Abstract
Double-strand breaks (DSBs) are dangerous chromosomal lesions that must be efficiently repaired in order to avoid loss of genetic information or cell death. In all organisms studied to date, two different mechanisms are used to repair DSBs: homologous recombination (HR) and non-homologous end joining (NHEJ). Previous studies have shown that during DSB repair, non-homologous exogenous DNA (also termed ‘filler DNA’) can be incorporated at the site of a DSB. We have created a genetic system in the yeast Saccharomyces cerevisiae to study the mechanism of fragment capture. Our yeast strains carry recognition sites for the HO endonuclease at a unique chromosomal site, and plasmids in which a LEU2 gene is flanked by HO cut sites. Upon induction of the HO endonuclease, a linear extrachromosomal fragment is generated in each cell and its incorporation at the chromosomal DSB site can be genetically monitored. Our results show that linear fragments are captured at the repaired DSB site at frequencies of 10−6 to 10−4 per plated cell depending on strain background and specific end sequences. The mechanism of fragment capture depends on the NHEJ machinery, but only partially on the homologous recombination proteins. More than one fragment can be used during repair, by a mechanism that relies on the annealing of small complementary sequences. We present a model to explain the basis for fragment capture.
INTRODUCTION
Double-strand breaks (DSBs) can be created by different DNA-damaging agents or can occur spontaneously during cell growth. If not properly repaired, DSBs have the potential to affect cell viability, or to cause the loss or modification of genetic information (1). In all organisms studied to date, two different mechanisms are used to repair DSBs: homologous recombination (HR) and non-homologous end joining (NHEJ) [reviewed in (2)]. In HR a DNA break or gap is repaired by copying similar information present on a sister chromatid, a homologous chromosome, or at an ectopic location. In contrast, in NHEJ broken ends are ligated together without the need for extensive homology.
Among eukaryotes, these processes have been best characterized at the genetic and molecular level in the yeast Saccharomyces cerevisiae. Homologous recombination is the major repair pathway in yeast. Yeast cells are able to repair broken chromosomes by carrying out a genome-wide search for homology, and using that information as a template to patch the broken chromosome. This usually results in the transfer of genetic information between the two loci, and may also lead to crossovers. If the information is present at different genomic locations, HR may result in genomic rearrangements, such as translocations, inversions and deletions [reviewed in (3)]. HR usually requires a set of genes termed the RAD52 epistasis group [reviewed in (4)]. These proteins help search for homologous sequences and carry out a strand exchange reaction between regions sharing sequence similarities.
NHEJ is a repair mechanism that is conserved from bacteria to higher eukaryotes [reviewed in (5)]. While NHEJ appears to be the major pathway for DSB repair in human cells (6), it represents a relatively minor pathway in Saccharomyces cerevisiae. A conserved set of proteins is required for NHEJ, including the Ku70/Ku80 heterodimer, DNA IV ligase and its associated factor Lif1/XRCC4. The MRX complex (MRN in humans) plays a role in promoting DNA joining both by HR and by NHEJ (7).
The relative contribution of HR and NHEJ varies depending on both the organism and the context of the DSB. In organisms with genomes rich in repeats, recombination between non-allelic sequences can potentially lead to crossover and genome rearrangements. In such a case, NHEJ might prove safer (8). However, NHEJ has also been linked to chromosomal rearrangements associated with the repair of a specific, induced DSB. These repair events consist of insertions, deletions, translocations and inversions (9–13).
Previous studies have shown that unrelated DNA fragments, sometimes termed ‘filler DNA’ can be inserted into junctions during the joining reaction. For example, in mammalian lymphoid cells, extra nucleotides of filler DNA are usually found at VDJ joints; only part of this filler DNA is generated by terminal transferase activity (14). DSB repair events in mammalian cells can also be associated with the capture of endogenous or exogenous DNA sequences up to several kilobases in length (15). Filler DNA is also commonly observed in the repair of DSBs in plant cells (16,17). The insertion of exogenous DNA has also been observed in yeast cells. cDNA sequences of the natural Ty1 yeast retrotransposon could be found at repaired DSB sites (9,11,18). Additionally, short mitochondrial DNA segments can be captured at break sites. This integration was not accompanied by modification of the junction sites, except for the loss or gain of 1–5 nucleotides (10,11).
A selectable assay system has been developed to study NHEJ-mediated chromosomal rearrangements associated with repair of a unique DSB in haploid yeast cells. Using this system Yu and Gabriel showed that extrachromosomal DNA fragments could be captured at the DSB site, in particular Ty1 cDNA and mitochondrial DNA sequences. In both wild type and rad52 cells, the inserted sequences were flanked by sequences with microhomology to the cut site, suggesting that NHEJ plays a central role in generating these events. Accordingly, the insertion process was shown to be RAD52-independent and YKU80 dependent (11,12).
In order to study in more detail the mechanism that captures filler DNA, we created yeast strains that allow the generation of a linear DNA fragment in each cell undergoing a chromosomal DSB. The experimental setup allowed us to directly identify events in which insertions had occurred at the break site. As there are no sequences in the genome of these strains sharing extensive homology to the region undergoing the DSB, survival becomes dependent on the NHEJ repair pathway. Here we use this selectable assay system to show that annealing of complementary sequences at the termini of linear fragments play an important role in linear fragment capture. We also show that more than one linear fragment can be used during a single repair event. Finally, we present a model for DSB repair by capture of a linear fragment.
MATERIALS AND METHODS
Yeast strains
All yeast strains were originally derived from YFP17 (ho, Δhml::ADE1, Δmata::hisG, Δhmr::ADE1, ade3::GAL-HO, ade1, lys5, trp1::hisG, ura3-52, leu2::HOcs) (19). Strains AGY670 and strain AGY673 carry an intronless copy of the ACT1 gene (ACT1Δi) and a URA3 allele into which the ACT1 intron was inserted (URA3::ACT1-i or URA3::ACT1-i::HOcs, respectively). Strain AHY22 is a leu2Δ derivative (YFP17, URA3::ACT1-i::HOcs, Δleu2::kanMX4, ACT1Δi).
Strains AHY52 (AHY117, URA3::ACT1-i::2dirHOcs) and AHY51 (AHY117, URA3::ACT1-i::2inv HOcs) were made in several steps. Strain AHY117 was streaked onto 5-FOA plates to obtain a spontaneous Ura-clone (AHY156). The non-functional ura3 allele on chromosome V was replaced with the URA3-containing BamHI fragment from either plasmid pAH146 (URA3::ACT1-i::2dir HOcs cassette) or plasmid pAH148 (URA3::ACT1-i::2inv HOcs cassette). Correct integration of the cassette at the URA3 locus was confirmed by PCR and sequencing.
Δ ku80::kanMX4, Δ rad51::kanMX4 and rad52::hisG derivatives of AHY119 were obtained by one-step gene replacement.
Plasmids
All plasmids were transformed into E. coli DH5alpha competent bacteria. E. coli SURE competent bacteria (Stratagene) were used for the palindrome-containing inverted orientation.
pAH150 (‘Direct’ plasmid) and pAH129 (‘Inverted’ plasmid) were derived from pLAY98 (20) and pRS414, a TRP1-CEN plasmid (21). Plasmid pAH174 (No-HOcs) was constructed by ligation of the SalI/SmaI LEU2 fragment into SalI and SmaI-digested pRS414.
pAH146 (URA3::ACT1-i::2dirHOcs cassette) and pAH148 (URA3::ACT1-i::2invHOcs) were constructed simultaneously. First, the 117-bp HOcs was amplified using primers pMH1, which contains an engineered MfeI site upstream of the HOcs, and primer pMNH2, containing both MfeI and NcoI sites downstream of HOcs. The HOcs from plasmid pAH129 was used as a PCR template. The amplified fragment was then cut with MfeI and ligated to MfeI-digested pAGE1658 (URA3::ACT1-i::HOcs) (11). Plasmids carrying both possible orientations were distinguished by digestion with NcoI and XhoI (a site unique to the plasmid). Candidates were subjected to sequencing to confirm the presence of the predicted configuration.
To construct plasmid pAH316 [referred to as ‘Inverted +2invHOcs’ plasmid, the double HOcs from plasmid pAH148 was amplified and ligated to a SnaB1-digested pAH129 (‘Inverted’ plasmid)].
To construct plasmid pAH256 (‘Inverted-Fit’), pLAY98 was modified by replacing the upstream HOcs with an HOcs engineered to contain KpnI and XhoI/NcoI ends, thus ensuring its orientation. The LEU2 fragment cut with SalI and SmaI was ligated into this modified pLAY98 plasmid that was cut with the same restriction enzymes. The resulting LEU2 gene flanked with inverted HO cut sites was cut with PvuII and ligated to pRS414 as described earlier.
Media and growth condition
Yeast cells were grown in yeast extract-peptone-dextrose (YP-dextrose) or synthetic complete media (SC) with appropriate amino acids missing (22). Yeast extract-peptone-galactose (YP-galactose) and Yeast extract-peptone-raffinose (YP-raffinose) contain 2% galactose and 1% raffinose (w/v), respectively. 5-fluoroorotic acid (5-FOA) plates are SC-glucose plates supplemented with 1 mg/ml of 5-FOA (23).
Induction of HO endonuclease, determination of DSB repair efficiency (survival frequency), and 5-FOA resistance frequencies
Twelve to 36 independent colonies of various yeast strains were inoculated into 3 ml liquid medium and grown at 30° to a final concentration of ∼3 × 107 cells/ml. Serial dilutions were then plated on YP-dextrose or YP-galactose plates and counted after 3–5 days. The survival frequency was calculated by determining the ratio of colonies growing on YP-galactose plates versus YP-dextrose plates (median values). Statistical analysis was carried out using a non-parametric Mann–Whitney rank test. Standard deviations were always lower than 20% of the median value.
YP-galactose plates were replica plated onto 5-FOA plates. The ratio of colonies growing on 5-FOA to those growing on galactose was used to calculate the frequency of 5-FOA resistance per survivor. The frequency of 5-FOAR per plated cells was calculated by multiplying the previous two terms.
Colonies growing on 5-FOA plates were replica plated to SC-leu and SC-trp plates. The frequency of 5-FOAR per plated cell with particular marker combinations was determined by multiplying the previous calculation by the proportion of the total 5-FOAR colonies shown to have or lack those markers (i.e. Leu+ Trp–, Leu+ Trp+ and Leu– Trp–).
Analysis of repaired chromosomes
5-FOA resistant colonies were purified and grown in liquid YP-dextrose at 30° over night before total genomic DNA was isolated (24). Genomic DNA samples were used as templates for PCR, using primers upstream and downstream of the HO cut site and the URA3::ACT1- i::HOcs cassette. The PCR products were analyzed by sequencing.
RESULTS
The experimental system
The experimental system used (Figure 1) is based on the strains developed by Yu and Gabriel (11). Haploid yeast cells contain a single HO recognition sequence (HO cut site or HOcs) placed into a non-essential portion of the ACT1 intron, which has itself been engineered into the coding domain of the URA3 gene on chromosome V. This URA3::ACT1-i::HOcs allele is efficiently spliced, resulting in uracil prototrophy. In addition, this strain (AGY117) has been deleted of all MAT-related sequences, and also contains an integrated, galactose-inducible HO endonuclease gene (11). When transferred to galactose-containing media, a persistent and lethal DSB is formed on chromosome V, unless the break is repaired in a way that eliminates the HO recognition sequence. Using this system Yu et al. captured extrachromosomal DNA fragments at the DSB site, by selecting for resistance to 5-fluoroorotic acid (5-FOA, i.e. loss of uracil prototrophy). In particular Ty1 cDNA (140 bp to 5.6 kb) and mitochondrial DNA sequences (33 to 219 bp) were identified.
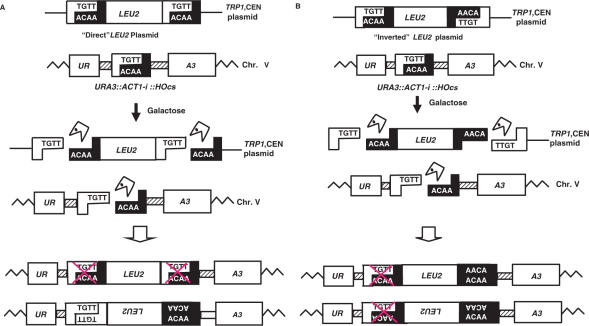
Figure 1.
Genetic system used. The URA3::ACT1-i::HOcs is located on Chromosome V. The LEU2 gene is carried by a TRP1-marked centromeric plasmid. (A) ‘Direct’ LEU2 orientation. Upon transfer of cells to galactose, the HO endonuclease cleaves the three HO cut sites. The ‘Direct’ LEU2 fragments can be inserted in either orientation. Crossed HOcs boxes represent additional mutations required to inactivate the HOcs and prevent it from being cut again. Note that only the upper orientation allows annealing of both ends. (B) The ‘Inverted’ LEU2 fragment. Note that in both orientations, only one end can undergo simple annealing.
To expand this work and study the mechanism by which filler DNA is incorporated into DSBs, we set up a related system to examine the fate of a defined extrachromosomal fragment in a cell with a defined DSB. We created a plasmid that upon induction of the HO endonuclease generates a linear DNA fragment carrying the LEU2 marker gene. In our strains, a centromeric TRP1 plasmid carries a selectable LEU2 marker that is flanked by two HOcs. The two HOcs sequences are present either in the same (referred to as ‘Direct’) or in opposite (referred to as ‘Inverted’) orientation relative to each other (Figure 1). These plasmids were independently transformed into AHY22, a strain carrying the URA3::ACT1-i::HOcs allele in which all the relevant homologous sequences (the ACT1 intron and the LEU2 gene) had been deleted. Upon transfer of cells to galactose-containing medium, the HO endonuclease is transcriptionally activated and cleaves the three HO cut sites, causing the release of the LEU2 fragment from the plasmid and creating at the same time a single DSB in the genome (Figure 1). The cell's ability to survive depends on successful repair of the genomic DSB in a way that modifies the HO recognition sequence and forms HO endonuclease-resistant colonies. In previous studies, carried out in the absence of a linearized DNA molecule, the most common form of repair was an imprecise end joining event within the intron, that does not interfere with splicing, and which therefore does not make the cells resistant to FOA (11,12). We reasoned that if the linear LEU2 fragment could participate in these repair events, its insertion at the DSB site should interfere with splicing and thus generate FOA resistant uracil auxotrophs. This experimental setup therefore allows screening for repair events in which the DSB has incorporated a defined linear fragment: these are detectable as colonies that, upon loss of the TRP1-marked plasmid, remain Ura− and Leu+. ‘Direct’ LEU2 fragments can in principle be joined with the DSB either through annealing of the complementary 3′ overhanging four bases at both ends and precise re-ligation or by imprecise end joining of the non-complementary 3′ overhanging four bases (Figure 1). However, if the break is repaired by precise re-ligation, it will be re-cut by the HO endonuclease. In order to stably insert a linear fragment at the DSB site, cells must use more complex mechanisms that modify the junction sequences, either by imprecisely joining complementary overhangs or by joining ends where the overhanging bases are non-complementary. For ‘Inverted’ fragments only one terminus can anneal, while the other must be joined in the absence of terminal complementarities. This can occur in either orientation of the LEU2 fragment relative to the URA3 gene (Figure 1). As a control we constructed a plasmid without any HO cut sites. In this case, there is no release of a linear fragment.
Repair of a DSB by insertion of a linear fragment
Wild-type cells carrying plasmids with the ‘Direct’, ‘Inverted’ or control ‘uncuttable’ fragment were plated on galactose-containing medium. Under these conditions, chromosome V is cut by the HO endonuclease, and only cells able to repair the broken chromosome will survive to form colonies. Southern blot analysis showed that the efficiency of DSB formation by HO was very similar in all strains (data not shown). Survival of the three strains was also very similar (∼1% survival). These results suggest that the availability of linear fragments does not significantly increase a cell's ability to survive a DSB.
Most of the surviving colonies were 5-FOA sensitive, as expected from previous studies (11). Our experimental system allows us to directly identify potential insertion events, which result in FOA resistance. The frequency of these events differed greatly among the various strains (Table 1). Except for cells carrying the ‘No-HOcs’ plasmids, all Ura− colonies recovered were Trp−, i.e. they had lost their plasmids. This result demonstrates that the HOcs in the plasmids were cut with high efficiency. The 5-FOA resistant colonies were then tested for leucine prototrophy, to identify LEU2 fragment insertions, and individual colonies were analyzed by PCR amplification of the genomic region surrounding the chromosomal DSB at the URA3::ACT-i::HOcs locus.
Table 1.
Quantitative analysis of epair events after a DSB at the URA3 cassette locus
| Plasmid | wt (HOcs) | Rad52 | rad51 | ku80 | 2inv HOcs | 2dir HOcs | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FOAR cells (10−4) | Leu+ among FOAR cells | FOAR cells (10−4) | Leu+ among FOAR cells | FOAR cells (10−4) | Leu+ among FOAR cells | FOAR cells (10−4) | Leu+ among FOAR cells | FOAR cells (10−4) | Leu+ among FOAR cells | FOAR cells (10−4) | Leu+ among FOAR cells | |
| ‘Direct’ | 2.05 | 94% | 0.15* | 7% | 0.49* | 64% | nd | – | 0.15* | 34% | 3.11 | 84% |
| ‘Inverted’ | 0.18* | 6% | 0.09* | nd | 0.17* | 7% | nd | – | 0.12* | 26% | 0.74* | 12% |
| ‘Inverted fit’ | – | – | – | – | – | – | – | – | 1.84 | 99% | 1.12 | 30% |
| ‘No HOcs’ | 0.31 | nd | 0.06 | nd | 0.24 | nd | nd | – | 0.30 | nd | 0.26 | nd |
Note: nd: not detected (<10−9). ‘–’ not applicable.
*Significantly different from the frequency obtained for the strain with a single HOcs and the ‘Direct’ plasmid (P < 0.001).
In wild-type cells carrying a plasmid that released a linear ‘Direct’ LEU2 fragment, the frequency of 5-FOAR survivors per plated cell was 2.05 × 10−4 and 94% of the 5-FOAR cells carried a LEU2 insertion (Table 1). This is in marked contrast to cells carrying the linear fragment with inverted termini. In this configuration, the frequency of Leu+ colonies per plated cell was 0.01 × 10−4, a 200-fold difference (Table 1). These results demonstrate that a linear fragment present in the cells can be captured and used to repair a broken chromosome, but that the termini of the linear fragments affect the frequency of recoverable insertional repair. As expected, in cells carrying the plasmid lacking HO cut sites, no LEU2 insertions were detected among the FOA resistant survivors (less than 10−9 Leu+ colonies per plated cell).
We carried out PCR analysis of the junctions of 52 insertion events. All ‘Direct’ linear fragments were inserted in the same orientation relative to the target locus (URA3 gene). This is the orientation expected from events in which the complementary ssDNA ends can undergo annealing (Figure 1A). This bias strongly suggests that annealing of the compatible ends plays an important role in directing the capturing of the linear fragment. We sequenced 10 of the insertions, and found that in 9 out of 10 survivors both ends of the inserted fragment exhibited an addition of 2 bp (+GT) (Figure 2A). Addition of GT appears to be the predominant modification of the HOcs that prevents the site from being re-cut by the HO endonuclease. The GT addition was detected in previous studies (9), and in more than 100 other sequences analyzed in this work (see further data later).
Figure 2.
Sequence of the repaired DSBs at the URA3::ACT1-i::HOcs locus. (A) DNA sequences of independent 5-FOAR, Leu+ colonies of a wild type strain. The HOcs at the URA3 cassette is colored red, the HOcs from the linear LEU2 fragments is colored black. The new sequences at the insertions are colored blue. The insertions are shown in bold. (B) Events obtained in the rad52 genetic background. (C) Events obtained in the rad51 genetic background.
Sequence analysis of the junctions of three insertion events obtained with the ‘Inverted’ plasmid revealed simple end-joining events at one terminus with addition of 2 bp (+GT) while the other terminus exhibited a complex event (Figure 1B). In one case the terminal region of the LEU2 fragment was deleted, and in one case a genomic sequence (part of the RNR1 gene) was inserted at the deleted end including the downstream URA3 gene. These results suggest that repair of a chromosomal end where simple end-joining cannot occur is associated with degradation of either the linear fragment, the chromosomal HOcs and associated URA3 sequences or both. As these results suggested that degradation of the ‘Inverted’ fragment could in many cases result in insertion of a partially deleted LEU2 gene, which could produce a Leu- phenotype, we analyzed 20 independent 5-FOA resistant Ura– Leu– colonies obtained from the strain with the ‘Inverted’ plasmid, by PCR analysis with primers on either side of the HOcs. In these cases, the repaired chromosome exhibited either Ty or mitochondrial sequences insertions, or deletion of the HO cut site (11). No truncated LEU2 fragment was detected suggesting that mechanistically, end-joining precedes degradation, since the compatible ends of the same ‘Inverted’ fragment are not degraded.
Genetic control of fragment capture
To investigate whether the homologous recombination repair system affects the ability of cells to assimilate linear fragments, we carried out similar experiments in cells deleted either for the RAD51 or for the RAD52 gene. RAD51 encodes a RecA homolog, and is known to be involved in strand exchange. In contrast, the function of Rad52 is less understood; it is, however, an essential component of the HR pathway and its inactivation affects most forms of recombination (4).
Rad52 cells carrying the linear fragment with direct or inverted termini exhibited a 2–4-fold decrease in viability and a frequency of FOAR colonies per plated cell that was 10–20-fold lower than the wild type (Table 1). Furthermore, only 7% of the FOA resistant surviving rad52 cells with the ‘Direct’ fragment carried LEU2 insertions (0.011 × 10−4 Leu+ colonies per plated cell, Table 1). This represents a 183-fold reduction in the frequency of LEU2 DNA capture events. None of the rad52 cells carrying the plasmid with the inverted configuration had a LEU2 insertion (Table 1). These results suggest that Rad52p plays a central role in the capture of linear fragments, despite the lack of extensive homology between the chromosomal ends and the captured fragment.
We carried out an analysis of the junctions of 13 insertion events obtained from rad52 colonies carrying the ‘Direct’ plasmid. In 12 out of the 13 samples the ‘Direct’ linear fragment was inserted in the same orientation to the target locus (URA3 gene).
We sequenced both junctions of 11 insertion events obtained from rad52 colonies carrying the ‘Direct’ plasmid. All survivors carried mutations in the upstream and downstream HO cut sites. However, in contrast to wild-type cells, only half of the junctions exhibited the common GT insertion. Additional junctions included insertion of TT, TGT or a simple G (Figure 2B). In the only case in which the ‘Direct’ linear fragment was inserted in the opposite orientation to the target locus, sequencing revealed a complex event: at the upstream HOcs, the terminal 3′ T nucleotide of the chromosomal HOcs was deleted, and ligated to the HOcs of the LEU2 fragment that lost its 6 terminal nucleotides. In addition, a 50-bp deletion of the HOcs of the LEU2 fragment and 43-bp deletion of the chromosomal HOcs sequences were seen at the downstream HOcs (Figure 2B).
Rad51 cells carrying the ‘Direct’ plasmid exhibited a 5-fold lower frequency of FOAR colonies per plated cell, compared to the wild type (Table 1). Furthermore, a 6-fold decrease in the frequency of cells that repaired the chromosomal break by capturing the ‘Direct’ LEU2 fragment was observed (0.31 × 10−4 Leu+ colonies per plated cell). In contrast, the number of FOA resistant rad51 cells carrying the ‘Inverted’ fragment that captured the LEU2 fragment was similar to that of the wild-type control (0.011 × 10−4 Leu+ colonies per plated cell).
Junction analysis revealed that in all samples the ‘Direct’ linear fragment was inserted in the same orientation to the target locus (URA3 gene). These results, similar to those observed in wild type and rad52 cells, suggest again that the cohesive orientation is preferred, and that this preference is independent of the main HR proteins. Sequence analysis revealed a pattern very similar to that observed in wild-type cells: GT or TGT additions at both ends for the ‘Direct’ insertions, and GT addition on one end and rearrangement in the other for the ‘Inverted’ fragment (Figure 2C).
These results show that, in contrast to the results obtained in the rad52 strain, lack of RAD51 activity lowers the efficiency of capture in the ‘Direct’ orientation, without affecting the mechanism of capture in the ‘Inverted’ orientation.
Yku80 is part of the Ku heterodimer (composed of Yku70 and Yku80 in yeast), required for accurate NHEJ. To investigate whether Ku plays a role in linear fragment assimilation, we repeated our experiments in yku80 cells. Survival of yku80 cells on galactose was severely impaired (1000-fold lower than wild type). We were unable to recover any survivor carrying a LEU2 fragment. These results indicate the importance of the Ku complex for survival after a DSB when homologous recombination is not an option, and further, its apparently absolute requirement for the incorporation of linear fragments into the broken chromosome.
Repair of a DSB by insertion of a linear fragment lacking compatible ends
In the vast majority of events examined, the ‘Direct’ linear fragment was inserted into the target locus in the same orientation. This suggests that simple annealing of the ends plays a dominant role in the capture of our linear fragments. To test this possibility, we changed the genomic target locus to an HO cut site that is incompatible with the fragment termini. Two new yeast strains were created. The first (AHY51, URA3::ACT1-i::2inv HOcs) carries two HOcs in an inverted orientation relative to one another (hereafter named ‘2inv’ strain). The second strain (AHY52, URA3::ACT1-i::2dir HOcs) contains two HOcs in direct orientation (‘2dir’ strain). Upon transfer of cells to galactose-containing medium, the HO endonuclease cleaves both HOcs simultaneously, leaving non-complementary 3′-overhanging termini on chromosome V in the ‘2inv’ strain and complementary 4-base 3′-overhanging termini in the ‘2dir’ strain. The ‘2dir’ strain exhibited a survival frequency similar to that observed in cells with a single HOcs. In contrast, the ‘2inv’ strain exhibited a 24-fold lower survival frequency in the presence of either plasmid (Table 1). These results indicate that the presence of complementary ends at the DSB site on chromosome V is the major determinant of survival, despite the additional requirement for a mutation that prevents re-digestion by the HO endonuclease.
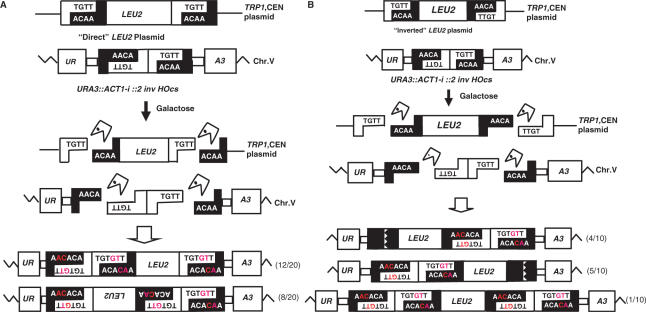
We carried out PCR analysis of insertion events of ‘Direct’ LEU2 fragments inserted at the 2inv locus, resulting in Leu+, 5-FOA resistant colonies. These were of particular interest, since cleavage of the HO cut sites on chromosome V should leave non-complementary ends, making the complementary overhang present at one end of the LEU2 fragment as the only likely joining molecule. The LEU2 fragments appeared in both orientations with respect to the URA3 target gene (12/20 insertions in the same orientation, and 8/20 insertions in the opposite one, Figure 3A). Surprisingly, all survivors, irrespective of the orientation of insertion, still carried two inverted HO cut sites upstream of LEU2 and a single HOcs downstream of the LEU2 insert. All the double HOcs analyzed carried inactivating mutations resulting from insertion of GT dinucleotides in each HOcs (Figure 3A). These results indicate that both HOcs at the chromosomal locus were cut and repaired, but that the 95 bp DNA fragment located between them was not lost or degraded. Our observations suggest a repair mechanism that keeps the broken ends together before the insertion of the linear fragment (see ‘Discussion’ section).
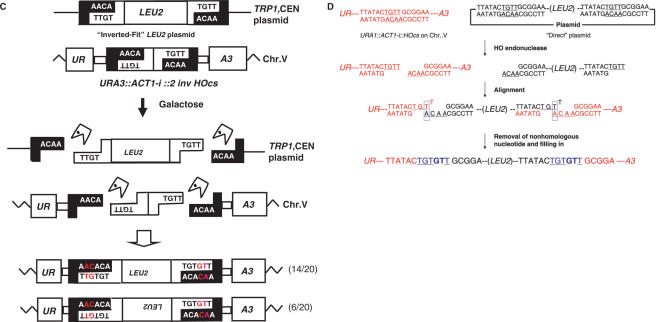
Figure 3.
Capture of linear LEU2 fragment into a DSB at the URA3::ACT1-i::2 inv HOcs locus. After HO endonuclease cleavage the termini of the broken chromosome V cannot anneal. (A) For the ‘Direct’ LEU2 fragment all samples carried two inverted HO cut sites with GT/AC insertions upstream of LEU2, and a single HOcs with a similar mutation downstream. (B) For the ‘Inverted’ LEU2 fragment all colonies analyzed carried two inverted HOcs exhibiting GT/AC insertions on one end of the fragment, and complex events on the other. (C) For the ‘Inverted fit’ LEU2 fragment, none of the events involved the use of the small inverted HOcs fragment. (D) Proposed mechanism of fragment capture (a single HOcs with a ‘Direct’ fragment are shown. The HO endonuclease cleaves both HO cut sites in the ‘Direct’ plasmid and the HOcs at the URA3 cassette, resulting in 4-bp 3′ overlapping sequences. Annealing between the terminal Adenine of the bottom strand and one T nucleotide prior to the terminal in the top strand, followed by DNA synthesis and ligation creates a GT insertion. DNA synthesis requires the removal of the terminal 3′ T nucleotide from the top strand.
For cells with the 2inv locus carrying the linear ‘Inverted’ LEU2 fragment, no complementary ends from the LEU2 fragment should be present to be captured at the DSB site, so insertions should require a complex NHEJ event at each end. In all samples analyzed (10 independent colonies) the linear fragment was inserted in the same orientation as the target locus. Sequence analysis revealed that the two inverted HOcs were retained either upstream (4/10 insertions) or downstream (5/10 insertions) of the LEU2 insert, mutated by GT/AC additions and in one case (1/10 insertion) the two inverted HOcs were retained in both upstream and downstream of the LEU2 insert. In all cases examined, the single HOcs at the other end of the insert contained a partial deletion (Figure 3B). Thus, as with the ‘Direct’ linear LEU2 fragment (Figure 3A) the chromosomal DSB is repaired by using two linear fragments, one released from the double-HOcs digestion, and a second one released from the cuts in the donor plasmid.
The ‘Inverted +2 invHOcs’ plasmid
The results presented earlier suggest that the small fragments generated by the endonuclease are utilized for DNA capture at the nearby break. This could suggest that the small fragment remains somehow associated with the break. In order to test whereas this adaptor molecule acts only in cis or represent the capture of an additional linear molecule that can be supplied in trans, we created a new plasmid carrying the ‘Inverted’ linear donor, plus, at a second location in the plasmid, a copy of the 2invHOcs construct. Upon expression of the HO endonuclease, five different DSBs are created: one at the URA3 locus on chromosome V, and two pairs of breaks that release a linear LEU2 fragment and a linear ‘adaptor’ fragment.
The presence of the 2invHOcs construct on the plasmid increased the efficiency of fragment capture: the frequency of 5-FOA resistant colonies was 8 × 10−4 per plated cell, a 44-fold increase with respect to the ‘Inverted’ linear fragments. Moreover, the frequency of Leu+ colonies was similar to that observed with the ‘Direct’ linear fragment (1.44 × 10−4 per plated cell). An analysis of insertion events revealed that in all cases, DNA capture involved the use of the small HOcs-derived fragments. In 13 out of 20 samples analyzed the LEU2 fragment was oriented as the target locus, whereas in the seven remaining samples it was oriented in the opposite direction. In all cases analyzed, all HOcs carried GT insertions. These results demonstrate that a broken chromosomal end can efficiently capture two independent linear fragments.
Fragment capture occurs through annealing
The highest frequency of capture was observed in strains carrying a single DSB at the URA3::ACT-i:: HOcs locus and ‘Direct’ plasmids. We reasoned that if this is due to its ability to carry out simple annealing of both ends (provided there is an alteration of the terminal sequences during the joining process), then similar combinations of chromosomal breaks and linear ends should result in similar high levels of capture. We created an additional linear substrate, which we call ‘Inverted-Fit’ (Figure 3C), which can be inserted into the DSB at the URA3::ACT-i:: 2inv HOcs locus by simple annealing. A similar case is found in cells with the 2dir locus carrying ‘Direct’ LEU2 fragment.
As predicted, in both cases the frequency of 5-FOAR colonies was similar to that of cells with a single HOcs carrying the ‘Direct’ plasmid (Table 1). Similarly, nearly all (99 and 84%) of the cells able to grow on 5-FOA had an insertion of the LEU2 fragment. An analysis of Leu+ colonies of these strains showed that the small excised inverted HOcs pair fragment was not used during repair. As before, both junctions carried single HOcs with GT insertions (Figure 3C and data not shown).
Thus, the small fragment containing the inverted HOcs was retained in cases where direct annealing between the linear ends was not possible, but was not utilized when annealing was readily carried out. These results once more point to annealing between complementary base pairs as a crucial step in the capture of linear fragments.
DISCUSSION
When a chromosome is broken, cells must repair the two pieces to reconstitute the genome's integrity. Extrachromosomal DNA has been found at the repaired junctions in several organisms, including yeast (9–11,18), plants (17) and mammalian cells (15, 25–28). In the present study, we have analyzed the mechanisms of DNA capture by using a single genomic DSB and defined linear fragments as the DNA to be captured. The presence of a linear LEU2 fragment (presumably in every cell of the population) enhanced the frequency of capture, but only when the ends were compatible with an annealing mechanism (see later). In cases in which at least one end had to undergo extensive processing, such as cells carrying the ‘Inverted’ plasmid, the percent of Leu+ colonies among those that contained an insert was relatively low. This suggests that DNA molecules that can serve as filler DNA are quite abundant, and it is end-processing that limits their utilization. Analysis of randomly picked 5FOAR Leu– colonies confirmed previous work that showed that Ty cDNA and random mitochondrial fragments constitute the main classes of ‘filler DNA’ in yeast (9,11,18).
Modification of the HOcs sequences
Insertion of GT appears to be the main HOcs modification that prevents the site from being recut by HO endonuclease. The GT insertion was detected in previous studies (9), and in more than 100 sequences analyzed in this work (in both wild type, rad51 and rad52 strains). A potential mechanism for this type of repair was previously proposed by Moore and Haber (9). In the case of a ‘Direct’ fragment, for example, the termini generated at the chromosome are complementary to those of the linear fragment. Simple annealing of such sequences results in the re-creation of HOcs that will be cut again by the enzyme. However, annealing misalignment of the terminal A nucleotide of the bottom strand with the penultimate T nucleotide on the top strand, followed by removal of the terminal 3′ T nucleotide in the top strand, DNA synthesis and ligation, results in a repair event that adds two nucleotides and is resistant to cleavage by HO (Figure 3D). In five cases (1 in wild type, 2 in rad52 and 2 in rad51) there was an insertion of 3 bp (+TGT) at the upstream or downstream HOcs, which can be most simply explained by annealing of the terminal nucleotides (terminal A and terminal T). Although the repair mechanism for the TGT insertion is simpler than that of the GT insertion (there is no need for removal of the terminal nucleotide) this insertion mutation was rare (5 out of >100 sequences in this work, and 1 out of 45 candidates in Moore and Haber (9). We have at present no explanation for this preference for the GT insertion over the TGT insertion, but it may imply that the intermediate shown in Figure 3D represents a particularly favorable conformation of ends for proteins such as the Ku heterodimer and the MRX complex to hold the broken chromosomes together and initiate the processing events that lead to an imprecise rejoining.
Annealing of homologous ssDNA sequences plays a role in fragment capture
All the results obtained in the present study point to the critical role played by annealing of the complementary ssDNA in fragment capture.
The frequency of insertion events differed greatly (200-fold) between wild-type cells carrying a plasmid that released a linear fragment with two complementary termini, compared to cells carrying a linear fragment with only a single complementary terminus (Table 1). In all cases analyzed (52 out of 52 cases), ‘Direct’ fragment insertion was carried out through simple complementary annealing/ligation. This lead to insertion in a preferred orientation. In contrast, ‘Inverted’ fragments are inserted by simple annealing of one terminus but the other end must be joined by NHEJ without annealing. Sequencing analysis of insertion events revealed that in all cases there were simple imprecise end-joining events at one terminus, whereas the other end exhibited a partial deletion event.
The cell can capture more then one fragment in order to repair a DSB
The excised small fragment generated by the double HO digestion, carrying complementary single-stranded DNA, was used for the repair of the DSB, but only if the LEU2 fragment did not end in compatible ends. For example in the case of the 2inv allele repaired by capturing the ‘Direct’ fragment, the two contiguous HOcs (inactivated by GT mutation) were always upstream of the LEU2 gene irrespective of its orientation relative to the 2inv allele, presumably because the upstream end in the LEU2 fragment could not anneal to any DSB end in 2inv (Figure 3A). Similarly, all the capture events involving the ‘Inverted + 2invHOcs’ plasmid carried double HOcs, however, this time the orientation of the LEU2 fragment was random, as expected from the fact that either end was equally likely to anneal to the broken chromosomal ends. In contrast, the small ‘adaptor’ fragment was lost in all cases in which the LEU2 linear molecule ended in compatible ends able to anneal (e.g. 2dir with the ‘Direct’ fragment or 2inv with the ‘Inverted-Fit’ fragment).
We showed that this small linear fragment could be supplied either in cis or in trans. Moreover, capture events involving the LEU2 fragment and either chromosomal or mitochondrial DNA as ‘adaptor molecules’ were also detected (Figure 2). DNA insertion events involving capture of more than one fragment were described in several studies (10, 11, 15). In a previous study of repair of the URA3::ACT1-i:: HOcs Yu et al. (11) found that in 3 out of 21 clones containing Ty1 cDNA sequences, two discontinuous fragments were inserted.
Capture is dependent on the Ku heterodimer and partially on HR
Survival of yku80 strains that underwent a DSB was extremely low, and we were unable to recover any survivor carrying a LEU2 fragment, implying that Ku-cells are incapable of incorporating extrachromosomal linear fragments into their chromosomes. Previous work by several laboratories has shown a role for Ku in the repair of DSBs in regions of the genome-lacking homology. However, the magnitude of the effect was smaller, and Ku-independent events were readily detected (12,29). This difference may be due in part to the absence, in our system, of flanking sequences that could allow repair by a pathway that has been named Microhomology-Mediated End Joining [MMEJ (30)]. This mechanism strongly depends on the MRX complex and on the Rad1/Rad10 endonuclease, but is independent of the Ku heterodimer (30). It seems that capture of linear fragments into a chromosomal DSB is particularly sensitive to defects in the NHEJ machinery, possibly because of its role in fragment stabilization. In contrast, mutations in the HR machinery had relatively mild effects on survival.
Deletion of the RAD52 gene, which abolishes most types of homologous recombination events, reduced the efficiency of capture of the ‘Direct’ fragment into a DSB by a factor of 180-fold and capture of the ‘Inverted’ fragment was below detection (Table 1). This suggests a role for Rad52p in capturing the linear fragment, probably by affecting either interaction with processed DNA ends necessary for capture, or the annealing efficiency. Rad52p has been shown to be able to promote strand annealing in vitro (31). Since sequence homology is extremely short, and repair seems to depend on both Ku and Rad52 activity the NHEJ and HR machineries may be required to cooperate in our system. Similar results have been observed in other systems (32,33). Notably, deletion of RAD51, the yeast RecA homolog, had a relatively mild effect on the capture of ‘Direct’ fragments, and no effect on the ‘Inverted’ system. These results are consistent with a repair mechanism that relays on annealing rather than strand invasion. Our results show that the use of homology, even a single base pair, is preferred over a mechanism that involves more complex processing of non-homologous DNA ends.
It is notable that the bias in orientation observed for integration of the ‘Direct’ linear fragment, which stresses the role of annealing in directing integration, is conserved in rad52 and rad51 strains. Thus, although Rad52 can promote annealing of homologous sequences in vivo, it is not completely essential for this step. In contrast, all capture is abolished in the absence of NHEJ.
Whereas annealing could drive insertion of complementary sequences, non-complementary 3′ overhangs should be first aligned by a DNA-binding protein to permit fill-in DNA synthesis on the opposite gapped strand and ligation. Sandoval and Labhart, using Ku-immunodepleted Xenopus egg extracts, showed that indeed the Ku heterodimer plays a role in the joining of fragments with overhangs. Moreover, overhangs carrying A:T base pairs, as in our system, were more affected by Ku depletion that those carrying G:C base pairs, suggesting that the requirement for Ku was dependent on the stability of the two cohesive DNA ends (34). Additional studies, using other eukaryotic systems, corroborate the role of Ku in stabilizing pairing overhangs (35).
The mechanism of linear fragment capture
Does the end-modification take place before or during the capture mechanism?
Sequencing of PCR products created by inverse PCR on the self-annealed ‘Direct’ fragment did not reveal any change in sequence. This suggests that before integration into the chromosomes, the ends remain unmodified. Modification is more likely to take place during capture, and not before.
Our results suggest that capture of a linear DNA fragment is a multi-step process that relies on one hand on the ability of the linear fragment to anneal to the broken ends, in a process that may require Rad52p but not Rad51p, and on the other hand on the NHEJ machinery, to join linear fragments. We therefore propose a model in which linear fragments are first synapsed with the broken ends by a process that involves the Rad52 homology recognition pathway. This process, which is akin to the homology recognition step of the single-strand annealing (SSA) mechanism (36), is Rad51-independent, as no strand invasion is necessary. In addition to its role in bringing the fragments together, Rad52 may play a role in catalyzing their annealing, similar to the one observed in vitro reactions (31). As only non-precise end-joining events can give rise to viable 5-FOAR colonies, the ends must be modified to introduce mutations that inactivate the chromosomal HOcs. This process is dependent on Ku, and probably on other members of the NHEJ machinery, and is likely to take place after a first recognition/annealing step has taken place. A local disengagement allows annealing of the penultimate chromosomal T residue with the internal adenine of the incoming fragment, as described in Figure 3D. An interesting possibility is that this repair event takes place only once, and is then copied during the repair of the second DSB. This could explain the high efficiency of repair of events containing several ends, all of which have to be modified, and the fact that usually the same mutation is found in the upstream and downstream HOcs. Notably, such a copying mechanism should be independent of Rad51p (as clearly demonstrated by the isolation of a capture event carrying identical copies of a mitochondrial sequences at both ends of the LEU2 insert) although it is apparently partially Rad52 dependent: about half of the capture events in rad52 strains carried different insertion mutations at the upstream and downstream HOcs. The Ku heterodimer and Rad52 may collaborate in this complex event that requires the recognition of homology and end modification.
Concluding remarks
Integration of linear fragments can have important genetic consequences, changing the genome in ways that may affect the future evolution of the organism. Genetic information from mitochondria has migrated to the nucleus of eukaryotic cells by a mechanism akin to the one we have described here (10). Similarly, linear DNA fragments of endogenous or exogenous origin can be incorporated into the genome to generate gene sequence duplications or to import useful traits from other organisms during horizontal transmission. On an evolutionary scale, these are events that create new opportunities for adaptation and speciation (29,37,38). We have shown that fragment capture is a complex event that involves both the NHEJ and HR machineries, and may utilize not only abundantly found DNA fragments, such as mitochondrial DNA or Ty cDNA, but also single copy sequences.
ACKNOWLEDGEMENTS
This work was supported from grants by the Israeli Science Foundation and the Israeli Ministry of Health to M.K. and from National Institute of General Medical Sciences, the American Cancer Society and the New Jersey Commission for Cancer Research to A.G. We thank all members of the Kupiec and Gabriel laboratory for encouragement and comments on the manuscript. Funding to pay the Open Access publication charges for this article was provided by the Israeli Science Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kupiec M. Damage-induced recombination in the yeast Saccharomyces cerevisiae. Mutat. Res. 2000;451:91–105. doi: 10.1016/s0027-5107(00)00042-7. [DOI] [PubMed] [Google Scholar]
- 2.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kupiec M, Steinlauf R. Damage-induced ectopic recombination in the yeast Saccharomyces cerevisiae. Mutat. Res. 1997;384:33–44. doi: 10.1016/s0921-8777(97)00012-8. [DOI] [PubMed] [Google Scholar]
- 4.Krogh BO, Symington LS. Recombination proteins in yeast. Annu. Rev. Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 5.Pastwa E, Blasiak J. Non-homologous DNA end joining. Acta Biochim. Pol. 2003;50:891–908. [PubMed] [Google Scholar]
- 6.Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat. Rev. Mol. Cell Biol. 2003;4:712–720. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- 7.Cahill D, Connor B, Carney JP. Mechanisms of eukaryotic DNA double strand break repair. Front Biosci. 2006;11:1958–1976. doi: 10.2741/1938. [DOI] [PubMed] [Google Scholar]
- 8.Gorbunova VV, Levy AA. How plants make ends meet: DNA double-strand break repair. Trends Plant Sci. 1999;4:263–269. doi: 10.1016/s1360-1385(99)01430-2. [DOI] [PubMed] [Google Scholar]
- 9.Moore JK, Haber JE. Capture of retrotransposon DNA at the sites of chromosomal double-strand breaks. Nature. 1996;383:644–646. doi: 10.1038/383644a0. [DOI] [PubMed] [Google Scholar]
- 10.Ricchetti M, Fairhead C, Dujon B. Mitochondrial DNA repairs double-strand breaks in yeast chromosomes. Nature. 1999;402:96–100. doi: 10.1038/47076. [DOI] [PubMed] [Google Scholar]
- 11.Yu X, Gabriel A. Patching broken chromosomes with extranuclear cellular DNA. Mol. Cell. 1999;4:873–881. doi: 10.1016/s1097-2765(00)80397-4. [DOI] [PubMed] [Google Scholar]
- 12.Yu X, Gabriel A. Ku-dependent and Ku-independent end-joining pathways lead to chromosomal rearrangements during double-strand break repair in Saccharomyces cerevisiae. Genetics. 2003;163:843–856. doi: 10.1093/genetics/163.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu X, Gabriel A. Reciprocal translocations in Saccharomyces cerevisiae formed by nonhomologous end joining. Genetics. 2004;166:741–751. doi: 10.1534/genetics.166.2.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roth DB, Chang XB, Wilson JH. Comparison of filler DNA at immune, nonimmune, and oncogenic rearrangements suggests multiple mechanisms of formation. Mol. Cell. Biol. 1989;9:3049–3057. doi: 10.1128/mcb.9.7.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin Y, Waldman AS. Promiscuous patching of broken chromosomes in mammalian cells with extrachromosomal DNA. Nucleic Acids Res. 2001;29:3975–3981. doi: 10.1093/nar/29.19.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorbunova V, Levy AA. Non-homologous DNA end joining in plant cells is associated with deletions and filler DNA insertions. Nucleic Acids Res. 1997;25:4650–4657. doi: 10.1093/nar/25.22.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salomon S, Puchta H. Capture of genomic and T-DNA sequences during double-strand break repair in somatic plant cells. Embo J. 1998;17:6086–6095. doi: 10.1093/emboj/17.20.6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teng SC, Kim B, Gabriel A. Retrotransposon reverse-transcriptase-mediated repair of chromosomal breaks. Nature. 1996;383:641–644. doi: 10.1038/383641a0. [DOI] [PubMed] [Google Scholar]
- 19.Paques F, Leung WY, Haber JE. Expansions and contractions in a tandem repeat induced by double-strand break repair. Mol. Cell. Biol. 1998;18:2045–2054. doi: 10.1128/mcb.18.4.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Negritto MT, Wu X, Kuo T, Chu S, Bailis AM. Influence of DNA sequence identity on efficiency of targeted gene replacement. Mol. Cell Biol. 1997;17:278–286. doi: 10.1128/mcb.17.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sherman F, Fink GR, Hicks JB. Methods in Yeast Genetics: A Laboratory Manual. Cold spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- 23.Boeke JD, LaCroute F, Fink GR. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 24.Hoffman CS, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 25.Sargent RG, Brenneman MA, Wilson JH. Repair of site-specific double-strand breaks in a mammalian chromosome by homologous and illegitimate recombination. Mol. Cell. Biol. 1997;17:267–277. doi: 10.1128/mcb.17.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang F, Han M, Romanienko PJ, Jasin M. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc. Natl Acad. Sci. USA. 1998;95:5172–5177. doi: 10.1073/pnas.95.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin Y, Waldman AS. Capture of DNA sequences at double-strand breaks in mammalian chromosomes. Genetics. 2001;158:1665–1674. doi: 10.1093/genetics/158.4.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Varga T, Aplan PD. Chromosomal aberrations induced by double strand DNA breaks. DNA Repair (Amst.) 2005;4:1038–1046. doi: 10.1016/j.dnarep.2005.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bergthorsson U, Adams KL, Thomason B, Palmer JD. Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature. 2003;424:197–201. doi: 10.1038/nature01743. [DOI] [PubMed] [Google Scholar]
- 30.Ma JL, Kim EM, Haber JE, Lee SE. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol. Cell. Biol. 2003;23:8820–8828. doi: 10.1128/MCB.23.23.8820-8828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mortensen UH, Bendixen C, Sunjevaric I, Rothstein R. DNA strand annealing is promoted by the yeast Rad52 protein. Proc. Natl Acad. Sci. USA. 1996;93:10729–10734. doi: 10.1073/pnas.93.20.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rapp A, Greulich KO. After double-strand break induction by UV-A, homologous recombination and nonhomologous end joining cooperate at the same DSB if both systems are available. J. Cell Sci. 2004;117:4935–4945. doi: 10.1242/jcs.01355. [DOI] [PubMed] [Google Scholar]
- 33.Couedel C, Mills KD, Barchi M, Shen L, Olshen A, Johnson RD, Nussenzweig A, Essers J, Kanaar R, et al. Collaboration of homologous recombination and nonhomologous end-joining factors for the survival and integrity of mice and cells. Genes Dev. 2004;18:1293–1304. doi: 10.1101/gad.1209204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sandoval A, Labhart P. High G/C content of cohesive overhangs renders DNA end joining Ku-independent. DNA Repair (Amst.) 2004;3:13–21. doi: 10.1016/j.dnarep.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 35.Feldmann E, Schmiemann V, Goedecke W, Reichenberger S, Pfeiffer P. DNA double-strand break repair in cell-free extracts from Ku80-deficient cells: implications for Ku serving as an alignment factor in non-homologous DNA end joining. Nucleic Acids Res. 2000;28:2585–2596. doi: 10.1093/nar/28.13.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ivanov EL, Sugawara N, Fishman-Lobell J, Haber JE. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics. 1996;142:693–704. doi: 10.1093/genetics/142.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, Lafontaine I, De Montigny J, Marck C, Neuveglise C, et al. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- 38.Lynch M, Conery JS. The evolutionary demography of duplicate genes. J. Struct. Funct. Genomics. 2003;3:35–44. [PubMed] [Google Scholar]