Abstract
Direct UV cross-linking combined with mass spectrometry (MS) is a powerful tool to identify hitherto non-characterized protein–RNA contact sites in native ribonucleoprotein particles (RNPs) such as the spliceosome. Identification of contact sites after cross-linking is restricted by: (i) the relatively low cross-linking yield and (ii) the amount of starting material available for cross-linking studies. Therefore, the most critical step in such analyses is the extensive purification of the cross-linked peptide–RNA heteroconjugates from the excess of non-crosslinked material before MS analysis. Here, we describe a strategy that combines small-scale reversed-phase liquid chromatography (RP-HPLC) of UV-irradiated and hydrolyzed RNPs, immobilized metal-ion affinity chromatography (IMAC) to enrich cross-linked species and their analysis by matrix-assisted laser desorption/ionisation (MALDI) MS(/MS). In cases where no MS/MS analysis can be performed, treatment of the enriched fractions with alkaline phosphatase leads to unambiguous identification of the cross-linked species.
We demonstrate the feasibility of this strategy by MS analysis of enriched peptide–RNA cross-links from UV-irradiated reconstituted [15.5K-61K-U4atac snRNA] snRNPs and native U1 snRNPs. Applying our approach to a partial complex of U2 snRNP allowed us to identify the contact site between the U2 snRNP-specific protein p14/SF3b14a and the branch-site interacting region (BSiR) of U2 snRNA.
INTRODUCTION
Protein–RNA complexes (ribonucleoprotein particles, RNPs) play a fundamental role in the control and regulation of gene expression in the eukaryotic cell. They participate in essential cellular processes such as pre-mRNA splicing, rRNA maturation, post-transcriptional control (mRNA stability), RNA export, translation and translational control. In the field of alternative splicing and of translational control by microRNAs, it was recently demonstrated that protein–RNA interactions and their dynamic changes provide a basis for the diverse and complex driving forces behind such processes (1–5).
There are various approaches to identifying the proteins involved in these processes. One is the overall analysis of the proteins associated with the complexes by mass spectrometry [MALDI-MS (6), Electrospray Ionisation (ESI)-MS (7)], as was recently demonstrated by several proteomic studies of RNP complexes that play fundamental roles in (alternative) splicing (8–10) and siRNA- and miRNA-mediated translational repression (11–14). However, in proteomic-driven studies, no information is gained regarding the question of which of the identified components interacts directly with RNA. A straightforward approach to identify proteins in direct contact to their cognate RNAs is protein–RNA cross-linking combined with MS (15). An alternative/additional method for mapping protein–RNA interactions using MS is the dissociation of intact protein–RNA complexes in the mass spectrometer and the analysis of components that are still associated with RNA (16,17).
One possibility for protein–RNA cross-linking is the direct UV-irradiation of RNPs at 254 nm (18), based on the natural UV-reactivity of the RNA nucleobases. Upon excitation, a covalent bond between a nucleobase and an amino-acid side chain of a protein is formed. This approach has several advantages over site-specific labelling (19,20) or over using heterobifunctional reagents [e.g. (21,22)]: (i) It can be applied directly to any native protein–RNA complex isolated from cells without reconstituting particles carrying site-specific cross-linkers (which can lead to a heterogeneous population and/or can reduce the yield of complexes for interaction studies). (ii) Zero-length cross-links have been proven to have a very high specificity, as demonstrated recently by the 3D structures of co-crystallized RNA–protein complexes (23,24). The site of contact identified in this way always reflects a structural (and functional) RNA interaction domain within or very close to the RNA-binding domain of the protein (25). (iii) It obviates extensive probing experiments, in contrast to comparable protein–RNA cross-linking studies using heterobifunctional reagents, in which the optimal probing conditions have to be carefully adjusted (21). (iv) No inter- or intramolecular protein–protein cross-links are generated (at least as reported so far), which reduces the number of putatively cross-linked species within the mass spectra and simplifies their interpretation.
However, combining protein–RNA cross-linking with mass spectrometry encounters several challenges: (i) because the yield in UV cross-linking is low(er), a purification strategy must be established that separates the cross-linked species from the excess of non-crosslinked species. (ii) MS per se must be adapted, since the peptide and RNA moieties of the cross-linked conjugates show divergent physico-chemical properties in the analysis. (iii) Enrichment and/or down-scaling strategies are required, both to reduce the amount of starting material (and thus to allow the study of material directly isolated from cells) and to increase the intensity of the peaks from (low-abundance) protein–RNA cross-links in MS.
In recent years, we have established a strategy for the purification and subsequent MALDI-Time-of-Flight (MALDI-ToF) MS analysis of cross-linked peptide–oligoribonucleotides derived from UV-irradiated native and reconstituted ribonucleoprotein particles (15,26 and below). It comprises: digestion of the protein moiety of cross-linked RNPs with endoproteinases, removal of the excess of non-crosslinked peptides by size-exclusion chromatography, hydrolysis of RNA-containing fractions with RNases and subsequent fractionation of the resulting mixtures on a microbore liquid-chromatography (LC) system. Fractions that showed an absorbance at 220 nm (peptide moiety) and 254 nm (RNA moiety) were considered to contain cross-linked species and were subsequently analysed by MALDI-MS and -MS/MS using 2,5-dihydroxybenzoic acid (DHB) and/or 2,4,6-trihydroxyacetophenone (THAP) as matrices (15).
On the basis of this work, we report here a down-scaling/enrichment strategy of cross-linked peptide–RNA oligonucleotide species from low amounts of starting material (≤50 pmol) obtained from UV-irradiated RNPs. The novel approach comprises enrichment of peptide–RNA cross-links by immobilized metal-ion affinity chromatography (IMAC) from capillary RP-HPLC fractions combined with treatment of the enriched species with calf intestinal alkaline phosphatase (CIP) to exclude false positives and the subsequent MS analysis by MALDI-ToF mass spectrometry.
In feasibility studies, we successfully applied this strategy to the detection of several peptide–RNA oligonucleotide heteroconjugates derived from (i) UV-irradiated partial complexes of the human minor spliceosome (26), i.e. [15.5K-61K-U4atac snRNA] complexes (24,25), and (ii) from UV-irradiated native U1 small nuclear ribonucleoprotein (snRNP) particles (27) of the human major spliceosome (28) that had been studied before (15,29,30). Moreover, we were able to enrich cross-links derived from a UV-irradiated [p14/SF3b14a-SF3b155282–424] protein complex bound to a U2 snRNA oligomer that mimics the branch-site interacting region (BSiR) of the U2 snRNA (31).
EXPERIMENTAL
Sample preparation
Reconstituted [15.5K-61K-U4atac snRNA] complexes (25) and native U1 snRNPs (27) were obtained and UV cross-linked as described previously (15). [p14/SF3b14a-SF3b155282–424-U2 snRNA BSiR] particles were reconstituted and UV-irradiated according to (31). The protein moiety of the particles was digested with endoproteinase trypsin (Promega) or chymotrypsin (Roche) (ratio 1:20 for both) and the RNA with RNase T1 (Ambion; ratio 1:15) or RNases T1 and A (Ambion; ratio 1:20 for both). The applied purification scheme for the cross-links was according to (15) with the exception for the [p14/SF3b14a-SF3b155282–424-U2 snRNA BSiR] particles. In this case, the non-crosslinked peptide moiety was separated from the non-crosslinked and cross-linked U2 snRNA BSiR oligomers by size exclusion chromatography on a Superdex Peptide column (300 mm × 3.2 mm) mounted on a SMART system (all GE Healthcare, Uppsala, Sweden). Peptide–RNA heteroconjugates were subsequently purified from the mixture by reversed-phase liquid chromatography (RP-LC) with an RP C18 column (150 mm × 0.3 mm; MicroTech Scientific, Vista, USA) coupled to a 140C microgradient system (Applied Biosystems, Foster City, USA) running at a flow rate of 2 µl/min. A gradient of water/0.1% trifluoroacetic acid (TFA) (solvent A) and 80% acetonitrile/0.085% TFA was used. Fractions of 8 µl volume were collected, evaporated to dryness and subjected to further analysis.
IMAC and CIP treatment of peptide–RNA heteroconjugates
Peptide–RNA oligonucleotide cross-links were enriched from one-half of the cap-LC fractions by IMAC using POROS 20 MC beads (Applied Biosystems) loaded with Fe(III) ions (32). For this, capillary LC fractions were redissolved in 50% acetonitrile (ACN) with 0.5% acetic acid (HOAc); one-half was incubated with the IMAC bead slurry for 30 min and the other half retained for hydrolysis (see below). The beads were subsequently washed with 25, 50 and 75% ACN in 100 mM HOAc. Bound cross-links were eluted from the beads with 1% phosphoric acid and subjected to MALDI-MS analysis. Peptide–RNA cross-links of the remaining half of the cap-LC fractions were IMAC-enriched as before and additionally incubated with 1 U of CIP (New England Biolabs) on Fe(III)-loaded POROS 20 MC beads. The beads were washed once with 50% ACN in 100 mM HOAc. Enriched and phosphatase-treated cross-linked species were subsequently eluted from the beads with 1% phosphoric acid as before and subjected to MALDI-MS analysis.
Mass spectrometry
Cross-linked samples were analysed on a Reflex IV mass spectrometer (Bruker Daltonik GmbH, Bremen, Germany) with 2,5-dihydroxybenzoic acid (DHB) as MALDI matrix in the positive reflectron mode for the U1 snRNP and the [15.5K-61K-U4atac snRNA] particles and in the negative reflectron mode for the [p14/SF3b14a-SF3b155282–424-U2 snRNA BSiR] complex, both under standard conditions, by summing up to 1000 laser shots.
0.5 µl of sample were mixed with the same volume of DHB (10 mg/ml in 50% ACN or water, respectively) on an AnchorChip™ MALDI target plate (Bruker Daltonik GmbH, Bremen, Germany) with spot sizes of 600 or 400 µm. The preparation was allowed to dry at room temperature and subjected to MS. Close external calibration with peptide standards was performed.
RESULTS AND DISCUSSION
Capillary LC
There are two critical steps in the entire analysis of isolated peptide–RNA cross-links from non-labelled UV-irradiated protein–RNA particles: (i) the yield achieved in cross-linking is usually very low, and (ii) the amount of starting material available is only small when native RNP particles are being investigated. As a consequence, the successful MS analysis of protein–RNA cross-links requires either an increase in the cross-linking yield or an improvement of the chromatographic separation and the enrichment of the heteroconjugates over the excess of non-crosslinked species.
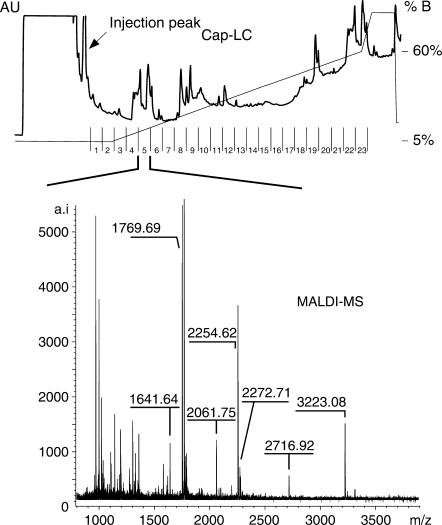
We therefore introduced a capillary liquid chromatography (cap-LC) system for the final purification step of the peptide–RNA heteroconjugates [instead of the microbore HPLC used in previous experiments (15)]. This allowed us to reduce drastically the amount of starting material of RNPs for cross-linking and for subsequent purification of the cross-linked heteroconjugates by LC to ≤50 pmol as compared to 1.5–3.0 nmol of starting material needed beforehand. An example of a cap-LC chromatogram derived from the purification of 50 pmol of native U1 snRNP particles after hydrolysis with trypsin and RNase T1 is given in Figure 1. In contrast to our previous experiments (15,29), defined collection of isolated peptide–RNA cross-links is not longer possible. Fractions contain a mixture of residual non-crosslinked peptides and cross-linked heteroconjugates and unambiguous identification of the putative cross-links (for subsequent MS/MS analysis) is hampered.
Figure 1.
Chromatogram of a capillary LC-run for the purification of peptide–RNA heteroconjugates derived from UV-irradiated U1 snRNP particles after hydrolysis with trypsin and RNase T1. The injection peak containing the moiety of non-crosslinked RNA oligonucleotides is indicated. A representative example of a MALDI-MS spectrum from a cap-LC fraction is shown.
IMAC enrichment of cross-links and CIP treatment
One possibility to overcome this problem is the introduction of an enrichment strategy for putative peptide–RNA heteroconjugates, employing a technique such as IMAC (33,34). In this context, we make use of a feature shared by protein–RNA cross-links and phosphopeptides, namely the phosphate groups that both carry. As phosphopeptides, cross-links and free RNA oligonucleotides can interact with the IMAC material through their negatively charged phosphate groups [as also demonstrated for cross-linked protein–DNA complexes (35–37)]. Since IMAC favours not only the enrichment of phosphate-containing species, but also a certain degree of unspecific binding of (acidic) peptides to the affinity matrix, the identity of enriched precursors must usually be verified by MS/MS experiments. Alternatively, additional treatment of the IMAC-enriched fractions with CIP can be used for the validation of the enriched species (32), as it allows the exclusion of false positives, i.e. acidic peptides, by a characteristic mass shift of 80 Da (=HPO3) in the corresponding MS spectra. CIP treatment together with highly accurate MALDI-ToF MS analysis in the reflectron mode and a computational database search (15) can reveal the cross-linked species in the spectrum without precursor selection and subsequent MS/MS analysis.
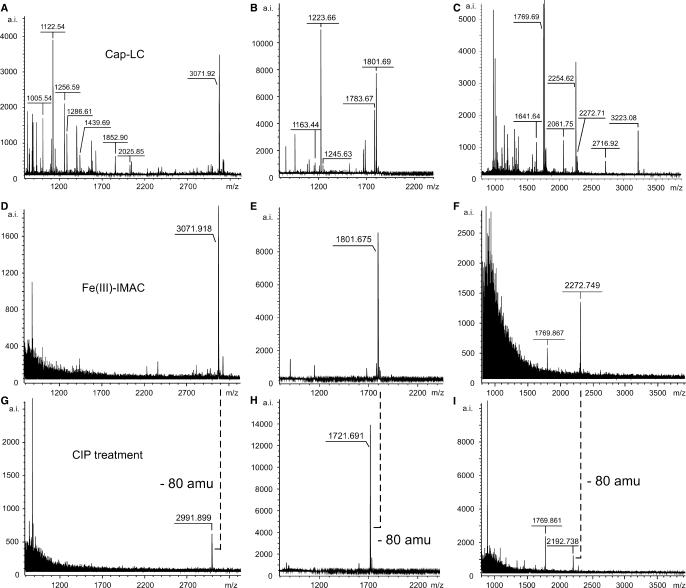
Figure 2A–C shows the MALDI-MS spectra of capillary-LC fractions of UV-irradiated [15.5K-61K-U4atac snRNA] complexes treated with chymotrypsin and RNase T1 (Figure 2A) or RNase T1/A (Figure 2B) and of UV-irradiated native U1 snRNPs hydrolyzed with trypsin and RNase T1 (Figure 2C) without IMAC treatment. Figure 2D–F shows the MALDI-MS spectra of the same fractions treated with IMAC. In all three cases, MALDI-MS revealed enriched species with m/z values of 3071.92, 1801.68 and 2272.75, respectively. MS/MS experiments of these putatively cross-linked precursors cause difficulties, as the amount of precursor is not sufficient (and precursor selection even in the non-enriched samples is restricted by the technical specifications of the MALDI instrument). To prove without MS/MS experiments that enriched precursors carry a phosphate moiety (32)—i.e. must contain a cross-linked RNA oligonucleotide with a 3′-phosphate (as derived from ribonuclease cleavage)—we treated the enriched precursors with CIP on the IMAC beads. Figure 2G–I shows the MALDI-MS spectra of the IMAC-enriched capillary-LC fractions additionally treated with CIP. Enriched precursors with m/z = 3071.918, 1801.675 and 2272.749 (Figure 2G–I) clearly show a shift of 80 amu, demonstrating that HPO3 was cleaved from the enriched species which thus must be a cross-linked peptide–RNA heteroconjugate. Comparison of the measured monoisotopic masses with calculated monoisotopic masses of all possible combinations of RNA oligonucleotides (http://library.med.utah.edu/masspec/compo.htm) revealed that the enriched species are no (modified) RNA oligonucleotides.
Figure 2.
Identification and validation of cross-linked peptide–RNA heteroconjugates by MALDI-ToF MS using IMAC and combined IMAC/CIP treatment. All spectra were recorded in positive ion-mode with DHB as matrix. (A–C) MALDI-ToF MS spectra of cap-LC fractions derived from UV-irradiated [15.5K-61K-U4atac snRNA] complexes after hydrolysis with chymotrypsin and RNase T1 (A), chymotrypsin and RNases A/T1 (B) and from UV-irradiated native U1 snRNP particles after hydrolysis with trypsin and RNase T1 (C). (D–F) MALDI-ToF MS spectra of the same fractions after treatment with IMAC. (G–I) MALDI-ToF MS spectra of the same fractions after combined IMAC/CIP treatment. The masses of the enriched precursor ions at m/z 3071.918 and 1801.675 correspond to a chymotryptic fragment of the 61K (hPrp31) protein encompassing positions 263–273 (SSTSVLPHTGY) cross-linked to a CAUAG pentanucleotide of the U4atac snRNA (positions 42–46) (A,D,G) and to an AU dinucleotide of the U4atac snRNA (positions 43–44) (B,E,H), respectively, with and without the 3′-phosphate. The enriched precursor ion at m/z 2272.749 corresponds to a tryptic fragment of the U1 snRNP-specific 70K protein comprising positions 173–180 (RVLVDVER) cross-linked to an AUCA tetranucleotide of the U1 snRNA with and without the 3′-phosphate.
An ambiguity of our strategy is the fact that enriched phosphopeptides (derived from native protein–RNA complexes) would also show the loss of HPO3 and thus a mass shift of 80 amu in MALDI-MS. However, enrichment of residual phosphopeptides in our experiments is not very likely since the vast majority of the non-crosslinked peptides is separated during size-exclusion chromatography. The probability that residual non-crosslinked peptides is still—to a certain degree—phosphopeptides is very low, because of the low abundance of phosphopeptides in protein–RNA complexes per se. Nonetheless, to exclude any ambiguity between cross-links and phosphopeptides we envisage the possibility of incubating the enriched species with nucleases (e.g. nuclease P1) on the beads instead of CIP. Nuclease P1 hydrolyzes both 3′-5′-phosphodiester bonds in single-stranded nucleic acids and 3'-phosphomonoester bonds in mono- and oligonucleotides terminated by a 3′-phosphate without base specificity. The corresponding mass shift could be observed in the MALDI-MS as for the CIP-treated samples.
Moreover, by applying this strategy additional sequence information about the cross-linked RNA moiety could be obtained. Alternatively, cross-linked and enriched species could be hydrolyzed with HF (37). Recent studies on protein–DNA cross-links demonstrated that HF treatment leaves only the nucleobase of the cross-linked nucleotides attached to the peptide (37).
Computer-aided cross-link search
As no structural information about the cross-linked peptide moiety could be obtained by MS/MS experiments, we set out to calculate the sequence of the cross-linked peptide moiety. We compared the experimental masses of the enriched precursors (i.e. putative cross-links) with the monoisotopic masses of proteolytic peptides of all the proteins within the RNP plus the monoisotopic masses of RNA oligonucleotides derived from in silico hydrolysis of the RNA molecules of the RNP (15). Assuming that the cross-linked precursor mass is additively composed of the molecular masses of the cross-linked peptide and RNA moieties, the following constraints were made for the calculation: (i) mass deviation for searching ≤0.1 Da; (ii) specificity of the endoproteinase (e.g. trypsin: R/K, not after P; chymotrypsin: Y/F/W/M/L), allowing two missed cleavages; (iii) highest priority for specificity of the endonuclease(s) (RNase T1, RNases T1 and A), but also consideration of 3′ and 5′ non-specific cleavage/hydrolysis products (15) and (iv) for each experiment/search, the scope of the database was restricted to the sequences of proteins and RNA(s) specific for the investigated RNP particle. A peptide hit was considered to be genuinely positive when the computational database search from both the experiments with RNase T1 and RNases A and T1 gave the same peptide sequence.
The results of the computational database searches for the enriched cross-linked species derived from reconstituted [15.5K-61K-U4atac snRNA] complexes and native U1 snRNPs are summarized in Table 1. Strikingly, enriched precursor masses derived after digestion of the RNA moiety of the two cross-linked complexes with RNase T1 and RNases T1/A showed only one specific peptide sequence for each RNP particle (Table 1, bold peptide sequences). The first of these is SSTSVLPHTGY, encompassing positions 263–273 of protein 61K (hPrp31), with the 61K protein cross-linked to an AU dinucleotide within the U4atac snRNA (in the stretch 42CAUAG46); and the second is RVLVDVER, positions 173–180 in the U1 70K protein cross-linked to an AU dinucleotide within the U1 snRNA (in A2C1U1).
Table 1.
Results of the computational database searches for IMAC-enriched cross-linked species derived from UV-irradiated [15.5K-61K-U4atac snRNA] complexes after hydrolysis with chymotrypsin and RNase T1 (m/z 3071.918) or RNases T1/A (m/z 1801.675) and from UV-irradiated native U1 snRNPs after hydrolysis with trypsin and RNase T1 (m/z 2272.749) or RNases T1/A (m/z 1638.671). Peptide sequences found after digestion of the RNA moiety with both RNase T1 and RNases T1/A are shown in bold. The abbreviations used are: Mexp—experimental monoisotopic molecular weight (MW); Mcal—calculated monoisotopic MW; EP—endoproteinase and chymo—chymotrypsin.
| m/z | Mexp | Mcal | Protein | EP | RNase | Composition/sequence | Stringency |
|---|---|---|---|---|---|---|---|
| 3071.918 | 3070.910 | 3070.899 | 61K | chymo | T1 | A2C1G1U1 + SGFSSTSVLPHTGY | 3′/5′ spec. |
| A1C3U1 + IYHSDIVQSL | 3′/5′ unspec. | ||||||
| C3G1U1 + PELESLVPNAL | |||||||
| A2C3 + TEIRKQANRMSF | |||||||
| A2C5 + RRKAARL | |||||||
| 3071.918 | 3070.910 | 3070.899 | 15.5K | chymo | T1 | A2C4G1 + TKKLLDL | 5′ unspec. |
| A2G1U2 + DLVQQSCNYKQL | 3′/5′ unspec. | ||||||
| A2C4G1 + TKKLLDL | |||||||
| 1801.675 | 1800.667 | 1800.667 | 61K | chymo | T1/A | A1U1 + SSTSVLPTGY | 3′/5′ spec. |
| G1U1 + QQILTNATIM | |||||||
| G1A1 + DEACDMALEL | 3′ unspec. | ||||||
| A1U2 + ESLVPNAL | 5′ unspec. | ||||||
| 2272.749 | 2271.741 | 2271.751 | 70K | trypsin | T1 | C1G2 + RQQEVETELK | 3′/5′ spec. |
| A2C1U1 + RVLVDVER | 3′ unspec. | ||||||
| C1G2 + RQQEVETELK | 5′ unspec. | ||||||
| C1G3 + RIHMVYSK | 3′/5′ unspec. | ||||||
| C1G3 + IHMVYSKR | |||||||
| C1G3 + ERSKDKDR | |||||||
| 2272.749 | 2271.741 | 2271.751 | U1C | trypsin | T1 | U1G1 + SMQGFPFYDKPMR | 3′/5′ spec. |
| 2272.749 | 2271.741 | 2271.751 | Sm D1 | trypsin | T1 | A2G2 + LVRFLMK | 3′/5′ spec. |
| A2G1U1 + VKSKKREA | 3′ unspec. | ||||||
| A2G1U1 + SKKREAVAG | |||||||
| 2272.749 | 2271.741 | 2271.751 | Sm D2 | trypsin | T1 | A1C1G2 + MSLLNKPK | 3′/5′ spec. |
| A2C1 + EMWTEVPKSGK | 3′/5′ unspec. | ||||||
| 2272.749 | 2271.741 | 2271.751 | Sm F | trypsin | T1 | C3G + CNNVLYIR | 5′ unspec. |
| A2C1G1 + PFLNGLTGK | 3′/5′ unspec. | ||||||
| 1638.671 | 1637.663 | 1637.660 | 70K | trypsin | T1/A | A1U1 + RVLVDVER | 3′/5′ spec. |
| 1638.671 | 1637.663 | 1637.660 | Sm D2 | trypsin | T1/A | A1C1 + IRFLILPD | 3′/5′ spec. |
| 1638.671 | 1637.663 | 1637.660 | Sm F | trypsin | T1/A | A1G1 + PFLNGLTGK | 3′/5′ spec. |
In general, unambiguous assignment of the cross-linking position on the RNA is more difficult. U4atac snRNA only contains a single specific RNase T1 fragment with the calculated composition of A2C1G1U1 (42AUCAG46). The U1 snRNP cross-link with m/z 2272.279 (corresponding to a cross-linked RNA with the composition A2C1U1) cannot be a specific RNase T1 fragment, as it does not contain a G (RNase T1 cleaves after G and RNase A after U and C). This fragment might result from the hydrolysis of a larger T1 fragment either during LC purification or from in-source gas-phase fragmentation during MS. However, the calculated nucleotide composition A2C1U1 is present in four RNase T1 fragments of the native U1 snRNA, namely nucleotide positions 21–28 (AUACCAUG), 29–33 (AUCACG), 111–117 (AAACUCG) and 122–130 (CAUAAUUUG). Without any further MS/MS experiments it is impossible to identify unambiguously the sequence of the cross-linked RNA.
Nonetheless, our enrichment/MS/computational database approach has proven—in this feasibility study—to be useful for the assignment of cross-linked proteins in RNPs and their cross-linked peptide region. Our calculations perfectly match the results obtained by MS/MS analysis of peptide–RNA heteroconjugates derived from the same UV-irradiated RNP particles in large-scale experiments (15).
Cross-linking sites in a partial complex of the U2 snRNP
We extended our approach to a partial complex of the U2 snRNP, i.e. the ternary [p14/SF3b14a-SF3b155282–424-U2 snRNA BSiR] complex, in order to identify the contact sites of the U2 snRNP-specific protein p14/SF3b14a and the so-called ‘branch-site interacting region’ (BSiR) on the U2 snRNA (31,38,39).
During pre-mRNA splicing—that is excision of intronic sequences and ligation of exons of a pre-mRNA in order to yield mature mRNA—sub-spliceosomal U1, U2 and [U4/U6.U5] tri-snRNP particles assemble on the pre-mRNA in a stepwise manner to generate the catalytically active spliceosome (28). A critical step in the assembly of the spliceosome is the recognition of conserved splicing motives of the pre-mRNA. Base-pairing of U2 snRNA (via its BSiR, nucleotide positions 33–38) with the branch-site sequence (BS) of the pre-mRNA forces an adenosine base (the so-called branch-point adenosine) to bulge out, defining a nucleophile for the first catalytic step of splicing. Both the p14/SF3b14a and the SF3b155 proteins are subunits of the U2 snRNP-associated splicing factor 3b (SF3b) that contacts pre-mRNA in the region of the branch-site, thus contributing to the recruitment of the U2 snRNP to the branch site (40). Cross-linking studies demonstrated that p14/SF3b14a is in direct contact with the branch-point adenosine (41,42) and with the U2 snRNA at nucleotide G31 next to the BSiR (nucleotides G33–A38) (43). In addition, p14/SF3b14a binds tightly to the SF3b155 protein between amino acids 255 and 424 (42) and its interaction facilitates binding to the BSiR on the U2 snRNA (31). Recent structural studies without RNA further demonstrated that p14/SF3b14a contains an RNA recognition motif (RRM) that is partly involved in the interaction with SF3b155 (31,39). Interestingly, Schellenberg and co-workers showed by cross-linking experiments with a site-specifically 32P-labelled RNA mimicking the pre-mRNA branch-point region that a conserved tyrosine residue in the RNP2 motif of the RRM is in direct contact with the bulged-out adenosine or its adjacent nucleotides (39).
A missing link in the overall picture in the absence of high-resolution structures is the identification of the contact sites between the p14/SF3b14a and its cognate U2 snRNA region, i.e. the branch-site interacting region. We therefore set out to identify the site(s) of such interaction(s) by applying our approach on reconstituted [p14/SF3b14a-SF3b155282–424-U2 snRNA BSiR] complexes (31).
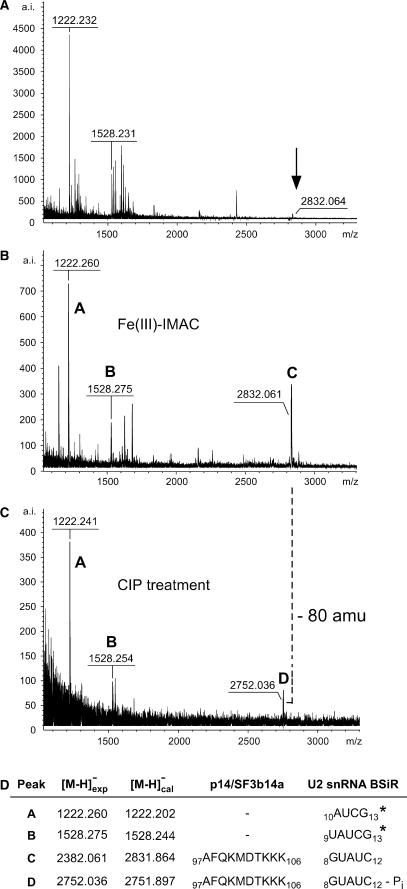
Figure 3 shows spectra of a cap-LC fraction before (Figure 3A) and after (Figure 3B) IMAC, revealing enrichment of a putative cross-link with m/z 2832.061 (labelled ‘C’). Peak C disappeared after treatment of the sample with alkaline phosphatase, and a new signal at m/z 2752.036 arose (‘D’ in Figure 3C). The characteristic mass difference of 80 amu between signals C and D was observed as before, consistent with an accessible phosphate residue.
Figure 3.
Identification and validation of a cross-linked peptide–RNA heteroconjugate derived from UV-irradiated [p14/SF3b14a-SF3b155282–424-U2 snRNA BSiR] complexes by MALDI-ToF MS using IMAC and combined IMAC/CIP treatment. All spectra were recorded in negative ion-mode with DHB as matrix. (A) MALDI-ToF spectrum of a cap-LC fraction derived from UV-irradiated [p14/SF3b14a-SF3b155282–424-U2 snRNA BSiR] complexes after hydrolysis with trypsin and RNase T1. The arrow indicates the signal of the cross-linked species (see panel B). (B) MALDI-ToF MS spectrum of the same fraction after treatment with IMAC. (C) MALDI-ToF MS spectrum of the same fraction after combined IMAC/CIP treatment. The masses of the enriched precursor ions (marked ‘C’ and ‘D’, respectively) correspond to a tryptic fragment of the p14/SF3b14a protein encompassing positions 97–106 (AFQKMDTKKK) cross-linked to a GUAUC pentanucleotide of the BSiR of U2 snRNA (positions 8–12) with and without the 3′-phosphate group. Ions marked ‘A’ and ‘B’ in panels B and C correspond to U2 snRNA BSiR oligonucleotides comprising positions 10–13 (AUCG) and 9–13 (UAUCG). Note that these oligonucleotides are located at the very 3′-end of the synthetic RNA oligomer and thus do not have a 3′-phosphate group accessible to CIP. (D) Assignment of the enriched precursor masses to peptide and RNA sequences. Asterisks indicate confirmation of RNA sequences by post-source decay (PSD) analysis.
We observed that MALDI-MS experiments performed in negative mode led to greater signal intensities, in particular when the cross-linked RNA moiety is larger and the cross-linking yield is low, which was the case in the latter experiment. A similar phenomenon has been described for the MALDI-MS analysis of phosphopeptides, where dramatically greater signal intensities are achieved in the negative mode (44). It is therefore feasible during MALDI-MS of enriched cross-links to switch between positive and negative mode to increase the signal intensities of the cross-linked precursors. Importantly, this can be executed within one experiment without consumption of noticeably greater quantities of sample material, so that the use of both modes does not incur any need for additional preparative experiments or a larger-scale preparation.
A computer database search revealed that the masses associated with peaks C and D did not match any combination of nucleotides derived from the RNA alone (http://library.med.utah.edu/masspec/compo.htm). However, it corresponded exactly to a p14/SF3b14a tryptic fragment encompassing positions 97–106 (AFQKMDTKKK) cross-linked to an RNA oligonucleotide with the composition A1C1G1U2. This composition matches two sequences within the oligonucleotide, namely 8GUAUC12 or 9UAUCG13; of these, the former 8GUAUC12 is not a specific RNase T1 fragment. Owing to the low abundance of the precursor ions C and D (after IMAC enrichment, 400 a.i. with 1000 laser shots; after CIP validation, 100 a.i. with 1000 laser shots), MS/MS was not possible. Nevertheless, a cross-link of the p14/SF3b14a fragment 97AFQKMDTKKK106 to the RNA oligonucleotide 8GUAUC12 most probably explains signals C and D (Figure 3) for the following reasons: (i) the alternative RNase T1 fragment (9UAUCG13) carries a 3′-hydroxyl group and could thus not have been dephosphorylated by alkaline phosphatase. Note that the ions marked with ‘A’ (m/z 1222.260) and ‘B’ (m/z 1528.275) in Figure 3B also show no loss of phosphate (although they are enriched by IMAC). Computational analysis of these mass peaks resulted in the oligonucleotides 10AUCG13 and 9UAUCG13 carrying a 3′-hydroxyl group, as G13 represents the extreme 3′ end of the oligonucleotide. Both these RNA sequences could be confirmed by MS/MS analysis of the pre-IMAC sample (Figure 3A and data not shown). (ii) Notably, mass peak A is a hydrolysis product of B (the actual RNase T1 fragment). Hydrolysis is presumably a consequence of cap-LC separation at pH 2, as RNA becomes labile below pH 3. Thus, the putatively cross-linked RNA oligonucleotide 8GUAUC12 can similarly be explained as a hydrolysis product of a larger T1 fragment.
In a similar manner, we identified a second peptide–RNA cross-link (data not shown). The m/z value of this second cross-link (2680.094) was again shifted by 80 amu after CIP treatment (to m/z 2600.056), thus again representing a putative peptide–RNA heteroconjugate. These masses exactly matched a p14/SF3b14a fragment encompassing positions 101–111 (MDTKKKEEQLK) cross-linked to an RNA oligonucleotide with the composition G1A1U2 that can be found at the RNA positions 4–7 (UGUA), 6–9 (UAGU) or 8–11 (GUAU). Strikingly, the deduced peptide fragment partially overlaps with that of the first cross-link, 97AFQKMDTKKK106. Taken together, these results argue that RNA can be cross-linked to p14/SF3b14a between positions 97 and 111 (AFQKMDTKKKEEQLK).
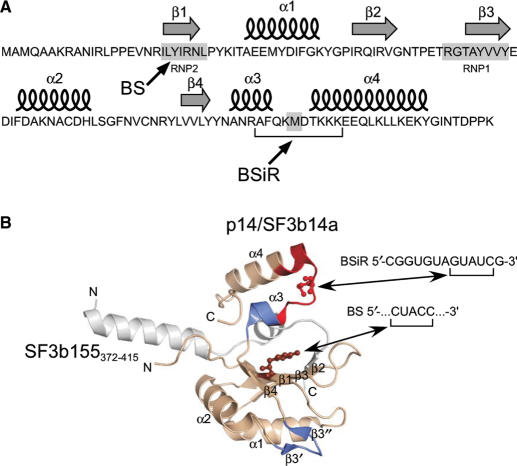
Importantly, the cross-linked peptide sequence encompasses parts of helix α3, helix α4 and the connecting loop between these helices (Figure 4A and B). It harbours the basic residue Lys100 that is postulated to be involved in interactions with RNA due to its close neighbourhood to Tyr22 (39). Furthermore, this particular residue is affected upon incubation with U2 BS-BSiR RNA duplex in NMR chemical shift experiments (31). On the basis of our former work, in which we found that methionine residues within loop regions of (ribosomal) proteins are highly susceptible to UV cross-linking (45), we postulate that Met101 in p14/SF3b14a is cross-linked to GUAUC (presumably to one of the uridine residues) within the BSiR (CGGUGUAGUAUCG). Our cross-linking data together with the data available from the 3D structures of p14/SF3b14a suggest a U2 BS/BSiR RNA binding interface on one site of the p14/SF3b14a encompassing β3′, β3″ and their connecting loop, β-sheet(s) of the RNP2 (and RNP1) and parts of the C-terminal helices α3 and α4 with their connecting loop (Figure 4B).
Figure 4.
Cross-linking sites in the 3D structure of p14/SF3b14a in complex with SF3b155373–415 according to ref. (39). (A) Secondary structure elements of p14/SF3b14a. Arrows indicate the amino acids cross-linked to the branch site [BS, (39)] and to the U2 branch-site interacting region (BSiR, this work). (B) Ribbon diagram of p14/SF3b14a in complex with SF3b155373–415. p14/SF3b14a is shown in beige, with Y22 in RNP2 (β1) that was cross-linked to the BS according to ref. (39) highlighted in balls-and-sticks, SF3b155373–415 is coloured grey. The peptide sequence (97AFQKMDTKKKEEQLK111) found to be cross-linked to the BSiR of U2 snRNA (this work) is marked in red with the putatively cross-linked amino acid M101 highlighted in balls-and-sticks. Regions marked in blue (β3′, β3″ and α3) were affected upon incubation with U2 BS/BSiR RNA duplex in NMR chemical shift experiments (31).
SUMMARY
We have developed an overall approach for the enrichment, validation and subsequent MALDI-ToF-MS-based identification of peptide–oligoribonucleotide heteroconjugates as obtained by UV cross-linking of RNP particles, both native and reconstituted in vitro—in particular when these are not available in large quantity.
Our approach can be applied to any protein–RNA complex for the identification of proteins in direct contact with RNA and for visualization of the site of interaction. Since it introduces capillary liquid chromatography for the final purification step of the cross-linked peptide–RNA heteroconjugates, it requires a relatively low amount of starting material for the protein–RNA cross-linking experiments (10–50 pmol of complex).
The strategy was first successfully applied to the purification of peptide–RNA oligonucleotide cross-links from two test systems that have been extensively studied before ([15.5K-61K-U4atac snRNA] complexes and U1 snRNP particles (15,30)). It further proved successful in the analysis of a contact site between the p14/SF3b14a protein and the U2 snRNA BSiR in partial U2 snRNP particles, i.e. [p14/SF3b14a-SF3b155282–424-U2 snRNA BSiR] complexes (31).
We assume that upon introduction of increasingly smaller chromatography systems, e.g. nano-LC, combined with enrichment strategies and MS(/MS)-based validation of peptide–RNA cross-links as presented here or elsewhere (30), a comprehensive analysis of protein–RNA contact sites in larger RNP particles [e.g. spliceosomal B (46) and C (47) complexes] is possible.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR Online.
ACKNOWLEDGEMENTS
We thank Marcus Wahl for his help in modelling the p14/SF3b14a-SF3b155373–415 structure. The excellent help of Monika Raabe and Uwe Pleßmann in cross-link purification is gratefully acknowledged. E.K.-H. is funded by the Peter und Traudl Engelhorn-Stiftung. This work is supported from a YIP grant from the EURASNET (within the 6th EU framework) to H.U. Funding to pay the Open Access publication charges for this article was provided by Max Planck Institute for Biophysical Chemistry.
Conflict of interest statement. None declared.
REFERENCES
- 1.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 2.Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 3.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 4.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev. Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Mallory AC, Vaucheret H. Functions of microRNAs and related small RNAs in plants. Nat. Genet. 2006;38:S31–S36. doi: 10.1038/ng1791. Erratum in: Nat. Genet., 38, 850. [DOI] [PubMed] [Google Scholar]
- 6.Karas M, Hillenkamp F. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal. Chem. 1988;60:2299–2301. doi: 10.1021/ac00171a028. [DOI] [PubMed] [Google Scholar]
- 7.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionisation for mass spectrometry of large biomolecules. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 8.Garcia-Blanco MA. Alternative splicing: therapeutic target and tool. Prog. Mol. Subcell. Biol. 2006;44:47–64. doi: 10.1007/978-3-540-34449-0_3. [DOI] [PubMed] [Google Scholar]
- 9.Horiuchi T, Aigaki T. Alternative trans-splicing: a novel mode of pre-mRNA processing. Biol. Cell. 2006;98:135–140. doi: 10.1042/BC20050002. [DOI] [PubMed] [Google Scholar]
- 10.Hui J, Bindereif A. Alternative pre-mRNA splicing in the human system: unexpected role of repetitive sequences as regulatory elements. Biol. Chem. 2005;386:1265–1271. doi: 10.1515/BC.2005.143. [DOI] [PubMed] [Google Scholar]
- 11.Nilsen TW. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 2007;23:243–249. doi: 10.1016/j.tig.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 12.Scherr M, Eder M. Gene silencing by small regulatory RNAs in mammalian cells. Cell Cycle. 2007;6:444–449. doi: 10.4161/cc.6.4.3807. [DOI] [PubMed] [Google Scholar]
- 13.Filipowicz W, Jaskiewicz L, Kolb FA, Pillai RS. Post-transcriptional gene silencing by siRNAs and miRNAs. Curr. Opin. Struct. Biol. 2005;15:331–341. doi: 10.1016/j.sbi.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 14.Tang G. siRNA and miRNA: an insight into RISCs. Trends Biochem. Sci. 2005;30:106–114. doi: 10.1016/j.tibs.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 15.Kühn-Hölsken E, Lenz C, Sander B, Lührmann R, Urlaub H. Complete MALDI-ToF MS analysis of cross-linked peptide-RNA oligonucleotides derived from non-labelled UV-irradiated ribonucleoprotein particles. RNA. 2005;11:1915–1930. doi: 10.1261/rna.2176605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hernandez H, Robinson CV. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nat. Protoc. 2007;2:715–726. doi: 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]
- 17.Sharon M, Robinson CV. The role of mass spectrometry in structure elucidation of dynamic protein complexes. Annu. Rev. Biochem. 2007;76:8.1–8.27. doi: 10.1146/annurev.biochem.76.061005.090816. [DOI] [PubMed] [Google Scholar]
- 18.Görner H. Photochemistry of DNA and related biomolecules: quantum yields and consequences of photoionization. J. Photochem. Photobiol. B. 1994;26:117–139. doi: 10.1016/1011-1344(94)07068-7. [DOI] [PubMed] [Google Scholar]
- 19.Konarska MM. Site-specific derivatization of RNA with photocrosslinkable groups. Methods. 1999;18:22–28. doi: 10.1006/meth.1999.0753. [DOI] [PubMed] [Google Scholar]
- 20.Rhode BM, Hartmuth K, Urlaub H, Lührmann R. Analysis of site-specific protein–RNA cross-links in isolated RNP complexes, combining affinity selection and mass spectrometry. RNA. 2003;9:1542–1551. doi: 10.1261/rna.5175703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Q, Yu ET, Kellersberger KA, Crosland E, Fabris D. Toward building a database of bifunctional probes for the MS3D investigation of nucleic acids structures. J. Am. Soc. Mass Spectrom. 2006;17:1570–1581. doi: 10.1016/j.jasms.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 22.Yu ET, Zhang Q, Fabris D. Untying the FIV frameshifting pseudoknot structure by MS3D. J. Mol. Biol. 2005;345:69–80. doi: 10.1016/j.jmb.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 23.Urlaub H, Raker VA, Kostka S, Lührmann R. Sm protein–Sm site RNA interactions within the inner ring of the spliceosomal snRNP core structure. EMBO J. 2001;20:187–196. doi: 10.1093/emboj/20.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu S, Li P, Dybkov O, Nottrott S, Hartmuth K, Lührmann R, Carlomagno T, Wahl MC. Binding of the human Prp31 Nop domain to a composite RNA-protein platform in U4 snRNP. Science. 2007;316:115–120. doi: 10.1126/science.1137924. [DOI] [PubMed] [Google Scholar]
- 25.Nottrott S, Urlaub H, Lührmann R. Hierarchical, clustered protein interactions with U4/U6 snRNA: a biochemical role for U4/U6 proteins. EMBO J. 2002;21:5527–5538. doi: 10.1093/emboj/cdf544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Will CL, Lührmann R. Splicing of a rare class of introns by the U12-dependent spliceosome. Biol. Chem. 2005;386:713–724. doi: 10.1515/BC.2005.084. [DOI] [PubMed] [Google Scholar]
- 27.Kastner B, Lührmann R. Purification of U small nuclear ribonucleoprotein particles. In: Haynes SR, editor. RNA–Protein Interaction Protocols, Methods in Molecular Biology. Vol. 118. Totowa, NY: Humana Press; 1999. pp. 289–298. [DOI] [PubMed] [Google Scholar]
- 28.Will CL, Lührmann R. Spliceosome structure and function. In: Gesteland RF, Gesteland TR, Atkins JF, editors. The RNA World III. 3rd. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2006. pp. 369–400. [Google Scholar]
- 29.Urlaub H, Hartmuth K, Kostka S, Grelle G, Lührmann R. A general approach for identification of RNA–protein cross-linking sites within native human spliceosomal small nuclear ribonucleoproteins (snRNPs). Analysis of RNA–protein contacts in native U1 and U4/U6.U5 snRNPs. J. Biol. Chem. 2000;275:41458–41468. doi: 10.1074/jbc.M007434200. [DOI] [PubMed] [Google Scholar]
- 30.Lenz C, Kühn-Hölsken E, Urlaub H. Detection of protein–RNA cross-links by nanoLC-ESI-MS/MS using precursor ion scanning and multiple reaction monitoring (MRM) experiments. J. Am. Soc. Mass Spectrom. 2007;18:869–881. doi: 10.1016/j.jasms.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 31.Spadaccini R, Reidt U, Dybkov O, Will CL, Frank R, Stier G, Corsini L, Wahl MC, Lührmann R, Sattler M. Biochemical and NMR analyses of an SF3b155-p14-U2AF-RNA interaction network involved in branch point definition during pre-mRNA splicing. RNA. 2006;12:410–425. doi: 10.1261/rna.2271406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stensballe A, Andersen S, Jensen ON. Characterization of phosphoproteins from electrophoretic gels by nanoscale Fe(III) affinity chromatography with off-line mass spectrometry analysis. Proteomics. 2001;1:207–222. doi: 10.1002/1615-9861(200102)1:2<207::AID-PROT207>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 33.Anderson L, Porath J. Isolation of phosphoproteins by immobilized metal (Fe3+) affinity chromatography. Anal. Biochem. 1986;154:250–254. doi: 10.1016/0003-2697(86)90523-3. [DOI] [PubMed] [Google Scholar]
- 34.Li S, Dass C. Iron(III)-immobilized metal ion affinity chromatography and mass spectrometry for the purification and characterization of synthetic phosphopeptides. Anal. Biochem. 1999;270:9–14. doi: 10.1006/abio.1999.4060. [DOI] [PubMed] [Google Scholar]
- 35.Steen H, Petersen J, Mann M, Jensen ON. Mass spectrometry analysis of a UV-cross-linked protein–DNA complex: tryptophans 54 and 88 of E. coli SSB cross-link to DNA. Protein Sci. 2001;10:1989–2001. doi: 10.1110/ps.07601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pingoud V, Geyer H, Geyer R, Kubareva E, Bujnicki JM, Pingoud A. Identification of base-specific contacts in protein-DNA complexes by photocrosslinking and mass spectrometry: a case study using the restriction endonuclease SsoII. Mol. Biosyst. 2005;1:135–141. doi: 10.1039/b503091a. [DOI] [PubMed] [Google Scholar]
- 37.Geyer H, Geyer R, Pingoud V. A novel strategy for the identification of protein-DNA contacts by photocrosslinking and mass spectrometry. Nucleic Acids Res. 2004;32:e132. doi: 10.1093/nar/gnh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Query CC, Strobel SA, Sharp PA. Three recognition events at the branch-site adenine. EMBO J. 1996;15:1392–1402. [PMC free article] [PubMed] [Google Scholar]
- 39.Schellenberg MJ, Edwards RA, Ritchie DB, Kent OA, Golas MM, Stark H, Lührmann R, Glover JN, MacMillan AM. Crystal structure of a core spliceosomal protein interface. Proc. Natl Acad. Sci. USA. 2006;103:1266–1271. doi: 10.1073/pnas.0508048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gozani O, Potashkin J, Reed R. A potential role for U2AF-SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol. Cell. Biol. 1998;18:4752–4760. doi: 10.1128/mcb.18.8.4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Query CC, McCaw PS, Sharp PA. A minimal spliceosomal complex A recognizes the branch site and polypyrimidine tract. Mol. Cell. Biol. 1997;17:2944–2953. doi: 10.1128/mcb.17.5.2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Will CL, Schneider C, MacMillan AM, Katopodis NF, Neubauer G, Wilm M, Lührmann R, Query CC. A novel U2 and U11/U12 snRNP protein that associates with the pre-mRNA branch site. EMBO J. 2001;20:4536–4546. doi: 10.1093/emboj/20.16.4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dybkov O, Will CL, Deckert J, Behzadnia N, Hartmuth K, Lührmann R. U2 snRNA-protein contacts in purified human 17S U2 snRNPs and in spliceosomal A and B complexes. Mol. Cell. Biol. 2006;26:2803–2816. doi: 10.1128/MCB.26.7.2803-2816.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Urlaub H, Kruft V, Bischof O, Müller EC, Wittmann-Liebold B. Protein-rRNA binding features and their structural and functional implications in ribosomes as determined by cross-linking studies. EMBO J. 1995;14:4578–4588. doi: 10.1002/j.1460-2075.1995.tb00137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Janek K, Wenschuh H, Bienert M, Krause E. Phosphopeptide analysis by positive and negative ion matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom. 2001;15:1593–1599. doi: 10.1002/rcm.417. [DOI] [PubMed] [Google Scholar]
- 46.Deckert J, Hartmuth K, Boehringer D, Behzadnia N, Will CL, Kastner B, Stark H, Urlaub H, Lührmann R. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol. Cell. Biol. 2006;26:5528–5543. doi: 10.1128/MCB.00582-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jurica MS, Sousa D, Moore MJ, Grigorieff N. Three-dimensional structure of C complex spliceosomes by electron microscopy. Nat. Struct. Mol. Biol. 2004;11:265–269. doi: 10.1038/nsmb728. [DOI] [PubMed] [Google Scholar]