Abstract
The impact of 2′-deoxy-2′-fluoroarabinonucleotide residues (2′F-araN) on different G-quadruplexes derived from a thrombin-binding DNA aptamer d(G2T2G2TGTG2T2G2), an anti-HIV phosphorothioate aptamer PS-d(T2G4T2) and a DNA telomeric sequence d(G4T4G4) via UV thermal melting (Tm) and circular dichroism (CD) experiments has been investigated. Generally, replacement of deoxyguanosines that adopt the anti conformation (anti-guanines) with 2′F-araG can stabilize G-quartets and maintain the quadruplex conformation, while replacement of syn-guanines with 2′F-araG is not favored and results in a dramatic switch to an alternative quadruplex conformation. It was found that incorporation of 2′F-araG or T residues into a thrombin-binding DNA G-quadruplex stabilizes the complex (ΔTm up to ∼+3°C/2′F-araN modification); 2′F-araN units also increased the half-life in 10% fetal bovine serum (FBS) up to 48-fold. Two modified thrombin-binding aptamers (PG13 and PG14) show an approximately 4-fold increase in binding affinity to thrombin, as assessed via a nitrocellulose filter binding assay, both with increased thermal stability (∼1°C/2′F-ANA modification increase in Tm) and nuclease resistance (4–7-fold) as well. Therefore, the 2′-deoxy-2′-fluoro-d-arabinonucleic acid (2′F-ANA) modification is well suited to tune (and improve) the physicochemical and biological properties of naturally occurring DNA G-quartets.
INTRODUCTION
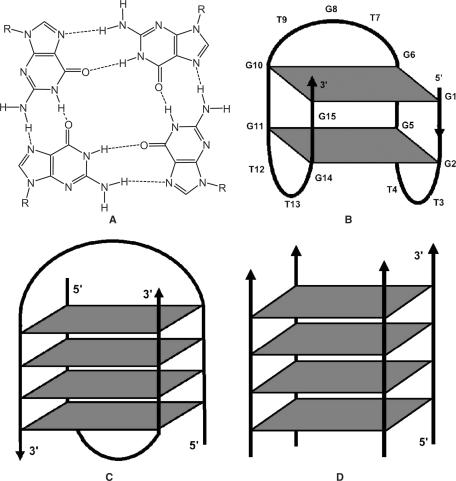
Guanine quartet (G-quartet) structures with four Hoogsteen-paired, coplanar guanines were first observed more than 40 years ago (1) and later demonstrated to be a unique structural motif of guanine-rich oligonucleotides (2) (Figure 1A). G-quartets are found in nature and also in sequences identified by screening techniques such as Systematic Evolution of Ligands by EXponential enrichment (SELEX) (3–5). They have also aroused interest as therapeutic agents to inhibit human thrombin (6,7), HIV infection (7–9), and as targets themselves to inhibit telomerase activity in anticancer drug design (10).
Figure 1.
Structure of (A) G-quartet with cyclic array of four guanines linked by Hoogsteen H-bonds (1); (B) thrombin-binding DNA d(G2T2G2TGTG2T2G2) with an edge-loop (chair-like) unimolecular G-quadruplex in K+ (15,16); (C) a telomeric DNA d(G4T4G4) with diagonal-loop dimeric hairpin complex [d(G4T4G4)]2 in Na+ (21,22); (D) a tetrameric G-quadruplex [d(PS-T2G4T2)]4 with four parallel strands (2,7).
G-quartets display extraordinary structural polymorphism (11–13). The 15-nt DNA sequence d(G2T2G2TGTG2T2G2) (6) binds and inactivate thrombin, a key enzyme in the blood clotting cascade. NMR spectroscopy reveals that it adopts a uniquely folded structure with two stacked G-quartets connected through edge-loops (one TGT and two TT loops) involving antiparallel alignment of adjacent strands (Figure 1B) (14–17). A seemly different crystal structure of this aptamer–thrombin complex (18,19) was later analyzed to be in agreement with the previous NMR structure (19,20). Potassium ions stabilize the G-quadruplex by coordination to the G residues (15). The G-quartet displays an alternating syn and anti conformation of guanine bases (syn- and anti-Gs) on the same plane and 5′-syn-G/3′-anti-G along each strand of the quadruplex (i.e. 5′-syn-anti-3′ connectivity where syn-G includes G1, G5, G10 and G14 and anti-G includes G2, G6, G11 and G15 in Figure 1B) and all the thymidines are in the anti orientation (15).
The telomeric sequence d(G4T4G4) adopts symmetrical dimeric quadruplexes comprising four G-quartets linked through a diagonal loop as analyzed by NMR in sodium (Na+) environment (Figure 1C) (21,22) or an edge-loop structure as revealed by X-crystallography in potassium (K+) environment (23). More recent studies re-examined the crystal structure of this sequence and suggested the crystal structure also adopts a diagonal conformation, consistent with the NMR solution structure (24). The diagonal loop configuration (Figure 1C) consists of 5′-syn-G/3′-anti-G along each G-strand, syn-syn-anti-anti conformations in a G-quartet and all sugar residues puckered in the south (C2′-endo) conformation (22,25).
Another interesting example is the phosphorothioate oligonucleotide PS-d(T2G4T2) identified as an inhibitor of HIV-1 infection in vitro by combinatorial screening of a library of DNA strands. This aptamer inhibits HIV envelope-mediated cell fusion in vitro and its structure consists of a parallel-stranded tetramer with all guanines in the anti-conformation (Figure 1D) (2,7).
Based on the above, the glycosidic conformations of guanines (i.e. syn- and anti-Gs) have a strong correlation with strand alignments. Parallel-stranded quadruplexes (Figure 1D) support only anti-G residues, while antiparallel-stranded quadruplexes favor alternating syn-anti Gs engaged in unimolecular (Figure 1B) or intermolecular complexes (Figure 1C) (11,26).
G-quadruplexes are sensitive to chemical modifications. Several studies aimed at modifying the thrombin-binding aptamer d(G2T2G2TGTG2T2G2) have been reported (27–31), but very few, if any, have led to an improvement over the original molecule. For example, Seela and coworkers recently reported the insertion of a hairpin-forming sequence GCGAAG into the position of the central loop (TGT) of the thrombin-binding aptamer. This construct formed a G-quadruplex fused to a mini-hairpin structure. According to the Tm data, the mini-hairpin induces a structural change in the aptamer section, leading to less stable G-quadruplex. Binding to thrombin was not investigated (30). Saccà et al. studied the effect of backbone charge and atom size, base substitutions as well as the effect of modification at the sugar 2′-position as analyzed by spectroscopy. Fully modified aptamers with sugar modifications (ribose, 2′-O-methylribose) and phosphate backbone modifications (methylphosphonate, phosphorothioate) led to a reduction in the thermal stability (31). In fact, the 2′-O-methylribose modification led not only to a destabilization of the structure but to a complete transformation of the G-tetrad conformation, as shown by spectroscopy in potassium buffer (31). 2′-O-methylribose was also shown to causes structural changes in RNA aptamers and often resulted in a loss of activity (32). Accordingly, there is a need for new chemical modifications to improve the nuclease stability of this and other aptamers. Ideally, these modifications will not alter the subtle binding interactions of the selected native aptamers and the thermal stability of G-quadruplexes.
2′-Deoxy-2′-fluoro-d-arabinonucleic acids (2′F-ANA) confer DNA-like (South/East) conformations (33) to oligonucleotides while rendering them more nuclease resistant (34). The incorporation of 2′F-araN units in oligonucleotides also raises the Tm of different systems, i.e. duplexes (∼+1°C/nt) (35), triplexes (∼+0.8°C/nt) (36) and C-rich quadruplexes (∼+1°C/nt, pH <4.0) (37). In light of this and other advantageous characteristics of 2F-ANA, such as synthetic accessibility through conventional solid-phase phosphoramidite chemistry, and promising antisense/siRNA properties (34,38–40), we have undertaken the first study concerning the ability of 2′F-ANA to form various G-quadruplex structures as analyzed by Tm and CD experiments. 2′F-ANA modified thrombin-binding aptamers were further evaluated by their nuclease resistance and binding affinity to thrombin. The outcome of these studies has opened new perspectives for the application of 2′F-ANA as aptamer oligonucleotides.
MATERIALS AND METHODS
Chemical synthesis of oligonucleotides
The sequence and composition of the oligomers prepared in this study are shown in Tables 1 and 2. Arabinose modified aptamer syntheses were carried out at a 1 μmol scale on an Applied Biosystems (ABI) 3400A synthesizer using standard β-cyanoethylphosphoramidite chemistry according to published protocols (41). Deoxyribonucleoside phosphoramidites were purchased from ChemGenes (Waltham, MA) and 2′F-arabinonucleoside 3′-O-phosphoramidites were provided by Topigen Pharmaceuticals Inc. (Montreal, Canada). The final concentrations of the monomers were 0.10 M for 2′-deoxyribonucleoside phosphoramidites and 0.125 M for the 2′F-arabinose phosphoramidites. The coupling time was extended to 150 s for the 2′-deoxyribonucleoside phosphoramidites dC and dG, and 15 min for the 2′F-araG and T phosphoramidites. These conditions gave about 99% average stepwise coupling yields. With the exception of PG17 through PG24, oligonucleotides were purified by anion-exchange HPLC (Waters Protein Pak DEAE-5PW column; 7.5 mm × 7.5 cm), desalted by size-exclusion chromatography on Sephadex G-25 resin, and characterized by MALDI-TOF mass spectrometry (Kratos Kompact-III Instrument; Kratos Analytical Inc., New York). Purity of the isolated oligonucleotides was >95% by HPLC. PG17–24 were used as obtained following deprotection and desalting.
Table 1.
Sequences, CD, Tm and binding data of 2′F-ANA modified thrombin-binding aptamers
| Code | Type | Sequencea | Tm (ΔTm)b (°C) | CD typec | Hysterisis in Tmd | Kd (nM)e | t1/2 (h)f |
|---|---|---|---|---|---|---|---|
| PG1 | All DNA | d(GGTTGGTGTGGTTGG) | 46.4 (42) | II | no | 210 | 0.5 |
| 47.4 | 200(6) | ||||||
| PG2 | All 2′F-ANA | d(GGTTGGTGTGGTTGG) | 54.1 (+0.4) | I | yes | 500 | >24 |
| PG3 | 2′F-ANA G-anti | d(GGTTGGTGTGGTTGG) | 53.3 (+1.5) | II | no | >700 | 4.8 |
| PG4 | 2′F-ANA G-anti& loop | d(GGTTGGTGTGGTTGG) | 61.6 (+1.3) | II | no | 500 | 9.4 |
| PG5 | 2′F-ANA G-syn | d(GGTTGGTGTGGTTGG) | 45.4 (– 0.5) | I | yes | >700 | 0.8 |
| PG6 | 2′F-ANA G-syn&loop | d(GGTTGGTGTGGTTGG) | 48.5 (+0.1) | I | yes | >700 | 0.6 |
| PG7 | 2′F-ANA G-quartet | d(GGTTGGTGTGGTTGG) | 50.2 (+0.4) | I | yes | 450 | 4.0 |
| PG8 | 2′F-ANA all-loop | d(GGTTGGTGTGGTTGG) | 56.3 (+1.3) | II | no | 280 | 5.9 |
| PG9 | 2′F-ANA loop | d(GGTTGGTGTGGTTGG) | 57.1 (+1.6) | II | no | 300 | 2.8 |
| PG10 | 2′F-ANA loop | d(GGTTGGTGTGGTTGG) | 51.2 (+0.8) | II | no | 250 | 2.7 |
| PG11 | 2′F-ANA loop | d(GGTTGGTGTGGTTGG) | 56.6 (+1.8) | II | no | 370 | 5.1 |
| PG12 | 2′F-ANA loop | d(GGTTGGTGTGGTTGG) | 59.1 (+2.9) | II | no | 310 | 3.5 |
| PG13 | 2′F-ANA loop | d(GGTTGGTGTGGTTGG) | 51.0 (+0.9) | II | no | 58 | 3.4 |
| PG14 | 2′F-ANA loop | d(GGTTGGTGTGGTTGG) | 50.6 (+0.8) | II | no | 40 | 2.0 |
| P8 | ssDNA control | d(GTCTCTTGTGTGACTCTGGTAAC) | NA | NA | NA | NC | 0.5 |
| H1 | Hairpin control (RNA) | r(GGACUUCGGUCC) | NA | NA | NA | NC | NA |
aCapital and bold letter: 2′F-ANA.
bΔTm is the Tm change per each 2′F-ANA residue between any modified aptamer with the unmodified DNA aptamer (PG1, Tm = 47.4°C); NA: not applicable.
c‘Type I’ CD spectrum refers to a positive CD band at ∼265 nm and a negative band at ∼240 nm that correlates with G-anti conformation in the G-quartet. ‘Type II’ CD refers to a CD spectrum with positive band at ∼295 nm and a negative band at ∼260 nm, which indicates a mixed anti-G and syn-G conformation in the G-quartet. CD was measured in the buffer of 10 mM Tris, pH 6.8, 25 mM KCl (12).
dHysterisis in Tm refers to the hysterisis existing between a heating and cooling process with 0.5°C/min temperature change during Tm measurements.
eKd was roughly estimated from the concentration (nM) where 50% of the maximum binding percentage was observed with a certain aptamer during the thrombin concentrations studied; NC: not calculated.
fHalf-life in 10% fetal bovine serum (FBS) as monitored by 20% polyacrylamide gel electrophoresis.
Table 2.
CD and Tm data of d(T2G4T2) and d(G4T4G4) and related oligonucleotides
| Code | Type | Sequencea | CD Typeb,c | Tmd (ΔTm)e (°C) |
|---|---|---|---|---|
| d(T2G4T2) and related sequences | ||||
| PG17 | All PO-DNA | PO-d(TTGGGGTT) | I | 66.0 |
| PG18 | All PS-DNA | PS-d(TTGGGGTT) | I | 73.5(43) |
| 74.0 | ||||
| PG19 | All PS-2′F-ANA | PS-d(TTGGGGTT) | I | 83.0 (+1.1) |
| PG20 | PS-2′F-ANA-G | PS-d(TTGGGGTT) | I | 87.0 (+3.3) |
| d(G4T4G4) and related sequences | ||||
| PG21 | All PO-DNA | PO-d(GGGGTTTTGGGG) | II | 65(44) |
| 64.4 | ||||
| PG22 | All PO-2′F-ANA | PO-d(GGGGTTTTGGGG) | I | 90.0 (+2.1) |
| PG23 | G-syn | PO-d(GGGGTTTTGGGG) | I | 72.5 (+2.0) |
| PG24 | G-anti | PO-d(GGGGTTTTGGGG) | II | 66.2 (+0.5) |
aCapital and bold letter: 2′F-ANA; PO: phosphate linkage; PS: phosphorothioate linkage.
bCD type I & II refer to the note in Table 1.
cdT2G4T2 and related sequences (PG17–20): phosphate-buffered saline (PBS buffer, pH 7.2 at 25°C), 137 mM NaCl, 2.7 mM KCl, 1.5 mM KH2PO4, 8 mM Na2HPO4; strand concentration: 20 µM for both CD and Tm experiments. dG4T4G4 and related sequences (PG21–24): 10 mM sodium phosphate buffer, 0.1 mM EDTA, pH 7 and 200 mM NaCl; strand concentration: 10 µM.
ddT2G4T2 and related sequences (PG17–20): phosphate-buffered saline (PBS buffer, pH 7.2 at 25°C), 137 mM NaCl, 2.7 mM KCl, 1.5 mM KH2PO4, 8 mM Na2HPO4; strand concentration: 20 µM; Tm data were generated from concentration-dependent CD spectra (Figure S5 and ‘Materials and methods’ section). dG4T4G4 and related sequences (PG21–24): 10 mM sodium phosphate buffer, 0.1 mM EDTA, pH 7 and 200 mM NaCl; strand concentration: 100 µM; Tm measurements were conducted at 295 nm wavelength.
eΔTm (°C) is the Tm change/2′F-ANA modification of PG18–20 or PG22–24 relative to the control PG18 (74.0°C) or PG21 (PG64.4°C), respectively.
UV thermal melting studies (Tm)
UV thermal dissociation data was obtained on a Varian CARY 1 spectrophotometer equipped with a Peltier temperature controller. Thrombin-binding aptamers (PG1–14) were dissolved in Tm buffer (10 mM Tris, pH 6.8, with and without 25 mM KCl) at a final concentration of 8 μM (42). Thrombin-binding aptamers were annealed in Tm buffer at 80°C for 10 min, allowed to cool to room temperature and refrigerated (4°C) overnight before measurements. dT2G4T2 and related sequences (PG17–20) were dissolved in phosphate-buffered saline (PBS buffer, pH 7.2) composed of 137 mM NaCl, 2.7 mM KCl, 1.5 mM KH2PO4, 8 mM Na2HPO4 at a final concentration of 20 µM (43). The telomeric DNA dG4T4G4 and related sequences (PG21–24) were dissolved in 10 mM sodium phosphate buffer (pH 7, 0.1 mM EDTA and 200 mM NaCl) at a final concentration of 100 µM (44). All samples (PG17–24) were annealed at 98°C for 5 min, naturally cooled down to room temperature and refrigerated (4°C) overnight before measurements. The annealed samples were transferred to pre-chilled Hellma QS-1.000 (Cat #114) quartz cell, sealed with a Teflon-wrapped stopper and degassed by placing them in an ultrasonic bath for 1 min. Extinction coefficients were obtained from the following internet site (http://www.idtdna.com/analyzer/applications/oligoanalyzer) based on the nearest-neighbor approach (45) and modified aptamers (phosphorothioates and 2′F-ANA) were assumed to have the same extinction coefficient as the natural DNA aptamer. Denaturation/cooling curves were acquired at either at 295 nm for d(G2T2G2TGTG2T2G2) and related sequences (PG1–14), dG4T4G4 and related sequences (PG21–24), or at 260 nm for dT2G4T2 and related sequences (PG17–20), at a heating/cooling rate of 0.5°C/min between 10 and 80°C (for PG1–14), 20 and 90°C (for PG17–20) or 40 and 98°C (for PG21–24). The data were analyzed with the software provided by Varian and converted to Microsoft Excel (Tables 1 and 2). The decreases in UV absorbance (hypochromicity) with increasing temperature were normalized between 1 and 0 by the formula: N = (At − Al)/(Ah − Al), where At is the absorbance at any given temperature (t), Al is the minimum absorbance reading at high temperature and Ah is the maximum absorbance reading at low temperature. Tm concentration dependence studies were also conducted in the same way at 295 nm using thrombin-binding aptamers (PG1–14) with different concentrations ranging from 4 to 76 μM. Starna quartz cells (Starna Cells, Inc., Cat. # 1-Q-1) with 1-mm path length were used at high concentrations to reduce the amount of aptamers required and to avoid exceeding the Absorbance range of the instrument.
Circular dichroism (CD) spectra
CD spectra (200–320 nm) were collected on a Jasco J-710 spectropolarimeter at a rate of 100 nm/min using fused quartz cells (Hellma, 165-QS). Measurements were carried out either in 10 mM Tris, pH 6.8 (with and without 25 mM KCl) at a concentration of 8 μM for thrombin-binding aptamers (PG1–14) (42); in PBS buffer (pH 7.2, 137 mM NaCl, 2.7 mM KCl, 1.5 mM KH2PO4, 8 mM Na2HPO4) for dT2G4T2 and related sequences (PG17–20) at a final concentration of 20 µM (43), or in sodium phosphate buffer (10 mM sodium phosphate buffer, pH 7, 0.1 mM EDTA and 200 mM NaCl) for dG4T4G4 and related sequences (PG21–24) at a final concentration of 100 µM (44). Temperature was controlled by an internal circulating bath (VWR Scientific) at constant temperature. The data was processed using J-700 Windows software supplied by the manufacturer (JASCO, Inc.). To facilitate comparisons, the CD spectra were background subtracted, smoothed and corrected for concentration so that molar ellipticities could be obtained. Temperature-dependent CD spectra were also conducted for dT2G4T2 and related sequences (PG17–20). A 10-min equilibration time was allowed at each temperature before CD scanning. The Tm profile was obtained by plotting the maximum molar ellipticities versus temperature and normalizing.
Nuclease stability assay
Nuclease stability of anti-thrombin aptamers was conducted in 10% fetal bovine serum (FBS, Wisent Inc., Cat. #080150) diluted with multicell Dulbecco's Modified Eagle's Medium (DMEM, Wisent Inc., Cat. #319005-CL) at 37°C. A single-strand DNA (ssDNA) 23mer (P-8), which is unable to form G-quadruplexes, was used as a control. Approximately 8 μmol of stock solution of aptamers and ssDNA control (∼1.2 O.D.U) was evaporated to dryness under reduced pressure and then incubated with 300 μl 10% FBS at 37°C. At 0, 0.25, 0.5, 1, 2, 6 and 24 h, 50 μl of samples were collected and stored at –20°C for at least 20 min. The samples were evaporated to dryness and then 10 μl of gel loading buffer and 10 μl of autoclaved water was added. 10 μl of the mixture was used for polyacrylamide gel electrophoresis (PAGE), which was carried out at room temperature using 20% polyacrylamide gel in 0.5× TBE buffer (Tris-borate-EDTA). The degradation patterns on the gels were visualized using Stains-All (Bio-Rad) according to the manufacturer's protocol.
5′-End labeling of synthetic oligonucleotides
Aptamers (PG1–14) were radiolabeled at the 5′-hydroxyl terminus with a radioactive 32P probe using a T4 polynucleotide kinase (T4 PNK) according to the manufacture's specifications (MBI Fermentas Life Sciences, Burlington, ON). Incorporation of the 32P label was accomplished in reaction mixtures consisting of DNA aptamers substrate (100 pmol), 2 μl 10 × reaction buffer (500 mM Tris–HCl, pH 7.6 at 25°C, 100 mM MgCl2, 50 mM DTT, 1 mM spermidine and 1 mM EDTA), 1 μl T4 PNK enzyme solution (10 U/1μl in a solution of 20 mM Tris–HCl, pH 7.5, 25 mM KCl, 0.1 mM EDTA, 2 mM DTT and 50% glycerol), 6 μl [γ-32P]-ATP solution (6000 Ci/mmol, 10 mCi/ml; Amersham Biosciences, Inc.) and autoclaved sterile water to a final volume of 20 μl. The reaction mixture was incubated for 30–45 min at 37°C, followed by a second incubation for 5 min at 95°C to thermally denature and deactivate the kinase enzyme. The solution was purified according to a standard protocol (46) and the isolated yield of 32P-5′-DNA following gel extraction averaged 50%. The pure labeled samples were kept at –20°C for future use.
Nitrocellulose filter binding assay
Labeled aptamers (1.25 pmol) were heated to 95°C for 5 min in the binding buffer (Tris–Ac, pH 7.4, 140 mM NaCl, 5 mM KCl, 1 mM CaCl2, 1 mM MgCl2) (6) and immediately placed on ice for 5 min before binding to increasing concentrations of thrombin protease (Amersham Biosciences, Inc.) ranging from 10 to 1000 nM in the binding buffer at 37°C in a final volume of 20 μl for 30 min. Mixtures were filtered through a nitrocellulose filter (13 mm Millipore, HAWP, 0.45 μm) pre-wetted with binding buffer in a Millipore filter binding apparatus, and immediately rinsed with 600 μl ice-cold washing buffer [Tris–Ac, pH 7.4, 140 mM NaCl, 5 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 1% sodium pyrophosphate (w/v)], then the filter was air-dried and the bound aptamer quantified by scintillation counting. The binding percentage (%) was calculated by the subtraction of the background from the counts in the microtube. Kd was roughly estimated from the concentration (nM) where 50% of the maximum binding percentage was observed with a certain aptamer during the thrombin concentrations studied.
RESULTS AND DISCUSSION
Oligonucleotide aptamers derived from SELEX have drawn great attention for their potential as therapeutic and diagnostic agents (47–50). The application of nucleic acid aptamers in vivo and their possible use in pharmacotherapy face the same key hurdles as siRNA and antisense based therapeutics, e.g. delivery, cellular uptake and biostability. To date, several methods have been devised to improve the stability of aptamers, most of which make use of SELEX, including a mirror-design of RNA aptamers (or ‘Spiegelmers’) (51); post-SELEX modification by 2′-OMe to increase the stabilization of RNA aptamers (32); and direct evolution in SELEX using modified dNTPs or rNTPs (e.g. 2′-F, 2′-NH2 pyrimidine 5′-triphosphate) (52–56). Macugen®, an aptamer recently approved by the FDA for the treatment of neovascular age-related macular degeneration, is a 2′F/2′-OMe ribose-modified oligomer (57). There still exists a great demand for new methods and more diverse range of chemistries to create aptamers with more favorable pharmacokinetic properties.
A fully 2′F-ANA modified thrombin-binding aptamer
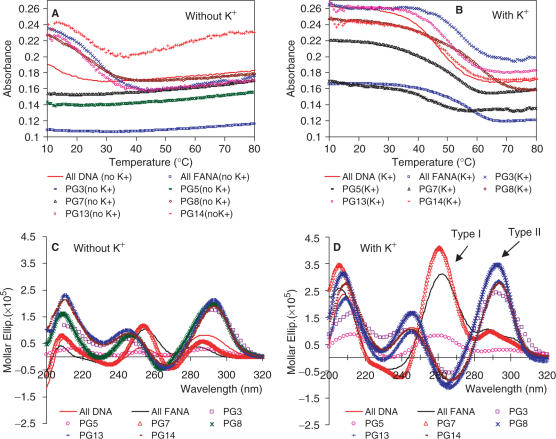
The study began with the replacement of all the deoxynucleotides in the thrombin-binding aptamer d(G2T2G2TGTG2T2G2) by 2′F-ANA units. Consistent with literature results (15,42), it was found through comparisons of the Tm profiles both with and without the presence of K+, that potassium ions stabilized the unimolecular G-quadruplex of the DNA aptamer (PG1) (Figure 2A and B). As shown in the literature, similar hypochromicity (UV absorbance decrease with increased temperature) was observed at 295 nm for all DNA sequence (42), which is an indication of G-quartet formation (12,58). The Tm of the all DNA aptamer obtained in this study was 47.4°C (Table 1 and Figure 2B), consistent with 46.4°C reported by Smirnov and Shafe (42) and the CD spectra also support the UV melting experiment. Previous studies have shown two typical CD spectra: a CD spectrum with a positive CD band at ∼265 nm and a negative band at ∼240 nm (Type I CD spectrum) is related to a parallel quadruplex with all anti-Gs, while one with a positive band at ∼295 nm and a negative band at ∼260 nm (Type II CD spectrum) corresponds to antiparallel quadruplex with alternating syn-anti Gs (12). The all DNA aptamer shows very low amplitude bands without K+, whereas it demonstrates characteristic Type II CD when K+ is present, indicating a G-quadruplex structure with alternating syn-anti Gs (Figure 2C and D) (11,12). The lack of concentration dependence in the Tm data (Figure 3) and the lack of hysteresis in the heating/cooling processes (Figure 4) support a unimolecular G-quadruplex structure (Figure 1B). Lack of hysteresis at a heating/cooling rate of 0.5°C/min indicates a fast kinetics for the formation of unimolecular quadruplexes (58).
Figure 2.
Tm profiles of selected 2′F-ANA-modified thrombin-binding aptamers (Table 1) measured at 295 nm in buffer 10 mM Tris, pH 6.8 (A) without KCl; (B) with 25 mM KCl, at a final strand concentration of 8 μM. The Tm data are provided in Table 1. CD spectra in the same buffer consisting of 10 mM Tris, pH 6.8 (C) without KCl; (D) with 25 mM KCl at 15°C at a final strand concentration of 8 μM. Type I and Type II CD types are shown in (D).
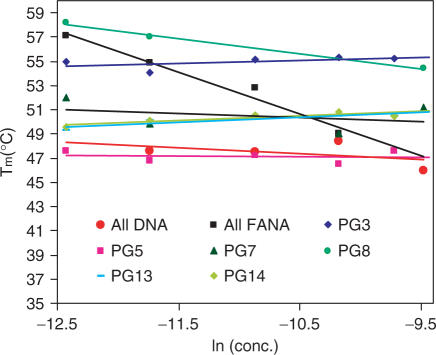
Figure 3.
Tm versus concentration dependence (15–20-folds) study measured at 295 nm in a buffer consisting of 10 mM Tris, pH 6.8, 25 mM KCl for selected 2′F-ANA modified thrombin-binding aptamers (Table 1).
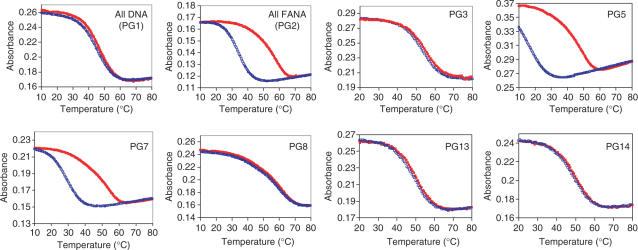
Figure 4.
Heating and cooling Tm transitions of 2′F-ANA modified thrombin-binding aptamers (Table 1) measured at 295 nm in a buffer of 10 mM Tris, 25 mM KCl, pH 6.8, at a final strand concentration of 8 μM in a heating/cooling rate of 0.5°C/min (red filled square: heating; blue empty square: cooling).
The all 2′F-ANA aptamer (PG2) also forms a defined G-quadruplex stabilized by K+ in the UV melting experiment (Figure 2A and B). The G-quadruplex of all 2′F-ANA aptamer (PG2) is more thermally stable than the all DNA aptamer (PG1) (ΔTm = 0.4°C/2′F-ANA modification in Table 1). Further characterization experiments indicated that the G-quadruplex PG2 formed in K+ is different from that of that of PG1. The effect of K+ on the CD spectra is consistent with the Tm experiment: very low absorbance in the absence of K+ and a strong positive absorbance peak at 260 nm in the presence of K+. Clearly this CD spectrum is Type I, which corresponds to a parallel quadruplex with all anti-Gs (11,12). A concentration-dependent Tm profile indicates the existence of intermolecular G-quadruplex formation (Figure 3). Even though we could not tell the existence of a dimeric or tetrameric G-quadruplex with all parallel strand alignments, it is clearly shown that all 2′F-ANA aptamer (PG2) could not maintain the unimolecular G-quadruplex topology (Figure 1B). An intermolecular G-quadruplex is thus formed to support a G-quartet structure with all anti-Gs. Hysteresis was observed in the heating and cooling processes, again supporting an intermolecular G-quadruplex adopted by PG2 (Figure 4).
Replacement of anti-Gs and syn-Gs with 2′F-ANA
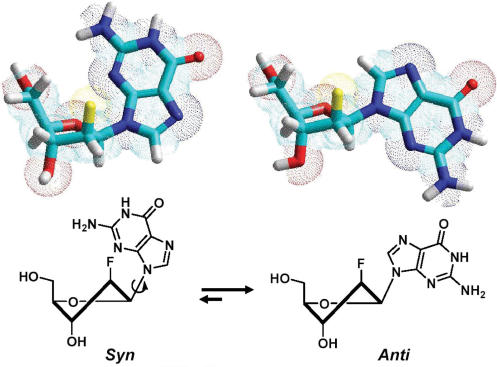
Nucleotides with preferred glycosidic conformations have been utilized to investigate their effect on the topology and molecularity of G-tetrads. For example, 8-methyldeoxyguanosine (8-Me-dG) and 8-bromodeoxyguanosine (8-Br-dG) which prefer syn conformation have demonstrated to play an important role in G-quadruplex conformation and thermal stability when they are used to modify a tetrameric G-quadruplex [d(TGGGT)]4 (59,60). Recent studies by Tang and Shafer (61) demonstrate that ribonucleic acids which favor an anti conformation provide enough driving force to change the topology and molecularity for the thrombin-binding aptamer d(G2T2G2TGTG2T2G2). A single locked nucleic acid (LNA) modification in an expected antiparallel folded quadruplex d(G4(T4G4)3) has also been shown to induce a significant topological change to a parallel quadruplex in the presence of K+ (62). 2′F-ANA unit which has been shown to have a conformationally biased sugar pucker (33) (i.e. more south/east than its DNA counterpart), is expected to favor an anti conformation in order to avoid steric interaction between guanine and the β-face fluorine (63) (Figure 5). Therefore, the next phase of the investigation was to study the effect of 2′F-araG on anti-Gs and syn-Gs observed in the different G-quadruplexes discussed earlier (see ‘Introduction’ section). We expected that the replacement of anti-Gs with 2′F-ANA would lead to thermal stabilization since preorganization of the anti glycosidic conformation favored by 2′F-araG (63) would reduce the loss of entropy during G-quartet assembly. Conversely, replacement of syn-Gs with 2′F-araGs is expected to cause destabilization or complete disruption of the G-quartet structure altogether.
Figure 5.
Glycosic conformation equilibrium of dG and 2′F-araG with south furanose sugar ring.
The obtained Tm and CD data are consistent with the expected results. Modifying the anti-Gs with 2′F-ANA (PG3) or modifying both the anti-Gs and the loops with 2′F-ANA (PG4) increases thermal stability in the presence of K+ (ΔTm = +1.3–1.5°C/2′F-ANA modification). The CD spectrum of PG3 was Type I CD, similar to the all DNA aptamer (PG1) (Figure 2C and D). Remarkably, PG4 shows a clear Type I CD both without and with K+, demonstrating that the G-quadruplex adopted by PG4 could form even in the absence of K+. In addition, it suggests that replacement of the loop with 2′F-ANA further benefits the G-quadruplex formation. This is consistent with previous work showing that all the thymidines in this thrombin-binding DNA aptamer adopt the anti orientation (15). A concentration-independent Tm profile (Figure 3) and lack of hysteresis in heating/cooling processes both support a unimolecular G-quadruplex structure like the DNA control (Figure 4 and Figure S1 in Supplementary Data). Replacement of anti-G with 2′F-ANA is therefore able to stabilize the G-quartet without changing the quadruplex topology.
The results of replacement of the syn-Gs with 2′F-ANA also match expectations. Modification of the syn-Gs with 2′F-ANA (PG5) and replacement of both syn-Gs and loops (PG6) with 2′F-ANA result in less stable complexes compared with PG3 and PG4. PG5 is even less stable than the original all DNA aptamer (ΔTm = –0.5°C/2′F-ANA modification, Table 1), and PG6 is only as stable as the all DNA aptamer control (ΔTm = +0.1°C/2′F-ANA modification, Table 1). CD experiments show that both PG5 and PG6 display very low amplitude peaks at 260 nm even in the presence of K+, indicating little structure formed (Figure 2C and D and Figure S4 in Supplementary Data). A Tm versus concentration study suggested that the Tm of PG5 and PG6 is independent of concentration so they likely still form unimolecular structures (Figure 3) but the formation of these structures seems kinetically slow based on the finding that major hysteresis is observed at a heating/cooling rate of 0.5°C/min (Figure 4 and Figure S1 in Supplementary Data). Taken together, if the unimolecular G-quadruplex is maintained in PG5 and PG6, the folding topology should be changed to tolerate a unimolecular all-parallel G-quadruplex structure with all anti-Gs indicated by a Type I CD profile. A parallel unimolecular quadruplex has been demonstrated in an X-ray crystal structure of the human telomere sequence d(AG3(TTAG3)3) by Neidle and coworkers (64) and in a LNA modified d(G4(T4G4)3) quadruplex by Dominick and Jarstfer (62). It seems that this atypical folding in PG5 and PG6 is not favored since the defined structure is not present in significant quantity in solution, as indicated by the previous CD experiments. The possible reason could be the loop size and sequences which are important factors in determining the folding topology (64). Furthermore, in the case where 2′F-araGs replace both syn-Gs and anti-Gs in the quartet (PG7), we found that 2′F-ANA brings minimal stabilization (ΔTm = +0.4°C/2′F-ANA modification, Table 1). Type I CD profile (i.e. a positive CD band at ∼260 nm and a negative band at ∼240 nm) was observed in the presence of K+ and not shown in the absence of K+, indicating a parallel quadruplex with all anti-Gs (12), as in PG2. Concentration-dependent Tm (Figure 3) and significant hysteresis observed in heating/cooling processes (Figure 4) support the idea that PG7 could adopt an intermolecular parallel G-quadruplex with all anti-Gs. Further non-denaturing shift gel analysis experiments are needed to confirm the molecularity of intermolecular G-quadruplexes studied here.
To verify whether anti-Gs can be stabilized by 2′F-ANA in other types of G-quadruplexes, we chose two other aptamers from the literature.
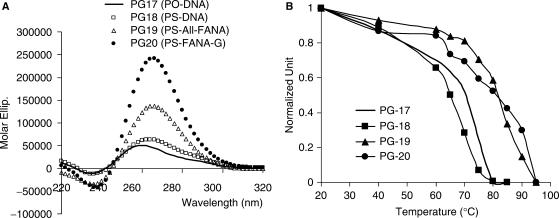
(A) Anti-HIV phosphorothioate PS-d(T2G4T2) modified with 2′F-ANA. Both the phosphodiester sequence PO-d(T2G4T2) and the phosphorothioate sequence PS-d(T2G4T2) are known to adopt a structure in which all G residues are in the anti conformation (7,43). As a result, this aptamer has a characteristic ‘Type I’ CD signature (Table 2 and Figure 6A) (12). Thermal denaturation studies at 260 nm yielded noisy curves. The reason for this could be the slow dissociation process of both phosphodiester and phosphorothioate as reported in the literature (43). Tm profiles shown in Figure 6B were obtained by plotting the maximum molar ellipticities versus temperature based on temperature-dependent CD spectra (Figure S5, Supplementary Data). Although it is impossible to get accurate Tm data for these kinetically slow G-quadruplexes, clearly our apparent Tm data help to confirm the significant thermal stabilization provided by the 2′F-ANA-G units (PG18–20, Table 2). Our results indicate that phosphorothioate modifications lead to stabilization, inconsistent with the literature finding (31). Compared with the controls PG17 (PO-DNA) and PG18 (PS-DNA), 2′F-ANA modified sequences PG19 and PG20 show large increases in molar ellipticities, indicating a better guanine–guanine interactions within the G-quartet structure.
Figure 6.
(A) CD spectra of dT2G4T2 and related sequences (PG17–20) obtained in PBS buffer at a strand concentration of 20 µM (single strand). (B) Tm curves generated by plotting the maximum absorbance at 265 nm wavelength (normalized) versus temperature in a series of CD-temperature-dependent measurements (Figure S5, Supplementary Data); the corresponding Tm data are shown in Table 2.
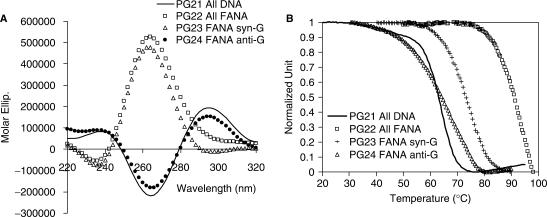
(B) A telomeric DNA d(G4T4G4) modified with 2′F-ANA. Structural analysis has shown that a telomeric DNA sequence d(G4T4G4) (PG21, Table 2) can form a symmetrical dimeric quadruplex with four G-quartets and a diagonal loop in the presence of Na+ (Figure 1C) (21,22). The guanine conformation is 5′-d(GsGaGsGa-TTTT-GsGaGsGa)-3′ where s denotes syn and a anti (22,25). Several sequences, including a fully 2′F-ANA modified sequence (PG22), a sequence with 2′F-ANA replacing all syn-Gs (PG23) and a sequence with 2′F-ANA modifications for all anti-Gs (PG24), were tested (Table 2). When the anti-G residues were replaced by 2′F-ANA G residues, a modest thermal stabilization of the G-quadruplex structure resulted, without disruption of the 3D structure (Table 2, and Figure 7A and B). When the syn-G residues were replaced (PG22 and PG23), a remarkable increase in thermal stabilization of the G-quadruplex structure resulted (up to 25°C; Table 2), with a concomitant Type II-to-Type I conformational change induced by the 2′F-ANA-G(anti) replacement (Figure 7A).
Figure 7.
(A) CD spectra of dT4G4T4 and related sequences (PG21–24) in 10 mM sodium phosphate buffer, 0.1 mM EDTA, pH 7 and 200 mM NaCl at a strand concentration of 10 µM. (B) Tm profile measured at 295 nm in the same sodium phosphate buffer at a strand concentration of 100 µM.
Overall, these two additional examples confirm the results obtained for the thrombin-binding aptamers: the modification of anti-Gs with 2′F-ANA can generally stabilize a G-quartet requiring anti-Gs, maintaining the overall quadruplex conformation, while the modification of syn-Gs with 2′F-ANA is not favored and results in a complete conformational switch to an alternative G-quadruplex structure, as indicated by the change of CD type from II to I for PG2, PG5, PG6, PG7, PG22 and PG23 (Tables 1 and 2, Figure 2D, Figure 7A and Figure S4 in Supplementary Data). The stabilizing effects of 2′F-ANA on G-quadruplexes is related to the conformational preorganization and the small steric effect of the fluorine atom. 2′-O-Methyl modification generates steric problems at the 2′ position and is often shown to destabilize a unimolecular G-quadruplex, resulting in significant conformational perturbation in both intra- and intermolecular G-quadruplex systems (31). The ability to switch G-quartet structures with a β-fluorine atom may be exploited to enrich the population of one of several interconverting G-quadruplex conformations (a β-fluorine “switch”).
Replacement of loops with 2′F-ANA
Thymine bases in the loop region of the thrombin-binding aptamer d(G1G2T3T4G5G6T7G8T9G10G11T12T13G14G15) (Figure 1B) exclusively prefers the anti orientation (15). The literature reports that G8 shows base stacking interactions with the first G-quartet but the conformation of the nucleotide was not mentioned (18). The replacement of G8 with 2′F-araG in this study resulted in an increase of 0.8°C in Tm (PG8 and PG9 in Table 1). It was expected that the replacement of anti thymines in the loops with anti conformationally biased 2′F-araT should increase thermal stability. Thermal melting studies support this hypothesis (PG8–14, Table 1). Different positions of thymidine contribute to the stability of the G-quadruplex to different degrees (Table 1 and Figure S2, Supplementary Data). 2′F-ANA replacements in the loops consisting of two thymidines (PG12) bring a greater thermal stabilization (ΔTm = 2.9°C/2′F-ANA modification, Table 1) than with modification in the TGT loop (PG9, ΔTm = 1.6°C/2′F-ANA modification, Table 1). T3 and T12 are the most sensitive positions to 2′F-araT modification resulting in an almost 6°C difference in Tm (PG9 versus PG10; PG11 versus PG14, Table 2), while the replacement of T13 and T4 with 2′F-araT is much less sensitive and results only in slight Tm increases of 1°C (PG9 versus PG11, Table 2) and 0.2°C (PG10 versus PG13, Table 2), respectively. The crystal structure of this DNA aptamer bound to thrombin suggests that T3 and T12 are different from T4 and T13. It appears that the bases of T3 and T12 do not interact with any other moieties within the aptamer, nor do they interact with thrombin, instead, they extend out into the solvent (18). Modifying T3 and T12 with 2′F-araT might change the conformation of the loops, bringing out stabilization to the whole G-quadruplex structure as suggested by the Tm increase. A detailed picture, however, is difficult to describe at this stage. The replacement of the central nucleotide G8 with a 2′F-araG is also less important for stabilization (PG8 versus PG9, only 0.8°C Tm difference). Overall, 2′F-ANA modifications of loop deoxynucleotides stabilizes the formation of a unimolecular G-quadruplex (Figure 1B), as supported by all CD and UV melting analysis. It was found that all loop-modified thrombin-binding aptamers (PG8–14) maintain Type II CD in the presence of K+, like the DNA aptamer control (Figure S4, Supplementary Data). Remarkably, in the absence of K+, a relatively strong positive peak at ∼295 nm is still observed, which indicates significant G-quartet formation. Therefore, replacement of the loop deoxynucleotides with 2′F-ANA significantly helps with the formation of G-quadruplex structure. They show concentration-independent Tm data, indicating a unimolecular structure, in agreement with a lack of hysteresis in heating/cooling processes (Figure 4 and Figure S1, Supplementary Data).
Nuclease resistance induced by 2′F-ANA
Incorporation of 2′F-ANA in oligonucleotides leads to an improvement in nuclease resistance (34). This study further evaluates the nuclease resistance of fully 2′F-ANA-modified thrombin-binding aptamers (PG1–14, Table 1). The data clearly shows that 2′F-ANA modified aptamers have enhanced half-lives in 10% FBS (in some cases >48-fold compared with PG1, Figure S6 in Supplementary Data and Table 1) but nuclease stability is found to be dependent on both the position (esp. syn-G and anti-G) and number of 2′F-ANA residues within the oligonucleotide backbone. For example, replacing anti-Gs with 2′F-ANA (PG3 and PG4) increases the half-lives to 4.8 and 9.4 h, respectively, while replacing syn-Gs with 2′F-ANA (PG5 and PG6) does not yield much gain in biostability with half-life increases of only 0.8 and 0.6 h, respectively, comparable to the DNA aptamer (PG1) (Figure S6 in Supplementary Data and Table 1). These differences could be justified by the previous finding that PG5 and PG6 only form insignificant G-quadruplex structures (Figure 2D) due to the unfavored conformation of the perturbed syn-Gs. It is expected that a complex structure should be less accessible for nuclease attack compared to dissociated or ‘open’ oligonucleotides. Furthermore, the nuclease stability of PG13 and PG14 is enhanced over PG1 by a factor of 4–7 with only four 2′F-ANA incorporated (Table 1). Modification of the loop residues of stable structures (e.g. PG4, PG8) may have a double effect since the increased stability of the duplex would be combined with protection of the nucleotides most exposed to endonuclease attack.
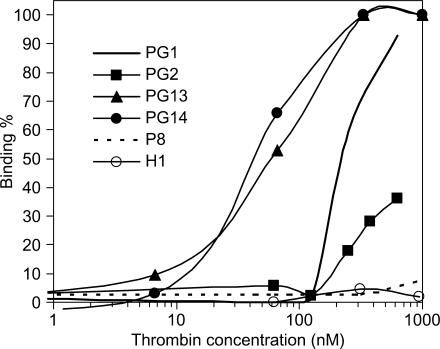
Binding affinity of 2′F-ANA-modified thrombin-binding aptamers with thrombin
As mentioned in the ‘Introduction’ section, the structure of the thrombin-binding aptamer is very sensitive to chemical modifications, which often undermine its thermal stability and/or binding activity (31). Thus, a final experiment in this study was conducted using nitrocellulose filter binding assays to assess the effect of 2′F-ANA modifications on the thrombin binding affinity. Data are presented in Table 1 and in Figure 8. ssDNA (P8) and hairpin (H1) controls show no binding to thrombin as expected (18). Binding of 2′F-ANA aptamers to thrombin is always adversely affected by 2′F-ANA modifications on G-quartets themselves, whether syn- or anti-Gs are modified (PG2-PG6). Some loop modifications with 2′F-ANA also disfavor thrombin binding (PG8–PG12). However, two loop-modified aptamers (PG13 and PG14, Table 1) show a 4–5-fold enhancement in thrombin-binding affinity. This conclusion should bear in mind that while these two aptamers may have enhanced binding affinity, the slope of the concentration response is not as sharp as that for the native PG1. Therefore, this study reveals the first 2′F-ANA modified thrombin-binding aptamers to combine enhanced thermal and nuclease stability with apparently slightly stronger binding affinity. Previous structural studies by NMR and crystallography suggest that the loop moieties would change conformation during binding to thrombin (18). The TGT loop is involved with the fibrinogen exosite and two TT loops could interact with the heparin exosite. It seems therefore that two 2′F-ANA-T residues when present in one of the TT loops and the TGT loop enhance thrombin binding.
Figure 8.
Binding affinity of selected aptamers with human thrombin assessed by nitrocellulose filter binding assays.
CONCLUSIONS
In the present study 2′F-ANA modified oligonucleotides were investigated based on a thrombin-binding DNA aptamer d(G2T2G2TGTG2T2G2), an anti-HIV phosphothioate PS-d(T2G4T2) and a DNA telomeric sequence d(G4T4G4) (Tables 1 and 2) by UV thermal denaturation and CD experiments. Generally, replacement of anti-Gs with 2′F-ANA can stabilize a G-quartet requiring anti-Gs and maintain the quadruplex conformation, while replacement of syn-Gs with 2′F-ANA is not favored and results in complete conformational change of the G-quadruplexes. The data shows that appropriate incorporation of 2′F-ANA residues into G-quadruplexes leads to an increase in the melting temperature of the complex formed (ΔTm up to ∼+3°C/2′F-ANA modification, Tables 1 and 2). The structure of thrombin-binding aptamers is stabilized by the presence of potassium ions. Nuclease resistance of 2′F-ANA modified thrombin-binding aptamers is increased up to 48-fold in 10% FBS. Two 2′F-ANA-modified thrombin-binding aptamers (PG13 and PG14) show a 4–5-fold enhancement in binding affinity to thrombin along with increased thermal stability and nuclease resistance. Therefore, the impact of 2′F-ANA modifications on G-quartets and loop regions has been demonstrated. The study suggests that 2′F-ANA may positively impact oligonucleotide-based therapeutics involving G-quadruplexes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We acknowledge financial support through a grant from the Canadian Institutes of Health Research (CIHR) and Topigen Pharmaceuticals, Inc. CGP acknowledges support from a FQRNT fellowship and a Clifford Wong McGill Major Fellowship. We thank J. Watts, R. Donga and J. Lackey for helpful feedback during the preparation of this manuscript. C. G. P. and M.J.D. are recipients of the Carl Winkler Dissertation Award (McGill University) and Bernard Belleau Award (Canadian Society for Chemistry), respectively. Funding to pay the Open Access publication charges for this article was provided by CIHR (Canadian Institutes of Health Research). Dedicated to Hermano Juan Antón on the occasion of his 61th birthday.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gellert M, Lipsett MN, Davies DR. Helix formation by guanylic acid. Proc. Natl Acad. Sci. USA. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sen D, Gilbert W. Formation of parallel 4-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 3.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 4.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 5.Joyce GF. Amplification, mutation and selection of catalytic RNA. Gene. 1989;82:83–87. doi: 10.1016/0378-1119(89)90033-4. [DOI] [PubMed] [Google Scholar]
- 6.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 7.Wyatt JR, Vickers TA, Roberson JL, Buckheit RW, Klimkait T, Debaets E, Davis PW, Rayner B, Imbach JL, et al. Combinatorially selected guanosine-quartet structure is a potent inhibitor of human-immunodeficiency-virus envelope-mediated cell-fusion. Proc. Natl Acad. Sci. USA. 1994;91:1356–1360. doi: 10.1073/pnas.91.4.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phan AT, Kuryavyi V, Ma JB, Faure A, Andreola ML, Patel DJ. An interlocked dimeric parallel-stranded DNA quadruplex: a potent inhibitor of HIV-1 integrase. Proc. Natl Acad. Sci. USA. 2005;102:634–639. doi: 10.1073/pnas.0406278102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jing NJ, Hogan ME. Structure-activity of tetrad-forming oligonucleotides as a potent anti-HIV therapeutic drug. J. Biol. Chem. 1998;273:34992–34999. doi: 10.1074/jbc.273.52.34992. [DOI] [PubMed] [Google Scholar]
- 10.Borman S. Targeting telomerase. Chem. Eng. News. 2006;84:32–33. [Google Scholar]
- 11.Keniry MA. Quadruplex structures in nucleic acids. Biopolymers. 2000;56:123–146. doi: 10.1002/1097-0282(2000/2001)56:3<123::AID-BIP10010>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 12.Williamson JR. G-quartet structures in telomeric DNA. Annu. Rev. Biophys. Biomol. Struct. 1994;23:703–730. doi: 10.1146/annurev.bb.23.060194.003415. [DOI] [PubMed] [Google Scholar]
- 13.Kerwin SM. G-quadruplex DNA as a target for drug design. Curr. Pharm. Des. 2000;6:441–471. doi: 10.2174/1381612003400849. [DOI] [PubMed] [Google Scholar]
- 14.Schultze P, Macaya RF, Feigon J. Three-dimensional solution structure of the thrombin-binding DNA aptamer d(GGTTGGTGTGGTTGG) J. Mol. Biol. 1994;235:1532–1547. doi: 10.1006/jmbi.1994.1105. [DOI] [PubMed] [Google Scholar]
- 15.Macaya RF, Schultze P, Smith FW, Roe JA, Feigon J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl Acad. Sci. USA. 1993;90:3745–3749. doi: 10.1073/pnas.90.8.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang KY, Mccurdy S, Shea RG, Swaminathan S, Bolton PH. A DNA aptamer which binds to and inhibits thrombin exhibits a new structural motif for DNA. Biochemistry. 1993;32:1899–1904. doi: 10.1021/bi00059a003. [DOI] [PubMed] [Google Scholar]
- 17.Wang KY, Krawczyk SH, Bischofberger N, Swaminathan S, Bolton PH. The tertiary structure of a DNA aptamer which binds to and inhibits thrombin determines activity. Biochemistry. 1993;32:11285–11292. doi: 10.1021/bi00093a004. [DOI] [PubMed] [Google Scholar]
- 18.Padmanabhan K, Padmanabhan KP, Ferrara JD, Sadler JE, Tulinsky A. The structure of alpha-thrombin inhibited by a 15-mer single-stranded DNA aptamer. J. Biol. Chem. 1993;268:17651–17654. doi: 10.2210/pdb1hut/pdb. [DOI] [PubMed] [Google Scholar]
- 19.Padmanabhan K, Tulinsky A. An ambiguous structure of a DNA 15-mer thrombin complex. Acta Crystallogr. D Biol. Crystallogr. 1996;D52:272–282. doi: 10.1107/S0907444995013977. [DOI] [PubMed] [Google Scholar]
- 20.Kelly JA, Feigon J, Yeates TO. Reconciliation of the x-ray and NMR structures of the thrombin-binding aptamer d(GGTTGGTGTGGTTGG) J. Mol. Biol. 1996;256:417–422. doi: 10.1006/jmbi.1996.0097. [DOI] [PubMed] [Google Scholar]
- 21.Schultze P, Smith FW, Feigon J. Refined solution structure of the dimeric quadruplex formed from the Oxytricha telomeric oligonucleotide d(GGGGTTTTGGGG) Structure. 1994;2:221–233. doi: 10.1016/s0969-2126(00)00023-x. [DOI] [PubMed] [Google Scholar]
- 22.Smith FW, Feigon J. Quadruplex structure of Oxytricha telomeric DNA oligonucleotides. Nature. 1992;356:164–168. doi: 10.1038/356164a0. [DOI] [PubMed] [Google Scholar]
- 23.Kang C, Zhang XH, Ratliff R, Moyzis R, Rich A. Crystal structure of 4 stranded Oxytricha telomeric DNA. Nature. 1992;356:126–131. doi: 10.1038/356126a0. [DOI] [PubMed] [Google Scholar]
- 24.Haider S, Parkinson GN, Neidle S. Crystal structure of the potassium form of an Oxytricha nova G-quadruplex. J. Mol. Biol. 2002;320:189–200. doi: 10.1016/S0022-2836(02)00428-X. [DOI] [PubMed] [Google Scholar]
- 25.Schultze P, Hud NV, Smith FW, Feigon J. The effect of sodium, potassium and ammonium ions on the conformation of the dimeric quadruplex formed by the Oxytricha nova telomere repeat oligonucleotide d(G4T4G4) Nucleic Acids Res. 1999;27:3018–3028. doi: 10.1093/nar/27.15.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hardin CC, Perry AG, White K. Thermodynamic and kinetic characterization of the dissociation and assembly of quadruplex nucleic acids. Biopolymers. 2000;56:147–194. doi: 10.1002/1097-0282(2000/2001)56:3<147::AID-BIP10011>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 27.Heckel A, Mayer G. Light regulation of aptamer activity: an anti-thrombin aptamer with caged thymidine nucleobases. J. Am. Chem. Soc. 2005;127:822–823. doi: 10.1021/ja043285e. [DOI] [PubMed] [Google Scholar]
- 28.Di Giusto DA, King GC. Construction, stability, and activity of multivalent circular anticoagulant aptamers. J. Biol. Chem. 2004;279:46483–46489. doi: 10.1074/jbc.M408037200. [DOI] [PubMed] [Google Scholar]
- 29.Buijsman RC, Schipperijn JWJ, Kuyl-Yeheskiely E, Van Der Marel GA, Van Boeckel CAA, Van Boom JH. Design and synthesis of a possible mimic of a thrombin-binding DNA aptamer. Bioorg. Med. Chem. Lett. 1997;7:2027–2032. [Google Scholar]
- 30.Rosemeyer H, Mokrosch V, Jawalekar A, Becker E-M, Seela F. Single-stranded DNA: replacement of canonical by base-modified nucleosides in the minihairpin 5′-d(GCGAAGC)-3′ and constructs with the aptamer 5′-d(GGTTGGTGTGGTTGG)-3′. Helv. Chim. Acta. 2004;87:536–553. [Google Scholar]
- 31.Sacca B, Lacroix L, Mergny J-L. The effect of chemical modifications on the thermal stability of different G-quadruplex-forming oligonucleotides. Nucleic Acids Res. 2005;33:1182–1192. doi: 10.1093/nar/gki257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lebruska LL, Maher LJ., III Selection and characterization of an RNA decoy for rranscription factor NF-kB. Biochemistry. 1999;38:3168–3174. doi: 10.1021/bi982515x. [DOI] [PubMed] [Google Scholar]
- 33.Trempe J-F, Wilds CJ, Denisov AY, Pon RT, Damha MJ, Gehring K. NMR solution structure of an oligonucleotide hairpin with a 2′F-ANA/RNA stem: implications for RNase H specificity toward DNA/RNA hybrid duplexes. J. Am. Chem. Soc. 2001;123:4896–4903. doi: 10.1021/ja003859p. [DOI] [PubMed] [Google Scholar]
- 34.Kalota A, Karabon L, Swider CR, Viazovkina E, Elzagheid M, Damha MJ, Gewirtz AM. 2′-Deoxy-2′-fluoro-beta-D-arabinonucleic acid (2′F-ANA) modified oligonucleotides (ON) effect highly efficient, and persistent, gene silencing. Nucleic Acids Res. 2006;34:451–461. doi: 10.1093/nar/gkj455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilds CJ, Damha MJ. 2′-Deoxy-2′-fluoro-β-D-arabinonucleosides and olignucleotides (2′F-ANA): synthesis and physicochemical studies. Nucleic Acids Res. 2000;28:3625–3635. doi: 10.1093/nar/28.18.3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilds CJ, Damha MJ. Duplex recognition by oligonucleotides containing 2′-deoxy-2′-fluoro-D-arabinose and 2′-deoxy-2′-fluoro-D-ribose. Intermolecular 2′-OH-phosphate contacts versus sugar puckering in the stabilization of triple-helical complexes. Bioconjug. Chem. 1999;10:299–305. doi: 10.1021/bc9801171. [DOI] [PubMed] [Google Scholar]
- 37.Wilds CJ. Ph.D. Thesis. Montreal: McGill University; 2000. [Google Scholar]
- 38.Lok C-N, Viazovkina E, Min K-L, Nagy E, Wilds CJ, Damha MJ, Parniak MA. Potent gene-specific inhibitory properties of mixed-backbone antisense oligonucleotides comprised of 2′-deoxy-2′-fluoro-D-arabinose and 2′-deoxyribose nucleotides. Biochemistry. 2002;41:3457–3467. doi: 10.1021/bi0115075. [DOI] [PubMed] [Google Scholar]
- 39.Dowler T, Bergeron D, Tedeschi AL, Paquet L, Ferrari N, Damha MJ. Improvements in siRNA properties mediated by 2′-deoxy-2′-fluoro-beta-D-arabinonucleic acid (FANA) Nucleic Acids Res. 2006;34:1669–1675. doi: 10.1093/nar/gkl033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watts JK, Choubdar N, Sadalapure K, Robert F, Wahba AS, Pelletier J, Pinto BM, Damha MJ. 2′-Fluoro-4′-thioarabino-modified oligonucleotides: conformational switches linked to siRNA activity. Nucleic Acids Res. 2007;35:1441–1451. doi: 10.1093/nar/gkl1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Viazovkina EV, Mangos MM, Elzagheid MI, Damha MJ. Current Protocols in Nucleic Acid Chemistry. John Wiley & Sons, Inc; 2002. pp. 4.15.11–22. [DOI] [PubMed] [Google Scholar]
- 42.Smirnov I, Shafer RH. Effect of loop sequence and size on DNA aptamer stability. Biochemistry. 2000;39:1462–1468. doi: 10.1021/bi9919044. [DOI] [PubMed] [Google Scholar]
- 43.Wyatt JR, Davis PW, Freier SM. Kinetics of G-quartet-mediated tetramer formation. Biochemistry. 1996;35:8002–8008. doi: 10.1021/bi960124h. [DOI] [PubMed] [Google Scholar]
- 44.Lu M, Guo Q, Kallenbach NR. Thermodynamics of G-tetraplex formation by telomeric DNAs. Biochemistry. 1993;32:598–601. doi: 10.1021/bi00053a027. [DOI] [PubMed] [Google Scholar]
- 45.Cantor CR, Warshaw MM, Shapiro H. Oligonucleotide interactions. 3. Circular dichroism studies of conformation of deoxyoligonucleotides. Biopolymers. 1970;9:1059–1077. doi: 10.1002/bip.1970.360090909. [DOI] [PubMed] [Google Scholar]
- 46.Galarneau A, Min K-L, Mangos MM, Damha MJ. Methods Mol. Biol. Vol. 288. NJ, United States: Totowa; 2005. pp. 65–80. [DOI] [PubMed] [Google Scholar]
- 47.Nimjee SM, Rusconi CP, Sullenger BA. Aptamers: an emerging class of therapeutics. Annu. Rev. Med. 2005;56:555–583. doi: 10.1146/annurev.med.56.062904.144915. [DOI] [PubMed] [Google Scholar]
- 48.Brody EN, Gold L. Aptamers as therapeutic and diagnostic agents. Rev. Mol. Biotechnol. 2000;74:5–13. doi: 10.1016/s1389-0352(99)00004-5. [DOI] [PubMed] [Google Scholar]
- 49.White RR, Sullenger BA, Rusconi CP. Developing aptamers into therapeutics. J. Clin. Invest. 2000;106:929–934. doi: 10.1172/JCI11325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pestourie C, Tavitian B, Duconge F. Aptamers against extracellular targets for in vivo applications. Biochimie. 2005;87:921–930. doi: 10.1016/j.biochi.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 51.Nolte A, Klussmann S, Bald R, Erdmann VA, Fuerste JP. Mirror-design of L-oligonucleotide ligands binding to L-arginine. Nat. Biotechnol. 1996;14:1116–1119. doi: 10.1038/nbt0996-1116. [DOI] [PubMed] [Google Scholar]
- 52.Ruckman J, Green LS, Beeson J, Waugh S, Gillette WL, Henninger DD, Claesson-Welsh L, Janjic N. 2′-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J. Biol. Chem. 1998;273:20556–20567. doi: 10.1074/jbc.273.32.20556. [DOI] [PubMed] [Google Scholar]
- 53.Lato SM, Ozerova NDS, He K, Sergueeva Z, Shaw BR, Burke DH. Boron-containing aptamers to ATP. Nucleic Acids Res. 2002;30:1401–1407. doi: 10.1093/nar/30.6.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kato Y, Minakawa N, Komatsu Y, Kamiya H, Ogawa N, Harashima H, Matsuda A. New NTP analogs: the synthesis of 4′-thioUTP and 4′-thioCTP and their utility for SELEX. Nucleic Acids Res. 2005;33:2942–2951. doi: 10.1093/nar/gki578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jhaveri S, Olwin B, Ellington AD. In vitro selection of phosphorothiolated aptamers. Bioorg. Med. Chem. Lett. 1998;8:2285–2290. doi: 10.1016/s0960-894x(98)00414-4. [DOI] [PubMed] [Google Scholar]
- 56.Pagratis NC, Bell C, Chang Y-F, Jennings S, Fitzwater T, Jellinek D, Dang C. Potent 2′-amino-, and 2′-fluoro-2′-deoxyribonucleotide RNA inhibitors of keratinocyte growth factor. Nat. Biotechnol. 1997;15:68–73. doi: 10.1038/nbt0197-68. [DOI] [PubMed] [Google Scholar]
- 57.Ng EWM, Shima DT, Calias P, Cunningham ET, Guyer DR, Adamis AP. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006;5:123–132. doi: 10.1038/nrd1955. [DOI] [PubMed] [Google Scholar]
- 58.Mergny JL, Phan AT, Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 59.Virgilio A, Esposito V, Randazzo A, Mayol L, Galeone A. 8-Methyl-2′-deoxyguanosine incorporation into parallel DNA quadruplex structures. Nucleic Acids Res. 2005;33:6188–6195. doi: 10.1093/nar/gki924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Esposito V, Randazzo A, Piccialli G, Petraccone L, Giancola C, Mayol L. Effects of an 8-bromodeoxyguanosine incorporation on the parallel quadruplex structure [d(TGGGT)](4) Org. Biomol. Chem. 2004;2:313–318. doi: 10.1039/b314672c. [DOI] [PubMed] [Google Scholar]
- 61.Tang CF, Shafer RH. Engineering the quadruplex fold: Nucleoside conformation determines both folding topology and molecularity in guanine quadruplexes. J. Am. Chem. Soc. 2006;128:5966–5973. doi: 10.1021/ja0603958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dominick PK, Jarstfer MB. A conformationally constrained nucleotide analogue controls the folding topology of a DNA G-quadruplex. J. Am. Chem. Soc. 2004;126:5050–5051. doi: 10.1021/ja039192z. [DOI] [PubMed] [Google Scholar]
- 63.Sapse AM, Snyder G. Ab initio studies of the antiviral drug 1-(2-fluoro-2-deoxy-beta-D-arabinofuranosyl) thymine. Cancer Invest. 1985;3:115–121. doi: 10.3109/07357908509017494. [DOI] [PubMed] [Google Scholar]
- 64.Parkinson GN, Lee MPH, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]