Abstract
Some selected amino acids, in particular l-aspartic acid (l-Asp) and l-histidine (l-His), can function as leaving group during polymerase-catalyzed incorporation of deoxyadenosine monophosphate (dAMP) in DNA. Although l-Asp-dAMP and l-His-dAMP bind, most probably, in a different way in the active site of the enzyme, aspartic acid and histidine can be considered as mimics of the pyrophosphate moiety of deoxyadenosine triphosphate. l-Aspartic acid is more efficient than d-aspartic acid as leaving group. Such P-N conjugates of amino acids and deoxynucleotides provide a novel experimental ground for diversifying nucleic acid metabolism in the field of synthetic biology.
INTRODUCTION
DNA and RNA molecules naturally occurring in living cells derive from nucleoside triphosphates, through an iterative catalytic process condensing the nucleoside 5′-phosphate moiety of these activated substrates to the 3′-hydroxyl moiety of the elongating strand and releasing pyrophosphate. A crucial aspect of this polymerization process is that the pyrophosphate leaving group undergoes hydrolysis under the action of pyrophosphatase, the essential enzyme in charge of the irreversibility of macromolecular biosynthesis (1). The energetics of protein synthesis is also based on the release and subsequent destruction of pyrophosphate through the transient formation of amino acyladenylates by amino acyl-tRNA synthetases (2). As an anhydride made up of two identical phosphoryl moieties, pyrophosphate is endowed with a compositional simplicity that provides the basis of straightforward recycling processes for reconstituting the pools of RNA and DNA precursors through P-O bond rearrangements catalyzed by nucleotide kinases and nucleoside diphosphate kinase (1). Altogether, cells have evolved an efficient network of enzymatic phospho-transfers so as to reload phosphoanhydride bonds in nucleotides using the potential energy of activated intermediates in metabolism, e.g. phosphoenolpyruvate, or using the chemo-osmotic potential of the cell membrane proton gradient (2). This coupling between energy storage and nucleic acid polymerization through phosphoanhydride formation and pyrophosphate release lies at the core of cells chemical, energetic and genetic design. If, for synthetic biology purposes, one attempted to diversify enzymatic polymerization of nucleic acids in vivo by condensing an additional category of activated nucleotides bearing no pyrophosphate leaving group, the design of such precursors would have to integrate features similar to those embodied in the economy and simplicity of phosphoanhydride metabolism. As a first step toward in vivo experiments along these lines, we set out to explore in vitro metabolic prototypes of activated DNA precursors such that: (i) they would serve as substrate for a DNA polymerase; (ii) their consumption in DNA biosynthesis would release as leaving group a metabolite common in cells; (iii) the leaving group could be actively degraded or recycled so as to enforce irreversibility of polymerization and (iv) the leaving group could facilitate uptake of activated DNA precursors through cell membranes.
Few successful attempts have been reported in the literature to substitute functionally the pyrophosphate leaving group of nucleoside triphosphates in nucleic acid polymerization. Phosphorimidazolides and their 2-methyl derivatives have been the topic of a systematic investigation of non-enzymatic polymerization of canonical and non-canonical nucleotides in prebiotic studies (3). Systematic substitution at the beta and gamma positions of deoxynucleoside triphosphates has been studied by Krayevsky and collaborators (4). Remarkably, dTTP analogues bearing bulky hydrophobic groups at the gamma position of dNTPs were shown to undergo polymerization catalyzed by HIV reverse transcriptase and other viral and cellular DNA polymerases (5). More recently, deoxynucleoside triphosphate analogues with a P-C-P distal bridge replacing the P-O-P phosphoanhydride have also been demonstrated to undergo condensation by DNA polymerase beta (6). Among conceivable P-O, P-S, P-C and P-N conjugates of deoxynucleoside monophosphates with physiological metabolites, phosphoramidate conjugates with amino acids (alias dNAP) seemed particularly worthy of interest because their structure lends itself to activation by the catalytic Mg2+ of DNA polymerases, much as deoxynucleoside triphosphates (7). Their condensation by polymerases is designed to release common amino acids, which can be recycled or destroyed in subsequent metabolic steps. In addition, they offer an easy synthetic access and a high enough stability toward spontaneous hydrolysis under physiological conditions (37°C, pH = 7.5).
The second aim of this research project is to bypass the kinase pathway for intracellular activation of modified nucleosides. Nucleoside reverse transcriptase inhibitors (NRTI) are designed to be recognized as substrates for RT and incorporated into a growing strand for further termination of chain elongation (8). Inhibition of reverse transcriptase activity and chain termination by NRTI's is achieved by introduction of structural modifications in the sugar moiety. These RT inhibitors are usually administered as biologically inactive free nucleosides or nucleoside phosphonates, or as nucleoside monophosphate/phosphonate prodrugs where the phosphate moiety is masked with a lipophilic group (9). In the case of nucleoside administration, three steps of kinase-mediated activation are needed to generate the triphosphate in the cell. In the prodrug concept, nucleoside 5′-monophosphate kinase and nucleoside 5′-diphosphate kinase activities are needed to provide the biological active congener (10). The efficiency of these enzymatic phosphorylation reactions depends on the substrate specificity of the different kinases. A nucleotide analogue that would not depend on activation by nucleoside/nucleotide kinases whilst serving as a natural substrate mimic, would be of great interest.
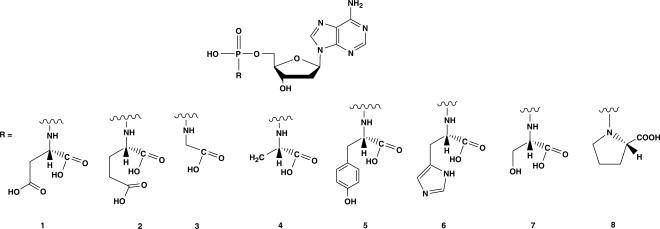
Here, we present a series of amino acids phosphoramidate analogues in which a natural l-amino acid moiety is linked through a P-N bond to 2′-deoxyadenosine 5′-O-monophosphate (Figure 1) and which might serve as potential leaving group in a nucleotidyl transfer reaction. Especially, the 5′-aspartyl-phosphoramidate and the 5′-histidyl-phosphoramidate can mimic a natural triphosphate moiety quite effectively for the incorporation of 2′-deoxyadenosine into a growing DNA strand by HIV RT and Therminator DNA polymerase. As demonstrated by modelling studies, the amino acid moiety of this deoxynucleotide analogue provides structural and electrostatic features essential for salt formation and/or metal coordination and assembly of the catalytic residues in the polymerase active site. In this respect, this study might contribute to a better understanding of the mechanism of biological polymerization reactions.
Figure 1.
Structure of l-Amino Acid Phosphoramidates used in this study.
RESULTS
Synthesis of amino acid phosphoramidates
The synthesis of methyl ester amino acid phosphoramidate nucleotides analogues was accomplished according to the method described by Wagner and colleagues starting from nucleoside monophosphate (11). The details of the synthesis of the compounds used in the study have been described in a preliminary communication of this work (7). l-amino acids were used for synthesis of the first series of phosphoramidate analogues. The deprotection of the amino acid moiety was carried out with 0.4 M sodium hydroxide in methanol-water solution. The series of phosphoramidate analogues coupled to a variety of natural l-amino acids, synthesized for the study, is shown in Figure 1.
Single nucleotide incorporation by HIV RT
HIV reverse transcriptase is involved in copying of the HIV genome and uses deoxynucleotides as substrates. HIV RT is an error-prone polymerase and has high mutation rate (12). The essential role of the HIV RT in viral replication and its flexibility and tolerance toward modified nucleotides renders this enzyme a primary target in treatment of HIV infection. In the presented study, the ability of HIV RT to incorporate a series of amino acid phosphoramidate nucleotide analogues was investigated by the gel-based single nucleotide incorporation assay (13,14).
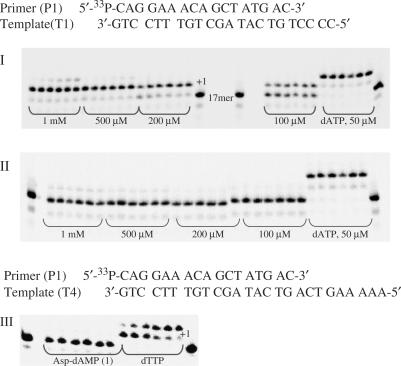
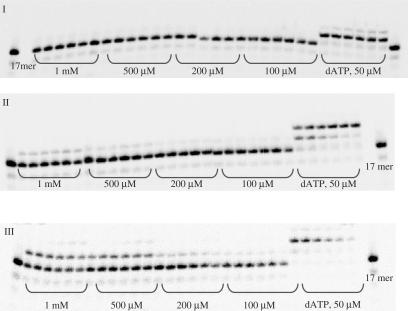
Among amino acid phosphoramidate nucleotides (1–8) remarkable results were observed with Asp-dAMP (1) (Figure 2).
Figure 2.
Panel I: incorporation of Asp-dAMP (1) by HIV RT. Panel II: incorporation of dimethyl ester Asp-dAMP (1a) by HIV RT. Panel III: control experiment for Asp-dAMP (1). Conditions. Panels I and II: primer (P1) was 5′-labelled with 33P followed by annealing to a template T1; 125 nM primer/template (P1/T1), [HIV RT] = 0.03 U/μl, time intervals: 5, 15, 30, 60, 90 and 120 min. Panel III: primer (P1) was 5′-labelled with 33P followed by annealing to a template T4; 125 nM primer/template (P1/T4), [L-Asp-dAMP] = 500 μM, [dTTP] = 10 μM, [HIV RT] = 0.03 U/μL, time intervals: 5, 15, 30, 60, 90 and 120 min.
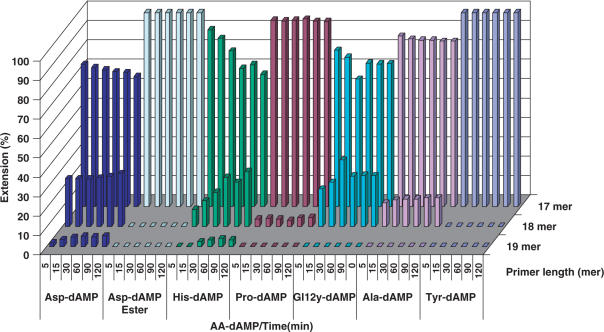
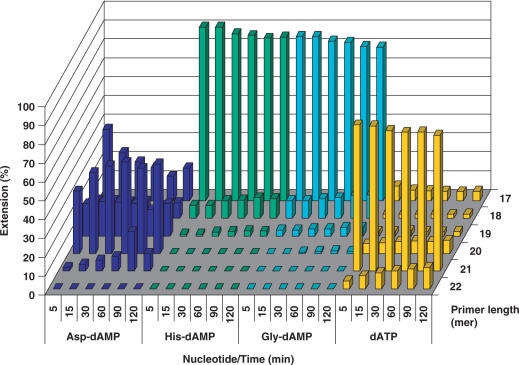
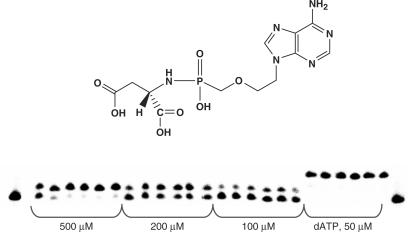
This phosphoramidate analogue was recognized by HIV RT and efficiently incorporated into a growing primer strand resulting in 90% conversion to an (n + 1) strand in 60 min (500 μM nucleotide concentration). At the same conditions, incorporation of His-dAMP(6), Gly-dAMP (3) and Pro-dAMP (7) were 1.5-, 6.5-, and 3.7-fold less efficient, respectively. Efficient incorporation of Asp-dAMP (24.1%) was also observed when the substrate concentration was decreased 10-fold. However, significantly lesser incorporation of amino acid phosphoramidate was detected for nucleotides coupled to non-polar, hydrophobic amino acids. Ala-dAMP and Tyr-dAMP behaved as poor substrates leading to merely 7- and 10-fold reduction in primer extension, respectively (Figure 3).
Figure 3.
Efficiency of phosphoramidate incorporation by HIV RT.
Interestingly, no incorporation occurred when respective methyl ester derivatives of 1–8 (dimethylester for 1 and 2 as also the carboxylic acid function in the side chain is methylated) were used as substrates in the polymerase reaction (Figure 3, Panel II). An unexpected result was observed with Glu-dAMP analogue (2) that also acted very poorly as an HIV RT substrate. These observations suggest that recognition and incorporation of amino acids (AA) dAMPs is a very specific process and is likely to be dictated by the chemical structure and electrostatics of the amino acid moiety.
Since it is known that polymerases have tendency to incorporate dAMP in a non-template manner, we investigated whether the former observations were due to a true base-pair extension. A control experiment with a mismatch sequence (A against A) was carried out (Figure 3, Panel III). As expected, Asp-dAMP (1) was not incorporated at all into the growing primer strand. After 2 h of the polymerase reaction at 500 μM substrate concentration 0% conversion was observed. The same results were observed when the substrate concentration was increased to 1 mM.
Single nucleotide incorporation by Therminator DNA polymerase
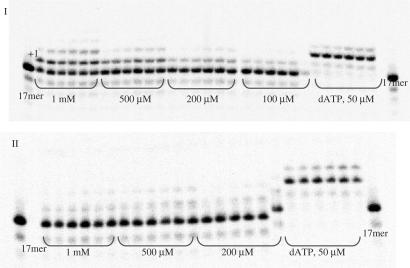
Another polymerase enzyme that demonstrates similar trends in recognition and utilization of AA-dAMPs is the thermostable Therminator DNA polymerase, a variant of (9°N−7) Thermococcus species DNA polymerase. This enzyme demonstrated effective recognition and incorporation of a number of nucleotides bearing unnatural nucleobase and sugar moieties (15–20). Likewise, probing of AA-dAMP incorporation directed by Therminator DNA polymerase revealed property of analogues 1, 3 and 6 to act effectively as alternative substrates in the DNA polymerization reaction (Figures 4 and 5).
Figure 4.
Panel I: incorporation of Asp-dAMP (1) by Therminator DNA pol. Panel II: incorporation of dimethyl ester Asp-dAMP by Therminator. Reaction conditions: primer (P1) was 5′-labelled with 33P followed by annealing to a template T1; 125 nM primer/template (P1/T1), [Therminator] = 0.03 U/μl, [primer] = 0.125 μM, time intervals: 5, 15, 30, 60, 90 and 120 min.
Figure 5.
Efficiency of phosphoramidate incorporation by Therminator DNA polymerase.
Yet again, the best results were obtained with Asp-dAMP, which led to 25.2% primer extension over 60 min at 500 μM nucleotide concentration. At the same conditions, similar results were obtained for Gly-dAMP and His-dAMP (26 and 25.4% primer extension, respectively). In the case of Glu-dAMP and methyl protected AA-dAMPs, Therminator DNA polymerase displays selectivity analogous to HIV reverse transcriptase and fails to direct incorporation of those phosphoramidate analogues (Figure 5).
Single nucleotide incorporation by other DNA polymerases
The remarkable property of Asp-dAMP encouraged further investigation and testing 1 as a substrate for other DNA polymerases. However, in the case of Taq, Vent (exo−), and KF (exo−) DNA polymerases, recognition and incorporation efficiency were significantly less appealing. Incorporation and primer extension were observed only in the case of KF (exo−) DNA pol demonstrating 32.5% conversion of the primer strand in 60 min. This is in contrast to Taq and Vent (exo−) DNA polymerases that failed to insert 1 into a growing primer strand. The diversity in incorporation selectivity that are observed among the polymerases [Therminator, Taq, Vent (exo−), KF (exo−) and HIV reverse transcriptase] could indicate the differences in the active site flexibility and tolerance to the triphosphate modifications (Figure 6). Although no data from human polymerases are yet available, the selectivity for HIV-RT points to the potential of this approach for the design of direct reverse transcriptase inhibitors as potential anti-HIV agents.
Figure 6.
Panel I: incorporation of Asp-dAMP (1) by Taq DNA polymerase. Panel II: incorporation of Asp-dAMP (1) by Vent (exo−) DNA polymerase. Panel III: Incorporation of Asp-dAMP (1) by Klenow Fragment (KF) (exo−) DNA polymerase. Reaction conditions: primer (P1) was 5′-labelled with 33P followed by annealing to a template T1; 125 nM primer/template (P1/T3), [Taq] = 0.005 U/μl, [Vent] = 0.005 U/μl, [KF] = 0.05 U/μl; time points: 5, 15, 30, 60, 90 and 120 min.
Primer extension by HIV RT
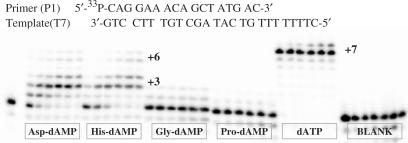
The further investigation of Asp-dAMP recognition by the reverse transcriptase focused on ability of HIV RT to direct template dependent incorporation of more than one phosphoramidate nucleotides. For this purpose, template T3 containing a string of three thymidine nucleobases flanked with cytidine nucleobases at the 3′ end and the template T7 that has an overhang of seven thymidine residues were used. Ability to HIV RT to synthesize a DNA sequence using phosphoramidate nucleotides as substrates was tested among Asp-dAMP, His-dAMP, Gly-dAMP and Pro-dAMP.
Among this series of phosphoramidate nucleotide, the most encouraging results using T3 template were observed with Asp-dAMP and His-dAMP which were used by HIV RT to extend a primer with three adenine nucleobases (n + 3 product). The results of these gel electrophoresis experiments are described in the preliminary communication (7).
However, after 60 min of the polymerase reaction the (n + 2) product predominates over the (n + 3) product (56.3% versus, 5.2% for Asp-dAMP and 67.1% versus 13.5%). Interestingly, efficiency of DNA synthesis with His-dAMP at 500 μM substrate concentration is similar or better to that when Asp-dAMP serves as the substrate (67.1% versus 56.2%, respectively, for the synthesis of the (n + 2) primer). This is in contrast to the single nucleotide incorporation results that indicate that His-dAMP is less good than Asp-dAMP as a substrate for HIV RT (Figure 7). Efficiency of chain elongation and single nucleotide incorporation is dependent on the structure of the ‘leaving group’.
Figure 7.
Extension efficiency of HIV reverse transcriptase.
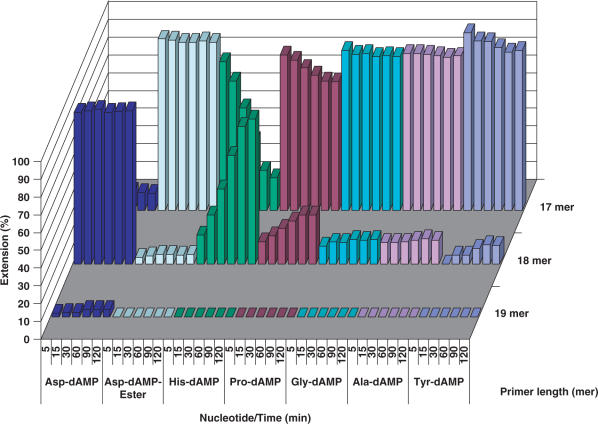
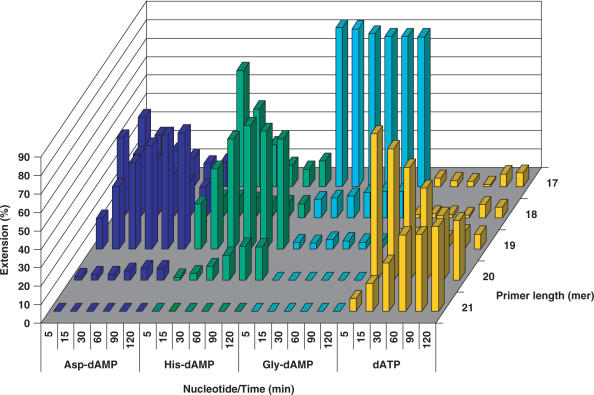
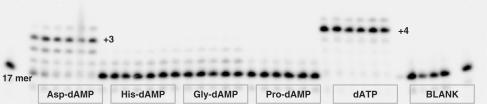
In the case of the T5 template with the overhang of seven thymidine nucleobases, HIV RT indeed generates (n + 6) and (n + 7) products at a very little extent while the (n + 2) and (n + 3) products are prevalent (Figure 8). The obvious stalling of the HIV RT polymerase after incorporation of two adenine nucleobases might indicate substrate inhibition or a template sequence effect. The primer strand extension for 1 h with 500 μM of Gly-dAMP or Pro-dAMP takes place with low efficiency and does not result in the formation of the full-length extension products.
Figure 8.
Primer extension with amino acid phosphoramidates by HIV RT. Reaction conditions: primer (P1) was 5′-labelled with 33P followed by annealing to a template T5; 125 nM primer/template (P1/T5), [AA-dAMP] = 500 μM; [dATP] = 50 μM; [HIV RT] = 0.03 U/μl; time points: 5, 15, 30, 60, 90 and 120 min. Blank: 125 nM primer/template (P1/T5), [HIV RT] = 0.03 U/µl, no nucleotide substrate.
Primer extension by Therminator DNA polymerase
The Therminator DNA pol mediated addition of amino acid phosphoramidate nucleotides instead of natural dNTPs at the 3′ terminal end was investigated for several AA-dAMPs. Similarly, to the case of HIV RT, the best results were observed with Asp dAMP phosphoramidate, which was successfully incorporated across from a string of thymidine residues (T3 template) to provide an (n + 3) product (Figure 9).
Figure 9.
Primer extension with amino acid phosphoramidates by Therminator DNA polymerase. Reactions conditions: primer (P1) was 5′-labelled with 33P followed by annealing to a template T3; 125 nM primer/template (P1/T3), [AA-dAMP] = 500 μM; [dATP] = 50 μM; [Therminator] = 8 × 10−4 U/μl; time points: 5, 15, 30, 60, 90 and 120 min. Blank: 125 nM primer/template (P1/T3), [Therminator] = 8 × 10−4 U/μl, no nucleotide substrate.
However, when His-dAMP and Gly-dAMP were used as substrates for Therminator DNA polymerase, the primer extension took place with significantly lesser efficiency (13.6 and 18.1% of primer extension, respectively) with the (n + 1) product being predominant and halted after addition of 2 nt phosphoramidate residues (Figure 10). The primer extension with Pro-dAMP was very ineffective and resulted only in addition of 1 nt phosphoramidate residue at the primer's end.
Figure 10.
Primer extension efficiency of Therminator DNA polymerase.
It is interesting to note that in the case of the T7 template with the overhang of seven thymidine residues the predominant product of the primer extension was the (n + 2) oligonucleotide. Nonetheless, Therminator DNA polymerase was able to carry out the extension of the T7 primer with Asp-dAMP phosphoramidate and incorporate up to five adenine residues.
Stereoselectivity of reverse transcriptase for the amino acid leaving group and chain termination using an antiviral nucleoside
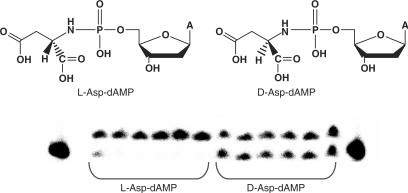
In order to evaluate the influence of chirality of the amino acid to function as a leaving group during the incorporation reaction, we have compared d-Asp-dAMP and l-Asp-dAMP as substrate for RT using template T1 and primer P1.
From the gel electrophoresis experiments it is clear that the natural l-Asp-dAMP is a better substrate for RT than d-Asp-dAMP (Figure 11). The stereoselectivity of the reaction further support the idea that appropriate binding and coordinating of the amino acid in the active site of the enzyme is needed, in order to function as substrate.
Figure 11.
Structure of l-Asp-dAMP and d-Asp-dAMP and primer extension reaction by HIV-RT. Reaction conditions: primer (P1) was labelled with 13P followed by annealing to a template T1; 125 nM primer/template (P1/T1); [HIV-RT] = 0.03 U/μl; [l-Asp-dAMP] =[d-Asp-dAMP] = 200 μM, time points: 5, 15, 30, 60, 90 and 120 min.
From the point of view of applicability in the antiviral field, we have evaluated the potential of l-Asp-PMEA (PhosphonoMethoxyEthylAdenine) to function as chain terminator in the RT assay. PMEA is a potent anti-HIV agent with a phosphonate moiety instead of a phosphate group and lacking a free hydroxyl in the nucleoside moiety, so that chain elongation is not possible (21). The incorporation of PMEA using l-Asp as leaving group is demonstrated in Figure 12, although high concentration of the substrate is needed (100 µM). It can be concluded that l-Asp-PMEA is a substrate for RT and incorporation of PMEA leads to chain termination. This experiment demonstrates the potential of the approach to use modified nucleosides having an alternative leaving group as potential anti-HIV agents.
Figure 12.
Chain terminating effect of l-Asp-PMEA using reverse transcriptase of HIV-1. Reaction conditions: primer (P1) was 5′-labeled with 33P followed by annealing to a template T1; 125 nM primer/template (P1/T1); [HIV-RT] = 0.03 U/μl; time points: 5, 15, 30, 60, 90 and 120 min.
Molecular modelling of l-Asp-dAMP and l-His-dAMP in the active site of HIV-RT
With the l-Asp-dAMP and l-His-dAMP molecules bound to reverse transcriptase, stable molecular dynamics (MD) trajectories were obtained.
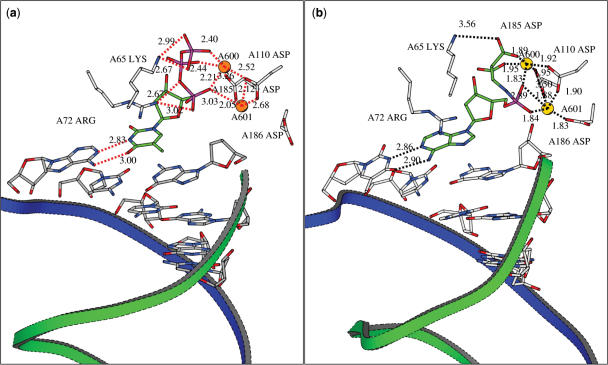
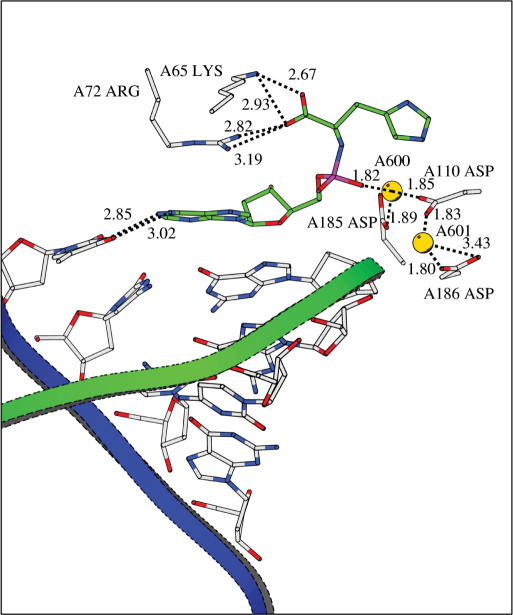
In the l-Asp-dAMP complex (Figure 13b), the 2 Mg2+ ions are comparable in position to their situation in the original TTP complex (Figure 13a). They are tightly bound to both COO− groups of the l-Asp-dAMP (mimicking the second and third phosphate groups), the phosphoramidite group and to three Asp groups in the enzyme: Asp110A and Asp185A which are widely conserved in the polymerases and Asp186A (22,23). In the original X-ray structure, Asp186A is not involved in an ionic bond with one of the Mg2+ ions. In the l-His-dAMP complex MD simulation reveals that the l-His-dAMP binds in a different manner (Figure 14).
Figure 13.
(a) X-ray structure of the RT dNTP pocket with bound Thymidine triphosphate (TTP). The primer strand is visualized by a green ribbon, the template strand has a blue ribbon. The stabilizing interactions of the TTP within the complex are shown: hydrogen bond distances between the bases are indicated in Å. The ligation of the 2 Mg2+ ions to the complex is also visualized by dashes and distances in Å. (b) Average structure of l-Asp-AMP in the RT dNTP pocket.
Figure 14.
Model of l-His-dAMP bound in the active site of RT. Stabilization of the structure is maintained by hydrogen bonding/ionic bonds (indicated by dotted lines) and a potential cation-π interaction between the neutral histidine group of l-His-dAMP and one of the Mg2+ ions. The distance of this Mg2+ to the plane of that histidine is 4.4 Å.
The Mg2+ ions shift during the simulation, i.e. Mg601A moves 3.44 Å to Asp186A, while Mg600A moves 2.51 Å in the direction of Asp185A. There is an ionic bond from one phosphate oxygen to Mg2+, and the coordination of the Mg2+ ions to three Asp residues is still present. The amino acid COO− of l-His-dAMP however is involved in a salt bridge with Lys65A. The neutral His group of l-His-dAMP is in an orientation facilitating a cationic-aromatic interaction with one of the Mg2+ ions (24).
Only a few dAMP residues are incorporated into the primer, when feeding the enzyme with l-Asp-dAMP. An explanation could be that the leaving group is bound too tight to the enzyme so that it stays in the enzyme, becoming an obstruction for new entering residues. When mutating the l-Asp-dAMP into l-Glu-dAMP, this glutamine side chain is too far from the Mg2+ ions to interact. Although the l-Glu-dAMP molecule fits into the NTP binding pocket (model not shown), no conclusion on why this molecule is not incorporated in the chain can be drawn based on that model.
Figures 13 and 14 are generated using Bobscript, Molscript and Raster3d (25–27).
Stability of l-Asp-dAMP
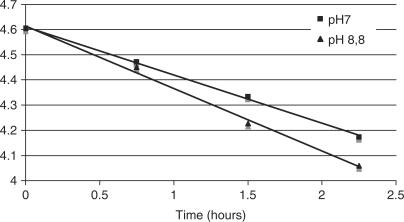
In order to evaluate whether the low incorporation level using some thermostable polymerases is not due to the instability of the compound at higher temperature, we have determined the chemical stability of dNAP in different conditions. The stability of l-Asp-dAMP was investigated using 1D 31P NMR spectra. Within a pH range of 6–8 and at a temperature of 25°C no degradation could be observed after a period of 2 days. At pH 7 and 8.8, at a temperature of 70°C, the product degraded following first order reaction kinetics with a half life of 3.3 h kdegr = and 2.8 h, respectively.
DISCUSSION
A considerable amount of work has been done in the last two decades exploring DNA polymerase mechanisms and the factors involved in nucleotide recognition and polymerase fidelity (28–31). Importantly, these studies have provided extremely valuable information for rational design of anti-viral, anti-cancer compounds as well as nucleoside probes for various biotechnological applications (32–35). The presented study, hence, demonstrates the successful and efficient use of the amino acid moiety linked to the α phosphate via a (P-N) phosphoramidate bond as a diphosphate (pyrophosphate) group mimic.
This study revealed that the most remarkable recognition and nucleotide analogue incorporation was achieved with 2′-deoxyadenosine-5′-aspartyl phosphoramidate nucleotide analogue. Gel-based polymerase assay shows that this analogue is successfully inserted into a growing DNA strand by HIV Reverse Transcriptase and Therminator DNA polymerase and that this incorporation is selective. It was also demonstrated that HIV RT is capable of incorporation of 2–3 consecutive residues of 1 in template-directed DNA synthesis. Likewise, Therminator DNA polymerase also efficiently extends a DNA primer by several nucleobases using the modified substrate. Notably, in the case of HIV RT, the observed stalling and termination of the DNA synthesis after incorporation of two Asp-dAMP residues could possibly indicate enzyme inhibition by either a competitive or a non-competitive mechanism. Incorporation of the modified substrate and the primer extension were observed with His-dAMP analogue. Although His-dAMP was inserted at a lesser extent than Asp-dAMP, primer extension was comparable to the extension with 1 and also resulted in a noticeable stalling at the +2 position. Thus, it would be of a great interest to explore the causes of the stalling and possible modes of substrate inhibition.
The kinetic analysis of Asp-dAMP incorporation shows that the specificity for incorporation of this modified substrate by HIV reverse transcriptase is ∼1300-fold lower than that for the natural substrate (dATP) (7). The significantly higher Km value for the amino acid phosphoramidate analogue than for the natural substrate, suggests that the phosphoramidate substrate dissociates from the active site more readily and faster. Thus, high Km value for Asp-dAMP implies weak binding to the polymerase active site. However, the measured Vmax is only 3-fold lower than this for the natural substrate suggesting fast and efficient nucleophilic displacement of amino acid moiety once the amino acid phosphoramidate substrate is bound at the active site and formation of a phosphodiester bond. Therefore, it might be feasible to design an amino acid phosphoramidate analogue with higher affinity for the polymerase active site while retaining structural features responsible for efficient recognition and nucleotidyl transfer.
To be useful in biological experiments, it is important that the amino acids-dAMP analogues are chemically and enzymatically stable. The enzymatic stability has not been evaluated, yet. However, the compounds have been proven to be stable in water between pH 6 and pH 8 for at least 2 days, their half life at pH 7 and pH 8.8 at 70°C is 3.3 and 2.8 h, respectively. These stability studies have been performed using NMR spectrometry to follow the potential degradation reactions (data not shown). It is expected that the phosphoramidate bond will become unstable at a pH lower than 5. Exploration of the substrate properties among a series of nucleotide phosphoramidates coupled to a wide spectrum of natural amino acids revealed several important structural and electrostatic features involved in triphosphate moiety binding, recognition and nucleotidyl transfer. Numerous structural studies indicate that the binding of the nucleoside triphosphate at the polymerase active site results in coordination of at least two catalytically essential metals (Mg2+) (36–40). Furthermore, several catalytic, highly conserved amino acid residues are also involved in chelation of metal ions and proper positioning of the triphosphate moiety for an ‘in-line’ nucleophilic attack at the α phosphorus. It was suggested previously that initial recognition of an incoming dNTP occurs through the binding of the triphosphate moiety (38). Structural and genetic analysis of a number of DNA polymerases and reverse transcriptase indicates that amino acid residues involved in the triphosphate binding are highly conserved (36,38,39). In the case of HIV RT, binding of the incoming dNTP is coordinated by Arg72 and Lys65 that make interactions with the α- and γ-phosphates, respectively (Figure 13a) (39). This dNTP is also accompanied by 2 Mg2+ ions, which are bound to the phosphates of the nucleotide and to the two residues Asp185 and Asp110 (Figure 13a) (22,23).
Importantly, studies have shown that the binding of the incoming dNTP and catalytic metal ions is responsible for further rearrangements of the catalytic amino acid residues as well as the relocation of the 3′ primer terminus in a position for the effective nucleotidyl transfer (39). Therefore, a proper geometric and spatial arrangement of all reacting residues and atoms are essential for the formation of the productive tertiary complex (36,37,40). Efficient incorporation of 1 might imply that the aspartyl amino acid effectively replaces the β and γ phosphate groups, likely. It can also be suggested that aspartate moiety acts as a leaving group and mimics of a pyrophosphate group. Another example of activated nucleotide and a use of a good leaving group is phosphorimidazolide nucleotides, in which the α-phosphorus atom is activated by imidazolide or methyl imidazolide moieties. Such phosphoimidazolides deoxy- and ribonucleotides were described as efficient substrates for non-enzymatic templated and non-templated oligomerization (41–43).
Furthermore, the presented study clearly demonstrates the requirement for the presence of the negative charge and electrostatic interactions for efficient binding of an incoming nucleotide. This is evident from the study with the aspartyl phosphoramidate (1) and bis-methoxy aspartyl phosphoramidate (1a) where the protection of a carboxylate groups brings drastic changes in ability of HIV RT or Therminator DNA pol to recognize and incorporate these two modified substrates. Although aspartyl phosphoramidate nucleotide behaves as a good nucleotide triphosphate analogue and substrate for HIV RT and Therminator DNA polymerase, its methyl protected derivative does not support DNA synthesis at all. Comparison among the series of synthesized amino acid phosphoramidates demonstrates that Asp-dAMP, which possesses an extra negative charge, displays superior properties as a polymerase substrate. The lower efficiency of DNA synthesis using l-Asp-dAMP (when compared with dATP) may also be attributed to these electrostatic effects (three negative charges for l-Asp-dAMP versus four negative charges for dATP, respectively), and the suggestion that the incoming dATP brings 1 Mg2+ in the active site, while the pyrophosphate leaving group takes 1 Mg2+ with it (44). Unexpectedly, Glu-dAMP analogue that also possesses an extra negative charge failed to serve as a substrate for both HIV RT and Therminator DNA polymerase. A possible explanation for this outcome could be steric clashing (crowding) due to a longer amino acid side chain (an extra methylene group) as compare to the Asp-dAMP analogue. However, a modelling study (data not shown) demonstrates that Glu-dAMP can also be accommodated in the active site of the enzyme complex. Apparently, the binding of Glu-dAMP is not optimal to function as substrate in the polymerization reaction. It can also lead to misalignment of the negative charge in the polymerase active site and disruption of the binding of the catalytic metal ions. HIV RT and Therminator DNA pol were effective in incorporation of His-dAMP suggesting that imidazole moiety of the histidine side chain might likewise be involved in binding interactions of metal ions and the triphosphate group. Interestingly, a modelling study demonstrates that His-dAMP is bound in a different way as Asp-dAMP and that the α-carboxylate group is involved in interactions with Lys-65 and Arg-72, while the imidazole ring of His-dAMP is involved in cation-π interactions.
Moreover, very little or no nucleotide incorporation occurred when phosphoramidate nucleotides coupled to non-polar amino acids were used as substrates for HIV RT or DNA polymerase. These observations provide further support for the rationale that the electrostatic interactions are indispensable for polymerase recognition, the assembly of the catalytic complex in the active site and for chain elongation. These suggestions, however, need to be confirmed by co-crystallization experiments and molecular dynamics.
In conclusion, aspartyl phosphoramidate moiety serves as a mimic of a pyrophosphate group and behaves as a good leaving group in a nucleotidyl transfer reaction. Incorporation of nucleotides, although to a lesser extent, was likewise observed for histidyl and glycinyl phosphoramidates, respectively. The fact that different AA-dAMP function as substrate for the polymerase reaction, does not mean that they bind to the enzyme in the same way and that their mode of action is identical.
Therefore, is seems feasible to use chain terminating nucleotide analogues coupled to newly designed leaving groups through a phosphoramidite or phosphodiester linkage for a direct inhibition of HIV-RT or other viral polymerase. Another application may be the enzymatic synthesis of DNA containing natural and unnatural nucleobases, avoiding at times cumbersome nucleoside triphosphate synthesis and purification. As we emphasized elsewhere (45), the propagation of certain genes and replicons that would require the exogenous supply of nucleic acid precursors absent from cells and natural food chains, such as deoxynucleotide-aspartate conjugates, stands as a promising option for preventing genetic pollution by nutritionally containing the dissemination of genetically engineered microbes. The logical next step toward implementing dNAPs, e.g. the set of deoxynucleotide-aspartate conjugates corresponding to the four bases A, C, G and T, as bona fide precursors of certain DNA plasmids in a bacterial cell will be to evolve a DNA polymerase variant with an enhanced efficiency and selectivity in condensing these substrates but having lost the capability to condense canonical dNTPs. Techniques of computational design (46) and of in vitro evolution (47) could be applied for accomplishing such a swap of substrate specificity in the active site of HIV reverse transcriptase and other DNA polymerases.
MATERIALS AND METHODS
Synthesis
The synthesis and analysis of all l-AA-dAMP compounds have been described in the Supplementary Data of the first communication of this research (7).
d-Asp-dAMP
2′-Deoxyadenosine 5′-monophosphate (100 mg, 0.30 mmol) and d-aspartic acid methyl ester (418 mg, 2.11 mmol) were dissolved in tBuOH (2.7 ml) and water (0.94 ml). Then, a solution of DCC (312 mg, 1.51 mmol) in tBuOH (2 ml) was added and the reaction mixture was heated in a boiling water bath for 4 h. The residue was resuspended in water (20 ml) and extracted with diethyl ether (4 × 15 ml). The aqueous phase was then lyophilized. The colourless solid that was obtained was subjected to column chromatography on silica gel using the following solvent gradient: CHCl3:MeOH (5:1), CHCl3:MeOH:H2O (5:2:0.25), CHCl3:MeOH:H2O (5:3:0.5) and finally CHCl3:MeOH:H2O (5:4:1). The product obtained was treated with 2 ml of 0.4 M NaOH in MeOH–H2O (1:1) and the reaction mixture was allowed to stir at room temperature for 4 h. The solvents were then removed under reduced pressure. The white solid that was obtained was subjected to column chromatography on silica gel using the following solvent gradient: iPrOH, iPrOH:NH3:H2O (7:1:1). The product was isolated as a colourless solid (94 mg, 70% for two steps). 1H NMR (300 MHz, D2O) δ 8.41 (1H, s), 8.12 (1H, s), 6.41 (1H, m), 4.71 (1H, m), 4.24 (1H, m), 3.95–3.89 (2H, m), 3.77 (1H, m), 2.41–2.84 (4H, m). 13C NMR (75 MHz, D2O) δ 177.5, 174.3, 155.1, 152.3, 148.4, 139.8, 118.3, 85.9 (d, J = 9.0), 83.5, 71.1, 64.0 (d, J = 5.2), 43.1, 38.9, 36.5. 31P NMR (121 MHz, D2O) δ 6.85. HRMS calcd for C14H18N6O9P (M-H+) 445.08729, found 445.08755.
l-Asp-PMEA
PMEA (50 mg, 0.18 mmol) was coupled with l-aspartic acid methyl ester (253 mg, 1.28 mmol) in the presence of DCC (189 mg, 0.92 mmol) and tBuOH (2.8 ml)–H2O (0.57 ml) as described above for d-Asp-dAMP. After purification by column chromatography, the white solid obtained was treated with 0.4 M NaOH in MeOH–H2O as described for d-Asp-dAMP. The product was isolated as a colourless solid (44 mg, 58% for two steps). 1H-NMR (D2O): δ 8.17 (2H, bs), 4.40–4.38 (2H, m), 4.93–4.88 (3H, m), 3.58–3.56 (2H, m), 2.81 (1H, dd, J = 3.9, J = 17.5), 2.69 (1H, dd, J = 8.2, J = 17.5). 13C NMR (75 MHz, D2O) δ 177.5, 174.2, 154.9, 151.8, 148.5, 142.8, 117.9, 70.2 (d, J = 11.1), 68.1, 52.2, 43.1, 36.5. 31P NMR (121 MHz, D2O) δ 15.5. HRMS calcd for C12H16N6O7P (M-H+) 387.08181, found 387.10119.
Polymerase assay
The assays for single nucleotide incorporation by HIV reverse transcriptase and the steady state kinetics has been described (7).
In the case of Therminator DNA polymerase, the enzyme was obtained from Westburg (NEB) (2 U/µl) and the reactions were carried out in a 10X Thermopol reaction buffer containing 20 mM TRIS-HCl, 10 mM KCl, 2 mM MgSO4, 0.1% Triton X-100, pH 8.8. The final concentration of the Therminator DNA pol in the reaction mixture was 8.33 × 10−4 U/µl. The polymerase reaction involving Therminator DNA polymerase or any other thermostable polymerase [Vent (exo−), Taq DNA polymerase] was carried out in a similar way as the HIV RT reaction with some modifications. The dNTP solutions and the primer/template/DNA polymerase mixture solutions were topped with mineral oil (30–60 µl) and pre-incubated at 70°C for 2 min. The polymerase reactions were performed at 70°C as well.
Molecular modelling
Electrostatic charges
Atomic electrostatic charges of the l-Asp-dAMP and l-His-dAMP molecules, to be used in the amber software package were calculated from the electrostatic potential at the 6-31G* level using the package Gamess (48) and the two-stage RESP fitting procedure (49) l-Asp-dAMP is supposed to carry an electric charge of −3 while l-His-dAMP is supposed to have a charge of −2.
Amber parameters
The force field parameters used in the amber simulations are those from the parm99 dataset (50). Missing bond and angle parameters were taken from comparable bonds, or angles from the AMBER force field. In particular, the parameters for the P-N bond were added.
Model building
The modelling is based on the crystal structure of RT in complex with a trapped entering triphosphate (pdb structure file 1RTD) (51). The geometry of the l-Asp-dAMP and l-His-dAMP molecules was optimized in gamess in the AM1 force field (48). A locally developed software was used to fit the l-Asp-dAMP structure on the entering triphosphate TTP in the 1RTD structure. This method uses flexible superposition by changing dihedral angles and optimizing the atomic overlap. In this 7 modelling work, the variable angles were the χ angle and the angles in the P-bound leaving group. The base was modified to an adenine and the complementary adenine base (E5) was changed into a thymine by an inverse fitting procedure using Quatfit (Quatfit program in CCL software archives). The same procedure was repeated for the l-His-dAMP molecule.
Molecular dynamics simulations
Solvated molecular dynamics was used to verify the stability (52). The complex was solvated in a truncated octahedron TIP3P water box (53). 31 Na+ counter-ions were then added to get an electrostatic neutral system. The water molecules and counter-ions were then allowed to relax their positions while keeping the solute fixed. Molecular dynamics simulations were then initiated with all restraints removed, with periodic boundary conditions and using a cutoff distance of 8 Å for the non-bonded interactions and the particle-mesh-Ewald method for the summation of the coulombic interactions (54), MD time step = 0.002 ps. Initially, for 20 ps, the system was heated up to 300 K with constant-T, constant-V conditions while restraining the position of the solute and using a Langevin temperature equilibration scheme. The MD was then continued for 200 ps at constant T and constant P. The simulation temperature was 300 K. The presence of the base pair hydrogen bonds between primer and template strand was taken as a valid indicator for the stability of the structures: nearly all H-bonds were still present in the final dynamic structures. An average structure of the last 50 ps was generated and analysed.
NMR Spectroscopy
NMR Spectra were recorded on a Bruker Avance II 500 NMR spectrometer. Chemical shifts δ are indicated in ppm relative to the solvent signals (1H and 13C) or H3PO4 as external standard (31P).
The 31P resonance of l-Asp-dAMP could be observed at 6.90 p.p.m. and the assignment was confirmed using an 1H detected 31P-1H COSY in which a clear scalar coupling could be observed between the 31P resonance and the α-proton of the l-Asp moiety and between the 31P resonance and the 5′ protons of the deoxyribose moiety.
Degradation of l-Asp-dAMP was monitored at pH 8.8 and pH 7 using the integral of the compound's 31P resonance peak. After each period of 45 min at 70°C the sample was cooled to 25°C and a 31P spectrum was recorded. The degradation could be fitted to Equation (1), which indicates that the degradation follows a first order kinetics. C represents the integral of the 31P resonance signal after one or several periods of 45 min at 70°C. C0 is the integral of the 31P resonance signal at the start of the degradation and was set to 100.
| 1 |
Plotting the natural logarithm of the integrals against time allowed us to extract the degradation constant and half-life at both pH conditions (Figure 15).
Figure 15.
Degradation of l-Asp-dAMP at 70°C and pH of 7 (blue dots) and 8.8 (red dots). C was expressed as the integrals of the l-Asp-dAMP 31P peaks with the integral before degradation equal to 100.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The research was financially supported by NEST project, Orthosome of the European Community. The authors thank Chantal Biernaux for editorial help. Funding to pay the Open Access publication charges for this article was provided by the NEST project.
Conflict of interest statement. None declared.
REFERENCE
- 1.Kornberg A, Baker TA. DNA replication. 2nd edn. University Science Books; 2005. ISBN 1-891389-44-0. [Google Scholar]
- 2.Gottschalk G. Bacterial Metabolism. Springer Verlag; 1992. [Google Scholar]
- 3.Kozlov IA, Politis PK, Van Aerschot A, Busson R, Herdewijn P, Orgel LE. Nonenzymatic synthesis of RNA and DNA oligomers on hexitol nucleic acid templates: the importance of the A structure. J. Am. Chem. Soc. 1999;121:2653–2656. doi: 10.1021/ja983958r. [DOI] [PubMed] [Google Scholar]
- 4.Alexandrova LA, Skoblov AY, Jasko MV, Victorova LS, Krayevsky AA. 2′-Deoxynucleoside 5′-triphosphates modified at alpha-, beta- and gamma-phosphates as substrates for DNA polymerases. Nucleic Acids Res. 1998;26:778–786. doi: 10.1093/nar/26.3.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arzumanov AA, Semizarov DG, Victorova LS, Dyatkina NB, Krayevsky AA. Gamma-phosphate-substituted 2′-deoxynucleoside 5′-triphosphates as substrates for DNA polymerases. J. Biol. Chem. 1996;271:24389–24394. doi: 10.1074/jbc.271.40.24389. [DOI] [PubMed] [Google Scholar]
- 6.Sucato CA, Upton TG, Kashemirov BA, Batra VK, Martinek V, Xiang Y, Beard WA, Pedersen LC, Wilson SH, et al. Modifying the beta,gamma leaving-group bridging oxygen alters nucleotide incorporation efficiency, fidelity, and the catalytic mechanism of DNA polymerase beta. Biochemistry. 2007;46:461–471. doi: 10.1021/bi061517b. [DOI] [PubMed] [Google Scholar]
- 7.Adelfinskaya O, Herdewijn P. Amino acid phosphoramidate nucleotides as alternative substrates for HIV-1 reverse transcriptase. Angew. Chem. Int. Ed. 2007;46:4386–4358. doi: 10.1002/anie.200605016. [DOI] [PubMed] [Google Scholar]
- 8.Arts EJ, Marois JP, Gu Z, Le Grice SF, Wainberg MA. Effects of 3′-deoxynucleoside 5′-triphosphate concentrations on chain termination by nucleoside analogues during human immunodeficiency virus type 1 reverse transcription of minus-strand strong-stop DNA. J. Virol. 1996;70:712–720. doi: 10.1128/jvi.70.2.712-720.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Clercq E. New approaches toward anti-HIV chemotherapy. J. Med. Chem. 2005;48:1297–1313. doi: 10.1021/jm040158k. [DOI] [PubMed] [Google Scholar]
- 10.Wagner CR, Iyer VV, McIntee EJ. Pronucleotides: toward the in vivo delivery of antiviral and anticancer nucleotides. Med. Res. Rev. 2000;20:417–451. doi: 10.1002/1098-1128(200011)20:6<417::aid-med1>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
-
11.Abraham TW, Kalman TI, McIntee EJ, Wagner CR. Synthesis and biological activity of aromatic amino acid phosphoramidates of 5-fluoro-2′-deoxyuridine and 1-
 arabinofuranosylcytosine: evidence of phosphoramidase activity. J. Med. Chem. 1996;39:4569–4575. doi: 10.1021/jm9603680. [DOI] [PubMed] [Google Scholar]
arabinofuranosylcytosine: evidence of phosphoramidase activity. J. Med. Chem. 1996;39:4569–4575. doi: 10.1021/jm9603680. [DOI] [PubMed] [Google Scholar] - 12.Bebenek K, Abbotts J, Wilson SH, Kunkel TA. Error-prone polymerization by HIV-1 reverse transcriptase. Contribution of template-primer misalignment, miscoding, and termination probability to mutational hot spots. J. Biol. Chem. 1993;268:10324–10334. [PubMed] [Google Scholar]
- 13.Boosalis MS, Petruska J, Goodman MF. DNA polymerase insertion fidelity. J. Biol. Chem. 1987;262:14689–14696. [PubMed] [Google Scholar]
- 14.Creighton S, Bloom LB, Goodman MF. Gel fidelity assay measuring nucleotide misinsertion, exonucleolytic proofreading, and lesion bypass efficiencies. Meth. Enzymol. 1995;262:232–256. doi: 10.1016/0076-6879(95)62021-4. [DOI] [PubMed] [Google Scholar]
- 15.Gardner AF, Jack WE. Determinants of nucleotide sugar recognition in an archaeon DNA polymerase. Nucleic Acids Res. 1999;27:2545–2553. doi: 10.1093/nar/27.12.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gardner AF, Jack WE. Acyclic and dideoxy terminator preferences denote divergent sugar recognition by archaeon and Taq DNA polymerases. Nucleic Acids Res. 2002;30:605–613. doi: 10.1093/nar/30.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adelfinskaya O, Nashine VC, Bergstrom DE, Davisson VJ. Efficient primer strand extension beyond oxadiazole carboxamide nucleobases. J. Am. Chem. Soc. 2005;127:16000–16001. doi: 10.1021/ja054226j. [DOI] [PubMed] [Google Scholar]
- 18.Chaput JC, Ichida JK, Szostak JW. DNA polymerase-mediated DNA synthesis on a TNA template. J. Am. Chem. Soc. 2003;125:856–857. doi: 10.1021/ja028589k. [DOI] [PubMed] [Google Scholar]
- 19.Ichida JK, Horhota A, Zou KY, McLaughlin LW, Szostak JW. High fidelity TNA synthesis by Therminator polymerase. Nucleic Acids Res. 2005;33:5219–5225. doi: 10.1093/nar/gki840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Renders M, Emmerechts G, Rozenski J, Krecmerová M, Holý A, Herdewijn P. Enzymatic synthesis of phosphonomethyl nucleosides by therminator polymerase. Angew. Chemie Int. Ed. 2007;46:2501–2504. doi: 10.1002/anie.200603435. [DOI] [PubMed] [Google Scholar]
- 21.Balzarini J, Naesens L, Herdewijn P, Rosenberg I, Holy A, Pauwels R, Baba M, Johns DG, De Clercq E. Marked in vivo antiretrovirus activity of 9-(2-phosphonylmethoxyethyl)adenine, a selective anti-human immunodeficiency virus agent. Proc. Natl Acad. Sci. USA. 1989;86:332–336. doi: 10.1073/pnas.86.1.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steitz TA. A mechanism for all polymerases. Nature. 1998;391:231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- 23.Yang L, Beard WA, Wilson SH, Broyde S, Schlick T. Highly organized but pliant active site of DNA polymerase β: compensatory mechanisms in mutant enzymes revealed by dynamics simulations and energy analyses. Biophys. J. 2004;86:3392–3408. doi: 10.1529/biophysj.103.036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallivan JP, Dougherty DA. Cation-π interactions in structural biology. Proc. Natl Acad. Sci. USA. 1999;96:9459–9464. doi: 10.1073/pnas.96.17.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Esnouf RM. Further additions to Molscript 1.4, including reading and contouring of electron-density maps. Acta Crystallogr. 1999;D55:938–940. doi: 10.1107/s0907444998017363. [DOI] [PubMed] [Google Scholar]
- 26.Kraulis PJ. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- 27.Merritt EA, Bacon DJ. Raster3D: photorealistic molecular graphics. Meth. Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 28.Bergstrom DE. Unnatural Nucleosides with Unusual Base Pairing Properties. Current Protocols in Nucleic Acids Chemistry. 2001. pp. 1.4.1–1.4.13. [DOI] [PubMed] [Google Scholar]
- 29.Kool ET. Hydrogen bonding, base stacking, and steric effects in DNA replication. Annu. Rev. Biophys. Biomol. Struct. 2001;30:1–22. doi: 10.1146/annurev.biophys.30.1.1. [DOI] [PubMed] [Google Scholar]
- 30.Kool ET. Active site tightness and substrate fit in DNA replication. Annu. Rev. Biochem. 2002;71:191–219. doi: 10.1146/annurev.biochem.71.110601.135453. [DOI] [PubMed] [Google Scholar]
- 31.Loakes D. The applications of universal DNA base analogues. Nucleic Acids Res. 2001;29:2437–2447. doi: 10.1093/nar/29.12.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henry AA, Romesberg FE. Beyond A, C, G and T: augmenting nature's alphabet. Curr. Opin. Chem. Biol. 2003;7:727–733. doi: 10.1016/j.cbpa.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 33.Hill F, Loakes D, Brown DM. Polymerase recognition of synthetic oligodeoxyribonucleotides incorporating degenerate pyrimidine and purine bases. Proc. Natl Acad. Sci. USA. 1998;95:4258–4263. doi: 10.1073/pnas.95.8.4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loeb LA, Essigmann JM, Kazazi F, Zhang J, Rose KD, Mullins JI. Lethal mutagenesis of HIV with mutagenic nucleoside analogs. Proc. Natl Acad. Sci. USA. 1999;96:1492–1497. doi: 10.1073/pnas.96.4.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogawa AK, Wu Y, McMinn DL, Liu J, Schultz PG, Romesberg FE. Efforts toward the expansion of the genetic alphabet: information storage and replication with unnatural hydrophobic base pairs. J. Am. Chem. Soc. 2000;122:3274–3287. [Google Scholar]
- 36.Beard WA, Wilson SH. Structural insights into the origins of DNA polymerase fidelity. Structure. 2003;11:489–496. doi: 10.1016/s0969-2126(03)00051-0. [DOI] [PubMed] [Google Scholar]
- 37.Brautigam CA, Steitz TA. Structural and functional insights provided by crystal structures of DNA polymerases and their substrate complexes. Curr. Opin. Struct. Biol. 1998;8:54–63. doi: 10.1016/s0959-440x(98)80010-9. [DOI] [PubMed] [Google Scholar]
- 38.Li Y, Kong Y, Korolev S, Waksman G. Crystal structures of the Klenow fragment of Thermus aquaticus DNA polymerase I complexed with deoxyribonucleoside triphosphates. Protein Sci. 1998;7:1116–1123. doi: 10.1002/pro.5560070505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sarafianos SG, Das K, Ding J, Boyer PL, Hughes SH, Arnold E. Touching the heart of HIV-1 drug resistance: the finger close down on the dNTP at the polymerase active site. Chem. Biol. 1999;6:R137–R146. doi: 10.1016/s1074-5521(99)80071-4. [DOI] [PubMed] [Google Scholar]
- 40.Joyce CM, Steitz TA. Function and structure relationships in DNA polymerases. Annu. Rev. Biochem. 1994;63:777–822. doi: 10.1146/annurev.bi.63.070194.004021. [DOI] [PubMed] [Google Scholar]
- 41.Kozlov IA, De Bouvere B, Van Aerschot A, Herdewijn P, Orgel LE. Efficient transfer of information from hexitol nucleic acids to RNA during nonenzymatic oligomerization. J. Am. Chem. Soc. 1999;121:5856–5859. doi: 10.1021/ja990440u. [DOI] [PubMed] [Google Scholar]
- 42.Wu T, Orgel LE. Nonenzymatic template-directed synthesis on oligodeoxycytidylate sequences in hairpin oligonucleotides. J. Am. Chem. Soc. 1992;114:317–322. doi: 10.1021/ja00027a040. [DOI] [PubMed] [Google Scholar]
- 43.Acevedo OL, Orgel LE. Template-directed oligonucleotide ligation on hydroxylapatite. Nature. 1986;321:790–792. doi: 10.1038/321790a0. [DOI] [PubMed] [Google Scholar]
- 44.Yang L, Arora K, Beard WA, Wilson SH, Schlick T. Critical role of magnesium ions in DNA polymerase β's closing and active site assembly. J. Am. Chem. Soc. 2004;126:8441–8453. doi: 10.1021/ja049412o. [DOI] [PubMed] [Google Scholar]
- 45.Pochet S, Kaminski PA, Van Aerschot A, Herdewijn P, Marlière P. Replication of hexitol oligonucleotides as a prelude to the propagation of a third type of nucleic acid in vivo. C. R. Biol. 2003;36:1175–1184. doi: 10.1016/j.crvi.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 46.Dwyer MA, Looger LL, Hellinga HW. Computational design of a biologically active enzyme. Science. 2004;304:1967–1971. doi: 10.1126/science.1098432. [DOI] [PubMed] [Google Scholar]
- 47.Ghadessy FJ, Ramsay N, Boudsocq F, Loakes D, Brown A, Iwai S, Vaisman A, Woodgate R, Holliger P. Generic expansion of the substrate spectrum of a DNA polymerase by directed evolution. Nature Biotechnol. 2004;22:755–759. doi: 10.1038/nbt974. [DOI] [PubMed] [Google Scholar]
- 48.Schmidt MW, Baldridge KK, Boatz JA, Elbert ST, Gordon MS, Jensen JH, Koseki S, Matsunaga N, Nguyen KA, et al. General atomic and molecular electronic structure system. J. Comput. Chem. 1993;14:1347–1363. [Google Scholar]
- 49.Bayly CI, Cieplak P, Cornell WD, Kollman PA. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J. Phys. Chem. 1993;97:10269–10280. [Google Scholar]
- 50.Wang J, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 2000;21:1049–1074. [Google Scholar]
- 51.Huang H, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-1 RT: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 52.Pearlman DA, Case DA, Caldwell JW, Ross WR, Cheatham TE, DeBolt S, Ferguson D, Seibel G, Kollman P. AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comput. Phys. Commun. 1995;91:1–41. [Google Scholar]
- 53.Jorgensen WL, Chandrasekhar J, Madura J, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 54.Cheatham TE, III, Miller JL, Fox T, Darden TA, Kollman PA. Molecular dynamics simulations on solvated biomolecular systems: the particle mesh Ewald method leads to stable trajectories of DNA, RNA, and proteins. J. Am. Chem. Soc. 1995;117:4193–4194. [Google Scholar]