrane protein often do not provide enough basis for a putein reconstruction. The inclusion of the EnvTracer module was intended to diminish false negativity in env gene detection. The lower frequency of env in the retroviral chains (Table 3) is probably due to mutational decay, occasional misses by EnvTracer or to env-less proviruses which may transpose without leaving the cell, e.g. the betaretroviruslike IAP elements in mice (26), and possibly the bulk of the HERVH elements in humans (27–30).
Table 2.
ReTe predicted genomic ERV contentsa
| Genome analyzed | Total detected elements | Elements with ReTe version 0.12, score ≥300b |
|---|---|---|
| Homo sapiens (hg16) | 18 213 | 3164 |
| Pan troglodytes (PanTro1) | 13 003 | 2117 |
| Gallus gallus (gg01) | 3921 | 262 |
aThe compilations are under improvement, depending on sequence draft qualities and ReTe optimization.
bReTe score ≥300 suggests true integrations of relatively intact elements. Modified from (28).
Table 3.
Proviral chains of score ≥300 detected in recent runs with ReTe version 1.0 and more complete genome assemblies
| Genus/Host | ERVs (all chains) | 2 × LTR | gag | pro | pol | env | Full lengtha |
|---|---|---|---|---|---|---|---|
| Alpha-like | |||||||
| galGal3 | 33 | 4 | 17 | 13 | 26 | 1 | 1 |
| AlphaBeta-likeb | |||||||
| galGal3 | 61 | 29 | 45 | 37 | 51 | 10 | 7 |
| Beta-like | |||||||
| hg18 | 770 | 299 | 424 | 498 | 694 | 328 | 142 |
| panTro2 | 768 | 287 | 423 | 511 | 683 | 307 | 137 |
| rheMac2 | 828 | 348 | 533 | 548 | 733 | 321 | 138 |
| canFam2 | 60 | 47 | 11 | 11 | 51 | 11 | 0 |
| musMus8 | 5928 | 2746 | 3446 | 3391 | 5316 | 1339 | 536 |
| galGal3 | 162 | 38 | 74 | 83 | 150 | 16 | 6 |
| Gamma-like | |||||||
| hg18 | 2713 | 1439 | 2070 | 1436 | 2491 | 1178 | 466 |
| panTro2 | 2055 | 1234 | 1195 | 976 | 1738 | 953 | 317 |
| rheMac2 | 1736 | 994 | 1086 | 875 | 1523 | 750 | 254 |
| canFam2 | 438 | 242 | 224 | 199 | 371 | 96 | 18 |
| musMus8 | 1461 | 808 | 974 | 921 | 1370 | 744 | 455 |
| galGal3 | 3 | 3 | 0 | 1 | 1 | 0 | 0 |
| Spuma-like | |||||||
| hg18 | 145 | 75 | 3 | 49 | 142 | 42 | 0 |
| panTro2 | 96 | 58 | 0 | 26 | 92 | 12 | 0 |
| rheMac2 | 96 | 58 | 0 | 26 | 92 | 12 | 0 |
| canFam2 | 5 | 5 | 0 | 0 | 5 | 2 | 0 |
| musMus8 | 438 | 307 | 2 | 428 | 436 | 285 | 1 |
| galGal3 | 3 | 3 | 0 | 1 | 1 | 0 | 0 |
As discussed below, a basic problem for sensitivity and specificity determination for an ERV detection algorithm is the absence of a generally recognized and curated HERV database. An established general mechanism for repeat detection, and a limited level of repeat characterization, is provided by RepeatMasker (7,11), based on RepBase (6). HERVd (12,13) represented an attempt to reduce the fragmentation of Repeatmasker output, and to amend its retroviral nomenclature. Unfortunately, it could not be maintained. Nevertheless, these sources are the best established references.
The ReTe cutoff score of 300 is motivated (i) by the lack of chains from random sequences above this limit, (ii) by the clear reduction of chains not overlapping RepeatMasker hits when this cutoff is used (Supplementary Data S10–S15), (iii) The relation between sensitivity and specificity versus Repeatmasker hits (receiver operating characteristic; ROC) curves for three genomes (Supplementary Data S10–S15).
ReTe-derived sequences were evaluated versus RepeatMasker output and HERVd annotations on the human genome hg15, see also supplementary Data S6 and S7, as well as (28–31). Using ≥300 as a chain score cutoff, 3373 retroviral chains were detected in hg15. Repeatmasker reported 457 600 ‘LTR’ elements, generally as fragments. A total of 2625 ReTe chains were colocalized with a Repeatmasker entry, using a criterion of overlap within 12 000 nt of the start point of the ReTe chain. The Supplementary Data S2 and S4 shows a chain missed in hg15. The remainder occurred in both data sets. A detailed comparison of these discrepancies requires a detailed ERV classification, which is out of scope for this article. The Supplementary Data (S6–S17) does however give a survey. Repeat-based recognition does not in itself identify retroviral sequences. RepeatMasker relies on the man-made assignments in RepBase to characterize elements as ‘LTR’ elements or not.
In a comparison of ReTe hg15 findings with HERVd, 255 515 partial or full elements in total were annotated in HERVd (which is based on the hg15 genome version). Of these, 3117 had a coincident ReTe chain. Thus, with the present motifs and distance constraints ReTe misses many HERV-L related sequences, which is an evolutionarily old and deviating group. According to RepeatMasker it constitutes ∼1.9% of the human genome. If they are subtracted from the abovementioned 3%, this leaves around 1% as detectable by ReTe. The 3373 chains scoring ≥300 found by ReTe version 0.10 in hg15 cover 24 487 571 nt, i.e. 0.79% of the human genome. A probable explanation for the discrepancy is that ReTe prefers relatively complete proviruses, and misses some fragmented ones. This is elaborated in Supplementary Data S8–S17. It is likely that by modifications in the distance model, and addition of more motifs, a greater proportion of HERV-L can be detected with ReTe.
On the other hand, ReTe is not dependent upon repetition for detection, and therefore could detect single or low-copy-number retroviral elements. Examples are ERV-FRD on chromosome 6p24.2 (32), and HERV-Fc1 (33) on chromosome Xq21.33, which are single or low-copy-number elements with unusually open reading frames (Supplementary Data S2 and S4). Both elements are however now fully or partially covered in an April 2007 version of the RM output of the human genome version hg18.
Accuracy
Reference retroviral genomes give chains with scores of; 3341 (RSV), 3792 (MMTV), 3439 (MPMV), 3022 (MLV), 2934 (FLV), 2814 (HIV-1), 2345 (HTLV2) and 879 (WDSV). The errantivirus elements ZAM and CER1 give 935 and 687, respectively. They are gypsy elements from Drosophila melanogaster and Caenorhabditis elegans, respectively. They are related to the Orthoretroviruses (Figure 7; and Supplementary Data S2), and have the same genome organization. The four major genes are predicted in all of the 10 viruses, except for the env gene in RSV. This may be due to deranged env distances due to the presence of src, and/or insufficient coverage of alpharetroviral SU and TM motifs. The start and stop positions of the respective ORFs were correct within 10% of the annotated position, see e.g. (1). Larger deviations are occasionally observed in complex retroviruses (lenti, delta and epsilon retroviruses), which have one or several additional regulatory protein genes. They can occur before gag and around env. Similar problems are caused by the sarcoma viruses, where oncogenes disrupt the retroviral structure (Supplementary Data, S2 and S5).
As mentioned in the discussion, ReTe has already been used in several studies on human ERVs (29,31,34–36).
Performance
ReTe was applied to the the human genome versions hg15, hg16, hg17 and hg18, the chimpanzee genome versions panTro1 and panTro2, the chicken genome versions galGal1 and galGal3, the dog genome version canFam2, the mouse genome version mm8 and the opossum genome monDom4. Depending on settings, a full analysis of the human and chimpanzee genomes takes 5–6 days, the chicken genome 3 days, using the computer set described in Systems and Methods. On the Uppmax Opteron Linux-based cluster, 1–2 days suffices.
DISCUSSION
Program design considerations
Solitary LTR detection is one of the most demanding aspects of retroviral sequence recognition. The principle of LTR selective split octamers used in ReTe is similar to the one used in the program Matinspector® (Genomatix Gmbh, Germany) (16–18). Despite a rather high LTR selectivity of the presented LTR recognition algorithm (LTRID), the number of false-positive hits is overwhelming when entire genomes are processed. Initially, we considered using hidden Markov models (HMMs) (21) for the detection of retroviral structures. This is computationally intensive, and we instead chose the faster algorithm ‘fragment threading’. We are currently attempting a limited introduction of HMMs for improved detection of solitary LTRs, and a few other motifs. The principle of ORF-selective hexamers used in ORFID is also used in gene-finding algorithms like GenScan (37). Clearly, certain binary triplet combinations are more likely to occur in retroviral ORFs compared to the two alternative reading frames. The functional basis for this selectivity is obscure.
We had the practising retrovirologist and geneticist in mind in the design. Although the modular design of ReTe leaves ample scope for future improvement, it has already proved useful in its present form (28–31,34–36,38–40). ReTe gives a rich basis for assessment of the functionality and taxonomy of a retroviral element. The ability to export protein and nucleic acid sequences of the four major retroviral genes (gag, pro, pol and env) from ERVs of an entire genome in FASTA format will aid phylogenetic studies, and promote the understanding of the ‘retroproteomes’ of these organisms. The usefulness of the ready availability of nucleic acid frequency, LTR divergence, Pol-based classification versus reference retroviral elements, and degree of nucleotide identity to RepBase elements for all detected ERVs in a genome has been demonstrated in studies on HERV-H (29,30), ERV3 (31), ERV9/HERV-W (36) and a comparison of ERVs unique to humans and chimpanzees (35). In addition, features such as splice prediction, prediction of additional ORFs besides Gag, Pro, Pol and Env, gag-pro-pol readthrough mechanisms and LTR structure aid further functional studies on ERVs.
ReTe, RepBase, RepeatMasker and HERVd
Repeat-based recognition of ERVs, using RepBase (6) and its corollaries RepeatMasker (7,11), Hubley,R. and Green,P., unpublished data (http://repeatmasker.org) and HERVd (12,13), has been conducted for over a decade. An elaborate classification (RepBase) based on nucleic acid identity to machine-generated consensus sequences, through the Censor program (10), exists for many repeated elements. It is gradually being supplemented by user contributions. This classification and detection procedure should be scrutinized against other alternatives. ReTe provides an independent route to ERV detection and classification. Only by using several approaches can a rational classification of ERVs be achieved. ReTe favors elements >1000-bp long due to its dependence on the presence of several retroviral fragments in the right order at approximate distances typical of retroviruses. RepeatMasker, however, can detect considerably shorter sequences but is limited by the need for a minimum number of repeats for recognition. It is also limited by a lack of internal retroviral structure interpretation. An exact appraisal is not possible due to the often fragmented nature of both HERVd and Repeatmasker outputs. Published methods for retrieval of retroviral sequences either center around detection of LTR pairs (41,42), specific conserved sequences, like TM (43,44), or RT, combined with an ORF search (45,46), or general repeat detection, collected in RepBase (6,10) and used in RepeatMasker (7,11) and HERVd (12,13). HESAS (HERVs Expression and Structure Analysis System) (47) merges dbEST information with Repeatmasker-based output. It yields information about the expression and structure of HERVs. However, none of them has the broad scope of ReTe.
Performance
Speed of analysis is essential as the sequencing and assembly of genomes becomes faster. Sequence lengths from 104 to 1010 nt can realistically be analyzed. As demonstrated, ReTe faithfully reconstructed many features both of the simple retrovirus MoMLV, and the complex retrovirus HIV (Figure 4; Supplementary Data S2 and S5). Many of the ReTe functions may be further optimized, e.g. splice site prediction based on canonical consensus sequences and LTR detection.
Limits of retroviral sequence detection
The limitation to the four different nucleotides in DNA imposes restrictions on the possibility for sequence recognition of mutated sequences. This is observed in multiple nucleic acid alignments, where identities <50% must be regarded with caution. The typical mutation frequency (here discussed as substitutions) for sequences without selection pressure is ∼0.2% per million years (48). Thus, 1% substitution corresponds to ∼5 Mya, and 50% corresponds to ∼250 Mya. Consequently, 200–300 Mya is a detection limit for selection neutral retroviral sequences, which probably are the majority of the ERVs. Selection for a functional protein, leading to persistence of conserved retroviral amino acid motifs, can push back the limits for recognition considerably. This is demonstrated by the ability of ReTe to recognize widely divergent retrovirus-related retrotransposons like Errantiviruses of invertebrates (Figure 7). Thus, using a collection of motifs (Supplementary Data S3) largely (but not exclusively) derived from retroviral sequences of higher vertebrates, ReTe can detect retroviruslike sequences in amphibians, insects and worms. In this situation, the model-based approach of ReTe surpasses the unbiassed recognition via the BLAST algorithm (Figures 5 and 6). This attests to the structural antiquity of retroviruses. However, the demonstrated ability to detect highly mutated ERVs requires a relatively intact structural backbone. Secondary integrations of a few large (e.g. LINEs) or many smaller (e.g. SINEs) elements into an ERV can derange structure beyond repair by the ‘broken chain’ function of ReTe, and masking of nonretroviral repeats. This problem is inherent to the systematic structural approach of ReTe. On the other hand, ReTe often provides an interpreted proviral structure, which can be used in further studies.
The structural model of ReTe thus allows recognition of many retroviral sequences. There are both minor and major obstacles to widening the scope of detection. Adjustments of the distance constraints and inclusion of more motifs, are simple measures which may lead to an enhanced recognition of retroviral sequences like HERV-L. However, the pol gene of copia has the gene order IN..RT instead of the usual RT..IN, which would require the use of alternative models for these elements. The MalR retrotransposons (11) are incomplete and very divergent from orthoretroviral gene structure, with very few recognizable conserved motifs. The latter two are major challenges for ReTe.
ReTe analysis of five vertebrate genomes (Figure 8) demonstrated that vertebrate lineages have a variable number and type of ERVs. The mouse had a high number of high-scoring chains, and the dog and chicken genome had a low number. The human, chimpanzee and rhesus genomes were intermediate. Especially complete proviruses were betaretroviruslike in mouse and gammaretroviruslike in humans (Table 3). The findings extend and confirm previous observations (28,49). Several of these species differences must have arisen relatively late in evolution, probably due to more or less successful modifications of antiretroviral restrictions or changes in habitat and habits (50–52). Further work with the extensive retroviral sequence data set provided by ReTe will undoubtedly shed light both on retroviral and vertebrate evolution.
CONCLUSION
ReTe is a rational tool for detection and annotation of retroviral sequences, alone, in contigs or in entire genome assemblies. It provides ERV detection based on retrovirological expert knowledge and is independent of RepBase and RepeatMasker. Further developments include improvements to the motifs, distance constraints and alignment library and to the database and presentation module. An extension to include more of HERV-L, gypsy and copia elements is considered.
SUPPLEMENTARY DATA
ReTe and documentation is available from the authors. Contact Jonas.Blomberg@medsci.uu.se. Supplementary information is given at http://www.kvir.uu.se/RetroTector%5CRetroTectorProject.html and at NAR online.
ACKNOWLEDGEMENTS
We thank Tore Eriksson, Alina Castell, and Björn Sperber for assistance during the long developmental process. The support from the Swedish scientific council (grant no. 521-2001-6520) and local funds at the Academic Hospital in Uppsala is gratefully acknowledged. Funding to pay the Open Access publication charges for this article was provided by ALF funds given to JB.
Conflict of interest statement. None declared.
REFERENCES
- 1.Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. New York, USA: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 2.IHGSC. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.CSAC. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 4.ICGSC. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 5.Kumar A, Bennetzen JL. Plant retrotransposons. Annu. Rev. Genet. 1999;33:479–532. doi: 10.1146/annurev.genet.33.1.479. [DOI] [PubMed] [Google Scholar]
- 6.Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 7.Smit AF. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- 8.Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 9.Larsson E, Kato N, Cohen M. Human endogenous proviruses. Curr. Top. Microbiol. Immunol. 1989;148:115–132. doi: 10.1007/978-3-642-74700-7_4. [DOI] [PubMed] [Google Scholar]
- 10.Jurka J, Klonowski P, Dagman V, Pelton P. CENSOR – a program for identification and elimination of repetitive elements from DNA sequences. Comput. Chem. 1996;20:119–121. doi: 10.1016/s0097-8485(96)80013-1. [DOI] [PubMed] [Google Scholar]
- 11.Smit AF. Identification of a new, abundant superfamily of mammalian LTR-transposons. Nucleic Acids Res. 1993;21:1863–1872. doi: 10.1093/nar/21.8.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paces J, Pavlicek A, Paces V. HERVd: database of human endogenous retroviruses. Nucleic Acids Res. 2002;30:205–206. doi: 10.1093/nar/30.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paces J, Pavlicek A, Zika R, Kapitonov VV, Jurka J, Paces V. HERVd: the Human Endogenous RetroViruses Database: update. Nucleic Acids Res. 2004;32:D50. doi: 10.1093/nar/gkh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 15.Baldi P, Brunak S. Bioinformatics: The Machine Learning Approach. 2nd. Cambridge, MA, USA: MIT Press; 2001. [Google Scholar]
- 16.Frech K, Danescu-Mayer J, Werner T. A novel method to develop highly specific models for regulatory units detects a new LTR in GenBank which contains a functional promoter. J. Mol. Biol. 1997;270:674–687. doi: 10.1006/jmbi.1997.1140. [DOI] [PubMed] [Google Scholar]
- 17.Frech K, Quandt K, Werner T. Software for the analysis of DNA sequence elements of transcription. Comput. Appl. Biosci. 1997;13:89–97. doi: 10.1093/bioinformatics/13.1.89. [DOI] [PubMed] [Google Scholar]
- 18.Frech K, Werner T. Specific modelling of regulatory units in DNA sequences. Pac. Symp. Biocomput. 1997:151–162. [PubMed] [Google Scholar]
- 19.Huang X. On global sequence alignment. Comput. Appl. Biosci. 1994;10:227–235. doi: 10.1093/bioinformatics/10.3.227. [DOI] [PubMed] [Google Scholar]
- 20.Haykin S. Neural Networks. 2nd. Upper Saddle River, NJ, USA: Prentice Hall; 1999. [Google Scholar]
- 21.Eddy SR. Hidden Markov models. Curr. Opin. Struct. Biol. 1996;6:361–365. doi: 10.1016/s0959-440x(96)80056-x. [DOI] [PubMed] [Google Scholar]
- 22.Kozak M. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc. Natl Acad. Sci. USA. 1986;83:2850–2854. doi: 10.1073/pnas.83.9.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pavlicek A, Paces J, Elleder D, Hejnar J. Processed pseudogenes of human endogenous retroviruses generated by LINEs: their integration, stability, and distribution. Genome Res. 2002;12:391–399. doi: 10.1101/gr.216902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blomberg J, Ushameckis D, Jern P. In: Retroviruses and Primate Genome Evolution. Sverdlov ED, editor. Georgetown, TX, USA: Eurekah.com/Landes Bioscience; 2004. pp. 227–262. [Google Scholar]
- 26.Dewannieux M, Dupressoir A, Harper F, Pierron G, Heidmann T. Identification of autonomous IAP LTR retrotransposons mobile in mammalian cells. Nat. Genet. 2004;36:534–539. doi: 10.1038/ng1353. [DOI] [PubMed] [Google Scholar]
- 27.Wilkinson DA, Goodchild NL, Saxton TM, Wood S, Mager DL. Evidence for a functional subclass of the RTVL-H family of human endogenous retrovirus-like sequences. J. Virol. 1993;67:2981–2989. doi: 10.1128/jvi.67.6.2981-2989.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jern P. Uppsala: Acta Universitatis Upsaliensis; 2005. Thesis. [Google Scholar]
- 29.Jern P, Sperber GO, Ahlsen G, Blomberg J. Sequence variability, gene structure, and expression of full-length human endogenous retrovirus h. J. Virol. 2005;79:6325–6337. doi: 10.1128/JVI.79.10.6325-6337.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jern P, Sperber GO, Blomberg J. Definition and variation of human endogenous retrovirus H. Virology. 2004;327:93–110. doi: 10.1016/j.virol.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 31.Andersson AC, Yun Z, Sperber GO, Larsson E, Blomberg J. ERV3 and related sequences in humans: structure and RNA expression. J. Virol. 2005;79:9270–9284. doi: 10.1128/JVI.79.14.9270-9284.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blaise S, de Parseval N, Benit L, Heidmann T. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc. Natl Acad. Sci. USA. 2003;100:13013–13018. doi: 10.1073/pnas.2132646100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benit L, Calteau A, Heidmann T. Characterization of the low-copy HERV-Fc family: evidence for recent integrations in primates of elements with coding envelope genes. Virology. 2003;312:159–168. doi: 10.1016/s0042-6822(03)00163-6. [DOI] [PubMed] [Google Scholar]
- 34.Jern P, Sperber GO, Blomberg J. Use of endogenous retroviral sequences (ERVs) and structural markers for retroviral phylogenetic inference and taxonomy. Retrovirology. 2005;2:50. doi: 10.1186/1742-4690-2-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jern P, Sperber GO, Blomberg J. Divergent patterns of recent retroviral integration in human and chimpanzee genomes; transmissions from other primates to chimpanzees. J. Virol. 2006;80:1367–1375. doi: 10.1128/JVI.80.3.1367-1375.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oja M, Sperber GO, Blomberg J, Kaski S. Self-organizing map-based discovery and visualization of human endogenous retroviral sequence groups. Int. J. Neural Syst. 2005;15:163–179. doi: 10.1142/S0129065705000177. [DOI] [PubMed] [Google Scholar]
- 37.Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 38.Forsman A, Yun Z, Hu L, Uzhameckis D, Jern P, Blomberg J. Development of broadly targeted human endogenous gammaretroviral pol-based real time PCRs quantitation of RNA expression in human tissues. J. Virol. Methods. 2005;129:16–30. doi: 10.1016/j.jviromet.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 39.Muradrasoli S, Forsman A, Hu L, Blikstad V, Blomberg J. Development of real-time PCRs for detection and quantitation of human MMTV-like (HML) sequences HML expression in human tissues. J. Virol. Methods. 2006;136:83–92. doi: 10.1016/j.jviromet.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 40.Schmidt P, Forsman A, Andersson G, Blomberg J, Korsgren O. Pig islet xenotransplantation: activation of porcine endogenous retrovirus in the immediate post-transplantation period. Xenotransplantation. 2005;12:450–456. doi: 10.1111/j.1399-3089.2005.00244.x. [DOI] [PubMed] [Google Scholar]
- 41.McCarthy EM, McDonald JF. LTR_STRUC: a novel search and identification program for LTR retrotransposons. Bioinformatics. 2003;19:362–367. doi: 10.1093/bioinformatics/btf878. [DOI] [PubMed] [Google Scholar]
- 42.Kalyanaraman A, Aluru S. Efficient algorithms and software for detection of full-length LTR retrotransposons. J. Bioinform. Comput. Biol. 2006;4:197–216. doi: 10.1142/s021972000600203x. [DOI] [PubMed] [Google Scholar]
- 43.Benit L, Dessen P, Heidmann T. Identification, phylogeny, and evolution of retroviral elements based on their envelope genes. J. Virol. 2001;75:11709–11719. doi: 10.1128/JVI.75.23.11709-11719.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Parseval N, Lazar V, Casella JF, Benit L, Heidmann T. Survey of human genes of retroviral origin: identification and transcriptome of the genes with coding capacity for complete envelope proteins. J. Virol. 2003;77:10414–10422. doi: 10.1128/JVI.77.19.10414-10422.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Villesen P, Aagaard L, Wiuf C, Pedersen FS. Identification of endogenous retroviral reading frames in the human genome. Retrovirology. 2004;1:32. doi: 10.1186/1742-4690-1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zdobnov EM, Campillos M, Harrington ED, Torrents D, Bork P. Protein coding potential of retroviruses and other transposable elements in vertebrate genomes. Nucleic Acids Res. 2005;33:946–954. doi: 10.1093/nar/gki236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim TH, Jeon YJ, Kim WY, Kim HS. HESAS: HERVs expression and structure analysis system. Bioinformatics. 2005;21:1699–1700. doi: 10.1093/bioinformatics/bti194. [DOI] [PubMed] [Google Scholar]
- 48.Li WH. Molecular Evolution. Sunderland, MA, USA: Sinauer Associates, Inc., Publishers; 1997. [Google Scholar]
- 49.Baillie GJ, van de Lagemaat LN, Baust C, Mager DL. Multiple groups of endogenous betaretroviruses in mice, rats, and other mammals. J. Virol. 2004;78:5784–5798. doi: 10.1128/JVI.78.11.5784-5798.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stoye JP. Koala retrovirus: a genome invasion in real time. Genome Biol. 2006;7:241. doi: 10.1186/gb-2006-7-11-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van der Kuyl AC, Dekker JT, Goudsmit J. Distribution of baboon endogenous virus among species of African monkeys suggests multiple ancient cross-species transmissions in shared habitats. J. Virol. 1995;69:7877–7887. doi: 10.1128/jvi.69.12.7877-7887.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tarlinton RE, Meers J, Young PR. Retroviral invasion of the koala genome. Nature. 2006;442:79–81. doi: 10.1038/nature04841. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.