Abstract
The Cockayne Syndrome group B (CSB) protein plays important roles in transcription, transcription-coupled nucleotide excision repair and base excision DNA repair. c-Abl kinase also plays a role in DNA repair as a regulator/coordinator of the DNA damage response. This study presents evidence that the N-terminal region of CSB interacts with the SH3 domain of c-Abl in vitro and in vivo. In addition, c-Abl kinase phosphorylates CSB at Tyr932. The subcellular localization of CSB to the nucleus and nucleolus is altered after phosphorylation by c-Abl. c-Abl-dependent phosphorylation of CSB increased in cells treated with hydrogen peroxide and decreased in cells pre-treated with STI-571, a c-Abl-specific protein kinase inhibitor. Activation of the c-Abl kinase in response to oxidative damage is not observed in CSB null cells. These results suggest that c-Abl and CSB may regulate each other in a reciprocal manner in response to oxidative stress.
INTRODUCTION
Cockayne Syndrome (CS) is a hereditary human disease characterized by profound deficiency of post-natal growth with heterogeneity of clinical symptoms and neural degeneration resembling premature aging. CS is associated with defective transcription-coupled nucleotide excision repair (1). At the cellular/molecular level, the phenotype of CS includes sensitivity to ultraviolet (UV)-irradiation and delayed recovery of RNA and DNA synthesis after exposure to UV (2–4) There are two CS complementation groups, A and B (5), corresponding to genetic defects in the CSA and CSB genes, respectively. The CSB gene encodes a 168-kDa protein which is a member of the SWI/SNF family of DNA-dependent ATPases (6). Previous studies show that primary cells from CS patients have a defect in initiating (i.e. incision) repair of oxidative DNA lesions (7). In addition, cells that are deficient in CSB-dependent ATPase (motif VI mutants) are sensitive to γ-irradiation and defective in general genome repair of oxidative DNA damage (8,9). These data suggest that CSB may play a role in repair of oxidative DNA damage via the base excision repair (BER) pathway.
c-Abl is a non-receptor protein tyrosine kinase whose activities are tightly regulated in the cell. For example, both nuclear and cytoplasmic c-Abl is activated in response to genotoxic or oxidative stress (10), DNA damage (11–14), or by a protein kinase Cδ (PKCδ)-dependent mechanism (15). Activated c-Abl plays a role in inducing apoptosis and/or necrosis (16,17) after exposure to oxidative and other types of cellular stress. c-Abl is phosphorylated by DNA-PK (18,19) and interacts with DNA repair associated proteins (20–23) including p53, p73 and Rad9 (24–26). Oxidative stress activates c-Abl (27,28).
Structure-function analysis of c-Abl indicates that it has a protein–protein interaction SH3 domain, which is likely to participate in interactions between c-Abl and its kinase substrates. SH3 domains bind preferentially to proline-rich motifs such as P-X-X-P (29,30) (Figure 2B). CSB is a phosphoprotein with an N-terminal proline-rich region (Figure 2E). No prior reports have indicated that CSB undergoes tyrosine phosphorylation.
Figure 2.
Identification of binding domains of CSB and c-Abl. c-Abl interacts with N-terminal of CSB via its SH3 domain. (A) GST pull-down assay was done using GST-SH3 or GST-SH2 domain of c-Abl and purified human CSB protein. Immunoblot was analyzed using anti-HA antibody (top). Amido black scan representing the equal loading of proteins (bottom). (B) Schematic diagram of c-Abl (top) and CSB (bottom). TK: tyrosine kinase, PxxP: proline-rich motif, DNA and Actin: DNA- and actin-binding domains, NLS: nuclear localization signal, NES: nuclear export signal, SH3 and SH2: Src Homology 3 and Src Homology 2 domains. CSB interacts with c-Abl via its N-terminal. S-protein pull-down products (D) of whole cell extracts obtained from CS1AN cells transfected with S-protein tagged fragments of CSB (B), incubated with human c-Abl protein, separated by SDS-PAGE and immunoblotted, either with anti-Abl (D, top panel) or anti-S-protein (D, bottom panel). The sequence panel shows the amino acids fragment of CSB containing proline-rich region (E).
Tyrosine phosphorylation, particularly by c-Abl tyrosine kinase, has been reported to play a role in the regulation of some DNA repair proteins. c-Abl-mediated phosphorylation of DNA-topoisomerase I (topo I) at Tyr268 in vitro and in cells conferred activation of the topo I function (31). Moreover, activation of c-Abl by treatment of cells with ionizing radiation was associated with c-Abl-dependent phosphorylation of topo I and induction of topo I activity (31). Rad51 is a key element of recombinational DNA repair and its activity is regulated by phosphorylation of the tyrosine residue at position 315 by c-Abl tyrosine kinase (32). This study explores the interactions between c-Abl and CSB and the role of this interaction in the response to oxidative DNA damage. Evidence is presented that CSB binds to c-Abl and is a substrate for c-Abl tyrosine kinase. We speculate that tyrosine phosphorylation of CSB plays an important role in regulating and/or coordinating the cellular response to oxidative stress.
EXPERIMENTAL PROCEDURES
Cell cultures
CS1AN.S3.G2 (CS1AN) cells, a SV40- transformed human CS-B fibroblast, either with empty vector or complemented with wild-type CSB, E646Q, Q942E point mutants, ECFP-CSB were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, 50 U of penicillin/ml, 50 µg of streptomycin/ml and 50 µg of genetectin/ml or neomycin (ECFP-CSB). K562 lymphoblasts were cultured in RPMI medium 1640 containing 10% fetal bovine serum and antibiotics. CS1AN cells, mouse wild-type and c-Abl-/- embryonic fibroblats were cultured in Dulbecco's modified Eagle medium containing 10% fetal bovine serum, 50 U of penicillin/ml and 50 µg of streptomycin/ml. GM00969, AG08470, AG01428 and AG05012 primary fibroblasts were grown to confluence in minimum essential medium containing 15% FBS, 2X MEM non-essential amino acids, 2X MEM vitamins and 1X MEM essential amino acids. The human cells were from the American Type Culture Collection (Manassas, VA) or Coriell Cell Repositories (Camden, NJ). In some experiments, CS1AN and CSB-WT complemented cells were incubated with hydrogen peroxide (Sigma, St. Louis, MO) in serum-free media at a concentration of 250 µM for 1 h, either in the presence or absence of STI-571 (5 µM).
Proteins and antibodies
The purified HA/His-tagged CSB protein, GST–c-Abl–Src homology domain 2 (SH2) and SH3 fragments and the purified histidine-tagged c-Abl from a baculovirus insect expression system were prepared as previously described (33,34). The c-Abl fragment (45 kDa, containing the SH2 and kinase domains, SH2-PTK) was purchased from New England BioLabs (Beverly, MA) and a human active almost full-length c-Abl was purchased from Upstate technologies (Beverly, MA).
Rabbit anti-human c-Abl was purchased from Sigma (St. Louis, MO). Cy3-conjugated secondary mAbs were purchased from Jackson Immunoresearch Laboratories (West Grove, PA). Mouse anti-GFP, Alexa 488-conjugated secondary mAbs and the DNA stain 4′,6-diamidino-2-phenylindole, dihydrochloride (DAPI) were purchased from Molecular Probes (Eugene, OR).
Immunoprecipitation and immunoblot analysis
Procedures for preparing cell lysates and conducting immunoprecipitation and immunoblot analysis were described previously (35). The pre-cleared lysates were immunoprecipitated with rabbit polyclonal anti-CSB (H-300; Santa Cruz biotechnology, Santa Cruz, CA) or mouse monoclonal anti-Abl (8E9; BD Pharmingen) antibodies. The protein G Sepharose-precipitated proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS–PAGE), transferred to polyvinylidene difluoride membranes, and analyzed by immunoblotting with anti-CSB, anti-Abl (8E9; BD Pharmingen), or anti-phosphotyrosine (Upstate Technology, Beverly, MA) monoclonal antibodies followed by the chemiluminescent method for detection with SuperSignal substrates (Pierce, Rockford, IL). Membranes were stripped by using the Restore western blot stripping buffer (Pierce, Rockford, IL) when necessary. Anti-CSB immunoprecipitates were used either to study the interactions or to analyze in vivo tyrosine phosphorylation. Anti-Abl immunoprecipitates were used either to study the interactions or to analyze in vivo tyrosine phosphorylation or in in vitro kinase assays using GST–Crk (aa 120–225) as c-Abl substrate.
GST pull-down experiments
The CSB and GST–c-Abl fragments and GST was incubated and the adsorbents were analyzed by SDS–PAGE. The membrane was analyzed by immunoblotting with the monoclonal anti-HA or anti-c-Abl antibody. Also, GST–c-Abl and GST was incubated with HeLa whole cell extracts (1 mg/ml) and the adsorbents were analyzed by SDS–PAGE. The membrane was analyzed by immunoblotting with the polyclonal anti-CSB or stained with amido black.
S-protein pull-down experiments
CS1AN cells were transfected with cDNA (6 µg) expressing various S-tagged fragments of CSB protein (Figure 2C) for 30 h with the PolyFect reagent (Qiagen, Valencia, CA). Whole cell extracts were prepared as described above. Equal amount of proteins were subjected to S-protein pull down using S-protein conjugated agarose beads (Novagen). The pull-down products were centrifuged at 500 × g for 2 min to collect the beads and washed five times with ice-cold lysis buffer. The final bead pellets were either used to incubate with full-length human c-Abl for binding studies or for in vitro kinase assay.
Microscopy and immunofluorescence
Cells were fractionated in situ to remove the cytoskeleton according to Mirzoeva and Petrini (36), and then were processed for indirect immunofluorescence and analyzed as described in Partridge et al. (37). For quantification, images were analyzed with co-localization software (Zeiss) using the exact same set of criteria for all cells.
In vitro kinase assay and in vivo phosphorylation
For in vitro kinase assays, the purified CSB was incubated with the c-Abl fragment, mouse full-length c-Abl or human full-length GST-c-Abl or GST-c-Abl (K-R) in a kinase buffer (50 mM Tris–HCl, 10 mM MgCl2, 1 mM EGTA, 2 mM dithiothreitol) for 15 min at 28°C. The c-Abl kinase inhibitor STI-571 (Novartis Pharma AG) (38) was used as a control. Phosphorylated proteins were separated by SDS–PAGE and analyzed by immunoblotting.
For in vivo phosphorylation, CSB-WT or CS1AN cells (2.4 × 106 cells/10 cm culture dish) were transfected with 6 µg of wild-type c-Abl or the dominant-negative c-Abl (K-R) mutant vectors (39) for 30 h with the PolyFect reagent (Qiagen, Valencia, CA). The cells were lysed as described above. Equal amount of protein from the whole cell extracts was used to immunoprecipitates CSB followed by phosphotyrosine analysis. Whole cell extracts were also immunoblotted to detect c-Abl, and CSB.
Tyrosine-phosphorylation mapping by MALDI-MS analysis
MALDI-MS analysis was used to identify the tyrosine-phosphorylation site of CSB. In brief, in vitro kinase assay was done as described above. The phosphorylated proteins were separated on 4–20% Tris–HCl gel by SDS–PAGE. The gel was then stained with Commassie blue stain. The bands were then excised and subjected to in-gel tryptic digestion (Pierce, Rickford, IL) according to the manufacturer's instructions.
The digests were then subjected to phosphopeptide isolation using a phosphopeptide isolation kit (Pierce, Rockford, IL). Elutes were subjected to MALDI-MS analysis. Mass spectra were collected on a Voyager MALDI-TOF mass spectrometer using delayed extraction parameter. The eluted phosphopeptide mixtures were analyzed in reflector mode. 70–200 shots from a nitrogen laser (337 nm) were averaged to yield each recorded mass spectrum. Spectra were externally calibrated with angiotensin I (MH+ = 1296.6853) and adrenocorticotropic hormone fragment (7–38; MH+ = 3657.93). The matrix solution was prepared immediately before use by mixing 20 mg of α-cyano-4-hydroxycinnamic acid and 4 mg of 2,5-dihydroxybenzoic acid in 500 µl of 0.1% trifluoroacetic acid in 50% CH3CN/dH2O, vortexing for 1 min, centrifuging for 1 min, and subsequently diluting the clear supernatant 1:4 with 0.1% TFA in 50% CH3CN/dH2O. Five microliters of the diluted matrix solution was mixed with the eluted tyrosine-phosphorylated peptides, and a 1 µl suspension was spotted on the target and allowed to air-dry completely before mass analysis.
For interpretation of the mass spectra, a list of predicted molecular weights was generated by theoretical cleavage with trypsin as a specific endoproteinases using the MS-Digest program available at the University of California, San Francisco (http://prospector.ucsf.edu/ucsfhtml3.2/msdigest.htm). The masses of the peaks recorded in the mass spectra were matched to the calculated masses within ∼0.1% or better.
RESULTS
Physical interaction between CSB and c-Abl
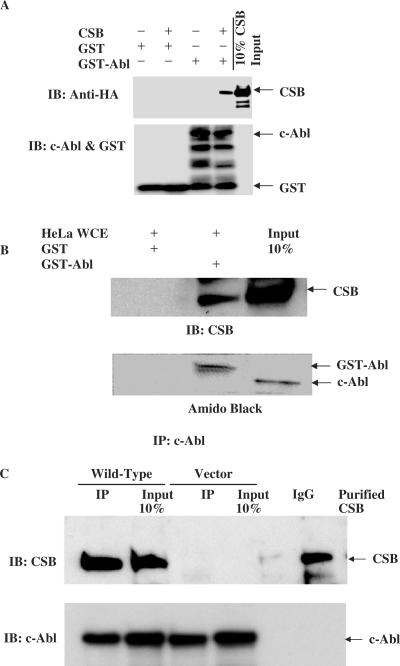
Physical interaction between purified CSB and c-Abl was examined in vitro using GST-pull-down or co-immunoprecipitation assays (Figure 1A, 1B and 1C). GST-pull-down products revealed that CSB and c-Abl interact with each other in vitro (Figure 1A). A GST-pull-down assay using GST-c-Abl and HeLa whole cell extracts (1 mg/ml) also revealed a potential interaction between c-Abl and endogenous CSB (Figure 1B). In vivo assays were also performed with extracts of CSB-deficient CS1AN cells or CS1AN cells complemented with wild-type CSB. Immunoprecipitation was carried out with anti-c-Abl antibody and the immunoprecipitates were blotted with anti-CSB. The results suggest that CSB interacts with c-Abl in vivo in CS1AN cells expressing wild-type CSB (Figure 1C).
Figure 1.
Physical interaction between CSB and c-Abl. (A) GST pull-down assay of purified HA-tagged CSB and GST-c-Abl. (B) GST pull-down assay of GST-c-Abl and HeLa whole cell extracts (WCE, 1 mg/ml). (C) Immunoprecipitation with anti-c-Abl antibody from the whole cell lysates (2-mg protein) obtained from CS1AN cells either complemented with wild-type CSB or an empty vector. Pull-down products or immunocomplex were separated by SDS–PAGE and the resulting blot was immunoblotted with respective antibodies as indicated in the figure.
The region of c-Abl that interacts with CSB was identified by incubating CSB with GST fusion proteins carrying the SH2 or the SH3 signaling domain of c-Abl. The results show that CSB binds to GST-c-Abl-SH3 (Figure 2A). Although we were not able to see any interaction of SH2 domain of c-Abl with CSB, there is a possibility of transient interaction of the SH2 domain with CSB. This interaction mapping was analyzed in more detail using S-protein tagged fragments of CSB. The results demonstrate that the N-terminal region of CSB, which includes proline-rich residues 301–304, interacts with the SH3 domain of c-Abl (Figure 2C, D and E). S-protein tagged CSB fragments (Figure 2C) were used in an alternate pull-down assay, which also indicated that the N-terminal region of CSB interacts with the SH3 domain of c-Abl (Figure 2C, D and E).
c-Abl phosphorylates CSB in vitro and in vivo
To determine whether CSB is a substrate for c-Abl, an in vitro kinase assay was performed by incubating CSB with a truncated form of c-Abl including its SH2 and protein tyrosine kinase domains (SH2-PTK). The results show that phosphotyrosine (p-Tyr) accumulates in CSB in the presence of active c-Abl kinase (SH2-PTK) but not in the presence of heat-inactivated c-Abl (SH2-PTK), a full-length kinase inactive c-Abl mutant (GST-c-Abl K-R), or after pre-incubation of either SH2-PTK fragment or full-length c-Abl with the kinase inhibitor STI-571 (Figure 3A and C). (Note that the amount of CSB protein was similar after phosphotyrosinylation; Figure 3A.) The kinase activities of full-length human c-Abl (Figure 3B) and human GST-c-Abl also phosphorylate CSB (Figure 3C). However, GST-c-Abl (K-R) failed to tyrosine phosphorylate CSB (Figure 3C). The location(s) of phosphotyrosine residues in SH2-PTK phosphorylated CSB were identified using an in vitro kinase assay with immunocomplexed S-protein fragments of CSB. The results indicate that phosphotyrosine is located only in CSB fragment aa 465–1056 (Figure 3D). The conserved ATPase and helicase motifs of CSB are also located in this region of CSB.
Figure 3.
CSB is phosphorylated by c-Abl in vitro and in vivo. Kinase assays were done by incubating purified CSB proteins with (A) increasing concentrations of SH2-PTK fragment of c-Abl (5, 10 and 20 ng), (B) full-length human-c-Abl protein (50 and 100 ng), and (C) full length human c-Abl protein or kinase-dead (K-R) c-Abl protein (50 and 100 ng). Phosphorylated proteins were separated by SDS–PAGE, transferred to a PVDF membrane, followed by immunoblotting for tyrosine phosphorylation using anti-p-Tyr. STI-571: kinase inhibitor (100 nM), +: present, −: absent, ▴: heat-inactivated c-Abl. (D) Kinase assays were done by incubating SH2-PTK fragment of c-Abl (20 ng) with S-protein pull-down products of whole cell extracts obtained from CS1AN cells transfected with S-protein tagged fragments of CSB. The resultant product was analyzed for phosphotyrosine (top panel), or c-Abl (middle panel) or S-protein (bottom panel). (E) c-Abl phosphorylates CSB in vivo. Wild-type CSB complemented CS1AN cells were either transfected with c-Abl or c-Abl (K-R) expression vectors. CSB was immunoprecipitated and anti-CSB immunoprecipitates were subjected to immunoblotting with anti-p-tyrosine antibody. Cell lysates were also immunoblotted with anti-c-Abl and anti-CSB antibodies. +: present, −: absent. (F) Anti-CSB immunoprecipitates from K562 CML lymphoblasts were analyzed for phosphotyrosine (top panel) or for CSB (bottom panel). (G) Anti-CSB immunoprecipitates either from wild-type or Abl−/− MEFs were analyzed for phosphotyrosine (top panel) or for c-Abl (bottom panel).
The ability of c-Abl to phosphorylate CSB in vivo was examined in CSB-deficient CS1AN cells expressing ectopic wild-type CSB. These cells were transfected with a plasmid expressing c-Abl or kinase-inactive c-Abl (K-R). CSB immunoprecipitates had significantly more phosphotyrosine in cells expressing c-Abl than in cells expressing c-Abl (K-R) or in cells that were not overexpressing c-Abl (Figure 3E). A low level of tyrosine phosphorylation was evident in cells expressing c-Abl (K-R), which could be due to endogenous c-Abl phosphorylation of CSB (Figure 3E). These results indicate that c-Abl tyrosine-phosphorylates CSB in vitro and in vivo and are consistent with the possibility that CSB is a physiologically important substrate of c-Abl.
CSB tyrosine-phosphorylation was also examined in chronic myeloid leukemia (CML) K562 lymphoblasts, which have a constitutive high level of Abl tyrosine kinase activity. These cells have been shown to express both cytoplasmic and nuclear c-Abl (40). CSB immunoprecipitates from whole cell extracts of these cells are significantly tyrosine-phosphorylated and the amount of phosphorylation is reduced if cells are pre-treated with STI-571 (Figure 3F). Similarly, CSB is tyrosine-phosphorylated in wild-type mouse embryonic fibroblasts (MEFs) but not in Abl-/- MEFs (Figure 3G). The amount of phosphotyrosine incorporated into CSB increased in MEFs exposed to oxidative stress and this increase was blocked by pre-treatment with STI-571 (Figure 3G). These data confirm the specificity of c-Abl for CSB and suggest that CSB may be a preferred substrate of c-Abl in cells exposed to oxidative stress.
c-Abl phosphorylates CSB at Tyr 932
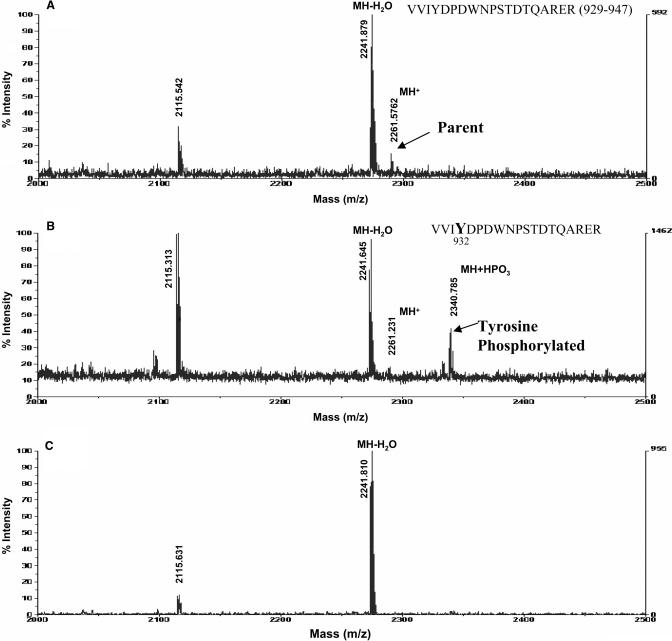
The tyrosine phosphorylation site of CSB was mapped at the amino acid level by MALDI-MS analysis of tryptic fragments of tyrosine-phosphorylated CSB. This experiment was carried out using the product of an in vitro kinase reaction containing c-Abl and CSB. The assay product was resolved by SDS–PAGE and digested by trypsin in situ. Phosphopeptides were eluted from the gel, concentrated using phosphopetide columns and analyzed by MALDI-MS (Figure 4). The peptides obtained from full-length CSB had a 95–99% match frequency with 59% coverage of full-length CSB. Mass analysis of each peptide indicated that a phosphate group was added to a 2262 kDa peptide VVIYDPDWNPSTDTQARER (929–947); this peptide contains Tyr932 (Figure 4B). Treatment with alkaline phosphatase released the phosphate moiety from the putative phosphotyrosine 932, suggesting correct identification of the peptide structure and correct mapping of tyrosine phosphorylation in CSB (Figure 4C). Tyr932 is located in motif VI of the putative ATPase/helicase domain of CSB.
Figure 4.
c-Abl phosphorylates CSB at Tyrosine 932. Purified CSB was phosphorylated by SH2-PTK domain on c-Abl in an in vitro kinase assay. Phosphorylated proteins were separated by SDS–PAGE. In-gel tryptic digestion was performed to digest phosphorylated CSB. Phosphopeptides were concentrated using phosphopeptide isolation column. The eluates were analyzed by MALDI-MS and the spectra were collected in reflector mode. Mass analysis of each peptide indicated that a phosphate group was added to a 2262 kDa peptide VVIYDPDWNPSTDTQARER (929–947); this peptide contains Tyr932 (A) CSB alone, (B) tyrosine-phosphorylated CSB and (C) tyrosine phosphorylated CSB after phosphatase treatment.
CSB and c-Abl co-localize after oxidative damage
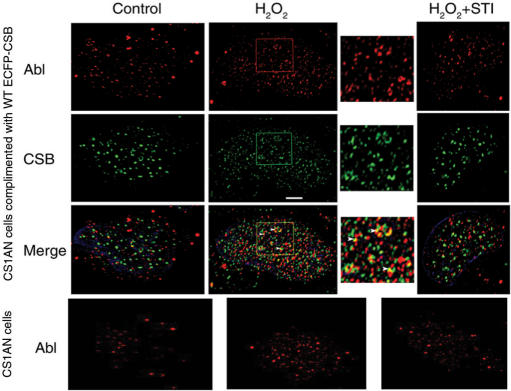
Indirect immunofluorescence of CSIAN cells expressing ECFP-CSB indicate that c-Abl (red, Figure 5) and CSB (green, Figure 5) are present throughout the nucleus in a punctate pattern in non-treated cells (Control). After exposure to hydrogen peroxide (H2O2), expression of ECFP-CSB and c-Abl increase and they re-distribute and co-localize in the nucleus and nucleolus (yellow in Merge, Figure 5). These effects can be inhibited with STI-571 (H2O2 + STI, Figure 5). These data raise the possibility that tyrosine phosphorylation of CSB by c-Abl promotes redistribution of CSB in the nucleus and enrichment of CSB in the nucleolus in response to oxidative stress; furthermore, these events may require c-Abl tyrosine kinase activity. Interestingly, when similar experiments were performed in CSB-deficient CS1AN cells (Bottom Panel, Figure 5), c-Abl redistribution to the nucleolus was not observed in response to oxidative stress. This result suggests that c-Abl and CSB might play complementary roles in a stress-response pathway that is activated by oxidative damage. Table 1 presents the quantitation of the data observed in Figure 5.
Figure 5.
CSB and c-Abl co-localize after H2O2 treatment. CSIAN cells complemented with ECFP-tagged WT CSB were processed for indirect immunofluorescence as described in Methods and stained with rabbit anti-c-Abl (1:40, red), anti-GFP1 mAb (1:200, green), and the DNA stain DAPI (blue). 0.1 µm Z-sections were obtained on an Axioplan 2i microscope (Carl Zeiss) and deconvolved with Openlab deconvolution software. Merge is the merged image of the three fluorescence channels examined of the same deconvolved Z-section. The bottom panel represents non-complemented CS1AN cells processed for indirect immunofluoresecence and stained with rabbit anti-c-Abl (1:40, red), serving as a comparative control. A representative cell from different experiments is shown. ×630; bar, 5 µm. An area of the H2O2 treated cells was enlarged (3×, square) and the arrowheads represent examples of co-localizing foci. Table 1 presents the quantitative analysis of images represented in this figure.
Table 1.
Quantitative analysis of images in Figure 5
| No of Pixels; NT = 100 | No of Foci; NT = 100 | |||||
|---|---|---|---|---|---|---|
| Groups | Green (CSB) % Increase | Red (c-Abl) % Increase | Yellow (co-localization) % Increase | % co-localizing yellow pixels/CSB (green) pixels | No. of co-localizing foci | % Increase |
| NT | 100.00 | 100.00 | 100.00 | 7.7 | 24.0 | 100.0 |
| H2O2 | 400.9 | 439.0 | 943.9 | 18.1 | 226.0 | 941.7 |
| STI + H2O2 | 125.0 | 108.1 | 100.0 | 6.1 | 25.0 | 104.2 |
Note: (i) There is a 4-fold increase over non-treated (NT) cells in both CSB (green) and c-Abl (red) signal after H2O2 treatment. A similar increase in signal was observed in CSB immunoprecipitates of H2O2 vs NT cells. (ii) There is an almost 10-fold increase in co-localization after H2O2 treatment, both in the number of co-localizing (yellow) pixels and in the number of co-localizing foci. (iii) STI-571 treated cells neither show an increase in signal intensity nor in co-localization, indicating the abrogation of the CSB-Abl interaction seen after H2O2 treatment, after use of the STI inhibitor. These results are similar to those obtained by immunoprecipitation.
Oxidative-stress induces c-Abl mediated tyrosine phosphorylation of CSB and CSB is necessary for activation of c-Abl after oxidative damage
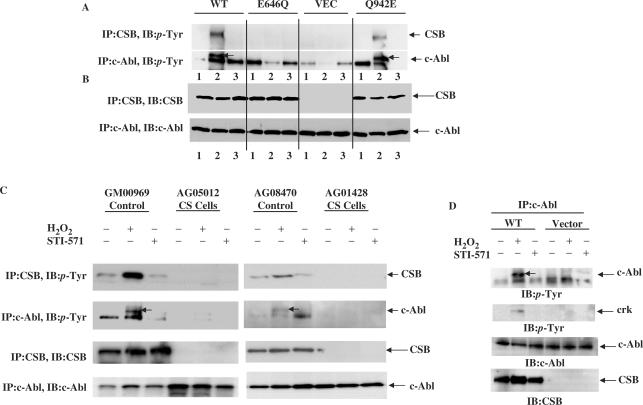
As stated above, we postulate that the interaction between CSB and c-Abl and the phosphorylation of CSB by c-Abl may play a role in the response to oxidative stress in vivo. This idea was tested by examining the efficiency of oxidative stress-induced c-Abl phosphorylation and autophosphorylation in the presence of wild-type and mutant forms of CSB. CS1AN cells expressing wild-type CSB, motif II E646Q CSB (ATPase-deficient) or motif VI Q942E CSB (BER-deficient) or no CSB were exposed to H2O2 and CSB phosphorylation and c-Abl autophosphorylation were examined. Autophosphorylation of c-Abl was significantly induced in cells expressing wild-type (WT) CSB and motif VI mutant CSB but not in cells expressing the CSB motif II mutant (Figure 6A). Tyrosine-phosphorylation increased significantly for WT CSB, but less for the motif VI mutant and not at all for the motif II mutant of CSB. Pre-treatment with STI-571 (5 µM) completely blocked c-Abl auto-phosphorylation and c-Abl mediated tyrosine phosphorylation of CSB (Figure 6A). c-Abl and CSB protein levels were similar in all cell extracts (Figure 6B). c-Abl auto-phosphorylation and tyrosine-phosphorylation of CSB was observed in normal human primary fibroblasts exposed to H2O2, but not in primary fibroblasts from CS patients (Figure 6C). Oxidative damage did not result in Abl auto-phosphorylation or tyrosine-phosphorylation of CSB in AG05012 or AG01428 primary fibroblasts obtained from CS patients. However, a significant auto-phosphorylation of c-Abl followed by a resultant tyrosine phosphorylation of CSB was observed in GM00969 and AG08470 fibroblasts derived from matched normal human subjects, after oxidative damage (Figure 6C). Pre-treatment with STI-571 inhibited auto-phosphorylation of c-Abl and tyrosine-phosphorylation of CSB (Figure 6C). These data suggest that CSB stimulates auto-phosphorylation of c-Abl and its activation in response to oxidative stress; this in turn results in tyrosine phosphorylation of CSB itself.
Figure 6.
Oxidative damage induces c-Abl activation and CSB phosphorylation by c-Abl. Oxidative damage induces activation of c-Abl and Abl-mediated phosphorylation of CSB, which is dependent on the ATPase function of CSB. Whole cell extracts were collected from CS1AN cells complemented with either wild-type CSB, or ATPase-deficient E646Q (motif II) mutant or motif VI (BER-deficient) Q942E mutant of CSB. 1. No treatment, 2. 250 µm H2O2, and 3. 24 h pre-treatment with STI-571 (5 µM) + 250 µM H2O2. Anti-CSB or anti-c-Abl immunoprecipitates were collected from 250 µg whole cell extracts and subjected to SDS page. The resultant membranes were either blotted with (A) anti-p-tyrosine or (B) anti-CSB (top panel) and anti-c-Abl (bottom panel). (C) Whole cell extracts were collected from primary fibroblasts from control subjects (GM00969 and AG08470) or from CS patients (AG05012 and AG01428) treated either with 250 µm H2O2 or 24 h pre-treatment with STI-571 (5 µM) + 250 µM H2O2. Anti-CSB or anti-c-Abl immunoprecipitates were collected from 250 µg whole cell extracts and subjected to SDS page. The resultant membranes were either blotted with anti-p-tyrosine, anti-CSB or anti-c-Abl antibodies. (D) Activation of c-Abl after H2O2. Whole cell extracts were collected from CS1AN cells complemented with either wild-type CSB or empty vector treated either with 250 µm H2O2 or 24 h pre-treatment with STI-571 (5 µM) + 250 µM H2O2. Kinase assay was done using immunocomplexed c-Abl and GST–Crk (aa 120–225). The product was analyzed either for phosphotyrosine (1st panel, c-Abl; 2nd panel, GST–Crk) or for c-Abl (3rd panel) or CSB (4th panel). The arrows on the p-Tyrosine panels of c-Abl represent 145 kDa c-Abl protein while the second band represents 119 kDa v-Abl isoform which also comes down with 8E9 antibody.
It is possible that auto-phosphorylation of c-Abl in response to oxidative damage influences c-Abl activity in a CSB-dependent manner. This idea was tested by measuring oxidative stress-induced c-Abl kinase activity towards a 15 amino acid fragment of Crk (aa 120–225) conjugated to GST in extracts of CS1AN cells with an ectopic plasmid expressing wild-type CSB. The results show that autophosphorylation of c-Abl increased in response to oxidative stress in extracts of CS1AN cells expressing wild type CSB but not in extracts of control cells carrying empty vector. Furthermore, activation/autophosphorylation of c-Abl stimulated tyrosine-phosphorylation of GST–Crk (aa 120–225) (Figure 6D). Pre-treatment with STI-571 inhibited c-Abl autophosphorylation and c-Abl-mediated tyrosine-phosphorylation of GST–Crk (aa 120–225) (Figure 6D). In order to decrease the possibility of phosphorylation by c-Abl immunoprecipitates from resting cells, low amounts of protein was used in these studies for immunoprecipitating c-Abl for kinase activity.
DISCUSSION
This study provides the first evidence of a physical interaction between CSB, a protein involved in the transcription-coupled DNA nucleotide excision DNA repair pathway, and the non-receptor tyrosine kinase c-Abl. CSB interacts with and is tyrosine phosphorylated by c-Abl in vitro and in vivo. Serine and threonine phosphorylation of CSB were reported previously (41), but this is the first report of tyrosine phosphorylation of CSB. Tyrosine-phosphorylated CSB is constitutive in CML K562 lymphoblasts. Also, wild-type MEFs exposed to hydrogen peroxide have increased CSB tyrosine-phosphorylation while it is lacking in Abl-/- MEFs. Tyrosine 932, which lies in motif VI of the ATPase domain of CSB, is the site of CSB phosphorylation by c-Abl. Oxidative stress stimulates c-Abl-mediated phosphorylation of CSB and this event is blocked by STI-571, a specific inhibitor of c-Abl kinase. Oxidative stress-induced c-Abl auto-phosphorylation and tyrosine phosphorylation of CSB appear to require CSB ATPase, because it was not observed in cells expressing an ATPase-deficient mutant of CSB. These data suggest that CSB and c-Abl may participate in and possibly regulate a common oxidative stress-response pathway.
c-Abl is a tightly regulated non-receptor tyrosine kinase that contains a Src homology domain 3 (SH3). The SH3 domain is a protein–protein interaction region which usually binds to proline-rich motifs (P-X-X-P) (42,43). This study shows that the interaction between c-Abl and CSB is mediated by the c-Abl SH3 domain and the N-terminal 355 amino acids of CSB (aa 1–355). This region of CSB includes a proline-rich motif at aa 301–304 (PVTP).
CSB cells and cell extracts are deficient in incision at 8-oxoguanine and 8-oxoadenine lesions (8–9,44) suggesting that CSB may play a role in the repair of these lesions. These lesions are believed to be removed by BER, and there is additional evidence for a role of CSB in the general genome BER pathway (45). Additional evidence comes from studies in mouse models (46) where increased accumulation of 8-oxoG is observed when mice are deficient in OGG1 in addition to lacking CSB. Also CSB is in protein complex with OGG1 (44) although 8-oxo-G appears to be removed by general genome BER, and there is no indication of transcription coupled repair of this lesion (47). This deficiency in 8-oxoguanine incision activity is complemented by ectopic expression of WT CSB; the deficiency is partially complemented by a motif II CSB mutant (ATPase deficient, E646Q) but not complemented by motif VI (Q942E) CSB mutants. (8,9,44). This result suggests that domain VI of CSB is required for repair of oxidative DNA lesions such as 8-oxoguanine. It is interesting to note that Tyr932, the site of c-Abl mediated tyrosine phosphorylation of CSB, also resides in domain VI of CSB protein.
In cells exposed to oxidative stress, c-Abl auto-phosphorylation and its phosphorylation of CSB increases. However, these effects are reduced and a lower extent of phosphorylation occurs in cells expressing the motif VI (Q942E) CSB mutant. CSB motif II is also required for these phosphorylation events, because they do not increase in response to oxidative damage in cells expressing a motif II CSB mutant or in cells lacking CSB. CSB is also required for oxidative stress-induced phosphorylation of Crk, a previously characterized substrate of c-Abl. This finding of the requirement of CSB in the activation of c-Abl during oxidative damage might have a greater role in the understanding of mechanisms of leukemia in CML patients. A constitutively active Abl and Bcr-abl feature the physiology of CML (48). Furthermore, a disruption of the CSB gene product has been reported as a possible anti-cancer target (49,50). It is possible that the disruption of CSB might help in the inactivation of Abl in case of CML. In addition, the lack of skin cancer in CSB patients might be due to the fact that because of lack of CSB in these patients, activation of c-Abl is hindered inhibiting the process of onset of carcinogenesis. These findings suggest that CSB may regulate auto-phosphorylation and activation of c-Abl in response to oxidative stress and it might play a role in the development of cancer. In addition, we also speculate that tyrosine phosphorylated CSB may play a specific role in regulating the response to oxidative DNA damage.
This study also shows that CSB and c-Abl co-localize in the nucleus and re-distribute in the nucleolus in cells treated with H2O2. This co-localization is inhibited by pre-treatment with STI-571. The redistribution of c-Abl and CSB may facilitate interaction between the two proteins and tyrosine phosphorylation of CSB. Alternatively, phosphorylation of CSB might promote its redistribution to sites of oxidative damage; this is consistent with the observation that redistribution of CSB and c-Abl is inhibited by STI-571. Furthermore, c-Abl and CSB do not co-localize to the nucleolus in unstressed cells, suggesting that activation of c-Abl may play a role in initiating redistribution. In future experiments, CSB tyrosine 932 mutants could be used to determine the role of c-Abl mediated tyrosine phosphorylation of CSB in the process of CSB redistribution and in the response to oxidative stress.
In conclusion, our results suggest that c-Abl interacts with and tyrosine phosphorylates CSB. This interaction may play an important role in the response to oxidative stress, resulting in activation of c-Abl, tyrosine phosphorylation of CSB and more efficient BER of oxidative DNA damage. Tyrosine-phosphorylated CSB may serve as a signal for repair proteins to localize to DNA damage and may help maintain active transcription in the nucleolus. These data also provide a first insight into a novel possible role of CSB as a signaling molecule in response to oxidative damage. This role can further be exploited to understand the phenotype of CS as well as in a general understanding of the role of CSB in response to various oxidative and DNA damage. Further studies are needed to understand the impact of tyrosine phosphorylation on the physiologically important functions of CSB in vivo.
ACKNOWLEDGEMENTS
We thank Drs Michael Seidman and Mohammed Hedayati for careful reading of the manuscript. We thank Glenn Quigley for excellent technical assistance for all the cell culture studies. This work was supported by funds from the National Institutes on Health/National Institute on Aging Intramural Program. Funding to pay the Open Access publication charges for this article was provided by the NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.van Hoffen A, Natarajan AT, Mayne LV, van Zeeland AA, Mullenders LH. Deficient repair of the transcribed strand of active genes in Cockayne's syndrome cells. Nucleic Acids Res. 1993;21:5890–5895. doi: 10.1093/nar/21.25.5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balajee AS, Bohr VA. Genomic heterogeneity of nucleotide excision repair. Gene. 2000;250:15–30. doi: 10.1016/s0378-1119(00)00172-4. [DOI] [PubMed] [Google Scholar]
- 3.Licht CL, Stevnsner T, Bohr VA. Cockayne syndrome group B cellular and biochemical functions. Am. J. Hum. Genet. 2004;6:1217–39. doi: 10.1086/380399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mayne LV, Lehmann AR. Failure of RNA synthesis to recover after UV irradiation: and early defect in cells from individuals with Cockayne's syndrome and xeroderma pigmentosum. Cancer Res. 1982;42:1473–1478. [PubMed] [Google Scholar]
- 5.Friedberg EC. Cockayne syndrome–a primary defect in DNA repair, transcription, both or neither? Bioessays. 1996;18:731–738. doi: 10.1002/bies.950180908. [DOI] [PubMed] [Google Scholar]
- 6.Troelstra C, van Gool A, De Wit J, Vermeulen W, Bootsma D, Hoeijmakers HJ. ERCC6 a member of a subfamily of putative helicases is involved in Cockaynes's syndrome and preferential repair of active genes. Cell. 1992;71:939–953. doi: 10.1016/0092-8674(92)90390-x. [DOI] [PubMed] [Google Scholar]
- 7.Tuo J, Jaruga P, Rodriguez H, Bohr VA, Dizdaroglu M. Primary fibroblasts of Cockayne syndrome patients are defective in cellular repair of 8-hydroxyguanine and 8-hydroxyadenine resulting from oxidative stress. FASEB J. 2003;17:668–674. doi: 10.1096/fj.02-0851com. [DOI] [PubMed] [Google Scholar]
- 8.Tuo J, Muftuoglu M, Chen C, Jaruga P, Selzer RR, Brosh R.M., Jr, Rodriguez H, Dizdaroglu M, Bohr VA. The Cockayne Syndrome group B gene product is involved in general genome base excision repair of 8-hydroxyguanine in DNA. J. Biol. Chem. 2001;276:45772–45779. doi: 10.1074/jbc.M107888200. [DOI] [PubMed] [Google Scholar]
- 9.Tuo J, Jaruga P, Rodriguez H, Dizdaroglu M, Bohr VA. The cockayne syndrome group B gene product is involved in cellular repair of 8-hydroxyadenine in DNA. J. Biol. Chem. 2002;277:30832–30837. doi: 10.1074/jbc.M204814200. [DOI] [PubMed] [Google Scholar]
- 10.Sun X, Majumder P, Shioya H, Wu F, Kumar S, Weichselbaum R, Kharbanda S, Kufe D. Activation of the cytoplasmic c-Abl tyrosine kinase by reactive oxygen species. J. Biol. Chem. 2000;275:17237–17240. doi: 10.1074/jbc.C000099200. [DOI] [PubMed] [Google Scholar]
- 11.Kharbanda S, Ren R, Pandey P, Shafman TD, Feller SM, Weichselbaum RR, Kufe DW. Activation of the c-Abl tyrosine kinase in the stress response to DNA-damaging agents. Nature. 1995;376:785–788. doi: 10.1038/376785a0. [DOI] [PubMed] [Google Scholar]
- 12.Jin S, Kharbanda S, Mayer B, Kufe D, Weaver DT. Binding of Ku and c-Abl at the kinase homology region of DNA-dependent protein kinase catalytic subunit. J Biol. Chem. 1997;272:24763–24766. doi: 10.1074/jbc.272.40.24763. [DOI] [PubMed] [Google Scholar]
- 13.Kharbanda S, Pandey P, Jin S, Inoue S, Bharti A, Yuan ZM, Weichselbaum R, Weaver D, Kufe D. Functional interaction between DNA-PK and c-Abl in response to DNA damage. Nature. 1997;386:732–735. doi: 10.1038/386732a0. [DOI] [PubMed] [Google Scholar]
- 14.Yuan ZM, Huang Y, Ishiko T, Nakada S, Utsugisawa T, Kharbanda S, Wang R, Sung P, Shinohara A, et al. Regulation of Rad51 function by c-Abl in response to DNA damage. J. Biol. Chem. 1998;273:3799–3802. doi: 10.1074/jbc.273.7.3799. [DOI] [PubMed] [Google Scholar]
- 15.Sun X, Wu F, Datta R, Kharbanda S, Kufe D. Interaction between protein kinase C delta and the c-Abl tyrosine kinase in the cellular response to oxidative stress. J. Biol. Chem. 2000;275:7470–7473. doi: 10.1074/jbc.275.11.7470. [DOI] [PubMed] [Google Scholar]
- 16.Sun X, Majumder P, Shioya H, Wu F, Kumar S, Weichselbaum R, Kharbanda S, Kufe D. Activation of the cytoplasmic c-Abl tyrosine kinase by reactive oxygen species. J. Biol. Chem. 2000;275:17237–17240. doi: 10.1074/jbc.C000099200. [DOI] [PubMed] [Google Scholar]
- 17.Sun X, Wu F, Datta R, Kharbanda S, Kufe D. Interaction between protein kinase C delta and the c-Abl tyrosine kinase in the cellular response to oxidative stress. J. Biol. Chem. 2000;275:7470–7473. doi: 10.1074/jbc.275.11.7470. [DOI] [PubMed] [Google Scholar]
- 18.Jin S, Kharbanda S, Mayer B, Kufe D, Weaver DT. Binding of Ku and c-Abl at the kinase homology region of DNA-dependent protein kinase catalytic subunit. J Biol. Chem. 1997;272:24763–24766. doi: 10.1074/jbc.272.40.24763. [DOI] [PubMed] [Google Scholar]
- 19.Kharbanda S, Pandey P, Jin S, Inoue S, Bharti A, Yuan ZM, Weichselbaum R, Weaver D, Kufe D. Functional interaction between DNA-PK and c-Abl in response to DNA damage. Nature. 1997;386:732–735. doi: 10.1038/386732a0. [DOI] [PubMed] [Google Scholar]
- 20.Agami R, Blandino G, Oren M, Shaul Y. Interaction of c-Abl and p73alpha and their collaboration to induce apoptosis. Nature. 1999;399:809–813. doi: 10.1038/21697. [DOI] [PubMed] [Google Scholar]
- 21.Gong JG, Costanzo A, Yang HQ, Melino G, Kaelin W.G., Jr, Levrero M, Wang JY. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature. 1999;399:806–809. doi: 10.1038/21690. [DOI] [PubMed] [Google Scholar]
- 22.Yoshida K, Komatsu K, Wang HG, Kufe D. c-Abl tyrosine kinase regulates the human Rad9 checkpoint protein in response to DNA damage. Mol. Cell Biol. 2002;22:3292–3300. doi: 10.1128/MCB.22.10.3292-3300.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan ZM, Huang Y, Fan MM, Sawyers C, Kharbanda S, Kufe D. Genotoxic drugs induce interaction of the c-Abl tyrosine kinase and the tumor suppressor protein p53. J. Biol. Chem. 1996;271:26457–26460. doi: 10.1074/jbc.271.43.26457. [DOI] [PubMed] [Google Scholar]
- 24.Yoshida K, Komatsu K, Wang HG, Kufe D. c-Abl tyrosine kinase regulates the human Rad9 checkpoint protein in response to DNA damage. Mol. Cell. Biol. 2002;22:3292–3300. doi: 10.1128/MCB.22.10.3292-3300.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan ZM, Huang Y, Fan MM, Sawyers C, Kharbanda S, Kufe D. Genotoxic drugs induce interaction of the c-Abl tyrosine kinase and the tumor suppressor protein p53. J. Biol. Chem. 1996;271:26457–26460. doi: 10.1074/jbc.271.43.26457. [DOI] [PubMed] [Google Scholar]
- 26.Yuan ZM, Shioya H, Ishiko T, Sun X, Gu J, Huang YY, Lu H, Kharbanda S, Weichselbaum R, et al. p73 is regulated by tyrosine kinase c-Abl in the apoptotic response to DNA damage. Nature. 1999;399:814–817. doi: 10.1038/21704. [DOI] [PubMed] [Google Scholar]
- 27.Sun X, Majumder P, Shioya H, Wu F, Kumar S, Weichselbaum R, Kharbanda S, Kufe D. Activation of the cytoplasmic c-Abl tyrosine kinase by reactive oxygen species. J. Biol. Chem. 2000;275:17237–17240. doi: 10.1074/jbc.C000099200. [DOI] [PubMed] [Google Scholar]
- 28.Sun X, Wu F, Datta R, Kharbanda S, Kufe D. Interaction between protein kinase C delta and the c-Abl tyrosine kinase in the cellular response to oxidative stress. J. Biol. Chem. 2000;275:7470–7473. doi: 10.1074/jbc.275.11.7470. [DOI] [PubMed] [Google Scholar]
- 29.Kay BK, Williamson MP, Sudol M. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 2000;14:231–241. [PubMed] [Google Scholar]
- 30.Mayer BJ. SH3 domains: complexity in moderation. J. Cell Sci. 2001;114:1253–1263. doi: 10.1242/jcs.114.7.1253. [DOI] [PubMed] [Google Scholar]
- 31.Yu D, Khan E, Khaleque MA, Lee J, Laco G, Kohlhagen G, Kharbanda S, Cheng YC, Pommier Y, et al. Phosphorylation of DNA topoisomerase I by the c-Abl tyrosine kinase confers camptothecin sensitivity. J. Biol. Chem. 2004;279:51851–51861. doi: 10.1074/jbc.M404396200. [DOI] [PubMed] [Google Scholar]
- 32.Conilleau S, Takizawa Y, Tachiwana H, Fleury F, Kurumizaka H, Takahashi M. Location of tyrosine 315, a target for phosphorylation by cAbl tyrosine kinase, at the edge of the subunit-subunit interface of the human Rad51 filament. J. Mol. Biol. 2004;339:797–804. doi: 10.1016/j.jmb.2004.04.017. [DOI] [PubMed] [Google Scholar]
- 33.Yoshida K, Komatsu K, Wang HG, Kufe D. c-Abl tyrosine kinase regulates the human Rad9 checkpoint protein in response to DNA damage. Mol. Cell. Biol. 2002;22:3292–3300. doi: 10.1128/MCB.22.10.3292-3300.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Christiansen M, Stevnsner T, Modin C, Martensen PM, Brosh R.M., Jr, Bohr VA. Functional consequences of mutations in the conserved SF2 motifs and post-translational phosphorylation of the CSB protein. Nucleic Acids Res. 2003;31:963–973. doi: 10.1093/nar/gkg164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng WH, von KC, Opresko PL, Fields KM, Ren J, Kufe D, Bohr VA. Werner syndrome protein phosphorylation by abl tyrosine kinase regulates its activity and distribution. Mol. Cell Biol. 2003;23:6385–6395. doi: 10.1128/MCB.23.18.6385-6395.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mirzoeva OK, Petrini JH. DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol. Cell Biol. 2001;21:281–288. doi: 10.1128/MCB.21.1.281-288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Partridge JJ, Lopreiato JO, Jr, Latterich M, Indig FE. DNA damage modulates nucleolar interaction of the Werner protein with the AAA ATPase p97/VCP. Mol. Cell Biol. 2003;14:4221–4229. doi: 10.1091/mbc.E03-02-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar S, Mishra N, Raina D, Saxena S, Kufe D. Abrogation of the cell death response to oxidative stress by the c-Abl tyrosine kinase inhibitor STI571. Mol. Pharmacol. 2003;63:276–282. doi: 10.1124/mol.63.2.276. [DOI] [PubMed] [Google Scholar]
- 39.Sawyers CL, McLaughlin J, Goga A, Havlik M, Witte O. The nuclear tyrosine kinase c-Abl negatively regulates cell growth. Cell. 1994;77:121–131. doi: 10.1016/0092-8674(94)90240-2. [DOI] [PubMed] [Google Scholar]
- 40.Bartova E, Harnicarova A, Pachernik J, Kozubek S. Nuclear topography and expression of the BCR/ABL fusion gene and its protein level influenced by cell differentiation and RNA interference. Leuk. Res. 2005;29:901–913. doi: 10.1016/j.leukres.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 41.Christiansen M, Stevnsner T, Modin C, Martensen PM, Brosh R.M., Jr, Bohr VA. Functional consequences of mutations in the conserved SF2 motifs and post-translational phosphorylation of the CSB protein. Nucleic Acids Res. 2003;31:963–973. doi: 10.1093/nar/gkg164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cao C, Leng Y, Huang W, Liu X, Kufe D. Glutathione peroxidase 1 is regulated by the c-Abl and Arg tyrosine kinases. J. Biol. Chem. 2003;278:39609–39614. doi: 10.1074/jbc.M305770200. [DOI] [PubMed] [Google Scholar]
- 43.Cao C, Leng Y, Kufe D. Catalase activity is regulated by c-Abl and Arg in the oxidative stress response. J. Biol. Chem. 2003;278:29667–29675. doi: 10.1074/jbc.M301292200. [DOI] [PubMed] [Google Scholar]
- 44.Tuo J, Jaruga P, Rodriguez H, Bohr VA, Dizdaroglu M. Primary fibroblasts of Cockayne syndrome patients are defective in cellular repair of 8-hydroxyguanine and 8-hydroxyadenine resulting from oxidative stress. FASEB J. 2003;17:668–674. doi: 10.1096/fj.02-0851com. [DOI] [PubMed] [Google Scholar]
- 45.Thorslund T, von KC, Harrigan JA, Indig FE, Christiansen M, Stevnsner T, Bohr VA. Cooperation of the Cockayne syndrome group B protein and poly(ADP-ribose) polymerase 1 in the response to oxidative stress. Mol. Cell Biol. 2005;25:7625–7636. doi: 10.1128/MCB.25.17.7625-7636.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trapp C, Reite K, Klungland A, Epe B. Deficiency of the Cockayne syndrome B (CSB) gene aggravates the genomic instability caused by endogenous oxidative DNA base damage in mice. Oncogene. 2007 doi: 10.1038/sj.onc.1210167. In Press. [DOI] [PubMed] [Google Scholar]
- 47.Thorslund T, Sunesen M, Bohr VA, Stevnsner T. Repair of 8-oxoG is slower in endogenous nuclear genes than in mitochondrial DNA and is without strand bias. DNA Repair (Amst.) 2002;1:261–273. doi: 10.1016/s1568-7864(02)00003-4. [DOI] [PubMed] [Google Scholar]
- 48.Ren R. Mechanisms of BCR-ABL in the pathogenesis of chronic myelogenous leukaemia. Nat. Rev. Cancer. 2005;5:172–183. doi: 10.1038/nrc1567. [DOI] [PubMed] [Google Scholar]
- 49.Lu Y, Mani S, Kandimalla ER, Yu D, Agrawal S, States JC, Bregman DB. The Cockayne syndrome group B DNA repair protein as an anti-cancer target. Int. J. Oncol. 2001;19:1089–1097. doi: 10.3892/ijo.19.6.1089. [DOI] [PubMed] [Google Scholar]
- 50.Lu Y, Lian H, Sharma P, Schreiber-Agus N, Russell RG, Chin L, van der Horst GT, Bregman DB. Disruption of the Cockayne syndrome B gene impairs spontaneous tumorigenesis in cancer-predisposed Ink4a/ARF knockout mice. Mol. Cell. Biol. 2001;21:1810–1818. doi: 10.1128/MCB.21.5.1810-1818.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]