Abstract
The efficient repair of double-strand breaks (DSBs) is crucial in maintaining genomic integrity. Sister chromatid cohesion is important for not only faithful chromosome segregation but also for proper DSB repair. During DSB repair, the Smc1–Smc3 cohesin complex is loaded onto chromatin around the DSB to support recombination-mediated DSB repair. In this study, we investigated whether Ctf18, a factor implicated in the establishment of sister chromatid cohesion, is involved in DSB repair in budding yeast. Ctf18 was recruited to HO-endonuclease induced DSB sites in an Mre11-dependent manner and to damaged chromatin in G2/M phase-arrested cells. The ctf18 mutant cells showed high sensitivity to DSB-inducible genotoxic agents and defects in DSB repair, as well as defects in damage-induced recombination between sister chromatids and between homologous chromosomes. These results suggest that Ctf18 is involved in damage-induced homologous recombination.
INTRODUCTION
DNA double-strand break (DSB) is probably the most dangerous type of DNA damage among the various types of DNA damage that can affect a cell. They are formed by exogenous agents, such as ionizing radiation (IR) and certain chemotherapeutic drugs and by endogenously generated reactive oxygen species and chromosomal stress. The inability to respond properly to DNA DSBs and repair the damage may lead to genomic instability, which in turn may either lead to cell death or increase the risk of pathological consequences such as the development of cancer (1).
Observations in yeast and mammalian cells suggest that sister chromatid cohesion is important for DNA repair as well as proper segregation of chromosomes. It has been proposed that cohesin facilitates DNA repair by holding sister chromatids locally at DSB sites to allow strand invasion during homologous recombination (HR) (2–5). The cohesin complex of budding yeast, which consists of Smc1, Smc3, Scc1 and Scc3, forms a ring-like structure (6–11). This holds the sister chromatids together by trapping the sister DNA molecules within its ring (11,12) and is essential for maintaining cohesion between sister chromatids until metaphase to assure equal segregation of sister chromatids (13). Loading of the cohesin complex onto chromatin requires the Scc2–Scc4 complex, whereas Eco1/Ctf7 is required to establish sister chromatid cohesion during S phase (14,15). The interaction between Eco1/Ctf7 and PCNA, which acts as a clamp for DNA polymerases, is essential for sister chromatid cohesion (16,17). However, Eco1/Ctf7 is neither required for the loading of cohesin onto chromatin nor for the maintenance of cohesion in G2/M phase (14,15).
Mutation of the CTF18 gene causes a decrease in the fidelity of chromosome transmission or chromosome loss (18). Mutations in CTF8 and DCC1 as well as CTF18, whose products form an alternative replication factor C (RFC) complex in conjunction with the four small subunits Rfc2-5 of RFC (Ctf18-RFC), cause a moderate non-lethal defect in sister chromatid cohesion (19–21). Ctf18-RFC has been shown to interact with PCNA and to load it onto DNA (22,23). Moreover, Ctf18-RFC is capable of unloading PCNA in vitro (23). Since Ctf18 physically associates with Eco1/Ctf7 (24,25), it seems likely that the moderate defect in sister chromatid cohesion of ctf18 mutant cells maybe related to the function of Eco1/Ctf7. Of note, Ctf18 and Eco1/Ctf7 are found at replication forks, and Ctf18 is required for the efficient recruitment of PCNA onto replication forks in HU-arrested cells (26).
Studies in yeast have revealed that normal loading of the cohesin complex onto chromatin during the progression of DNA replication is insufficient to hold DSB ends in close proximity. This suggests that the cohesin complex must be loaded within the vicinity of the DSBs following replication to facilitate the repair of the DSBs through sister chromatid recombination (SCR) (2,5,27). Until now, the function of Ctf18 in homologous recombination has not been considered because of the synthetic sick or lethal interaction between the mutation of the CTF18 gene and RAD52 (28) which plays a major role in homologous recombination repair. In this study, we present evidence that Ctf18 is involved either directly or indirectly in recombination-mediated DSB repair.
MATERIALS AND METHODS
Yeast strains
The yeast strains used in this study are listed in Supplementary Table S1. Null mutants and Myc- or HA-tagged alleles were made using standard PCR-based gene disruption and insertion methods, as previously described (29–31). Deletion mutants were replaced by KANMX6, HPHMX4 and CgTRP1, which were amplified from pFA6aKANMX6, pAG32 and SHB1805, respectively, by using gene-specific primers consisting of 40–45 nt. The resulting PCR fragments were then transformed into yeast cells and colonies that appeared on G418-, hygromycin B-containing YPAD plates or SC-Trp plates were isolated. Gene disruption was confirmed by PCR of genomic DNA. The sequences of the primers used to generate either the DNA constructs used for gene disruption or for checking disruption of the genes as well as details on the yeast strains will be provided upon request.
Sensitivity to DNA-damaging agents
For the analysis of sensitivity to continuous exposure to DNA-damaging agents, 10-fold serial dilutions of logarithmically growing cells in distilled water were spotted onto YPAD plates, or YPAD plates containing the indicated concentrations of methyl methanesulfonate (MMS), hydroxyurea (HU), phleomycin or camptothecin (Sigma-Aldrich). The plates were incubated for 3 days at 30°C and then photographed. For NHEJ analysis, strains containing galactose-inducible HO endonuclease were grown overnight to 1 × 107 cells/ml in YP medium containing 2% raffinose. After washing once with water, aliquots of 10-fold serial dilutions of cells were plated on YP medium, which contained 2% raffinose and was supplemented with either 2% galactose or 2% glucose. The plates were incubated for 3 days at 30°C and then photographed. For the analysis of sensitivity to phleomycin, logarithmically growing cells were diluted to 107 cells/ml and cultured in the presence of 15 μg/ml nocodazole for 3 h to induce G2/M phase arrest. G2/M-arrested cells were exposed to 100 μg/ml phleomycin for 0.5, 1, 1.5 and 2 h. The cells were washed to remove phleomycin and nocodazole, diluted and inoculated onto YPAD plates. After 3 days incubation at 30°C, colonies were counted.
Detection of Rad53 modification
To detect the modification of Rad53-13Myc in response to DNA damage, G2/M-arrested cells were exposed to 100 μg/ml phleomycin or 0.1% MMS for 2 h at 30°C, and then the cells were harvested for immunoblotting analysis. The cells were incubated at 4°C for 15 min in 230 μl 1.5 M NaCl, followed by incubation at 4°C for 10 min after addition of 30 μl 55% Trichloroacetic acid, and spun at 14 000 r.p.m. for 1 min at 4°C. The cell pellet was resuspended in 20 μl of HU buffer (200 mM Tris-HCl (pH 6.8), 1 mM EDTA, 5% SDS, 8 M Urea, 1% BPB (Bromo-phenol blue) and 20 μl of 5× SDS–polyacrylamide sample buffer. Samples were boiled for 5 min, and spun at 10 000 r.p.m. before loading to SDS-PAGE gels. The proteins were detected by immunoblotting with an anti-Myc antibody (9E10) or an anti-histone H3 antibody (Abcam).
Analysis of the incidence of sister chromatid recombination (SCR) and interchromosomal homologous recombination between heteroalleles
The strains constructed for detecting unequal SCR were previously described (32). Diploid strains with the MR101 background were constructed such that recombination between the heteroalleles his1-1 and his1-7 could be detected by the restoration of histidine prototrophy (33). The number of His+ colonies was scored for each of the 12 plates and the median number of His+ colonies for all 12 plates was determined. The rate of spontaneous recombination was then calculated by the median method (34,35). For detection of damage-induced recombination, logarithmically growing cells were inoculated onto SC-His plates and YPAD plates with or without MMS or phleomycin to evaluate the incidence of damage-induced recombination and colony forming cells, respectively. Alternatively, the logarithmically growing cells were diluted and arrested in G2/M phase in the presence of 15 μg/ml nocodazole for 3 h at 30°C and were exposed to 100 μg/ml phleomycin for the indicated time at 30°C. The cells were subsequently washed to remove the phleomycin as well as nocodazole and plated on YPAD plates and SC-His plates. The recombination frequency after treatment with MMS or phleomycin was determined by dividing the total number of recombinants in the culture by the total corresponding number of surviving cells following treatment with MMS or phleomycin.
Pulsed-field gel electrophoresis
Logarithmically growing cells were diluted to 8 × 106 cells/ml, exposed to 0.1% MMS for 1 h, and then cultured in MMS-free medium for the indicated periods of time. In the case of phleomycin treatment on G2/M-arrested cells, logarithmically growing cells were diluted to 8 × 106 cells/ml and cultured in the presence of 15 μg/ml nocodazole for 3 h at 30°C to induce G2/M phase arrest. G2/M-arrested cells were exposed to 100 μg/ml phleomycin for 2 h at 30°C, washed to remove the phleomycin, and then cultured at 30°C in YPAD medium containing 15 μg/ml nocodazole for the indicated periods of time.
Agarose plugs containing chromosomal DNA were prepared as previously described, with minor modifications (36). All plugs were subsequently treated with Zymolyase 100T (0.15 mg/ml) and proteinase K (1 mg/ml) at 30°C for 24 h. After electrophoresis, gels were stained with 0.5 μg/ml ethidium bromide for 30 min, destained in deionized water for 20 min, and then photographed.
Cell fractionation
Whole-cell extracts (WCEs) and chromatin pellets (ChP) were prepared as previously described (37) with modification. A total of 4 × 107 cells were harvested and sodium azide was added to 0.1%. Cells were incubated at room temperature for 4.5 min in 0.5 ml of prespheroplasting buffer [100 mM PIPES (pH 9.4), 10 mM DTT], followed by incubation in 0.5 ml of spheroplasting buffer [50 mM KH2PO4/K2HPO4 (pH 7.5), 0.6 M Sorbitol, 10 mM DTT, 0.125 mg/ml Zymolyase 100T] at 30°C for 4.5 min with occasional mixing. Sphereoplasts were washed with 1 ml of ice-chilled wash buffer [100 mM KCl, 50 mM HEPES-KOH (pH 7.5), 2.5 mM MgCl2 and 0.4 M Sorbitol, 1 mM PMSF], pelleted at 4000 r.p.m. for 1 min in a microcentrifuge at 4°C, and resuspended in 200 μl of extraction buffer [EBX; 100 mM KCl, 50 mM HEPES-KOH (pH 7.5), 2.5 mM MgCl2, 50 mM NaF, 5 mM Na4P2O7, 0.1 mM NaVO3], and protease inhibitors (1 mM PMSF, 20 μg/ml of leupeptin, 2 μg/ml of pepstatin, 2 mM benzamidine HCl and 0.2 mg/ml of bacitracin or 1× Complete (Roche), 0.25% Triton X-100). Spheroplasts were lyzed and incubating on ice for 5 min with gentle mixing. Lysate was underlayered with 100 μl of 30% sucrose, and spun at 14 000 r.p.m. for 3 min at 4°C. Pellet was washed with 100 μl of EBX, and spun again at 12 000 r.p.m. for 1 min at 4°C. All pellet fractions were resuspended in 20 μl EBX. Equal volumes of mixture of HU buffer [200 mM Tris-HCl (pH 6.8), 1 mM EDTA, 5% SDS, 8 M Urea, 1% Bromo-phenol blue] and 5× SDS–polyacrylamide sample buffer were added to each fraction. Samples were boiled for 5 min, and spun at 10 000 r.p.m. for 1 min before loading to SDS-PAGE gels. Histone H3 was used as a loading control for protein levels in WCEs and chromatin pellet (ChP) fractions. The proteins were detected by immunoblotting with an anti-Myc antibody (9E10; Santa Cruz) or anti-histone H3 antibody (Abcam).
Chromatin immunoprecipitation (ChIP)
ChIP was carried out as previously described, with minor modifications (33). Briefly, asynchronous cultures were grown overnight at 30°C in YP medium containing 2% raffinose. Nocodazole (15 μg/ml) was added to the cultures when they reached 6 × 106 cells/ml, and they were incubated at 30°C for 5 h to induce G2/M phase arrest. Expression of HO endonuclease was then induced by adding 2% galactose. The cultures were washed for 1 h after the addition of galactose and then treated with 2% glucose to repress the expression of HO endonuclease. Cells were harvested and incubated in 1% formaldehyde for 15 min to cross-link proteins to DNA, then the reaction was quenched by incubating the cells in 125 mM glycine for 5 min. Cells were lyzed with glass beads and extracts were sonicated to shear DNA to an average size of 1 kb. Extracts were then divided into two aliquots: Input DNA and immunoprecipitated (IP) DNA (1:20, respectively). Immunoprecipitation was carried out using a monoclonal anti-Myc antibody (9E10) (Santa Cruz Biotechnology, Inc.) or a monoclonal anti-HA antibody (12CA5) (Roche), and immune complexes were captured using Dynabeads Protein G (Dynal Biotech) for 4 h at 4°C. After a series of washes, proteins were released from the beads by incubation for 6 h at 65°C, and were treated with proteinase K. IP DNA was purified for PCR analysis using phenol extraction followed by ethanol precipitation.
PCR amplification
Primers used for ChIP assay are listed in Supplementary Table S2. Amplified PCR products from IP DNA were separated by agarose gel electrophoresis, and quantitated using Scion Image software. To correct for different levels of IP efficiency at different loci, the signal from the target locus was first normalized to the signal from an independent locus (SMC2) on chromosome VI by dividing each target signal by the corresponding SMC2 IP signal. For analysis of different time points, MATα or HML IP signals were normalized to the IP signal at 0 h, which was designated as 1.
In vivo physical monitoring of DSB induction, strand invasion and repair completion during recombination repair
DSB induction and strand invasion of the input DNA were detected by PCR. Primers are listed in Supplementary Table S2. The primer extension and ligation assays were carried out according to a previously described PCR-based method (38). Amplified PCR products were separated by agarose gel electrophoresis and quantitated using Scion Image software. The signal from the target locus was normalized to the amplified signal from an independent locus (SMC2) on chromosome VI. Primer extension and ligation were arbitrarily set at 100% for the highest wild-type level.
RESULTS
Ctf18 is required for damage-induced SCR but not for NHEJ-mediated DSB repair
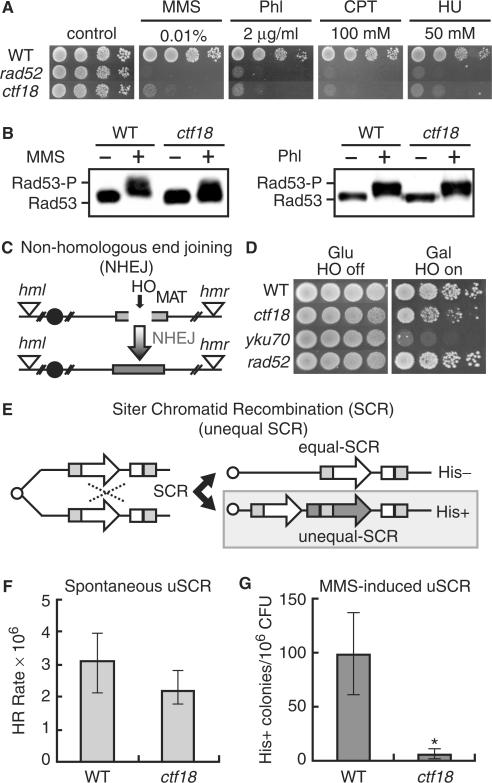
The cohesin complex is required for efficient repair of DSBs (5,27,39). Ctf18 is involved in establishing sister chromatid cohesion (26). However, the proteins that are involved in establishing cohesion for DNA repair have not been fully determined. In this study, we investigated the role of Ctf18 in DNA repair. We first examined the sensitivity of ctf18 deletion mutants to DNA-damaging agents, methyl methanesulfonate (MMS), phleomycin, camptothecin and hydroxyurea (HU), which reportedly induce DSBs both directly and indirectly (40–43). The ctf18 mutant cells, like rad52 cells, were highly sensitive to these DNA-damaging agents compared with wild-type cells (Figure 1A), which suggests that ctf18 cells have defects in their DNA damage checkpoints or DNA repair mechanism. To test the former possibility, we examined the phosphorylation of Rad53, which is a hallmark of activation of the damage checkpoint (44). Rad53 was phosphorylated in both ctf18 and wild-type cells following exposure to MMS and phleomycin (Figure 1B), suggesting that Ctf18 is not essential for the activation of DNA damage checkpoint. This is consistent with a previous report that ctf18 cells showed only a slight defect in checkpoint function unless Rad24 was also absent (21). The sensitivity of the ctf18 cells to these DNA-damaging agents suggests that Ctf18 is involved in DSB repair.
Figure 1.
Ctf18 is involved in DSB repair by HR, but not in NHEJ. (A) Sensitivity to various DNA-damaging agents. Wild-type (SCRL), rad52 (YHO602) and ctf18 (YHO605) cells were inoculated on YPAD plates containing the indicated concentrations of methyl methanesulfonate (MMS), phleomycin, camptothecin or hydroxyurea (HU). The plates were incubated for 3 days at 30°C and then photographed. (B) Modification of Rad53 in response to DNA damage. Wild-type (YHO302) and ctf18 (YHO308) cells expressing Rad53-13Myc were arrested in G2/M phase and then exposed to 0.1% MMS or 100 μg/ml phleomycin for 2 h at 30°C. Aliquots of the cultures were taken before (−) and after (+) exposure to MMS or phleomycin for 2 h. Proteins prepared from these cells were subjected to SDS-PAGE. The Rad53 protein was detected by western blotting using an anti-Myc antibody. (C) Schematic representation of the process and outcome of NHEJ at the galactose-inducible HO endonuclease-specific DSB site on chromosome III in yeast strain JKM179. (D) The NHEJ DSB repair pathway is intact in ctf18 mutants. The viability of wild-type (JKM179), yku70 (YHO505), rad52 (YHO506) and ctf18 (YHO511) mutants on a background of an HR-repair defective yeast strain was determined by growing the cells on either glucose (HO off) or galactose (HO on). A galactose-inducible HO endonuclease was integrated at the ADE3 locus of a haploid yeast strain that was defective in HR-mediated DSB repair. Upon switching the cells to galactose, the HO endonuclease is expressed that induces a single DSB at the MATα locus. Functional DSB repair can occur only through the NHEJ pathway in this strain, since the HM donor loci, HML and HMR, are deleted. (E) A schematic representation of the process and outcome of unequal sister chromatid (USCR) recombination. (F) Spontaneous unequal sister chromatid recombination rates. Wild-type (SCRL) and ctf18 (YHO605) cells were inoculated onto YPAD plates or SC plates lacking His and then incubated at 30°C for 3 days. Spontaneous unequal sister chromatid recombination rates were determined by the median method (34,35). The average number of viable cells (CFU) in the twelve independent colonies and the median number of His+ colonies for all twelve independent colonies was determined after incubation at 30°C for 3 days. The rate of production of recombinants was then calculated by the method of the median HR rates (34,35). The data present the mean and confidential interval (95%<) is shown and (G) MMS-induced sister chromatid recombination (SCR). Wild-type (SCRL) and ctf18 (YHO605) cells were inoculated onto YPAD plates or SC plates lacking His with 0.008% MMS and then incubated at 30°C for 3 days. The viability of the cells cultured with 0.008% MMS was 98%, and 34% for wild type (SCRL) and ctf18 (YHO605), respectively. The frequency of SCR is presented as the number of His colonies per 106 colony forming units (CFU). The columns and error bars represent the means and SDs of eight samples. Statistical difference in the recombination frequency was evaluated between WT and ctf18 mutant cells by Student's t-test (P < 0.001).
DSBs can be repaired by either non-homologous end-joining (NHEJ), which requires Yku70, or homologous recombination (HR), which requires Rad52 (45–47). First, we investigated whether Ctf18 was required for NHEJ-mediated repair by inducing a single DSB in the MATα locus using ctf18 cells that express HO endonuclease. The HR-mediated repair pathway was precluded in these cells by deleting the homologous donor loci, HML and HMR (Figure 1C). Sustained expression of the HO endonuclease leads to a continuous cycle of DNA cleavage and ligation. Cells that are capable of error-prone repair by NHEJ grow in the presence of the HO endonuclease, while cells deficient in NHEJ do not survive (48). Unlike yku70 cells, we found that ctf18 and rad52 mutants grew as well as wild-type cells under sustained expression of HO endonuclease (Figure 1D).
Next, we investigated if damage-induced homologous recombination was impaired in ctf18 cells. We examined damage-induced unequal sister chromatid recombination (uSCR) in haploid cells (Figure 1E). As shown in Figure 1F, the mean spontaneous uSCR rate was slightly lower in ctf18 cells than in wild-type cells; however there was no statistical difference in the rates between the wild-type and ctf18 cells. In contrast, the frequency of MMS-induced uSCR was very low in ctf18 cells compared with wild-type cells (Figure 1G). This suggests that Ctf18 is involved in the recombination between sister chromatids following exposure to MMS.
Ctf18 associates with recombination intermediates
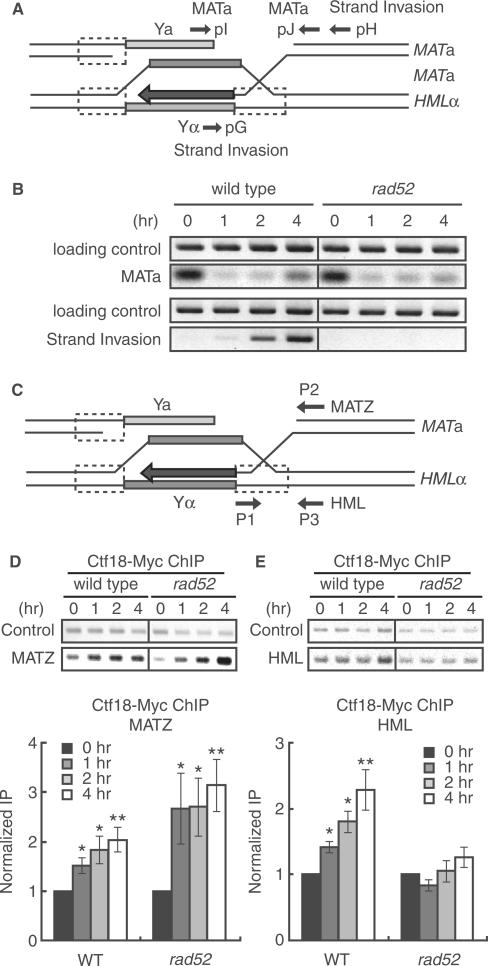
HR-mediated DSB repair is a multiple-step process that includes the recruitment of proteins to sites of DSBs, resection of DNA, strand invasion of ssDNA on the homologous template, DNA synthesis and ligation. One of the best characterized systems for studying homologous recombination is HO endonuclease-induced MAT switching, using HML as the donor template (49). In this system, repair of HO endonuclease-induced DSB at the MATa locus is accomplished through recombination with the homologous HML locus. We investigated the role of Ctf18 in DSB repair by HR using the yeast strain JKM161, which contains MATa and HML as the donor template. DSB formation at the HO recognition site and invasion of the MATa recipient strand into the donor strand (HML) were monitored by PCR using the primers shown in Figure 2A. The amount of PCR product corresponding to the MATa locus was considerably reduced after 1 h following induction of the HO endonuclease in wild-type and rad52 cells. This indicated that cleavage at the MATa locus occurred with high efficiency. In addition, strand invasion was detected 2 and 4 h following induction of the HO endonuclease in wild-type cells, but not in rad52 mutants (Figure 2B). To examine whether Ctf18 was recruited to both DSB sites in the MAT and the HML donor loci during the process of recombination, we performed ChIP analysis using primers for either MAT- or HML-specific sequences (Figure 2C). We observed an association of Ctf18 with the DSB site in the MAT locus 1 h after induction of the HO endonuclease in both wild-type and rad52 cells (Figure 2D). In addition, Ctf18 was also associated with the HML donor strand 4 h after HO endonuclease induction (Figure 2E). In contrast, an association between Ctf18 and the HML donor strand was not observed in rad52 cells (Figure 2E). This data indicates that Ctf18 associates with recombination intermediates.
Figure 2.
Ctf18 associated with HR intermediate. DSBs were induced in JKM161 cells that express Ctf18-13Myc and carry the HML donor sequence by treating cells with 2% galactose for 1 h, which induces the expression of the HO endonuclease, The cells were subsequently incubated in 2% glucose to repress the expression of the HO endonuclease. Genomic DNA was isolated at the indicated time points after the addition of galactose. (A) A schematic representation of the HR intermediates formed during mating-type switching. The approximate location of the primers used to monitor DSBs (pI and pJ primers) and strand invasion (pG and pH primers) are shown. (B) Time course of the formation of DSBs and the formation of strand-invasion intermediates following induction of HO-endonuclease expression. DNA was purified from cells at the indicated time points after transient expression of HO endonuclease. PCR was carried out using primers that flanked the MATa locus: pI and pJ primers were used to detect the formation of the DSB (upper panel) or pG and pH primers were used to detect products of strand invasion and primer extension (lower panel). The SMC2 locus was amplified as a loading control. The data are representative of three independent experiments. (C) A schematic representation of the MATa and HML loci showing the location of the primers used to detect an association of Ctf18 with sequences at the DSB site (P1 and P2 primers for the CEN-distal MATZ locus) or the homologous donor region (P1 and P3 primers for the HML locus) and (D and E) Wild type (YHO411) and rad52 mutant (YHO412) cells expressing Ctf18-13Myc were treated as described for B, and chromatin from the cells at the indicated time points after addition of galactose was subjected to immunoprecipitation using anti-Myc antibodies. DNA present in anti-Myc immune complexes was analyzed by PCR using primers specific for sequences at the DSB site (P1 and P2 primers for the CEN-distal MATZ locus; D) or the homologous donor region (P1 and P3 primers for the HML locus; E), as indicated. Representative images of agarose gels are shown (upper panels). Data from each target locus was normalized to the control SMC2 product. Fold increase was calculated by normalizing the data from each time point to the zero time point value (Bottom panel). The columns and error bars represent the means and SDs (n = 4). The asterisks indicate statistical significances of the results compared to that of 0 h evaluated by Student's t-test (*; P < 0.05; **; P < 0.005).
Mre11 is required for the recruitment of Ctf18 to DSB sites
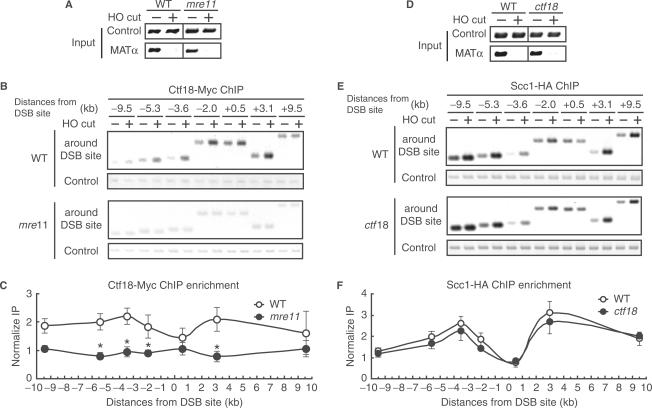
It was reported that recruitment of cohesin complex is dependent on Mre11. Therefore, we investigated whether or not the function of the MRX complex, a heterotrimeric protein assembly of Mre11, Rad50 and Xrs2, is required for the recruitment Ctf18 on the DSB site at MAT locus. In response to DSBs, the MRX complex arrives first at the sites of DSBs and performs nucleolytic resection to generate single-stranded DNA (ssDNA). The ssDNA is recognized by the ssDNA-binding protein Rpa, and proteins involved in DNA damage checkpoint are recruited at DSB sites in an Rpa-dependent manner. Subsequently, proteins that are involved in HR are recruited. The assembly of Rad51 on ssDNA is initiated by Rad52, which in turn causes strand invasion. We found that the induction of DSBs occurred in a similar manner in both wild-type cells and mre11 cells arrested in the G2/M phase (Figure 3A). However, binding of Ctf18 around the DSB site was not observed in the mre11 cells compared with wild-type cells (Figure 3B and C), suggesting that the generation of ssDNA by Mre11 is essential for the recruitment of Ctf18 to the DSB site. These findings indicate that Ctf18 is recruited to the DSB site in a manner dependent on the function of Mre11.
Figure 3.
Ctf18 enrichment to the DSB site requires Mre11. Cells expressing Ctf18-13Myc (A, B, C: wild type; YHO512, mre11; YHO513) or Scc1-3HA (D, E, F: wild type; YHO516, ctf18; YHO517) in JKM179 were arrested in G2/M phase in the presence of 15 µg/ml nocodazole and 2% raffinose at 30°C and then transferred into medium containing 2% galactose to induce the expression of the HO endonuclease and the formation of DSBs. Samples were processed 2 h after HO induction. (A and D) The kinetics of DSB induction. PCR was carried using ‘input DNA’ (see Materials and Methods section) and DSB primers (MATα), which flank the HO cleavage site. Amplification of the SMC2 locus on chromosome VI was used as a control. (B) Binding of Ctf18-13Myc in wild-type and mre11 cells or (E) binding of Scc1-3HA in wild-type and ctf18 cells at a DSB induced at the MATα site on chromosome III. Genomic DNA from cells at the indicated time points after HO endonuclease induction was subjected to ChIP analysis using an anti-Myc antibody. DNA that co-immunoprecipitated with the anti-Myc antibody (ChIP) was amplified using primer sets corresponding to sequences at different distances from the MATα cut. Data show representative photographs of the agarose gels and (C) ChIP signals of Ctf18-13Myc in wild-type and mre11 cells or (F) ChIP signals of Scc1-3HA in wild-type and ctf18 cells were quantified, and normalized to the SMC2 control fragment. Each time point was normalized to the zero time point value and was plotted on the y-axis versus coordinates (in kb) relative to the MATα cut site in chromosome III on the x-axis. The columns and error bars represent the means and SDs (n = 3). The asterisks indicate statistics significant differences determined by Student's t-test (P < 0.01).
Ctf18 is not required for the recruitment of Scc1 to DSB sites
Since Ctf18 is required for the establishment of sister chromatid cohesion, and cohesin is recruited to sites of DSBs, we examined whether a defect in Ctf18 alters the recruitment of cohesin to the DSB sites. Cohesin is composed of Smc1, Smc3, Scc1 and Scc3. We investigate whether the recruitment of Scc1 onto DSB sites required Ctf18. To characterize the association of these proteins with DSB sites, we used the JKM179 strain (50) in which the HML and HMR loci are deleted to prevent HO endonuclease-induced DSB repair by homologous recombination. In addition, to avoid any complication due to S-phase events, we used the cells arrested in G2/M phase. Following induction of the HO endonuclease, DSBs at the MATα locus were induced rapidly and efficiently (Figure 3D). Using ChIP analysis, we investigated the binding of HA-tagged Scc1 to sites around the DSB in wild-type and ctf18 cells. As reported previously, the binding of Scc1 to the MATα locus was increased by 2- to 3-fold following induction of the DSBs, and Scc1 was localized to regions 2–10 kb away from the DNA break on both sides of the break in wild-type cells [Figure 3E and F; (5,27)]. No difference was observed between ctf18 cells and wild-type cells in the localization of Scc1 around the DSB site. This suggests that Ctf18 is not required for cohesin enrichment around the DSB site.
Deletion of CTF18 does not influence HO-induced intrachromosomal recombination
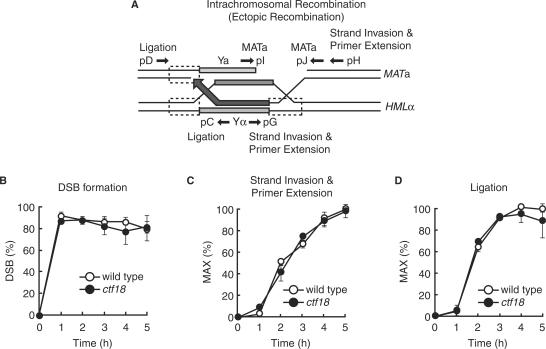
Recruitment of Ctf18 onto sites of DSBs and the association of Ctf18 with recombination intermediates prompted us to examine whether or not Ctf18 was required for the HO endonuclease-induced intrachromosomal recombination. We monitored the formation of DSBs at the HO endonuclease recognition site, primer extension following strand invasion on the donor strand, and ligation of synthesized DNA in the presence of PCR using the primers shown in Figure 4A. We observed that the induction of DSBs, the extension of the invading DNA strand, and ligation occurred with similar kinetics in ctf18 cells compared with wild type cells (Figure 4B, C and D). This suggests that Ctf18 is not required for HO endonuclease-induced intrachromosomal recombination.
Figure 4.
Ctf18 is not involved in intrachromosomal recombination. JKM161 cells carrying the HML donor sequence were treated as described for Figure 2. (A) A schematic representation of the HR intermediates formed during mating-type switching. The approximate location of the primers used to monitor DSBs (pI and pJ; MATa primers), strand invasion and primer extension (pG and pH; strand invasion and primer extension primers) and completion of repair (pC and pD; ligation primers) are shown. (B, C and D) Genomic DNA was isolated from wild-type (JKM161) and ctf18 mutant (YHO409) cells at the indicated time points after transient expression of HO endonuclease and subjected to PCR analysis to determine the efficiency of DSB formation (B), strand invasion and primer extension (C), and completion of repair (ligation) (C). Amplified PCR products were run on agarose gels and stained with ethidium bromide. DNA signals were quantified and then normalized to the control SMC2 product. A PCR product using primers specific for detecting strand invasion and primer extension will only be generated if invasion of the 3′ strand and replication of at least 222 nt of the donor template sequence occurs. The highest level of strand invasion-primer extension and completion of repair (ligation) in wild-type cells was designated as 100%. The data represent the mean and SD of two independent experiments.
Ctf18 may be involved in post-replicative DSB repair
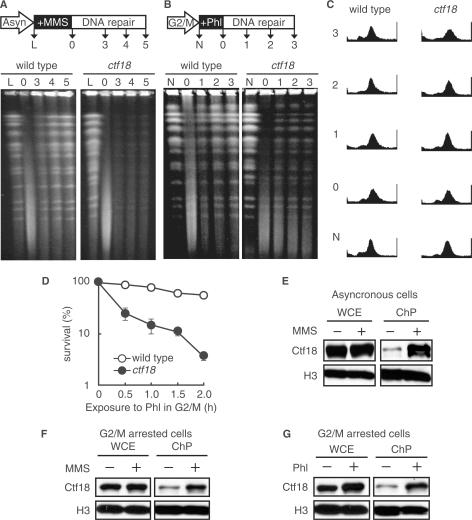
Although Ctf18 was associated with recombination intermediates (Figure 2), no apparent defect was observed in HO endonuclease-induced DSB repair by either NHEJ or HR in ctf18 cells (Figures 1D and 4). Therefore, we investigated whether or not Ctf18 was involved in DSB repair at the chromosomal level. Logarithmically growing cells were treated with MMS and then cultured in MMS-free medium. Chromosomal DNA was isolated from these cells and subjected to pulsed-field gel electrophoresis (PFGE). Immediately following the administration of MMS, the distinct chromosomal DNA bands were replaced by a low-molecular-weight DNA smear in both wild-type and ctf18 cells (Figure 5A). After culturing the cells in MMS-free medium, the chromosome-sized DNA bands were restored in wild-type cells, whereas the restoration of the DNA bands was impaired in ctf18 cells. The amount of fragmented DNA might not necessarily reflect the level of DSBs because strand breaks can occur in MMS-treated DNA during the heat treatment of PFGE plugs (51). However, the defect in restoring chromosome-sized DNA bands was only observed in ctf18 cells and suggests a defect in post-replicative repair in these cells.
Figure 5.
Association of Ctf18 with the damaged chromatin is required for post-replicative DSB repair. (A) A defect in DNA repair in ctf18 cells was monitored by PFGE. A schematic representation of the process used to prepare the cells in pulsed-field gel electrophoresis (PFGE) is shown (upper panel). Logarithmically growing wild-type (MR101) and ctf18 (YHO218) cells were exposed to 0.1% MMS for 1 h, and then cultured in MMS-free medium for the indicated periods of time. The cells were harvested, and their DNA was analyzed by PFGE as described in the Materials and Methods section. Lane L: untreated cells; Lane 0: cells treated with MMS for 1 h; Lanes 3, 4 and 5: cells treated with MMS for 1 h and then cultured in MMS-free medium for 3, 4 and 5 h, respectively. A representative gel image is shown (bottom panel). (B) Phleomycin-induced post-replicative DSB repair. A schematic representation of the process used to prepare the cells in pulsed-field gel electrophoresis (PFGE) is shown (upper panel). G2/M-arrested cells (wild type; MR101, ctf18; YHO218) were exposed to 100 μg/ml phleomycin at 30°C for 2 h and then cultured in phleomycin-free medium containing 15 μg/ml nocodazole for the indicated periods of time. The cells were harvested and their DNA was analyzed by PFGE as described in the Materials and Methods section. Lane N: untreated cells; Lane 0: cells treated with phleomycin for 2 h; Lanes 1, 2, 3: cells treated with phleomycin for 2 h, then cultured in phleomycin-free medium for 1, 2 and 3 h, respectively. A representative gel image is shown (bottom panel). (C) G2/M-arrested state of (B) confirmed by flow cytometry. To confirm the fact that cells are arrested during the course of above experiment described in (B), we monitored the cell cycle status of cells by FACS. (D) Sensitivity to phleomycin of cells arrested in G2/M. Cells (wild type; MR101, ctf18; YHO218) arrested in the G2/M phase with 15 μg/ml nocodazole were exposed to phleomycin (100 μg/ml) at 30°C for the indicated periods of time, washed to remove phleomycin, inoculated onto YPAD plates containing neither phleomycin nor nocodazole and incubated for 3 days at 30°C before counting the colonies. To measure the sensitivity to phleomycin, at least four cultures were taken for each point. The data represent the means and SD. (E and F) Chromatin binding of Ctf18 after exposure to MMS. Logarithmically growing cells (E) or G2/M-arrested cells (F) expressing Ctf18-13Myc (YHO512) were exposed to 0.1% MMS for 2 h. Aliquots of the cultures were taken before (−) and after (+) exposure to MMS. Whole-cell extracts (WCE) and chromatin containing pellets (ChP) were prepared and analyzed by immunoblotting for the presence of the indicated proteins. Histone H3 was used as a loading control for protein levels. The Myc-tagged proteins and histone H3 were detected using an anti-Myc monoclonal antibody and a specific anti-histone H3 antibody, respectively. (G) Chromatin binding of Ctf18 after exposure to phleomycin. G2/M-arrested cells expressing Ctf18-13Myc (YHO512) were exposed to 100 μg/ml phleomycin for 2 h. Cell aliquots were taken before exposure (−) and 2 h after phleomycin (Phl) exposure (+). Whole-cell extract (WCE) and chromatin containing pellets (ChP) were prepared and analyzed by immunoblot for the presence of the indicated proteins. Histone H3 was used as a loading control for protein levels in WCE and ChP fractions. Ctf18-13myc and Histone H3 were detected using an anti-Myc monoclonal antibody and anti-Histone H3-specific antibody, respectively. To confirm the fact that cells are arrested at G2/M during the course of experiments (F), and (G), we monitored the cell cycle status of cells by FACS (Supplementary Figure S1).
To further test the involvement of Ctf18 in DSB repair, we investigated the repair of DSBs induced by phleomycin in G2/M phase-arrested cells. Cells arrested in G2/M were treated with phleomycin and then cultured in phleomycin-free medium containing nocodazole to maintain G2/M arrest. Chromosomal DNA was isolated and subjected to PFGE (Figure 5B), and the cell cycle status of the cells was monitored by fluorescence activated cell sorting (FACS) (Figure 5C). Distinct chromosomal DNA bands were absent just after exposure to phleomycin in both wild-type and ctf18 cells and a low-molecular-weight DNA smear appeared, reflecting the occurrence of DSBs. The chromosome-sized DNA bands were restored in both wild-type and ctf18 cells after culturing cells in phleomycin-free medium. However, the restoration of these bands was less efficient in ctf18 cells compared to wild-type cells, which was reflected in the decrease in the viability of the ctf18 cells compared with wild-type cells after a brief exposure of G2/M phase-arrested cells to phleomycin (Figure 5D).
We also investigated the association of Ctf18 with chromatin following exposure to either MMS or phleomycin. We observed an increase in the amount of Ctf18 in the chromatin-containing fraction after exposure of logarithmically growing cells to MMS (Figure 5E). Moreover, the association of Ctf18 with chromatin was observed even in G2/M phase-arrested cells (Supplementary Figure S1) after exposure to MMS and phleomycin (Figure 5F and G). Taken together, these results suggest that Ctf18 plays a role in post-replicative DSB repair.
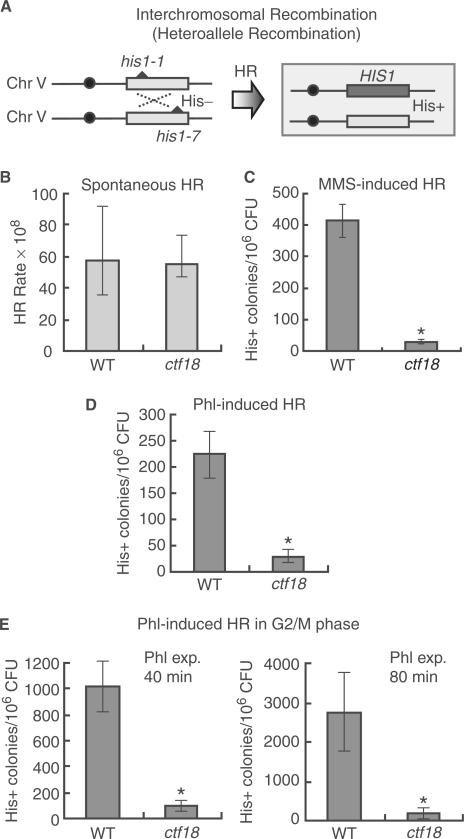
Damage-induced interchromosomal recombination is defective in ctf18 cells
Ctf18 is involved in the establishment of sister chromatid cohesion, which is important for sister chromatid-based homologous recombination repair. Therefore, the defect of uSCR in ctf18 cells (Figure 1G) may be due to a defect in sister chromatid cohesion, even if Ctf18 is not required for cohesin recruitment around the DSB site (Figure 3). In our previous study, we found that rad50 mutant cells show defects in damage-induced recombination between not only sister chromatids (data not shown) but also between homologous chromosomes (52). Thus, we investigated whether Ctf18 is required for interchromosomal recombination between the heteroalleles his1-1 and his1-7 in diploid cells (Figure 6A). The spontaneous interchromosomal recombination rate of ctf18 cells was comparable to that of wild-type cells (Figure 6B). In the presence of either MMS or phleomycin, the interchromosomal recombination frequency in wild-type cells dramatically increased (Figure 6C and D), whereas the interchromosomal recombination frequency in ctf18 cells remained low. The interchromosomal recombination frequency of ctf18 cells in G2/M phase was also very low compared to that of wild-type cells after temporal exposure to phleomycin (Figure 6E).
Figure 6.
Ctf18 is required for damage-induced recombination between homologous chromosomes. (A) A schematic representation of the process and outcome of heteroallele recombination between interchromosomes at the HIS1 locus. (B) Spontaneous heteroallelic recombination rates at the HIS1 locus were determined by the median method (34,35). Wild-type (MR101) and ctf18 (YHO218) cells were used for this assay. The average number of viable cells (CFU) in the twelve independent colonies and the median number of His+ colonies for all twelve independent colonies was determined after incubation at 30°C for 3 days. The rate of production of recombinants was then calculated by the method of the median HR rates (34,35). The data present the mean and confidential interval (95%<) is shown. (C and D) Frequency of MMS- or phleomycin-induced interchromosomal recombination at the HIS1 locus in wild-type and ctf18 cells. Wild-type (MR101) and ctf18 (YHO218) mutant cells were inoculated onto YPAD plates or SC plates lacking His with 0.003% MMS (C; The viability of the cells cultured with 0.003% MMS was 101%, and 87% for wild type (MR101) and ctf18 (YHO218), respectively.) or 0.4 μg/ml phleomycin (D; The viability of the cells cultured with 0.4 μg/ml phleomycin was 96%, and 80% for wild type (MR101) and ctf18 (YHO218), respectively.) and then incubated at 30°C for 3 days. The HR frequency is presented as the number of His+ colonies per 106 colony forming units (CFU). To measure recombination frequency, four (C) or twelve (D) independent cultures were taken for each point. (E) Interchromosomal recombination induced by phleomycin of cells arrested in G2/M phase. Cells (wild type; MR101, ctf18; YHO218) arrested in the G2/M phase with 15 μg/ml nocodazole were temporarily exposed to 100 μg/ml phleomycin at 30°C for 40 min (The viability of the cells treated with 100 μg/ml phleomycin for 40 min was 40%, and 8% for wild type (MR101) and ctf18 (YHO218), respectively.) or 80 min (The viability of the cells treated with 100 μg/ml for 80 min was 19%, and 2% for wild type (MR101) and ctf18 (YHO218), respectively.), washed to remove phleomycin, inoculated onto YPAD plates containing neither phleomycin nor nocodazole, and incubated for 3 days at 30°C before counting colonies. The HR frequency is presented as the number of His+ colonies per 106 colony forming units (CFU). To measure recombination frequency, at least four cultures were taken for each point. Statistical significances (*; P < 0.01) are indicated for the differences between the results of wild-type and ctf18 cells in each graph shown in C, D and E by Student's t-test.
DISCUSSION
The CTF18 gene was originally identified as the gene whose mutation causes a decrease in chromosome transmission fidelity or chromosome loss (18). The function of Ctf18 in homologous recombination repair has not been considered because of the synthetic sick or lethal interaction between the CTF18 mutation and the RAD52 mutation, which plays a major role in homologous recombination repair (28). In this study, we found that repair of DSBs induced by phleomycin (Figure 5), recombination of sister chromatids induced by DNA damage (Figure 1) and interchromosomal recombination between heteroalleles (Figure 6) were defective in ctf18 cells. This indicates that Ctf18 is involved in recombination repair. Although ctf18 cells were nearly normal for HO-endonuclease-based NHEJ, this result does not necessarily rule out the possibility of Ctf18 involvement in the NHEJ pathway under other assay conditions, such as those used by Schar et al. (4) who reported that smc1-2 mutants were much less efficient in plasmid end-joining mediated by the NHEJ pathway.
Although Ctf18 was recruited to the DSB site at the MAT locus (Figure 3) even in G2/M phase-arrested cells, we could not detect an obvious defect in the processes of HO endonuclease-induced recombination, such as primer extension and ligation (Figure 4). The discrepancy between the results obtained after inducing a single DSB with HO-endonuclease and those obtained after inducing DNA damage with phleomycin or MMS may be explained by the different mechanisms of recombination involved in repairing these two types of DNA damage; HO endonuclease induces intrachromosome recombination, whereas MMS and phleomycin induces interchromosomal or sister chromatid recombination. Alternatively, this discrepancy may be due to differences in the amount of DNA damage induced by these agents. For example, the function of Ctf18 maybe more important following the induction of multiple sites of DNA damage when the demands on the cell to execute HR-mediated DNA double-strand break repair are greater. Finally, the discrepancy may be due to the differences in the DNA ends created by HO endonuclease, MMS and phleomycin. Ctf18 could be required for the repair of DNA lesions induced by MMS and phleomycin but not for the repair of DSB induced by HO endonuclease.
How does Ctf18 contribute to recombination repair?
Ctf18 may affect recombination repair through the function of the cohesin complex. Cohesin complexes normally associate with distinct loci on chromosomes (53–56), but associate with damaged DNA regardless of whether cohesin-binding sites are present or not (5,27). During normal cell cycling, cohesion is established during S-phase, but not during G2-phase (57,58). Following DNA damage, cohesion is also established de novo in G2/M phase (5). Cohesin mutant also leads to an impairment of coordination of homologous recombination and NHEJ processes (4). In the current study, we showed that the loading of cohesin (Scc1) onto the DSB site in the MAT locus was not affected by deletion of the CTF18 gene (Figure 3). However, we observed that the function of Ctf18 is required for both recombination between sister chromatids and for recombination between homologous chromosomes (Figures 1 and 6). These results do not necessarily eliminate the possibility that Ctf18 is involved in establishing de novo cohesion of sister chromatids during recombination repair in G2/M phase-arrested cells or the possibility that the defect in homologous recombination repair of ctf18 cells is due to the inefficient establishment of cohesion during replication. In addition, the cohesin complex is composed of multiple subunits, including Scc1 and the Smc1/Smc3 heterodimer, and there exist at least two forms of cohesin, one tightly bound and the other loosely associated (59). Since a large cellular pool of the Smc1/Smc3 heterodimer in cells is apparently not in a complex with Scc1, the ChIP data for Scc1 presented here does not necessarily represent the behavior of the cohesin complex. In a future study, we will examine the loading of the Smc1/Smc3 heterodimer onto damaged sites to shed further light to this issue.
Interestingly, the recruitment of Ctf18 onto the DSB site in the MAT locus and the donor site, the HML locus, was dependent on Mre11 and Rad52, respectively. Therefore, Ctf18 may be recruited onto the DSB site via the function of Mre11 and thereafter function together with Rad52 in DSB repair. Moreover, Ctf18 is recruited onto chromatin even after the induction of DNA damage in G2/M phase-arrested cells. These data suggest that the Ctf18 may play a novel role other than damage-induced cohesion establishment in recombination repair.
Finally, genetic recombination not only promotes genetic diversity in a population, but also insures the integrity of an organism's genome. Inappropriate or inefficient recombination drives tumor formation and underlies certain premature aging diseases in humans. Mutations in the human CTF18 complex could lead to the chromosomal instability known as a hallmark of most cancers.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank J.E. Haber for yeast strains used in the study. We thank all members of the Enomoto lab for their support. This work was supported by Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan, and by Health Sciences Research Grants from the Ministry of Health and Welfare of Japan. Funding to pay the Open Access publication charges for this article was provided by Grants-in-Aid for Scientific Research on Priority Areas from The Ministry of Education, Science, Sports and Culture of Japan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Scott SP, Pandita TK. The cellular control of DNA double-strand breaks. J. Cell. Biochem. 2006;99:1463–1475. doi: 10.1002/jcb.21067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sjögren C, Nasmyth K. Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr. Biol. 2001;11:991–995. doi: 10.1016/s0960-9822(01)00271-8. [DOI] [PubMed] [Google Scholar]
- 3.Sonoda E, Matsusaka T, Morrison C, Vagnarelli P, Hoshi O, Ushiki T, Nojima K, Fukagawa T, Waizenegger IC, et al. Scc1/Rad21/Mcd1 is required for sister chromatid cohesion and kinetochore function in vertebrate cells. Dev. Cell. 2001;1:759–770. doi: 10.1016/s1534-5807(01)00088-0. [DOI] [PubMed] [Google Scholar]
- 4.Schar P, Fasi M, Jessberger R. SMC1 coordinates DNA double-strand break repair pathways. Nucleic Acids Res. 2004;32:3921–3929. doi: 10.1093/nar/gkh716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ström L, Lindroos HB, Shirahige K, Sjögren C. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol. Cell. 2004;16:1003–1015. doi: 10.1016/j.molcel.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 6.Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in. S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- 8.Orr-Weaver TL. The ties that bind: localization of the sister-chromatid cohesin complex on yeast chromosomes. Cell. 1999;99:1–4. doi: 10.1016/s0092-8674(00)80055-0. [DOI] [PubMed] [Google Scholar]
- 9.Anderson DE, Losada A, Erickson HP, Hirano T. Condensin and cohesin display different arm conformations with characteristic hinge angles. J. Cell. Biol. 2002;156:419–424. doi: 10.1083/jcb.200111002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gruber S, Haering CH, Nasmyth K. Chromosomal cohesin forms a ring. Cell. 2003;112:765–777. doi: 10.1016/s0092-8674(03)00162-4. [DOI] [PubMed] [Google Scholar]
- 11.Haering CH, Lowe J, Hochwagen A, Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell. 2002;9:773–788. doi: 10.1016/s1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- 12.Ivanov D, Nasmyth K. A topological interaction between cohesin rings and a circular minichromosome. Cell. 2005;122:849–860. doi: 10.1016/j.cell.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 13.Nasmyth K. Segregating sister genomes: the molecular biology of chromosome separation. Science. 2002;297:559–565. doi: 10.1126/science.1074757. [DOI] [PubMed] [Google Scholar]
- 14.Skibbens RV, Corson LB, Koshland D, Hieter P. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 1999;13:307–319. doi: 10.1101/gad.13.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toth A, Ciosk R, Uhlmann F, Galova M, Schleiffer A, Nasmyth K. Yeast cohesin complex requires a conserved protein, Eco1p (Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 1999;13:320–333. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skibbens RV. Unzipped and loaded: the role of DNA helicases and RFC clamp-loading complexes in sister chromatid cohesion. J. Cell. Biol. 2005;169:841–846. doi: 10.1083/jcb.200503129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moldovan GL, Pfander B, Jentsch S. PCNA controls establishment of sister chromatid cohesion during S phase. Mol. Cell. 2006;23:723–732. doi: 10.1016/j.molcel.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 18.Kouprina N, Tsouladze A, Koryabin M, Hieter P, Spencer F, Larionov V. Identification and genetic mapping of CHL genes controlling mitotic chromosome transmission in yeast. Yeast. 1993;9:11–19. doi: 10.1002/yea.320090103. [DOI] [PubMed] [Google Scholar]
- 19.Hanna JS, Kroll ES, Lundblad V, Spencer FA. Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol. Cell. Biol. 2001;21:3144–3158. doi: 10.1128/MCB.21.9.3144-3158.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayer ML, Gygi SP, Aebersold R, Hieter P. Identification of RFC (Ctf18p, Ctf8p, Dcc1p): an alternative RFC complex required for sister chromatid cohesion in. S. cerevisiae. Mol. Cell. 2001;7:959–970. doi: 10.1016/s1097-2765(01)00254-4. [DOI] [PubMed] [Google Scholar]
- 21.Naiki T, Kondo T, Nakada D, Matsumoto K, Sugimoto K. Chl12 (Ctf18) forms a novel replication factor C-related complex and functions redundantly with Rad24 in the DNA replication checkpoint pathway. Mol. Cell. Biol. 2001;21:5838–5845. doi: 10.1128/MCB.21.17.5838-5845.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bermudez VP, Maniwa Y, Tappin I, Ozato K, Yokomori K, Hurwitz J. The alternative Ctf18-Dcc1-Ctf8-replication factor C complex required for sister chromatid cohesion loads proliferating cell nuclear antigen onto DNA. Proc. Natl Acad. Sci. USA. 2003;100:10237–10242. doi: 10.1073/pnas.1434308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bylund GO, Burgers PM. Replication protein A-directed unloading of PCNA by the Ctf18 cohesion establishment complex. Mol. Cell. Biol. 2005;25:5445–5455. doi: 10.1128/MCB.25.13.5445-5455.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kenna MA, Skibbens RV. Mechanical link between cohesion establishment and DNA replication: Ctf7p/Eco1p, a cohesion establishment factor, associates with three different replication factor C complexes. Mol. Cell. Biol. 2003;23:2999–3007. doi: 10.1128/MCB.23.8.2999-3007.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skibbens RV. Chl1p, a DNA helicase-like protein in budding yeast, functions in sister-chromatid cohesion. Genetics. 2004;166:33–42. doi: 10.1534/genetics.166.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lengronne A, McIntyre J, Katou Y, Kanoh Y, Hopfner KP, Shirahige K, Uhlmann F. Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol. Cell. 2006;23:787–799. doi: 10.1016/j.molcel.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 27.Unal E, Arbel-Eden A, Sattler U, Shroff R, Lichten M, Haber JE, Koshland D. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell. 2004;16:991–1002. doi: 10.1016/j.molcel.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 28.Pan X, Ye P, Yuan DS, Wang X, Bader JS, Boeke JD. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–1081. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- 29.Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 30.Kitada K, Yamaguchi E, Arisawa M. Cloning of the Candida glabrata TRP1 and HIS3 genes, and construction of their disruptant strains by sequential integrative transformation. Gene. 1995;165:203–206. doi: 10.1016/0378-1119(95)00552-h. [DOI] [PubMed] [Google Scholar]
- 31.Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast. 1996;12:259–265. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C259::AID-YEA901%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 32.Ui A, Seki M, Ogiwara H, Onodera R, Fukushige S, Onoda F, Enomoto T. The ability of Sgs1 to interact with DNA topoisomerase III is essential for damage-induced recombination. DNA Repair. 2005;4:191–201. doi: 10.1016/j.dnarep.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 33.Ogiwara H, Ui A, Onoda F, Tada S, Enomoto T, Seki M. Dpb11, the budding yeast homolog of TopBP1, functions with the checkpoint clamp in recombination repair. Nucleic Acids Res. 2006;34:3389–3398. doi: 10.1093/nar/gkl411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lea DE, Coulson CA. The distribution of numbers of mutants in bacterial populations. J. Genet. 1949;49:264–284. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 35.Paulovich AG, Armour CD, Hartwell LH. The Saccharomyces cerevisiae RAD9, RAD17, RAD24 and MEC3 Genes are required for tolerating irreparable, ultraviolet-induced DNA damage. Genetics. 1998;150:75–93. doi: 10.1093/genetics/150.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogiwara H, Ui A, Enomoto T, Seki M. Role of Elg1 protein in double strand break repair. Nucleic Acids Res. 2007;35:353–362. doi: 10.1093/nar/gkl1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang C, Stillman B. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev. 1997;11:3375–3386. doi: 10.1101/gad.11.24.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.White CI, Haber JE. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 1990;9:663–673. doi: 10.1002/j.1460-2075.1990.tb08158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ström L, Sjogren C. DNA damage-induced cohesion. Cell Cycle. 2005;4:536–539. doi: 10.4161/cc.4.4.1613. [DOI] [PubMed] [Google Scholar]
- 40.Beranek DT. Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat. Res. 1990;231:11–30. doi: 10.1016/0027-5107(90)90173-2. [DOI] [PubMed] [Google Scholar]
- 41.Lundin C, Erixon K, Arnaudeau C, Schultz N, Jenssen D, Meuth M, Helleday T. Different roles for nonhomologous end joining and homologous recombination following replication arrest in mammalian cells. Mol. Cell. Biol. 2002;22:5869–5878. doi: 10.1128/MCB.22.16.5869-5878.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moore CW. Internucleosomal cleavage and chromosomal degradation by bleomycin and phleomycin in yeast. Cancer Res. 1988;23:6837–6843. [PubMed] [Google Scholar]
- 43.Koy JF, Pleninger P, Wall L, Pramanik A, Martinez M, Moore CW. Genetic changes and bioassays in bleomycin- and phleomycin-treated cells and their relationship to chromosomal breaks. Mutat. Res. 1995;336:19–27. doi: 10.1016/0921-8777(94)00040-d. [DOI] [PubMed] [Google Scholar]
- 44.Sanchez Y, Bachant J, Wang H, Hu F, Liu D, Tetzlaff M, Elledge SJ. Control of the DNA damage checkpoint by Chk1 and Rad53 protein kinases through distinct mechanisms. Science. 1999;286:1166–1171. doi: 10.1126/science.286.5442.1166. [DOI] [PubMed] [Google Scholar]
- 45.Van Dyck E, Stasiak AZ, Stasiak A, West SC. Binding of double-strand breaks in DNA by human Rad52 protein. Nature. 1999;398:728–731. doi: 10.1038/19560. [DOI] [PubMed] [Google Scholar]
- 46.Hiom K. DNA repair: Rad52-the means to an end. Curr. Biol. 1999;9:446–448. doi: 10.1016/s0960-9822(99)80278-4. [DOI] [PubMed] [Google Scholar]
- 47.Lundblad V. DNA ends: maintenance of chromosome termini versus repair of double strand breaks. Mutat. Res. 2000;451:227–240. doi: 10.1016/s0027-5107(00)00052-x. [DOI] [PubMed] [Google Scholar]
- 48.Valencia M, Bentele M, Vaze MB, Herrmann G, Kraus E, Lee SE, Schar P, Haber JE. NEJ1 controls non-homologous end joining in Saccharomyces cerevisiae. Nature. 2001;414:666–669. doi: 10.1038/414666a. [DOI] [PubMed] [Google Scholar]
- 49.Haber JE. Switching of Saccharomyces cerevisiae mating-type genes. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. Washington, DC: ASM Press; 2002. pp. 927–952. [Google Scholar]
- 50.Moore JK, Haber JE. Capture of retrotransposon DNA at the sites of chromosomal double-strand breaks. Nature. 1996;383:644–646. doi: 10.1038/383644a0. [DOI] [PubMed] [Google Scholar]
- 51.Lundin C, North M, Erixon K, Walters K, Jenssen D, Goldman AS, Helleday T. Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res. 2005;33:3799–3811. doi: 10.1093/nar/gki681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tomizawa Y, Ui A, Onoda F, Ogiwara H, Tada S, Enomoto T, Seki M. Rad50 is involved in MMS-induced recombination between homologous chromosomes in mitotic cells. Genes Genet. Syst. 2007;82:157–160. doi: 10.1266/ggs.82.157. [DOI] [PubMed] [Google Scholar]
- 53.Blat Y, Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- 54.Tanaka T, Cosma MP, Wirt KH, Nasmyth K. Identification of cohesin association sites at centromeres and along chromosome arms. Cell. 1999;98:847–858. doi: 10.1016/s0092-8674(00)81518-4. [DOI] [PubMed] [Google Scholar]
- 55.Laloraya S, Guacci V, Koshland D. Chromosomal addresses of the cohesin component Mcd1p. J. Cell. Biol. 2000;151:1047–1056. doi: 10.1083/jcb.151.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Glynn EF, Megee PC, Yu HG, Mistrot C, Unal E, Koshland DE, DeRisi JL, Gerton JL. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2004;2:E259. doi: 10.1371/journal.pbio.0020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Uhlmann F, Nasmyth K. Cohesion between sister chromatids must be established during DNA replication. Curr. Biol. 1998;8:1095–1101. doi: 10.1016/s0960-9822(98)70463-4. [DOI] [PubMed] [Google Scholar]
- 58.Haering CH, Schoffnegger D, Nishino T, Helmhart W, Nasmyth K, Lowe J. Structure and stability of cohesin's Smc1-kleisin interaction. Mol. Cell. 2004;15:951–964. doi: 10.1016/j.molcel.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 59.Gerlich D, Koch B, Dupeux F, Peters JM, Ellenberg J. Live-cell imaging reveals a stable cohesin-chromatin interaction after but not before DNA replication. Curr. Biol. 2006;16:1571–1578. doi: 10.1016/j.cub.2006.06.068. [DOI] [PubMed] [Google Scholar]