Abstract
Episomal gene expression vectors offer a safe and attractive alternative to integrating vectors. Here we describe the development of a high capacity episomal vector system exploiting human episomal retention sequences to provide efficient vector maintenance and regulated gene expression through the delivery of a genomic DNA locus. The iBAC-S/MAR vector is capable of the infectious delivery and retention of large genomic DNA transgenes by exploiting the high transgene capacity of herpes simplex virus type 1 (HSV-1) and the episomal retention properties of the scaffold/matrix attachment region (S/MAR). The iBAC-S/MAR vector was used to deliver and maintain a 135 kb genomic DNA insert carrying the human low density lipoprotein receptor (LDLR) genomic DNA locus at high efficiency in CHO ldlr−/− a7 cells. Long-term studies on CHO ldlr−/− a7 clonal cell lines carrying iBAC-S/MAR-LDLR demonstrated low copy episomal stability of the vector for >100 cell generations without selection. Expression studies demonstrated that iBAC-S/MAR-LDLR completely restored LDLR function in CHO ldlr−/− a7 cells to physiological levels and that this expression can be repressed by ∼70% by high sterol levels, recapitulating the same feedback regulation seen at the endogenous LDLR locus. This vector overcomes the major problems of vector integration and unregulated transgene expression.
INTRODUCTION
An ideal vector for functional genetics or for gene therapy applications should allow long-term expression of the delivered transgene at close to physiological levels. Many features that regulate gene expression have been characterized in detail but other contributory factors are poorly understood. It is known, for example, that expression of a gene can be influenced by several factors such as its replication timing during the cell cycle (1) and its localization within the nucleus (2,3). Complexity of gene expression also includes alternative splicing mechanisms, alternative promoter usage and effects of DNA polymorphisms, which could explain the production of a large number of proteins from a relatively small number of human genes (around 22 000 known genes, see http://www.ensembl.org/Homo_sapiens/index.html). To achieve physiological conditions of expression and retention, an efficient vector system for gene therapy or expression studies should be based on sequences derived from the human genome.
The delivery of a complete genomic locus has proven to be an excellent means to express a transgene at physiological levels, in contrast to cDNA-based expression cassettes (4–8). cDNA-based constructs have shown many limitations due to the silencing of the heterologous promoter or transgene overexpression, which can be toxic for the cell. The use of an entire genomic DNA locus, in which the native promoter of a gene and all regulatory sequences are included in the vector, has demonstrated to be effective in rescuing deficiency phenotypes by providing physiological levels of expression and correct alternative splicing and promoter usage mechanisms (5–8).
Long-term transgene retention can be achieved through the vector integration into the host genome or through the replication and persistence of the vector in the nucleus as an extrachromosomal unit (episome). Episomal vectors are capable of long-term persistence in mammalian cells requiring the two main features of replication and segregation into daughter cells. Episomal maintenance systems offer many advantages over integrating vectors as they avoid unpredictable integration into the host genome and the risk of cellular transformation.
Many episomal systems described in the past have been derived from viral genomes (9). To be maintained extrachromosomally, most episomal vector systems rely on the expression of viral products, which may possibly confer immunogenic or oncogenic properties. Recently, a new episomal system has been described, named pEPI-1, which possesses replication and episomal retention features through the function of a scaffold/matrix attachment region (S/MAR) isolated from the human β-interferon gene (10,11).
In this project, we describe the development of a high-capacity episomal vector system based on the above-described human S/MAR sequence. S/MARs are 70% AT-rich sequences, which are believed to play many important roles in chromatin function. The human genome is organized in a complex structure within the nucleus where DNA interacts with histones and chromatin proteins to form a 30-nm fibre, which appears to be organized into loops by interaction of S/MARs with the nuclear matrix (12). Besides their structural function, S/MARs also play an important role in temporal and spatial organization of gene expression (13,14).
The episomal features of an S/MAR-based self-replicating vector can be explained by its interaction with the nuclear matrix (11), since cellular processes such as DNA replication and transcription are closely associated with this proteinaceous structure (14). Close involvement in DNA replication has also been confirmed by the finding of S/MAR sequences close to replication sites (15,16). The inclusion of an S/MAR sequence in an expression vector can help increase the level of expression and avoid silencing of the transgene (17,18).
Here we have developed an episomal vector system based on an S/MAR element able to deliver, maintain and express a genomic DNA transgene at physiological levels. To investigate regulated expression from a genomic locus, we used a previously characterized BAC clone carrying the 45-kb human low density lipoprotein receptor locus (LDLR) within a 135-kb genomic DNA insert (6) since this gene is an excellent example of finely regulated expression (19,20). Delivery of such large genomic DNA loci by non-viral means can be technically challenging and exhibit poor efficiency. Therefore, to achieve a high level of delivery in cells we used the herpes simplex type 1 (HSV-1) amplicon vector exploiting its high transgene capacity, unique amongst viral vectors, in a system we have called the infectious bacterial artificial chromosome, or iBAC.
Here we describe the construction and analysis of the 156-kb iBAC-S/MAR-LDLR vector and demonstrate intact delivery to cells by HSV-1 amplicons. We generated stable clonal cell lines carrying the iBAC-S/MAR-LDLR vector as a low copy, stable episome. Functional expression studies on two independent clones demonstrated a full functional recovery of LDLR activity in a deficient cell line. LDLR transgene expression retained physiological regulation and was repressed by high sterol levels. Finally, we showed the high mitotic stability of iBAC-S/MAR-LDLR by demonstrating long-term episomal retention in both clones grown in the absence of selection.
Overall, these data describe the development of a new episomal vector system lacking viral coding sequences and able to provide efficient delivery and long-term regulated expression of a large genomic DNA transgene.
MATERIALS AND METHODS
Vector construction
The vector pEPHZ was obtained by modifying pEPI (a gift from H. Lipps, University of Witten/Herdecke, Germany). The resulting vector contains the HSV-1 amplicon elements (oris and pac), the LacZ gene under IE4/5 promoter for titration on G16.9 cells, the R6Kγ bacterial conditional replication origin, a loxP site and a 5875-bp fragment purified from pEPI PciI-EcoO109I digestion, which contains the EGFP-S/MAR cassette under CMV promoter and the Kan/Neo resistance cassette under SV40 promoter. pEPHZ DNA was produced using π-expressing bacteria to allow replication from the R6Kγ conditional replication origin. pEPHZ-LDLR was obtained by incubating 2 ng of pEPHZ and 500 ng of LDLR BAC (164O19 clone, GenBank AC011485) with 1 unit of purified Cre enzyme at 37°C for 1 h followed by inactivation at 65°C for 10 min. The reaction was drop-dialyzed against water for 2 h after which time the whole reaction was electroporated in DH10B (ElectroMAX DH10B Cells, Invitrogen). All enzymes used have been purchased from New England Biolabs (NEB), UK.
Tissue culture
CHO wild-type and CHO ldlr−/− a7 cells (gifts from M. Krieger, Massachusetts Institute of Technology, Cambridge, MA, USA) were cultured in Ham's F12 Nutrient Mix supplemented with 10% fetal bovine serum (FBS), 20 mM l-glutamine (l-glut), penicillin (100 U/ml), and streptomycin (100 μg/ml) (P/S). In all experiments, we used CHO ldlr−/− a7 cells expressing the herpes virus entry protein C (HveC) to enhance their transduction (21). CHO ldlr−/− a7 HveC+ cells (referred throughout simply as CHO ldlr−/− a7) (6) were grown in the same medium as CHO wild-type cells but supplemented with puromycin at a final concentration of 4 μg/ml. G16.9 cells (a derivative of the human Gli-36 glioma cell line expressing the HSV-1 VP-16 protein to enhance the expression from the amplicon pIE4/5-lacZ promoter cassette; a kind gift of Y. Saeki and E.A. Chiocca) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS, P/S, l-glut and hygromycin-B (200 μg/ml, Invitrogen). Vero 2-2 cells (a derivative of Vero African green monkey kidney cells constitutively expressing the HSV-1 ICP27 protein, kindly provided by Dr Rozanne Sandri-Goldin, University of California, Irvine, CA, USA) were grown in DMEM, 10% FBS, P/S, l-glut and G418 (Geneticin, Gibco) at 500 μg/ml. For the generation of pEPHZ-LDLR stable clones, 4 × 105 CHO ldlr−/− a7 cells were plated per well in a 6-well plate and infected with pEPHZ-LDLR amplicons 24 h later at a multiplicity of infection (MOI) of 0.1–0.5. Infected cells were diluted after 48 h in medium containing G418 450 μg/ml and single colonies isolated after 10–15 days. All cells were cultured at 37°C in 5% (v/v) CO2.
HSV-1 amplicons production
HSV-1 amplicons were produced using an improved HSV-1 helper virus-free system as previously described (22). Typically, the supernatant from three 6-cm dishes was concentrated by ultracentrifugation at 22 000 r.p.m. for 3 h in an SW41 rotor (Beckman) and resuspended in 250 μl of DMEM, 10% FBS, P/S, l-glut, to give a stock of 1–2 × 107 transducing units/ml. For amplicon titration 4 × 105 G16.9 cells were seeded per well in a 24-well plate and infected 24 h later with pEPHZ-LDLR amplicons. Twenty-four hours after infection, amplicons were removed and cells fixed with fixative solution (PBS containing 2% formaldehyde and 0.05% glutaraldehyde), washed with PBS and incubated with X-gal staining solution (PBS containing 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 2 mM MgCl2 and X-gal 1 mg/ml) at 37°C overnight to determine LacZ expression.
Plasmid rescue assay
Episomal DNA was prepared by an alkaline lysis method from a confluent 10-cm dish as follows. Cells were scraped into 1.5 ml of PBS and centrifuged at 5000 r.p.m. for 3 min. The cell pellet was resuspended in 60 μl STET buffer (8% sucrose, 5% Triton X-100, 50 mM EDTA, 50 mM Tris-Cl pH 8). The cells were lysed with 130 μl of alkaline SDS (1% SDS, 0.2 N NaOH), neutralized with 110 μl 7.5 M ammonium acetate, incubated on ice for 5 min and spun down at 13 000 r.p.m. for 30 min at 4°C. The supernatant was extracted twice with 1:1 phenol:chloroform and twice with chloroform, and the nucleic acids were precipitated and resuspended in 20 μl of TE (10 mM Tris, 1 mM EDTA) with RNAse A 50 μg/ml. Ten microlitres of the episomal preparation were used to transform by electroporation DH10B bacteria, which were plated on LB agar with antibiotics. Plasmid DNA was prepared from the resulting bacterial colonies, digested with NotI and the digests were resolved by pulsed-field gel electrophoresis.
LDLR expression analysis
Lipoprotein-deficient FBS (LPDS) was purchased from Biomedical Technologies, Inc. (Stoughton, MA, USA), human LDL and DiI-LDL were purchased from AbD Serotec (Kidlington, Oxford). Qualitative analysis of LDLR function by measuring DiI-LDL fluorescence was performed as follows. 3 × 104 CHO cells were seeded per well in a 24-well plate and the day after the medium was replaced with Ham's F12 medium supplemented by 5% LPDS. Forty-eight hours later, cells were incubated in medium containing DiI-LDL (10 μg/ml) for 5 h at 37°C. The incubation mix was then removed, the cells were washed once in Ham's F12 medium, and fluorescence was observed on a Nikon Eclipse TE2000-U inverted microscope and photographed. For quantitative analysis, the cells were then washed twice with PBS with Ca2+ and Mg2+ containing BSA 0.4% and three times with PBS alone and lysed with 0.1% SDS/0.1N NaOH (23). Fluorescence in the cell lysate was analysed with a Shimadzu RF-1501 spectrofluorophotometer at excitation and emission wavelengths of 520 and 580 nm, respectively and total protein content determined using Bicinchoninic acid solution (BCA, Sigma). Non-specific binding was determined in the presence of a 50-fold excess of unlabelled LDL and subtracted from total binding to give specific binding. To investigate the effect of sterols on LDLR expression, the medium was supplemented using stock solutions of 12 mg/ml cholesterol and 1.2 mg/ml 25-hydroxycholesterol dissolved in ethanol, to achieve final concentrations of 12 μg/ml cholesterol and 0.6 μg/ml 25-hydroxycholesterol. An equivalent amount of ethanol was added to the medium in control incubations.
Genomic analysis of stable pEPHZ-LDLR clones
The Southern blot analyses were performed as previously described (24). Five micrograms of genomic DNA samples were digested overnight with BamHI and digestion fragments resolved on a 1% agarose gel together with a set of 3-fold serial dilutions of BamHI digests of quantified pEPHZ, each dilution mixed with 5 μg of CHO ldlr−/− a7 genomic DNA prior to gel loading. The gel was blotted on Hybond N+ membrane (Amersham Biosciences) and hybridized with a 32P radio-labelled Kanr probe obtained purifying a 1015-bp fragment from AvrII pEPHZ digest. The labelling kit (Random Primers DNA Labeling System) was purchased from Invitrogen. Band intensity was analysed on the blot using a Bio-Rad FX personal imager and on the films using a Fluor Chem 8800 Imaging system and copy number calculated comparing band intensity of genomic DNA samples with pEPHZ known quantities. Rate of loss data have been calculated using a first order rate of loss model (λ = (−1/t)(ln Nt/N0) where N0 is the number of copies at the start of the experiment and Nt the number of copies after t generations), considering CHO replication time to be 17 h.
RESULTS
Construction of pEPHZ-LDLR
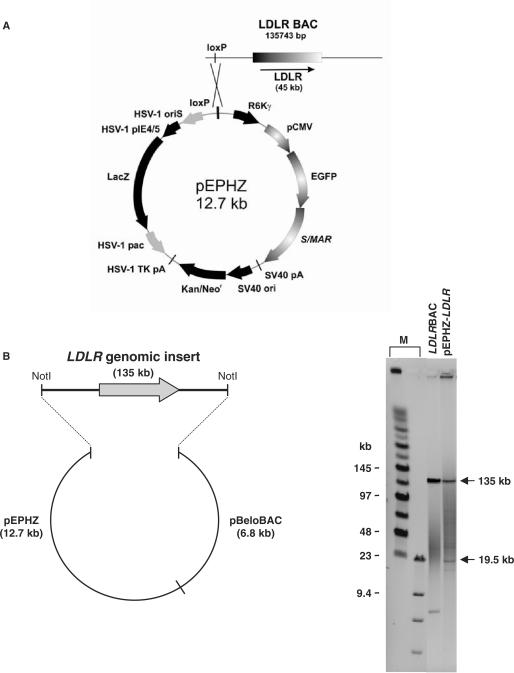
To construct pEPHZ-LDLR, we first modified the S/MAR-containing plasmid pEPI to create pEPHZ, by inserting a loxP site and the sequences necessary for vector packaging into HSV-1 amplicons (Figure 1A). To incorporate the human LDLR locus into this vector we used a previously identified BAC clone containing the complete 45 kb genomic DNA locus within a 135 kb insert (CIT-B 164O19, GenBank AC011485) (6). This LDLR BAC clone was retrofitted with pEPHZ using an in vitro reaction mediated by purified Cre enzyme, able to recognize the loxP sites present on both plasmids (Figure 1A).
Figure 1.
Construction of pEPHZ-LDLR. (A) Schematic representation of Cre-loxP recombination between pEPHZ and LDLR-BAC. (B) NotI restriction analysis of the retrofitted pEPHZ-LDLR gives the expected bands of 135.7 and 19.5 kb. M: size markers lanes.
Analysis of the resulting vector by NotI restriction digestion followed by pulsed-field gel electrophoresis showed the integrity of the 135 kb genomic insert and the presence of only one copy of pEPHZ in pEPHZ-LDLR. The presence of the expected bands confirmed the successful construction of pEPHZ-LDLR through a Cre-loxP-based recombination (Figure 1B).
Infectious delivery of intact pEPHZ-LDLR into CHO ldlr−/− a7 cells
We packaged pEPHZ-LDLR amplicons in Vero 2–2 cells using an improved HSV-1 amplicon helper-virus-free system (22). Typically amplicon titers of ∼1–2 × 107 transducing units (t.u.)/ml, measured on G16.9 cells by using X-gal staining, were achieved after concentration by ultracentrifugation.
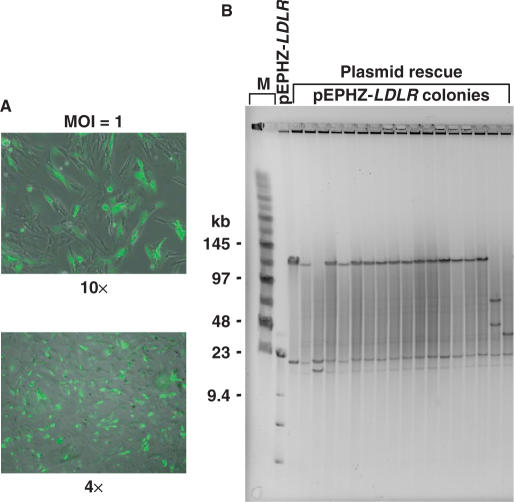
To demonstrate intact delivery of the 135-kb LDLR genomic DNA insert, we infected CHO ldlr−/− a7 cells with pEPHZ-LDLR amplicons at a multiplicity of infection (MOI) of 1. In all experiments, we used CHO ldlr−/− a7 cells expressing the herpesvirus entry protein C (HveC) to enhance their transduction (21). Forty-eight hours post-infection we prepared episomal DNA and performed a plasmid rescue assay. DNA inside HSV-1 amplicons is linear but it re-circularizes after entry into the nucleus. The plasmid rescue assay will shuttle re-circularized pEPHZ-LDLR molecules, from infected CHO ldlr−/− a7 cells back into bacteria where the vector can be analysed. Plasmid DNA was prepared from bacterial colonies grown on kanamycin/chloramphenicol plates, digested with NotI and analysed by pulsed-field gel electrophoresis. pEPHZ-LDLR was shown to be delivered intact by HSV-1 amplicons and re-circularize upon infection unrearranged, with an efficiency of 82% (Figure 2).
Figure 2.
Intact delivery of pEPHZ-LDLR by HSV-1 amplicons. (A) Infection is confirmed by fluorescence microscopy analysis showing expression of the vector reporter gene EGFP in the infected CHO ldlr−/− a7 cells. (B) Plasmid rescue 48 h after infection shows pEPHZ-LDLR to be delivered intact with an efficiency of ∼82%. M: size marker lanes.
This experiment confirmed the high capacity of HSV-1 amplicons demonstrating the high efficiency of packaging and delivery of the intact 156-kb pEPHZ-LDLR vector.
Generation of pEPHZ-LDLR episomal clonal cell lines
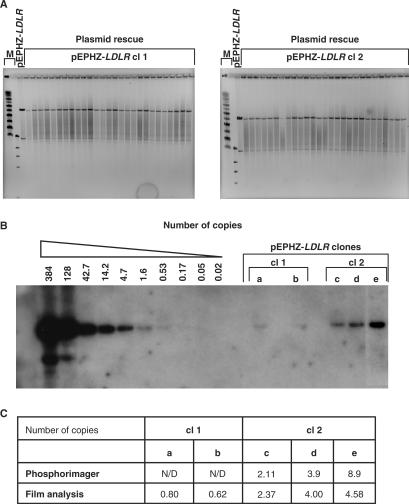
In order to assess whether pEPHZ-LDLR can persist in cells as an episome, we generated CHO ldlr−/− a7 stable clonal cell lines carrying this vector. CHO ldlr−/− a7 cells were seeded and then infected 24 h later with pEPHZ-LDLR amplicons at an MOI of 0.1–0.5. Infected cells were diluted 48 h later in medium supplemented with G418 (450 μg/ml) and left to grow for about 10–14 days. We isolated a total number of 108 early stage single clones. In most cases the clones did not survive prolonged culture, which we interpret as being due to the episome only providing transient antibiotic resistance and not successfully becoming permanently established. We obtained ten established growing clonal lines and screened them for episomal status by plasmid rescue, which we consider the optimal test for the presence of established episome. Plasmid rescue demonstrated the vector to be present as an episome in three of the ten (30%) established clones. Figure 3A shows NotI restriction analysis followed by pulsed-field gel electrophoresis of DNA rescued from two of the established episomal clones. These data demonstrate the replication/retention features of pEPHZ-LDLR since episomal plasmid DNA extracted from clones after several weeks of growth under antibiotic selection (∼70 cell generations) showed a restriction digest pattern identical to the one of the original starting DNA preparation used for packaging (Figure 3A). These two clones named pEPHZ-LDLR clones 1 and 2 (abbreviated to cl 1 and cl 2) were used for subsequent work. Plasmid rescue analysis was repeated every two weeks throughout the duration of the experiment to confirm continuing episomal retention.
Figure 3.
Characterization of pEPHZ-LDLR clonal cell lines. (A) Plasmid rescue analysis from the two pEPHZ-LDLR clones, cl 1 and cl 2, demonstrates the episomal retention and replication of pEPHZ-LDLR. M: size marker lanes. (B) pEPHZ-LDLR is retained in cl 1 and cl 2 at a number of ∼1 and ∼4 copies/cell, respectively. (C) pEPHZ-LDLR copy number has been assessed using two independent systems. N/D: not determined.
These results clearly show that pEPHZ-LDLR is replicated and retained episomally when delivered by HSV-1 amplicon infection and confirm the presence of an intact genomic locus as vector rearrangements were not observed.
Copy number estimation in pEPHZ-LDLR stable clones
To assess the number of copies of pEPHZ-LDLR, total genomic DNA was prepared from pEPHZ-LDLR clones 1 and 2, continuously grown in the presence of G418 selection. Southern blotting hybridization with a radiolabelled Kanr gene probe revealed the expected 8.5 kb band in the pEPHZ controls and in the pEPHZ-LDLR clones 1 and 2 (Figure 3B). Band intensity was quantified using two independent systems (Figure 3C) and pEPHZ-LDLR was estimated to be present in clones 1 and 2 at ∼1 and ∼4 copies/cell, respectively.
Gene expression from LDLR genomic locus
To assess gene expression from the genomic insert carried by pEPHZ-LDLR, episomal CHO ldlr−/− a7 clones 1 and 2 were analysed for the presence of specific LDLR activity. Fluorescently labelled low density lipoproteins (DiI-LDL), routinely used for qualitative microscopy studies on LDLR, can also be used to quantify LDLR expression when the samples are processed and analysed on a spectrofluorometer (23).
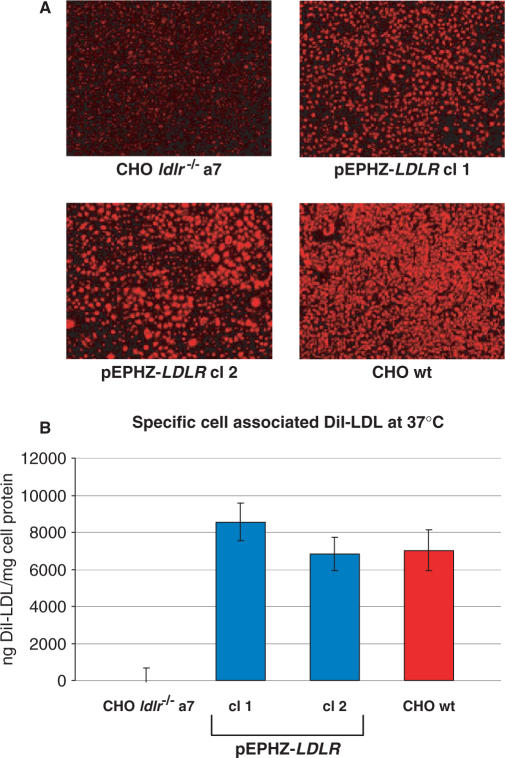
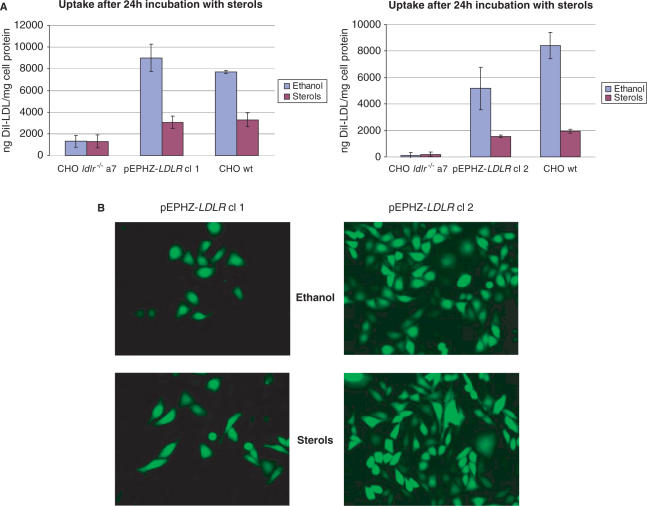
The two pEPHZ-LDLR clones were incubated in lipoprotein-deficient serum (LPDS) for 48 h to enhance LDLR expression. After incubation with LPDS, cell monolayers were incubated with DiI-LDL at a concentration of 10 μg/ml for 5 h after which time cells were washed and pictures were taken using an inverted fluorescence microscope. From these pictures LDLR expression similar to the wild-type cell line can be observed for both clones (Figure 4A). The same samples were then lysed and analysed on a spectrofluorometer using excitation and emission wavelengths of 520 and 580 nm, respectively. The data described in Figure 4B confirm the complete restoration of LDLR expression in pEPHZ-LDLR clones 1 and 2 and correction of the CHO ldlr−/− a7 deficiency phenotype.
Figure 4.
Functional complementation of the low density lipoprotein receptor deficiency in CHO ldlr−/− a7 pEPHZ-LDLR clones, cl 1 and cl 2. (A) Qualitative photomicrograph analysis of cell-associated DiI-LDL in CHO ldlr−/− a7, CHO ldlr−/− a7 pEPHZ-LDLR cl 1, CHO ldlr−/− a7 pEPHZ-LDLR cl 2 and CHO wt cells. (B) Quantitative fluorometric analysis of cell-associated DiI-LDL in CHO ldlr−/− a7, CHO ldlr−/− a7 pEPHZ-LDLR cl 1, CHO ldlr−/− a7 pEPHZ-LDLR cl 2 and CHO wt cells.
Inhibition of LDLR expression levels by elevated sterol levels
Expression from the endogenous LDLR locus is regulated to control the intracellular cholesterol concentration through a negative feedback mechanism. The pEPHZ-LDLR genomic DNA transgene contains the sterol response elements (SREs), necessary for transcriptional regulation in response to sterols. To investigate regulation of expression from the 135-kb genomic DNA insert delivered by pEPHZ-LDLR, CHO ldlr−/− a7 pEPHZ-LDLR stable clones were first incubated in LPDS for 24 h, after which time cholesterol and 25-hydroxycholesterol at concentrations of 12 and 0.6 μg/ml, respectively, were added. After a further 24 h in the presence of cholesterol and 25-hydroxycholesterol, cell monolayers were washed and the DiI-LDL assay was performed. Average reductions of LDLR expression in the presence of sterols by 66% in pEPHZ-LDLR clone 1, and 70% in clone 2, demonstrate that the classical mechanism of sterol inhibition acting on the endogenous site also applies to the iBAC-S/MAR-LDLR transgene (Figure 5A). Demonstration that only LDLR gene expression is affected by the presence of high sterol levels comes from fluorescence microscopy pictures of the same cell samples. No difference in reporter EGFP expression is found in control samples or samples treated with sterols (Figure 5B).
Figure 5.
Repression of LDLR expression by incubation with high sterol levels. (A) DiI-LDL assay on pEPHZ-LDLR clones, cl 1 and cl 2, in the presence of sterols shows a reduction of expression by 65 and 70%, respectively, when compared to the negative control. (B) Fluorescence microscopy analysis of the reporter gene EGFP shows that reduction of expression in the presence of sterol is LDLR specific as EGFP expression is not affected.
Vector retention in the absence of selection
Episomal vectors are slowly lost in rapidly dividing cells when selective pressure is removed and this occurs with a rate depending on the episomal retention efficiency of the vector. We used two different methods to calculate the efficiency of pEPHZ-LDLR retention. First, pEPHZ-LDLR clones 1 and 2 were grown in the absence of G418 selection and maintained in actively growing state. At each passage, episomal DNA was extracted and plasmid rescue assay was performed. DNA extracted from kanamycin/chloramphenicol-resistant bacterial colonies obtained by plasmid rescue was digested with NotI and compared with the original DNA preparation used for pEPHZ-LDLR packaging. It was possible to rescue the vector until 11 weeks in the absence of selection, demonstrating the very high retention efficiency of this episomal S/MAR vector (data not shown).
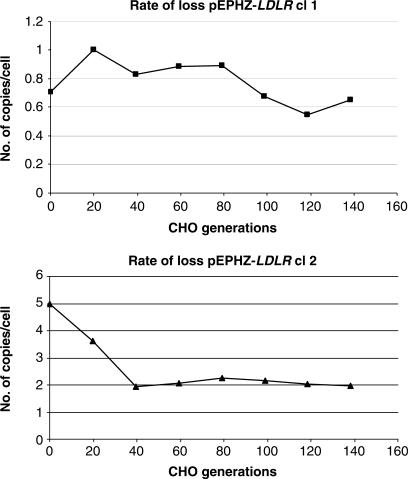
Second, from the same two pEPHZ-LDLR clones grown without G418, genomic DNA was also extracted every two weeks. After 3 months (at the end of the experiment) all samples were analysed by Southern blot hybridization for copy number assessment and band intensity calculated by Phosphorimager quantitative analysis. The data shown in Figure 6 demonstrate a rate of loss of 0.2 and 2.4% per cell generation for pEPHZ-LDLR clone 1 and clone 2, respectively, similar to that reported for pEPI, the basic S/MAR vector (25,26).
Figure 6.
Rate of loss analysis of pEPHZ-LDLR. pEPHZ-LDLR clones, cl 1 and cl 2, grown in the absence of selection, show a rate of loss of 0.2 and 2.4% respectively. These values have been calculated using a first order rate of loss model (λ = (−1/t)(ln Nt/N0) between time points zero and 84 days (118.6 generations) for cl 1 and zero and 28 days (39.5 generations) for cl 2) considering CHO replication time to be 17 h.
DISCUSSION
In this work we have developed an episomal vector for the efficient delivery and maintenance of genomic DNA loci. Many recent studies have demonstrated the advantages in gene expression that come from the delivery of a genomic DNA locus over expression cassettes based on cDNA driven by strong constitutive promoters (4–8). These advantages include physiologically regulated levels of expression, long-term vector retention and transgene expression, and correct alternative splicing and promoter usage (5,7,8,27). The importance of physiological regulation of transgene expression can be exemplified by the LDLR gene which is mutated in familial hypercholesterolemia (FH), a disorder characterized by high plasma levels of low density lipoproteins, which form atherosclerotic plaques leading to premature cardiovascular disease (28,29). Previous gene therapy vectors developed for the treatment of FH have focused on the use of cassettes carrying the LDLR cDNA driven by strong promoters (30). These small constructs were characterized, in vitro and in vivo, by induction of non-physiological overexpression of LDLR, which led to the formation of intracellular deposits of cholesterol followed by death of the transduced cells and, finally, loss of transgene expression (31,32). This does not occur in physiological conditions where levels of expression are controlled by regulatory elements present throughout the LDLR locus.
Long-term gene expression is dependent on efficient vector retention, which can be achieved by using a replicating episomal vector. Such vectors offer several advantages over integrating vectors by maintaining the transgene in an extrachromosomal state (33). The transgene of interest will not be disrupted and subject to regulatory constraints (silencing), a phenomenon referred to as ‘position effect’, which can often occur in the event of integration into cellular DNA. Moreover an episomal transgene will not lead to the interruption of important cellular genes or to cell transformation by insertional mutagenesis, the latter of which can occur if the integration site is near to growth-promoting genes. It has been demonstrated for integrating vectors that these vectors tend to integrate in coding or regulatory regions of expressed genes, making the integration event even more likely to induce oncogenesis (34,35). In 2003, three years into a clinical trial using an MLV vector for the therapy of human severe combined immunodeficiency X-linked (SCID-X1), after an initial success in developing a functional immune system, three of the eleven patients developed leukaemia caused by vector integration and activation of the proto-oncogene LMO2 (36,37). The important side effect observed in this clinical trial has raised important questions of safety, regarding the use of integrating vectors, which can be overcome by the use of episomal systems.
Delivery of vectors containing large fragments of DNA by non-viral means has proven to be inefficient. The obvious alternative to non-viral gene transfer is represented by the use of viral vectors, however the majority of them have a limited capacity. The HSV-1 amplicon system combines high capacity and efficient delivery with ease of construction and offers a capacity of up to ∼150 kb which can allow the delivery of ∼95% of human genomic DNA loci (see http://www.ensembl.org/Homo_sapiens/index.html). The recent development of an improved packaging system provides vector packaging into ‘gutless’ HSV-1 amplicons without helper virus contamination (22).
In this work we describe the construction of a vector that achieves all the above-mentioned features that an efficient vector for gene therapy or expression studies should possess. We developed an iBAC-S/MAR hybrid system for delivery and expression of a genomic DNA locus. In order to assess regulated levels of expression we chose to deliver the LDLR genomic locus since LDLR expression is tightly regulated by intracellular levels of cholesterol through a complex negative feedback mechanism, mediated by sterol response elements present in the promoter region (38,39). Therefore, we constructed the 156-kb vector pEPHZ-LDLR by recombining a modified pEPI vector, pEPHZ, with a BAC carrying the LDLR genomic locus. Infection of CHO ldlr−/− a7 cells with HSV-1 amplicons carrying pEPHZ-LDLR and rescue of episomal DNA after 48 h showed that this vector can be delivered and re-circularizes upon entry into cells with high efficiency, demonstrating the feasibility of this approach. In order to investigate the episomal stability of the iBAC-S/MAR-LDLR system, we infected CHO ldlr−/− a7 cells and isolated stable clonal cell lines carrying pEPHZ-LDLR. The plasmid rescue assay performed on established dividing clones revealed the vector to be present in an episomal state in three out of ten pEPHZ-LDLR clones. Since no episomal vector could be rescued from the remaining seven clones we assume the vector to be present in an integrated form in these clones. We therefore observe a final percentage of 30% (3/10) of episomal clones out of the total number of isolated dividing clones. We note, first, that we did not observe a long-term clone to contain a re-arranged episome suggesting that once established the episome retains structural integrity, and second, that an efficiency of 30% for obtaining episomal retention is similar to that observed for an earlier HSV-1/EBV system when 3/9 episomal colonies of mammalian cells were obtained following infection with a similar large >100 kb episomal construct (7). Plasmid rescue analysis carried out on pEPHZ-LDLR clones proved that this vector can replicate and be retained stably in an episomal state, and no rearrangements were observed. Moreover, Southern blot studies revealed the vector to be present at a low copy number, which allows pEPHZ-LDLR expression properties to be at physiological levels. Studies of vector retention demonstrated a very high retention efficiency for iBAC-S/MAR-LDLR as the vector could be rescued after more than 100 cell generations in the absence of selection. Southern blot analyses confirmed these results by showing a pEPHZ-LDLR mitotic stability of ∼97.6–99.8%, in line with observations made on pEPI in other research studies (25). To assess if pEPHZ-LDLR is able to express functional low density lipoprotein receptors on cell surface, we analysed pEPHZ-LDLR episomal stable clones for their ability to bind and internalize fluorescent-labelled LDLs. Qualitative and quantitative studies demonstrated a complete restoration of LDLR expression, at levels comparable to the wild-type cell line. Moreover incubation of pEPHZ-LDLR clonal cell lines with high sterol levels shows a typical reduction of ∼65–70% of LDLR expression in clones incubated with cholesterol and 25-hydroxycholesterol compared to the same clones in the absence of sterols, exhibiting an essentially identical response to that shown by endogenous LDLR expression from wild-type CHO cells.
The vector system described in this work represents a broadly applicable expression system for human genomic DNA loci. iBAC-S/MAR is a novel episomal vector which is based on human sequences, producing efficient episomal retention in the absence of virally encoded proteins. Moreover, the presence of the whole genomic DNA locus allows regulated levels of transgene expression. In this study, we have shown the iBAC-S/MAR system to be an efficient tool for long-term vector retention and regulated transgene expression, which are major targets of vector development.
ACKNOWLEDGEMENTS
The authors wish to thank Dr Olivia Hibbitt, Dr Sebastian J. Arnold and Dr Richard Harbottle for helpful advice and suggestions. The vector pEPI was kindly provided by H. Lipps, University of Witten/Herdecke, Germany. The work was supported by the Medical Research Council and the Wellcome Trust. R.W.-M. is a Wellcome Trust Research Career Development Fellow. Funding to pay the Open Access publication charges for this article was provided by The Wellcome Trust.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gilbert DM. Replication timing and transcriptional control: beyond cause and effect. Curr. Opin. Cell. Biol. 2002;14:377–383. doi: 10.1016/s0955-0674(02)00326-5. [DOI] [PubMed] [Google Scholar]
- 2.Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- 3.Verschure PJ, van Der Kraan I, Manders EM, van Driel R. Spatial relationship between transcription sites and chromosome territories. J. Cell Biol. 1999;147:13–24. doi: 10.1083/jcb.147.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schiedner G, Morral N, Parks RJ, Wu Y, Koopmans SC, Langston C, Graham FL, Beaudet AL, Kochanek S. Genomic DNA transfer with a high-capacity adenovirus vector results in improved in vivo gene expression and decreased toxicity. Nat. Genet. 1998;18:180–183. doi: 10.1038/ng0298-180. [DOI] [PubMed] [Google Scholar]
- 5.Inoue R, Moghaddam KA, Ranasinghe M, Saeki Y, Chiocca EA, Wade-Martins R. Infectious delivery of the 132 kb CDKN2A/CDKN2B genomic DNA region results in correctly spliced gene expression and growth suppression in glioma cells. Gene Ther. 2004;11:1195–1204. doi: 10.1038/sj.gt.3302284. [DOI] [PubMed] [Google Scholar]
- 6.Wade-Martins R, Saeki Y, Chiocca EA. Infectious delivery of a 135-kb LDLR genomic locus leads to regulated complementation of low-density lipoprotein receptor deficiency in human cells. Mol. Ther. 2003;7:604–612. doi: 10.1016/s1525-0016(03)00060-1. [DOI] [PubMed] [Google Scholar]
- 7.Wade-Martins R, Smith ER, Tyminski E, Chiocca EA, Saeki Y. An infectious transfer and expression system for genomic DNA loci in human and mouse cells. Nat. Biotechnol. 2001;19:1067–1070. doi: 10.1038/nbt1101-1067. [DOI] [PubMed] [Google Scholar]
- 8.Wade-Martins R, White RE, Kimura H, Cook PR, James MR. Stable correction of a genetic deficiency in human cells by an episome carrying a 115 kb genomic transgene. Nat. Biotechnol. 2000;18:1311–1314. doi: 10.1038/82444. [DOI] [PubMed] [Google Scholar]
- 9.Van Craenenbroeck K, Vanhoenacker P, Haegeman G. Episomal vectors for gene expression in mammalian cells. Eur. J. Biochem. 2000;267:5665–5678. doi: 10.1046/j.1432-1327.2000.01645.x. [DOI] [PubMed] [Google Scholar]
- 10.Piechaczek C, Fetzer C, Baiker A, Bode J, Lipps HJ. A vector based on the SV40 origin of replication and chromosomal S/MARs replicates episomally in CHO cells. Nucleic Acids Res. 1999;27:426–428. doi: 10.1093/nar/27.2.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baiker A, Maercker C, Piechaczek C, Schmidt SB, Bode J, Benham C, Lipps HJ. Mitotic stability of an episomal vector containing a human scaffold/matrix-attached region is provided by association with nuclear matrix. Nat. Cell Biol. 2000;2:182–184. doi: 10.1038/35004061. [DOI] [PubMed] [Google Scholar]
- 12.Heng HH, Goetze S, Ye CJ, Liu G, Stevens JB, Bremer SW, Wykes SM, Bode J, Krawetz SA. Chromatin loops are selectively anchored using scaffold/matrix-attachment regions. J. Cell Sci. 2004;117:999–1008. doi: 10.1242/jcs.00976. [DOI] [PubMed] [Google Scholar]
- 13.Alvarez JD, Yasui DH, Niida H, Joh T, Loh DY, Kohwi-Shigematsu T. The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev. 2000;14:521–535. [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J, Bramblett D, Zhu Q, Lozano M, Kobayashi R, Ross SR, Dudley JP. The matrix attachment region-binding protein SATB1 participates in negative regulation of tissue-specific gene expression. Mol. Cell. Biol. 1997;17:5275–5287. doi: 10.1128/mcb.17.9.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brylawski BP, Tsongalis GJ, Cordeiro-Stone M, May WT, Comeau LD, Kaufman DG. Association of putative origins of replication with the nuclear matrix in normal human fibroblasts. Cancer Res. 1993;53:3865–3868. [PubMed] [Google Scholar]
- 16.Dijkwel PA, Hamlin JL. Matrix attachment regions are positioned near replication initiation sites, genes, and an interamplicon junction in the amplified dihydrofolate reductase domain of Chinese hamster ovary cells. Mol. Cell. Biol. 1988;8:5398–5409. doi: 10.1128/mcb.8.12.5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phi-Van L, von Kries JP, Ostertag W, Stratling WH. The chicken lysozyme 5′ matrix attachment region increases transcription from a heterologous promoter in heterologous cells and dampens position effects on the expression of transfected genes. Mol. Cell. Biol. 1990;10:2302–2307. doi: 10.1128/mcb.10.5.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jenke AC, Scinteie MF, Stehle IM, Lipps HJ. Expression of a transgene encoded on a non-viral episomal vector is not subject to epigenetic silencing by cytosine methylation. Mol. Biol. Rep. 2004;31:85–90. doi: 10.1023/b:mole.0000031363.35839.46. [DOI] [PubMed] [Google Scholar]
- 19.Brown MS, Goldstein JL. Regulation of the activity of the low density lipoprotein receptor in human fibroblasts. Cell. 1975;6:307–316. doi: 10.1016/0092-8674(75)90182-8. [DOI] [PubMed] [Google Scholar]
- 20.Russell DW, Yamamoto T, Schneider WJ, Slaughter CJ, Brown MS, Goldstein JL. cDNA cloning of the bovine low density lipoprotein receptor: feedback regulation of a receptor mRNA. Proc. Natl Acad. Sci. USA. 1983;80:7501–7505. doi: 10.1073/pnas.80.24.7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geraghty RJ, Krummenacher C, Cohen GH, Eisenberg RJ, Spear PG. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science. 1998;280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 22.Saeki Y, Fraefel C, Ichikawa T, Breakefield XO, Chiocca EA. Improved helper virus-free packaging system for HSV amplicon vectors using an ICP27-deleted, oversized HSV-1 DNA in a bacterial artificial chromosome. Mol. Ther. 2001;3:591–601. doi: 10.1006/mthe.2001.0294. [DOI] [PubMed] [Google Scholar]
- 23.Teupser D, Thiery J, Walli AK, Seidel D. Determination of LDL- and scavenger-receptor activity in adherent and non-adherent cultured cells with a new single-step fluorometric assay. Biochim. Biophys. Acta. 1996;1303:193–198. doi: 10.1016/0005-2760(96)00094-x. [DOI] [PubMed] [Google Scholar]
- 24.Wade-Martins R, Frampton J, James MR. Long-term stability of large insert genomic DNA episomal shuttle vectors in human cells. Nucleic Acids Res. 1999;27:1674–1682. doi: 10.1093/nar/27.7.1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jenke AC, Stehle IM, Herrmann F, Eisenberger T, Baiker A, Bode J, Fackelmayer FO, Lipps HJ. Nuclear scaffold/matrix attached region modules linked to a transcription unit are sufficient for replication and maintenance of a mammalian episome. Proc. Natl Acad. Sci. USA. 2004;101:11322–11327. doi: 10.1073/pnas.0401355101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaarschmidt D, Baltin J, Stehle IM, Lipps HJ, Knippers R. An episomal mammalian replicon: sequence-independent binding of the origin recognition complex. EMBO J. 2004;23:191–201. doi: 10.1038/sj.emboj.7600029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xing W, Baylink D, Kesavan C, Mohan S. HSV-1 amplicon-mediated transfer of 128-kb BMP-2 genomic locus stimulates osteoblast differentiation in vitro. Biochem. Biophys. Res. Commun. 2004;319:781–786. doi: 10.1016/j.bbrc.2004.05.053. [DOI] [PubMed] [Google Scholar]
- 28.Goldstein JL, Basu SK, Brown MS. Receptor-mediated endocytosis of low-density lipoprotein in cultured cells. Methods Enzymol. 1983;98:241–260. doi: 10.1016/0076-6879(83)98152-1. [DOI] [PubMed] [Google Scholar]
- 29.Goldstein JL, Kita T, Brown MS. Defective lipoprotein receptors and atherosclerosis. Lessons from an animal counterpart of familial hypercholesterolemia. N. Engl. J. Med. 1983;309:288–296. doi: 10.1056/NEJM198308043090507. [DOI] [PubMed] [Google Scholar]
- 30.Wilson JM, Johnston DE, Jefferson DM, Mulligan RC. Correction of the genetic defect in hepatocytes from the Watanabe heritable hyperlipidemic rabbit. Proc. Natl Acad. Sci. USA. 1988;85:4421–4425. doi: 10.1073/pnas.85.12.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cichon G, Willnow T, Herwig S, Uckert W, Loser P, Schmidt HH, Benhidjeb T, Schlag PM, Schnieders F, et al. Non-physiological overexpression of the low density lipoprotein receptor (LDLr) gene in the liver induces pathological intracellular lipid and cholesterol storage. J. Gene Med. 2004;6:166–175. doi: 10.1002/jgm.473. [DOI] [PubMed] [Google Scholar]
- 32.Heeren J, Steinwaerder DS, Schnieders F, Cichon G, Strauss M, Beisiegel U. Nonphysiological overexpression of low-density lipoprotein receptors causes pathological intracellular lipid accumulation and the formation of cholesterol and cholesteryl ester crystals in vitro. J. Mol. Med. 1999;77:735–743. doi: 10.1007/s001099900045. [DOI] [PubMed] [Google Scholar]
- 33.Conese M, Auriche C, Ascenzioni F. Gene therapy progress and prospects: episomally maintained self-replicating systems. Gene Ther. 2004;11:1735–1741. doi: 10.1038/sj.gt.3302362. [DOI] [PubMed] [Google Scholar]
- 34.Bushman F, Lewinski M, Ciuffi A, Barr S, Leipzig J, Hannenhalli S, Hoffmann C. Genome-wide analysis of retroviral DNA integration. Nat. Rev. Microbiol. 2005;3:848–858. doi: 10.1038/nrmicro1263. [DOI] [PubMed] [Google Scholar]
- 35.Schroder ARW, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 36.Hacein-Bey-Abina S, von Kalle C, Schmidt M, Le Deist F, Wulffraat N, McIntyre E, Radford I, Villeval JL, Fraser CC, et al. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N. Engl. J. Med. 2003;348:255–256. doi: 10.1056/NEJM200301163480314. [DOI] [PubMed] [Google Scholar]
- 37.Hacein-Bey-Abina S, von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, Lim A, Osborne CS, Pawliuk R, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 38.Sudhof TC, Russell DW, Brown MS, Goldstein JL. 42 bp element from LDL receptor gene confers end-product repression by sterols when inserted into viral TK promoter. Cell. 1987;48:1061–1069. doi: 10.1016/0092-8674(87)90713-6. [DOI] [PubMed] [Google Scholar]
- 39.Sudhof TC, Van der Westhuyzen DR, Goldstein JL, Brown MS, Russell DW. Three direct repeats and a TATA-like sequence are required for regulated expression of the human low density lipoprotein receptor gene. J. Biol. Chem. 1987;262:10773–10779. [PubMed] [Google Scholar]