Abstract
DNA topoisomerase IIα (topo IIα) is an essential nuclear enzyme and its unique decatenation activity has been implicated in many aspects of chromosome dynamics such as chromosome replication and segregation during mitosis. Here we show that chromatin-associated protein HMGB1 (a member of the large family of HMG-box proteins with possible functions in DNA replication, transcription, recombination and DNA repair) promotes topo IIα-mediated catenation of circular DNA, relaxation of negatively supercoiled DNA and decatenation of kinetoplast DNA. HMGB1 interacts with topo IIα and this interaction, like the stimulation of the catalytic activity of the enzyme, requires both HMG-box domains of HMGB1. A mutant of HMGB1, which cannot change DNA topology stimulates DNA decatenation by topo IIα indistinguishably from the wild-type protein. Although HMGB1 stimulates ATP hydrolysis by topo IIα, the DNA cleavage is much more enhanced. The observed abilities of HMGB1 to interact with topo IIα and promote topo IIα binding to DNA suggest a mechanism by which HMGB1 stimulates the catalytic activity of the enzyme via enhancement of DNA cleavage.
INTRODUCTION
DNA topoisomerase II (topo II) is an essential and ubiquitous enzyme for proliferation of eukaryotic cells (1). It can alter the topological state of DNA and untangle DNA knots and catenanes (interlocked rings) via ATP-dependent passing of an intact double helix through a transient double-stranded break generated in a separate DNA segment, followed by religation and enzyme turnover (2). In mammalian cells, topo II exists in two isoforms, α (170 kDa) and β (180 kDa), both having similar primary structure and almost identical catalytic properties, but differing in their production during the cell cycle (1,3).
Topo II is the primary target of a number of active agents currently used in the treatment of human cancers, such as epipodophyllotoxins (etoposide and teniposide), anthracyclines (doxorubicin and daunorubicin) and mitoxantrone (3). These drugs (also termed topo II poisons) can stabilize the covalent enzyme-associated complexes and shift the DNA cleavage/religation equilibrium of the enzyme reaction toward the cleavage state, converting biological intermediates of topo II activity into lethal ones ultimately leading to triggering of programmed cell death pathways (1,3,4).
HMGB1 is an abundant, ubiquitous and evolutionarily highly conserved non-histone chromatin-associated protein in mammals, which functions in a number of fundamental cellular processes such as transcription, replication, DNA repair and recombination (5–8). HMGB1 is associated with chromosomes in mitosis and due to its extreme mobility in the cell, the protein is continuously exchanged between nucleus and cytoplasm (5 and references therein). HMGB1 also exhibits an important extracellular function in mediation of inflammation mechanisms, tumor growth and metastasis (5,6). HMGB1 binds relatively weakly to B-form DNA, but displays a high affinity for distorted DNA conformations [e.g. four-way DNA junction, DNA minicircles, hemicatenated DNA loops and cisplatin-modified DNA; (7–12)]. Binding of HMGB1 to DNA causes local distortions by bending/looping or changes of DNA topology (7,13,14). HMGB1 also interacts weakly with a number of proteins, including transcription factors, site-specific recombination and DNA repair proteins (5). The importance of HMGB1 for life is supported by the phenotype of the HMGB1 knockout mice, which die 24 h after birth due to hypoglycemia and exhibit a defect in the transcriptional function of the glucocorticoid receptor (15).
In the present study, we report a physical interaction between HMGB1 and human topo IIα in vitro. We show that HMGB1 promotes topo IIα-mediated catenation of circular DNA, enhances relaxation of negatively supercoiled DNA and decatenation of kinetoplast DNA. Stimulation of the catalytic activity of topo IIα by HMGB1 is mainly due to enhanced DNA binding and cleavage by the enzyme. Possible functioning of HMGB1 as a modulator of the cellular activity of topo IIα is discussed.
MATERIALS AND METHODS
Enzymes and antibodies
Experiments were carried out either with human topoisomerase IIα (topo IIα) purchased from Topogen, or with recombinant human topo IIα isolated from yeast strain JEL1Δtop1 (the strain was kindly provided by John L. Nitiss, Department of Molecular Pharmacology, St. Jude Children's Research Hospital, Memphis, USA) harboring plasmid YEpWOBalphaHT (kindly provided by Anni H. Andersen, Department of Molecular Biology, University of Aarhus, Aarhus, Denmark) as detailed in references (16–18). Escherichia coli topoisomerase IV (10 U/μl) and wheat germ topoisomerase I (2–10 U/μl) were purchased from Topogen and Promega, respectively. Kinetoplast DNA (kDNA) was isolated from Crithidia fasciculata (the cells were kindly provided by Paul T. Englund, Haskins Laboratories and Biology Department, Pace University, New York, USA) as detailed in (19). kDNA was also kindly provided by Julius Lukeš (Institute of Parasitology, České Budějovice, Czech Republic). Antibodies against the following proteins were used: anti-HMGB1 (affinity purified rabbit polyclonal, BD Pharmingen), and anti-topo IIα (rabbit polyclonal, Topogen).
Plasmids
DNA plasmids were isolated by alkaline lysis method, followed by purification by two rounds of cesium chloride gradient or by the Qiagen plasmid kits. All purified plasmids exhibited ratios A260/A280 higher than 1.85. In some cases (catenation assays), supercoiled plasmids pTZ19R or pBR322 were relaxed by wheat germ topoisomerase I, followed by deproteinization of the relaxed plasmid as detailed earlier (20).
Cloning of HMGB1 and site-directed mutagenesis
HMGB1 (residues 1–215), HMGB1 domain A (residues 1–88), HMGB1 domain B (residues 85–180) and HMGB1 di-domain A+B (residues 1–180) were derived from rat HMGB1 cDNA (the amino acid sequence of the rat HMGB1 protein is identical to that of the human HMGB1 protein). Alanine mutagenesis of intercalating residues Phe38 (domain A), Phe103 and I122 (domain B) of the individual HMGB1 domains or the full-length HMGB1 was carried out using PCR-based protocol generating ‘chimeric proteins’ (Štros, unpublished data). The introduced mutations were verified by dideoxi-sequencing of both strands. The DNA sequences coding for the HMGB1 and truncated forms were inserted into BamHI and SalI sites of the vector pQE-80L (Qiagen) which allows tightly regulated N-terminal 6xHis-tagged protein expression in E. coli. The N-terminally GST (Glutatione S-Transferase) tagged HMGB1 protein and its truncated forms were also synthetized from the pGEM-4T1 vector (GE Healthcare). Purification of HMGB1 proteins by FPLC-chromatography, and SDS–18% polyacrylamide gel electrophoresis was performed as described earlier (11,21,22).
DNA supercoiling assay
DNA supercoiling assay was carried out as previously described (20) with the following modifications. Negatively supercoiled plasmid pBR322 DNA (0.5 μg or ∼9 nM) was relaxed in the relaxation buffer (40 mM NaCl, 50 mM Tris-HCl pH 7.5, 1 mM EDTA, 20% glycerol, 1 mM DTT) by wheat germ topoisomerase I (2U, Promega) at 37°C for 90 min. Then a new portion of the enzyme was added and the relaxed DNA was subsequently mixed either with wild-type HMGB1 or mutated HMGB1 (0–6 μM) in a final volume of 20 μl. The reactions were allowed to proceed for 1 h at 37°C after which 5 μl of the termination mix (5× TBE, 5% SDS, 15% sucrose, 0.1% bromphenol blue, 0.1% xylene cyanol, 0.2 μg/μl proteinase K in 20 mM Tris-HCl pH 8.0,1 mM CaCl2) was added and the samples were incubated at 45°C for 30 min, followed by phenol/chloroform extraction (11). The DNA topoisomers were then resolved on a 1% agarose gel in 0.5× TBE buffer at ∼3 V/cm for 17 h, and the DNA samples were visualized by UV-illumination of the ethidium bromide-stained gel (0.5 μg/ml).
Enhancement of intermolecular DNA ligation by HMGB1
Supercoiled plasmid pTZ19R (∼15 nM) was linearized by HindIII digestion, the deproteinized linear DNA was mixed with HMGB1 (2 or 6 μM), and pre-incubated on ice for 20 min. The DNA was then ligated with 0.2 U of T4 DNA ligase in a final volume of 20 μl at 30°C for 30 min in the presence or absence of 5% (w/v) polyethylene glycol (PEG 8000, Sigma). Deproteinized DNA samples were resolved on 1% agarose gels, followed by staining with ethidium bromide (0.5 μg/ml).
Human topoisomerase IIα catalytic activity assays
The following assays were used to assess topo IIα activity: catenation, decatenation, relaxation, cleavage and religation. Most experiments were carried out both with human topo IIα purchased from Topogen and with recombinant human topo IIα isolated from yeast (see ‘Materials and Methods section’). ‘Catenation assay’ was carried out with either negatively supercoiled or relaxed closed-circular plasmids. The plasmid pTZ19R (∼15 nM) was pre-incubated on ice in a total volume of 20 μl of topo IIα assay buffer (50 mM Tris-HC1 pH 8, 85 mM KCl, 10 mM MgCl2, 1 mM ATP, 0.5 mM dithiothreitol, 30 μg/ml BSA) containing 0–10% PEG (w/v) with different amounts of HMGB1 (as indicated in the figure legends) for 20 min, followed by addition of topo IIα (typically 5 nM). The mixture was finally incubated at 37°C for 40–60 min. ‘Decatenation assay’ was performed in the topo IIα assay buffer by incubation of 0.2 μg of kinetoplast DNA (kDNA) with topo IIα (as indicated in the figure legends). No PEG was present in the decatenation assay buffer. ‘Relaxation assay’ was carried out in the topo IIα assay buffer by treatment of negatively supercoiled plasmid pBR322 (∼8 nM) with either a fixed amount of topo IIα and increasing amounts of HMGB1 or with increasing amounts of topo IIα and a fixed amount of HMGB1 as indicated in the figure legends. No PEG was present in the relaxation assay buffer. All other reaction conditions of the relaxation assay were identical to the catenation assay. The above topo IIα activity assays were terminated by rapid addition of 460 μl of 1 M NaCl/1% SDS/10 mM EDTA and 2 μl of 2.5% linear polyacrylamide as carrier, followed by vortexing with an equal volume of chloroform–isoamylalcohol (24:1, v/v). The mixtures were then centrifuged at room temperature at 10 000 g for 10 min, and the clarified upper layers were precipitated with 1 ml of absolute ethanol at −70°C for 20 min. The DNA precipitates were washed with 70% ethanol, air-dried and dissolved in 0.1× TE buffer. ‘DNA cleavage assay’ was carried out by incubation of plasmid pBR322 (∼4 nM) with topo IIα in DNA cleavage buffer (10 mM Tris-HCl pH 8, 50 mM KCl, 50 mM NaCl, 5 mM MgCl2, 2.5% glycerol, 0.1 mM EDTA, containing either 1 mM ATP or 1 mM non-hydrolyzable analog ANP-PNP, Sigma) in a total volume of 20 μl at 37°C for 20 min. Some reactions also contained HMGB1 (1–3 μM) and/or etoposide (10–100 μM, Sigma) as indicated in the figure legends. The cleavage reactions were terminated by trapping the DNA cleavage complexes by addition of 2 μl of 10% SDS, followed by addition of 1.5 μl of 0.25 M EDTA and 2 μl of proteinase K (0.8 μg/μl in 50 mM Tris-HCl pH 8, and 1 mM CaCl2) and incubation at 45°C for 60 min. ‘Religation assay’ was carried out in DNA cleavage buffer containing 30 ng BSA (bovine serum albumin)/μl in a DNA mixture with negatively supercoiled plasmid pBR322 (4 nM), 50 μM etoposide, and topo IIα (8 nM). In some cases, HMGB1 (1–3 μM) was pre-incubated with DNA for 20 min on ice before addition of the enzyme. The reactions were started by incubation at 37°C for 15 min, followed by shifting the incubation temperature to 65°C or 75°C (to promote only the topo IIα-mediated religation and not the cleavage). Aliquots (20 μl) were then withdrawn at different times (typically 0–40 min), immediately mixed with 2.2 μl of 10% SDS, and finally digested with proteinase K (see ‘DNA cleavage assay’). The deproteinized DNA samples were finally resolved on 1% agarose gels containing 0.5× TBE. DNA was visualized by ethidium bromide staining which was either present in the gels prior to electrophoresis (DNA decatenation, cleavage and religation assays), or gels were stained after electrophoresis (DNA relaxation and catenation assays). DNA was quantified either using the ImageQuant TL software (GE Healthcare) or Multi Gauge software using imaging system LAS-3000 (Fuji).
Decatenation of kDNA by E. coli topoisomerase IV
Decatenation assay was carried out with 0.2 μg of kDNA in 40 mM HEPES-KOH (pH 7.6), 100 mM potassium glutamate, 10 mM magnesium acetate, 2 mM ATP, 10 mM DTT and 50 μg BSA/ml with E. coli topoisomerase IV (typically 0.1–1 U) in the presence or absence of HMGB1 (1 or 4 μM) in a final volume 20 μl at 37°C for 30 min, according to the manufacturer's instructions (Topogen). Analysis of decatenated products was performed as detailed for decatenation by human topo IIα.
Electrophoretic mobility shift assay
The 36-mer oligonucleotides for EMSA containing a high-affinity topo IIα-binding site (47,48) were (5′ to 3′): oligo_1, ATGAAATCTAACAATGCGCTCATCGTCATCCTCGGC; oligo_2, GCCGAGGATGACGATGAGCGCATTGTTAGATTTCAT.
The oligo_1 was 32P-labeled at its 5′-terminus by T4 DNA kinase and [γ-32P]ATP (specific activity 3000 Ci/mmol, GE Healthcare) and annealed with its corresponding complementary strand (oligo_2) to form a DNA duplex. Reaction mixtures for EMSA contained isolated topo IIα (75 nM) and increasing amounts of HMGB1 proteins (0.4–1.6 μM), ∼35 ng of 32P-labeled DNA duplex in EMSA buffer (20 mM HEPES, pH 7.6, 4% Ficoll, 0.02% Nonidet P-40, 1.5 mM spermidine, 0.1 mM EDTA, 0.125 M KCl and 0.5 mM DTT) in a total volume of 20 μl. In some EMSA assays, 0.2 μg of homopolymer poly (dI-dC) was used as a non-specific competitor DNA to reduce non-sequence specific binding of HMGB1. However, we have noticed that the presence of the competitor DNA significantly reduced the binding of topo IIα to the linear 36-bp DNA duplex as previously reported (23). The proteins were pre-incubated with DNA on ice for 20 min (order of mixing had no effect on the outcome of the EMSA assays) and finally resolved on 5% polyacrylamide gels in 0.5× TBE buffer at 250 V at 4°C until the bromphenol blue reached the bottom of the gel. The gels were dried and the labeled DNA was imaged by Storm PhosphorImager (Molecular Probes).
ATP hydrolysis by topoisomerase IIα
ATP hydrolysis was studied in reactions containing 1 mM cold ATP plus 20 × 106 c.p.m. of [γ-32P]ATP (specific activity 3000 Ci/mmol, GE Healthcare) and negatively supercoiled plasmid pBR322 (50 nM). Some reactions also contained HMGB1 (0.2–3 μM). ATP hydrolysis was initiated by addition of topo IIα (5 nM). Termination of ATP hydrolysis was accomplished by spotting the mixtures on TLC cellulose plates (20 × 20 cm; Baker-flex, Phillipsburg) at different times as previously described (24). Products of ATP hydrolysis were separated by thin layer chromatography and the free phosphate was quantified by PhosphorImager.
Electron microscopy
Following incubation of the relaxed circular plasmid DNA with topo IIα and HMGB1, the DNA was deproteinized by chloroform:isoamylalcohol (24:1, v/v) extraction. RecA coating of DNA for EM was carried out by the protocols detailed in (25). Briefly, for single-strand DNA coating with RecA (∼9.1 kb plasmid pAK-9.1), DNA was denatured at 63°C in the presence of glyoxal and purified on a Sephacryl S-500 minicolumn (0.4 × 4 cm) (GE Healthcare). Denatured DNA was then mixed with RecA protein at a molar ratio of 70:1 (∼2.8-kb plasmid pTZ19R) or 30:1 (∼9.1-kb plasmid pAK-9.1), cross-linked with 0.2% (v/v) glutaraldehyde, and purified on a Sephacryl S-500 minicolumn. Coating of double-stranded DNA (∼2.8 kb plasmid): following DNA coating with RecA, 0.5 mM ATP-γS was added to stabilize the complexes. DNA cross-linking with glutaraldehyde and purification of the RecA-coated dsDNA was carried out as for single-stranded DNA. RecA-coated DNA samples (15 μl) were adsorbed to a freshly glow-discharged (26) carbon-coated parlodion film on EM grids. Specimens were washed with 100 mM ammonium acetate, stained on drops of 5% (w/v) uranyl acetate and washed on the surface of 10 mM ammonium acetate (25). The grids were both rotary and unidirectionally shadowed with platinum/palladium at an angle of ∼7° and observed with a JEOL JEM 1200EX electron microscope operating at 60 kV. The photographs were taken at 50 000 magnification.
GST pull-down assay
GST alone or GST fused with full-length HMGB1 or its HMG-box domains at their N-termini were isolated as described (21,22). The purifed proteins (typically 1–2 μg) were bound to glutathione Sepharose 4B beads (40 μl of 1:1 v/v slurry), and incubated with purified human topo IIα (0.5 μg) in buffer PD [20 mM Tris-HCl pH 7.6, 0.2 M NaCl, 10 mM DTT, 0.2% Triton X-100, 0.2% Tween-20, 20% glycerol (v/v), protease inhibitors: 1 μg/ul aprotinin, 10 μg/ul leupeptin, 1 μg/ul pepstatin A, 100 μg/ul trypsin inhibitor, 0.1 mM TLCK and 20 mM benzamidine] in the presence or absence of 10 U of DNAse I for 15 min at 25°C, followed by rocking the samples for 2 h at 4°C. Subsequently, the beads were washed five times in 1 ml of 1× PD buffer. The proteins associated with Sepharose beads were finally eluted from the beads by addition of 20 μl of 10× SDS loading buffer and boiling the samples for 3–4 min. The beads-associated proteins were resolved on an SDS–7.5% polyacrylamide gel, transferred onto the PVDF membrane by western blotting and detected using polyclonal human topo IIα antibody (1:1000 dilution, Topogen), followed by incubation of the membranes with horseradish peroxidase-conjugated anti-rabbit antibodies (IgG-HRP) (1:2000 dilution, GE Healthcare) and ECL detection (GE Healthcare).
RESULTS
HMGB1 stimulates interlocking of DNA by human topoisomerase IIα as revealed by gel electrophoresis
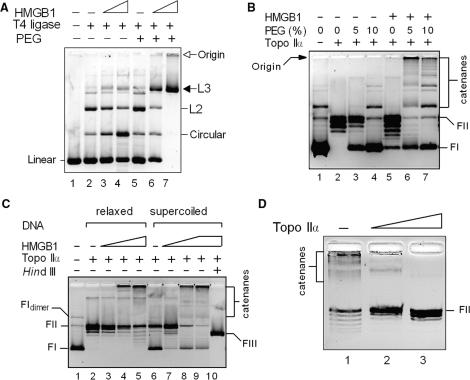
We have previously shown that chromatin architectural protein HMGB1 could stimulate T4 DNA ligase-mediated end-to-end joining of linear DNA molecules by promoting intramolecular association of DNA molecules via their ends (21,27). Ligation of diluted solutions of linearized plasmid DNA in the presence of HMGB1 resulted in preferential intramolecular ligation into closed-circular DNA (Figure 1A, lanes 2–4). However, as both DNA and HMGB1 are present in eukaryotic nuclei in relatively high concentrations, the effect of the protein on DNA end-joining was re-investigated in the presence of a macromolecular crowding (volume excluding) agent, polyethyleneglycol (PEG), to mimic crowding in vitro. In the absence of HMGB1, PEG alone had only little effect on DNA ligation using limiting amounts of the enzyme (Figure 1A, compare lanes 2 and 5). However, the simultaneous presence of HMGB1 and PEG suppressed intramolecular ligation and markedly promoted intermolecular DNA end-joining. Under these conditions, all monomeric DNA molecules were converted into linear DNA multimers, the majority of which migrating on agarose gels with the mobility of trimers or higher multimers (designated as L3 in Figure 1A), and into a minor fraction of high molecular mass linear multimers (which were susceptible, like L3, to ExoIII digestion, data not shown) not entering the gel (Figure 1A, compare lanes 5 and 7).
Figure 1.
HMGB1 promotes intermolecular association of DNA. (A) Macromolecular crowding favors intermolecular ligase-mediated DNA end-joining by HMGB1. Linearized plasmid pTZ19R (∼15 nM) was pre-incubated with 0.5 μM (lanes 3 and 6) or 1.5 μM (lanes 4 and 7) HMGB1, and then treated with 0.2 U of T4 DNA ligase in the presence (lanes 6 and 7) or absence (lanes 3 and 4) of 5% polyethyleneglycol (PEG). L2, dimers; L3 trimers or higher multimers. Linear, linearized plasmid pBR322; circular, closed-circular plasmid pBR322. (B) HMGB1 promotes topo IIα-catalyzed interlocking of DNA into multimers (catenanes) in the presence of PEG. Supercoiled plasmid pTZ19R (∼15 nM, lane 1) was pre-incubated with HMGB1 (4.5 μM) in the absence or presence of PEG (as indicated), and treated with topo IIα (∼7 nM). (C) Both relaxed and supercoiled plasmid DNAs form multimers with HMGB1 and topo IIα. Relaxed or supercoiled plasmids pTZ19R (∼15 nM) were pre-incubated with 0.5 μM (lanes 3 and 7), 1.5 μM (lanes 4 and 8) and 4.5 μM HMGB1 (lanes 5 and 9) in the presence of 5% PEG, followed by treatment with topo IIα (∼7 nM). (D) DNA multimers formed by topo IIα and HMGB1 are catenanes. Reactions from (C) (lane 4) were deproteinized and treated with increasing amounts of topo IIα (10 and 20 nM, left to right) for 30 min at 37°C. Deproteinized samples in (A–D) were separated on 1% agarose gels, and the resolved DNA samples were visualized by ethidium bromide staining as detailed in Materials and Methods section. The gels are presented as negatives. FI, supercoiled plasmid DNA; FII, relaxed closed-circular plasmid DNA; FIII, linearized plasmid DNA (HindIII).
The ability of HMGB1 to promote intermolecular association of DNA prompted us to determine whether HMGB1 could also promote interlocking (catenation) of covalently closed DNA by human topoisomerase IIα (topo IIα). As shown in Figure 1B (lanes 2 and 5), incubation of supercoiled DNA and HMGB1 with limiting amounts of topo IIα resulted in no formation of DNA multimers unless 5% PEG was included in the reaction buffer (Figure 1B, lane 6). Most of the complex DNA multimers did not enter the agarose gel and remained at its origin. The apparent size of the complex DNA multimers was visibly decreased at 10% PEG, and a significant fraction of the DNA multimers was no longer trapped in the wells and migrated rather as discrete bands of lower mobility (Figure 1B, lane 7).
Stimulation of topo IIα-mediated formation of DNA multimers by HMGB1 was independent of the topological state of the initial DNA (Figure 1C). Interestingly, HMGB1 and topo IIα could induce small changes in the linking number of relaxed closed-circular DNA at the highest molar ratio HMGB1-to-DNA studied as evidenced by the formation of faster migrated DNA topoisomers (Figure 1C, lane 5). The latter finding is reminiscent of the effect of HMGB1 on DNA supercoiling of closed-circular plasmid DNA by topoisomerase I (13,14).
In order to characterize the DNA multimers formed by plasmid DNA, HMGB1 and topo IIα in the presence of PEG, the deproteinized DNA multimers were treated with restriction nuclease HindIII or topo IIα. Digestion of the DNA multimers by HindIII resulted in a single band corresponding to the linearized plasmid DNA (Figure 1C, lane 10). The DNA multimers could be resolved into relaxed circular DNA and a minor fraction of circular plasmid dimers upon treatment with topo IIα (Figure 1D). The above results suggested that the DNA multimers formed by plasmid DNA, HMGB1 and topo IIα in the presence of PEG represented catenated DNA, i.e. double-stranded DNA molecules interlocked by a double-stranded DNA pass, rather than hemicatenated DNA molecules.
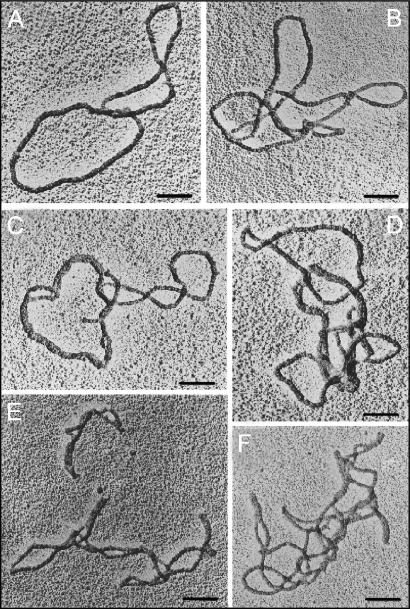
Electron microscopy reveals that DNA catenanes are formed by HMGB1 and topoisomerase IIα
In order to further verify that HMGB1 promotes formation of fully inter-locked DNA molecules by topo IIα, and to assess the complexity of these multimers, the DNA complexes were visualized by electron microscopy (EM). Prior to EM, the DNA multimers (originating either from a 2.8-kb or a 9.1-kb plasmid for double-stranded or single-stranded DNA coating, respectively) were deproteinized, followed by coating with RecA protein for better imaging of the DNA at cross-over points. Examination of the samples by EM revealed that the DNA multimers formed by HMGB1 and topo IIα contained many linked DNA molecules (>60% of all visualized DNA samples), and that the majority of these catenated molecules formed heterogeneous population of DNA molecules inter-linked with one or several DNA molecules (Figure 2; no DNA multimers were observed when DNA was incubated with HMGB1 in the absence of topo IIα, data not shown). The complex nature of these DNA multimers revealed by EM (Figure 2) corresponded to their migration in the course of agarose gel electrophoresis: they either remained at the origin of the gel or migrated with very low mobility (Figure 1), suggesting their high-molecular mass.
Figure 2.
The complex multimers formed by topoisomerase IIα and HMGB1 consist of fully catenated DNA molecules. Multimers were formed with plasmid DNA of 9.1 kb (panels A–D) or 2.8 kb (panels E–F), HMGB1 and human topoisomerase IIα. The samples were deproteinized and coated with RecA protein as single-stranded (panels A–D) or double-stranded DNA (panels E–F) before visualization by electron microscopy. All negatives were shot at 50 000 magnification. The bars represent 0.1 μm.
HMGB1 stimulates decatenation of kinetoplast DNA and relaxation of supercoiled DNA by topoisomerase IIα
To find out whether other catalytic activity of topo IIα than DNA catenation are stimulated by HMGB1, the effect of HMGB1 on topo IIα-mediated decatenation of kinetoplast (kDNA) or relaxation of negatively supercoiled DNA was studied.
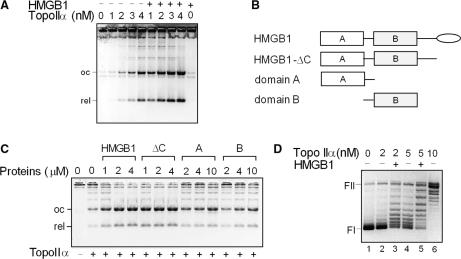
To study the effect of HMGB1 on DNA decatenation, increasing amounts of the topo IIα were added to kinetoplast DNA (kDNA) in the presence or absence of HMGB1. As shown in Figure 3A, the amount of decatenated DNA products increased proportionally with the amount of added topo IIα, and the decatenation activity of the enzyme was up to ∼10-fold higher (depending on the amount of the enzyme) in the presence of HMGB1 as revealed by the densitometry of the band intensities of the decatenated kDNA minicircles. No decatenation of kDNA was observed by incubation with HMGB1 alone in the absence of topo IIα (Figure 3A).
Figure 3.
HMGB1 stimulates decatenation of kinetoplast DNA and relaxation of supercoiled DNA by topoisomerase IIα. (A) Decatenation of kinetoplast DNA (kDNA) by topo IIα is stimulated by HMGB1. kDNA (0.2 μg) was incubated with increasing concentrations of topo IIα (as indicated) in the absence or presence of HMGB1 (1 μM). (B) Domain structure of HMGB1, HMGB1-ΔC, HMGB1 lacking the acidic C-tail (depicted as oval); A, domain A; B, domain B. (C) HMGB1 stimulates decatenation of kDNA by topo IIα via the HMG-box domains. kDNA (0.2 μg) was incubated with topo IIα (2 nM) in the presence or absence of increasing amounts of HMGB1, HMGB1-ΔC, domain A or domain B (as indicated). ΔC, HMGB1-ΔC. (D) HMGB1 stimulates relaxation of negatively supercoiled DNA by HMGB1. Relaxation of negatively supercoiled pBR322 plasmid DNA (∼8 nM) by different concentrations of topo IIα (as indicated) in the presence or absence of recombinant HMGB1 (1 μM), 37°C for 45 min. Decatenation and relaxation assays were carried out in the absence of PEG. Deproteinized DNA samples were resolved on 1% agarose gels containing ethidium bromide (A and C) or without ethidium bromide (panel D). The gels are presented as negatives. FI, supercoiled plasmid DNA; FII, relaxed closed-circular plasmid DNA. oc, nicked closed-circular DNA minicircle; rel, relaxed closed-circular DNA minicircle.
HMGB1 protein consists of two HMG-boxes, domains A and B, and a highly acidic C-terminus (Figure 3B). To find out which part of HMGB1 is responsible for stimulation of decatenation of kDNA by topo IIα, decatenation experiments were carried out with HMGB1 and its truncated forms. As shown in Figure 3C, both HMGB1 and HMGB1-ΔC could enhance decatenation of kDNA to a similar extent, suggesting that the HMG-boxes of HMGB1, and ‘not’ the acidic C-tail, were responsible for the enhancement of the decatenation activity of topo IIα. Although both isolated HMGB1 domains, A and B, could decatenate kDNA, the observed stimulatory effect was significantly lower (up to ∼10-fold) than that observed with the full-length HMGB1 or the A+B di-domain (HMGB1-ΔC), Figure 3C.
The fact that complex DNA multimers were formed under conditions when the relaxation activity of the enzyme was severely compromised by higher concentrations of PEG, suggested that HMGB1 could also stimulate the relaxation activity of the enzyme (Figure 1B, compare lanes 6 and 7). As shown in Figure 3D, when increasing amounts of topo IIα were incubated with negatively supercoiled plasmid DNA, DNA was relaxed more efficiently when HMGB1 was present in the reactions.
The above experiments provided evidence that HMGB1 could stimulate both DNA catenation/decatenation and relaxation of supercoiled DNA by topo IIα. While PEG was necessary to observe DNA catenation by low amounts of topo IIα in the presence of HMGB1 (Figure 1), it was clearly dispensable for HMGB1-mediated stimulation of kDNA decatenation and relaxation of supercoiled DNA (Figure 3), suggesting that PEG itself could not stimulate topo IIα activity.
The possibility that HMGB1 might act as a chaperone for topo IIα protecting it from inactivation during the course of catalytic assays was excluded based on results from control experiments in which the enzyme was incubated in reaction buffer in the presence or absence of HMGB1 prior to addition of DNA (data not shown).
HMGB1-mediated stimulation of DNA decatenation by topoisomerase IIα is not due to the effect of HMGB1 on DNA topology
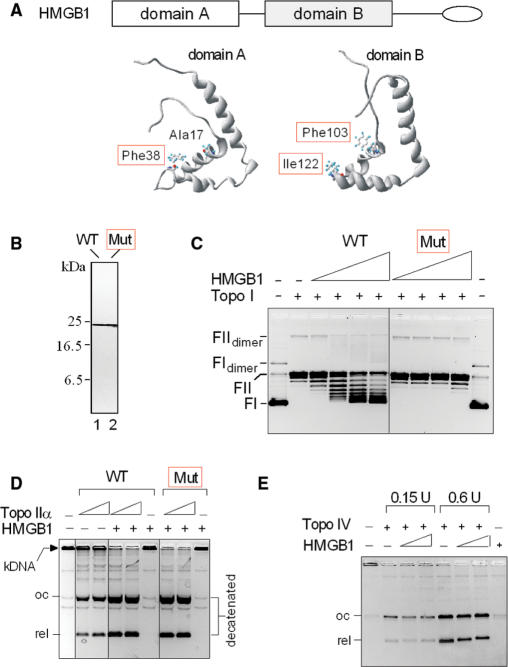
HMGB1 has been known to affect DNA topology by unwinding or by inducing negative supercoiling (13,14,20,28). Recently we demonstrated that alanine mutagenesis of intercalating residues of HMGB1 domains, A (Phe 38) and B (Phe 103 and Ile122) (Figure 4A and B), abrogated high-affinity binding of HMGB1 to bent DNA (minicircles) without affecting significantly its affinity to linear DNA (12). Here we show that the HMGB1 mutant also lost the ability to supercoil DNA in a topo I-mediated supercoiling assay (Figure 4C). To find out whether the mutated HMGB1 could still promote the catalytic activity of topo IIα, decatenation of kDNA was carried out. As shown in Figure 4D, mutated HMGB1 could promote decatenation of kDNA by topo IIα indistinguishably from the wild-type protein, suggesting that the observed enhancement of catalytic activity of topo IIα by HMGB1 was not affected by the HMGB1-mediated changes in DNA topology.
Figure 4.
Stimulation of topo IIα activity by HMGB1 is not due to the effect of HMGB1 on DNA topology. (A) HMGB1 protein and structure of HMGB1 domains A and B indicating positions of the mutated intercalating amino acid residues (in red frames). (B) Purity of HMGB1 and mutant as revealed by electrophoresis on an SDS–18% polyacrylamide gel and Coomassie blue R-250 staining. Lane 1, wild-type HMGB1; lane 2, HMGB1 mutant (F38A/F103A/I122A). A, alanine; F, phenylalanine; I, isoleucine. (C) Alanine mutagenesis of intercalating residues of HMGB1 abrogated the ability of HMGB1 to introduce supercoils into DNA by topoisomerase I. Closed-circular pBR322 plasmid DNA (∼9 nM) was incubated with wheat germ topoisomerase I in the absence or presence of HMGB1 or mutant (1, 2, 3 and 6 μM, left to right). FI, supercoiled plasmid DNA; FII, relaxed closed-circular plasmid DNA. (D) Both wild-type HMGB1 and HMGB1 mutant can enhance decatenation of kDNA by human topo IIα. Decatenation of kDNA was carried out as in Figure 3A. HMGB1 and mutant were at 1 μM, topo IIα was 7.5 and 15 nM. (E) HMGB1 does not stimulate decatenation of kDNA by E. coli topoisomerase IV. Decatenation of kDNA by E. coli topoisomerase IV (0.15 and 0.6 U) was carried out as detailed in the Materials and Methods section using HMGB1 or mutant (1 and 4 μM, left to right). The DNA samples in (C–E) were separated on 0.8% agarose gels without (C) or with (D–E) ethidium bromide in the course of electrophoresis as detailed in the Materials and Methods section. The gels are presented as negatives. WT, wild-type HMGB1; Mut, triple-mutant (F38A/F103A/I122A) of HMGB1.
The specificity of HMGB1 on the catalytic activity of human topo IIα was further investigated using E. coli type-II topoisomerase IV (29). As shown in Figure 4E, addition of increasing amounts of HMGB1 to the decatenation reactions did not enhance the activity of the prokaryotic enzyme. Thus, while the above data ruled out the effect of HMGB1-mediated changes in DNA topology on the stimulation of topo IIα activity (and also E. coli type-II topoisomerase IV), they may suggest an involvement of protein–protein interactions in the stimulatory effect of HMGB1 protein on the catalytic activity of human topo IIα.
HMGB1 physically interacts with topoisomerase IIα
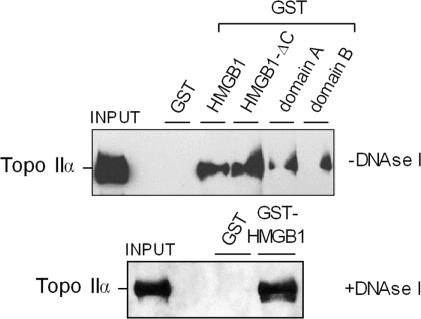
To find out whether the stimulatory effect of HMGB1 on the catalytic activity of topo IIα could be due to a direct physical interaction of HMGB1 with the enzyme, the pull-down assay was used to investigate the possible existence of the HMGB1–topo IIα interactions. To address this question, recombinant GST alone or GST-HMGB1 were immobilized on glutathione-agarose and incubated with isolated human topo IIα. After extensive washing of the beads, the bound proteins were resolved by SDS-PAGE and subjected to western blotting and immunological detection using an antibody specific to topo IIα. While no topo IIα was found to be associated with GST alone, a significant fraction of topo IIα was bound to GST-HMGB1 (Figure 5), suggesting that HMGB1 could interact with topo IIα. Using a similar approach we found that approximately similar amounts of topo IIα were associated with HMGB1 or HMGB1-ΔC, indicating that the HMG-boxes, and not the polyanionic C-tail, of HMGB1 were responsible for the interaction with topo IIα (Figure 5). From Figure 5 (upper panel), it also follows that both HMG-box domains, A and B, could interact with topo IIα. Similar results were obtained when DNAse I was included in the incubation buffer (Figure 5, lower panel), demonstrating that the interaction of HMGB1 with topo IIα was independent of DNA that might have been associated with the isolated proteins. Specificity of the ‘pull-down’ assay was verified with other proteins known to interact with HMGB1 [p53, pRb, (5,11,30)] as compared to those not binding to HMGB1 (E2F1, data not shown). Thus, the above ‘pull-down’ binding experiments strongly suggest that the interaction of HMGB1 with topo IIα is specific.
Figure 5.
HMGB1 physically interacts with topoisomerase IIα. HMGB1, HMGB1-ΔC, domains A and B (expressed in E. coli and fused proteins with GST at their N-termini) were bound to glutathione-Sepharose beads (GST was used as a negative control). The beads were then incubated with isolated human topo IIα, followed by extensive washing of the beads, and boiling them in SDS-containing buffer. The released proteins were resolved by electrophoresis on an SDS–7.5% polyacrylamide gel, followed by western blotting and immunological detection using polyclonal antibody specific to topo IIα. Input, 10% of the total topo IIα used for the pull-down assay. Upper and lower PD experiments were performed in the absence or presence of 10 U of DNAse I in the incubation buffer, respectively.
HMGB1 stimulates binding of topoisomerase IIα to DNA
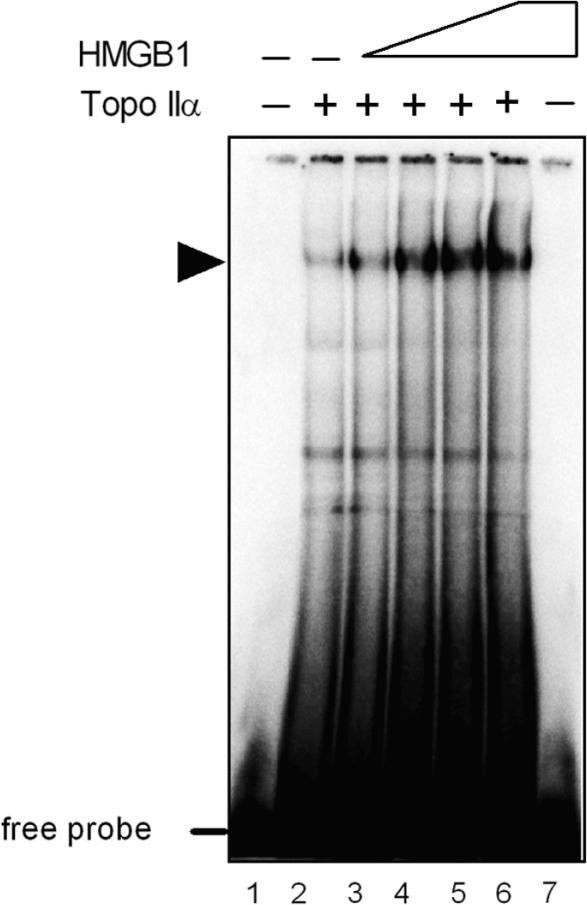
HMGB1 has previously been reported to promote binding of a plethora of transcription factors and other sequence-specific proteins to their cognate sites (5). To find out whether the stimulatory effect of HMGB1 on the catalytic activity of topo IIα might have originated from enhanced binding of the enzyme to DNA, the EMSA technique (Electrophoretic Mobility Shift Assay) was used to study the effect of HMGB1 on topo IIα binding to DNA containing a strong topo IIα recognition sequence. A gradual increase in topo IIα binding to DNA was observed when increasing amounts of HMGB1 were added to a mixture of DNA and topo IIα (Figure 6, arrow). Densitometric analysis revealed that the topo IIα binding to DNA was up to ∼10-fold enhanced by HMGB1. HMGB1 promoted topo IIα binding to DNA (Step 1 in Figure 10) without formation of a ternary complex topo IIα-HMGB1-DNA as no super-shift of the topo IIα–DNA complex was detected by specific anti-HMGB1 antibody (data not shown).
Figure 6.
HMGB1 stimulates binding of topoisomerase IIα to DNA. 32P-labeled 36-bp DNA duplex (∼75 nM) containing a high-affinity binding site for topo IIα was mixed with topo IIα (75 nM) and increasing amounts of HMGB1 protein (0, 0.4, 0.6, 0.8 and 1.6 μM, lanes 2–6, respectively), followed by separation of unbound DNA (free probe) and DNA–protein complexes on a 5% non-denaturing polyacrylamide gel (EMSA) as detailed in Materials and Methods section. The dried gel was subjected to PhosphorImaging. Lane 7, HMGB1 at 1.6 μM. Arrowhead indicates mobility of the topo IIα–DNA complex.
Figure 10.
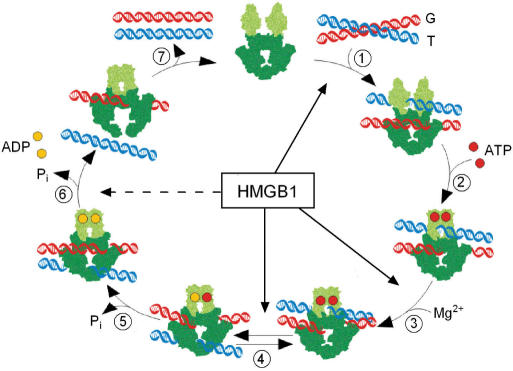
HMGB1 and catalytic cycle of DNA topoisomerase IIα. The drawing is adapted from ref. (46). The ATPase domains of topo IIα (light green) and the core domains (dark green) are depicted (the C-terminal domain which is not required for the enzymatic activity of topo IIα, is not shown). Binding of two double-stranded DNA duplexes G (in red) and T (in blue) initiates the catalytic cycle (Step 1). Upon binding of two ATP molecules (red filled circles), the two ATPase domains dimerize (Step 2). Cleavage of G duplex in Step 3 (‘pre-strand passage DNA cleavage’) is followed by passage of the T duplex through the double-stranded break in the G duplex (‘Gpost-strand passage DNA cleavage’) (Step 4). Religation of the G duplex and hydrolysis of ATP proceeds in Step 5. Two molecules of ADP dissociate in Step 6, followed by release of the T duplex through the C-terminal part of topo IIα. Reopening of the ATPase domains allows dissociation of the G duplex (Step 7) to regenerate the enzyme for the next catalytic cycle. Individual steps of the topo II catalytic cycle which are stimulated the most by HMGB1 are indicated by arrows: Step 1 (DNA binding, Figure 6), Steps 3 and 4 (DNA cleavage, Figure 7). The dashed arrow indicates the HMGB1-mediated partial relief of topo IIα inhibition by ICRF-193 in Step 6 (Figure 9).
HMGB1 promotes DNA cleavage activity by topoisomerase IIα
All catalytic activities of topo IIα are associated with the ability of the enzyme to form transient double-stranded breaks in the DNA duplex and to form covalent enzyme–DNA intermediates, referred as to the ‘cleavable complexes’. Topoisomerase IIα establishes two distinct DNA cleavage/religation equilibria: a ‘pre-strand passage DNA cleavage’ (corresponding to the ATP-free form of the enzyme), and a ‘post-strand passage DNA cleavage’ (corresponding to the ATP-bound form) (31). To monitor a ‘pre-strand passage DNA cleavage’, ATP is omitted in the assays (Step 3 in Figure 10). A ‘post-strand passage DNA cleavage’ is measured in the assays employing a non-hydrolyzable ATP analog, adenyl-5′-yl imidodiphosphate (AMP-PNP) (Step 4 in Figure 10). When ATP is included in the cleavage assays, a mixture of the two equilibria (Steps 3 and 4 in Figure 10) is measured (31).
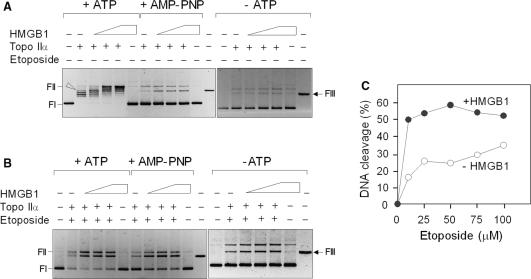
To find out whether the observed stimulation of the catalytic activity of topo IIα by HMGB1 is due to enhanced cleavage of the DNA substrate, the topo IIα-mediated cleavage of the supercoiled plasmid DNA was carried out. This assay allows one to quantify the topo IIα-mediated double-stranded DNA cleavage by detecting the conversion of covalently closed-circular DNA to linear molecules. In the absence of drugs stabilizing the ‘cleavable complexes’, the evaluation of the DNA scission by topo IIα using ATP as a co-factor was complicated by the appearance of individual topoisomers in the course of relaxation of supercoiled plasmid DNA (Figure 7A, +ATP). This was not the case when DNA cleavage assay was performed either in the absence of any cofactor (Figure 7A, −ATP) or using the non-hydrolyzable ATP analog (Figure 7A, AMP-PNP). The latter DNA cleavage experiments detected a significant (∼3-fold) HMGB1-mediated enhancement of DNA scission by topo IIα, while no visible DNA scission was observed when HMGB1 was incubated with supercoiled DNA in the absence of topo IIα (Figure 7A).
Figure 7.
HMGB1 stimulates DNA cleavage activity of topoisomerase IIα. (A) Effect of HMGB1 on DNA cleavage activity of topo IIα in the absence of etoposide. DNA cleavage reactions contained negatively supercoiled plasmid pBR322 (∼4 nM), human topo IIα (∼8 nM) and increasing concentrations of HMGB1 (1, 2 and 3 μM, left to right). ATP or a non-hydrolyzable ATP analog (AMP-PNP) were at 1 mM. Arrowhead indicates position of linearized DNA. (B) Effect of HMGB1 on DNA cleavage activity of topo IIα in the presence of etoposide. The molar concentrations of topo IIα and plasmid pBR322 were identical as in panel A. HMGB1 protein was at 1 μM and etoposide at 50 μM. (C) Effect of HMGB1 on DNA cleavage by topo IIα in the presence of increasing concentrations of etoposide. Negatively supercoiled plasmid DNA (4 nM) was pre-incubated on ice either with buffer (control) or HMGB1 (1 μM) in the presence of 0–100 μM etoposide. Cleavage reactions were initiated by addition of topo IIα (8 nM) and incubation was carried out at 37°C for 15 min. The cleavage complexes in A–C reactions were trapped by 1% SDS, followed by digestion with proteinase K as detailed in Materials and Methods section. Deproteinized DNA samples were resolved on 1% agarose gels and subsequently stained with ethidium bromide. FIII, HindIII-digested plasmid pBR322 (last right lanes in A and B) indicates mobility of linearized plasmid DNA. FI, supercoiled plasmid DNA; FII, relaxed closed-circular plasmid DNA. The gels are presented as negatives. The percentage of cleaved plasmid DNA was quantified from three independent cleavage experiments, each in duplicates, by ImageQuant TL (GE Healthcare). The average SD for the data was <7%. DNA cleavage is expressed as percentage of initial uncleaved supercoiled plasmid pBR322. (open circle) In the absence (control) or in the presence of HMGB1 (Filled circle).
The percentage of ‘cleavable complexes’ is usually very low, but the levels of the cleavable complexes are significantly enhanced in the presence of topo II poisons (such as etoposide) that affect the DNA cleavage/religation equilibrium (3). Similarly to the DNA cleavage experiments in the absence of etoposide (Figure 7A), HMGB1 could enhance >3-fold DNA cleavage by topo IIα when etoposide was included in the reactions (Figure 7B). Densitometric analysis of the percentage of linearized DNA revealed that HMGB1 could stimulate more efficiently (>3-fold) the ‘post-strand passage DNA cleavage’ than the ‘pre-strand passage DNA cleavage’ (∼2-fold), regardless of the presence or absence of etoposide.
The effect of HMGB1 on DNA cleavage by topo IIα was also studied in reactions with increasing concentrations of etoposide (0–100 μM) and a fixed amount of the enzyme. Approximately ∼2–4 times higher percentage of linear DNA was observed in the presence of HMGB1, with the most prominent effect at lower concentrations of the drug (10–50 μM), Figure 7C. The stimulatory effect of HMGB1 on formation of topo IIα–DNA cleavage complexes was detectable over a range of enzyme:plasmid ratios at fixed concentrations of the drug and HMGB1 (data not shown).
HMGB1 does not promote the ability of topoisomerase IIα to religate cleaved DNA
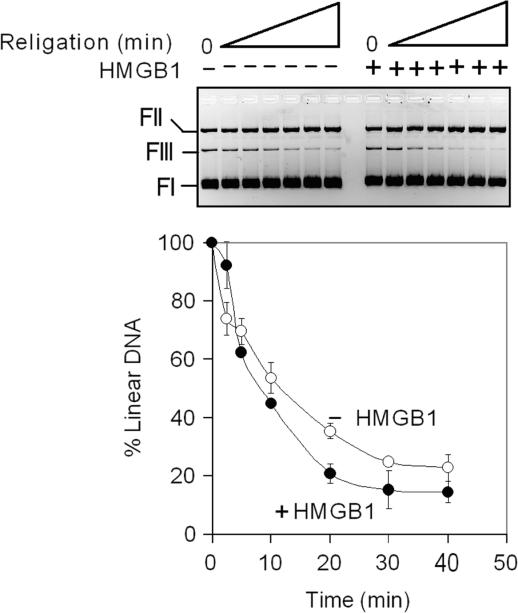
The ability of topo IIα to cleave and religate double-stranded DNA is essential for the regulation of DNA topology by the enzyme. As HMGB1 stimulates the DNA cleavage by topo IIα (Steps 3 and 4 in Figure 10), we have asked whether HMGB1 could also influence the ability of topo IIα to religate DNA using a heat-induced religation assay (Step 5 in Figure 10). This assay is based on the finding that religation activity of topo IIα is less sensitive to variations in temperature than DNA cleavage activity (32). By shifting the temperature from 37 to 65°C before termination with SDS, linearized DNA plasmid (generated by topo IIα-mediated DNA cleavage in the presence of etoposide) is re-circularized in a time-dependent manner. As shown in Figure 8, religation of DNA (as revealed by the decrease of linearized DNA) was very little, if any, enhanced by HMGB1, indicating that HMGB1 could not promote religation of the cleaved DNA by topoisomerase IIα. This was even more apparent when religation assay was carried out at 75°C (data not shown), a temperature which has been recently shown to inhibit more efficiently DNA cleavage by human topo IIα without significantly affecting the ability of the enzyme to religate DNA (33).
Figure 8.
HMGB1 does not significantly affect DNA religation by topoisomerase IIα. DNA cleavage reactions contained negatively supercoiled plasmid DNA (4 nM), 50 μM etoposide in the absence (control) or presence of HMGB1 (1 μM). Cleavage reactions were initiated by addition of topo IIα (8 nM) and incubation at 37°C for 15 min. The incubation temperature was then shifted to 65°C to suppress cleavage and to promote religation as described in Materials and Methods section. A representative ethidium bromide-stained agarose gel of the DNA religation assay is shown (religation times were 0, 2.5, 5, 10, 20, 30 and 40 min, left to right). The percentage of linear DNA was estimated by the Multi Gauge software using imaging system LAS-3000 (Fuji). Each curve represents the average of 2–3 independent assays. The average SD for the data was <8%. The amount of linearized plasmid molecules in the cleavage reaction containing or lacking HMGB1 was expressed as 100%. (open circle) in the absence (control) or in the presence of HMGB1 (filled circle). FI, supercoiled plasmid DNA; FII, relaxed closed-circular plasmid DNA; FIII, linear plasmid DNA.
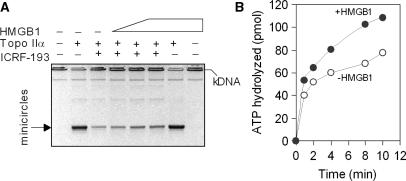
HMGB1 reduces inhibition of catalytic activity of topoisomerase IIα by ICRF-193 and enhances the rate of ATP hydrolysis
Topoisomerase IIα changes the topology of DNA by coupling binding and hydrolysis of two ATP molecules to the passage of one DNA duplex through a transient double-stranded break in another DNA duplex. The ‘catalytic inhibitor’ ICRF-193 traps the topo IIα in the closed-clamp intermediate form in the presence of ATP (Step 6 in Figure 10), resulting in the inhibition of ATPase activity, strand passage and catalytic turnover of the enzyme (34). As shown in Figure 9A, HMGB1 could partially relieve (<2-fold) the inhibitory effect of ICRF-193 on decatenation of kDNA by topo IIα. A slight, but reproducible, decrease in the inhibitory effect of ICRF-193 on topo IIα-mediated decatenation of kDNA was also detected over a wide range of the drug concentrations (data not shown). The latter results prompted us to examine whether HMGB1 could modulate the rate of ATP hydrolysis by topo IIα using thin layer chromatography enabling to separate released phosphate from ATP upon incubation of [γ-32P]ATP with topo IIα and negatively supercoiled plasmid DNA (24). Quantification of the released phosphate revealed that HMGB1 could enhance (<2-fold) the rate of ATP hydrolysis by topo IIα (Figure 9B). As the stimulatory effect of HMGB1 on ATPase activity of topo IIα is relatively modest, the DNA binding and cleavage are most likely the major control steps by which HMGB1 modulates the catalytic activity of the enzyme.
Figure 9.
HMGB1 enhances ATP hydrolysis by topoisomerase IIα. (A) HMGB1 reduces inhibition of catalytic activity of topo IIα by ICRF-193. kDNA (0.2 μg) was decatenated with topo IIα (2 nM) in the absence or presence of 10 μM ICRF-193. Some decatenation reactions contained HMGB1 (0.5, 1.5 and 3 μM, left to right). Decatenation was carried out as detailed in the Materials and Methods section. Due to the absence of ethidium bromide in the agarose gel in the course of electrophoresis, the decatenated minicircles migrated as single bands comprising both nicked and closed-circular DNA (designated as oc and rel in Figures 3 and 4). A representative ethidium bromide-stained 1% agarose gel (presented as a negative) is shown. (B) HMGB1 enhances the rate of ATP hydrolysis by topo IIα. ATP hydrolysis by topo IIα (5 nM) was studied in reactions containing 1 mM cold ATP and [γ-32P]ATP and negatively supercoiled plasmid pBR322 (50 nM). The ATP hydrolysis (as determined by a time-dependent release of free PO4) was measured by thin layer chromatography (24) and quantified by PhosporImaging. Data represent the average of two independent experiments.
DISCUSSION
A distinctive property of living systems is that biochemical processes proceed in a medium which is highly crowded with macromolecules (35). It has previously been shown that molecular crowding has a strong effect both on equilibrium and on the rate of reactions involving macromolecules (35). Polyethylene glycol (PEG) as a volume excluding agent may imitate the conditions under which HMGB1 is able to facilitate (non-covalent) intermolecular associations of DNA helices with subsequent ligation by limiting amounts of enzymes (21,27,36,37 and this study). The observed interlinking of circular, covalently closed DNA molecules by topo IIα in the presence of HMGB1 is likely a consequence of the PEG-mediated increase of local DNA and protein concentrations. Apart from increase of the DNA concentration, the presence of PEG obviously increases the concentration of HMGB1 in the catenation reaction (typically ∼1 μM in the test tube) which may then approach the expected actual concentration of HMGB1 in the cell nucleus, ∼25 μM (assuming ∼106 copies of HMGB1 per cell, the volume of the cell nucleus 6.5 × 10−11 ml, and disregarding the contribution of nuclear crowding agents which would otherwise likely lead to even higher concentration of HMGB1 in the nucleus). As higher amounts of topo IIα can promote DNA catenation in the absence of other factors (38), there has been a possibility that the observed DNA catenation in the presence of HMGB1 might have had arisen solely as a consequence of a PEG-induced increase in the topo IIα concentration. However, this seems unlikely as PEG itself does not promote DNA catenation by low amounts of topo IIα when HMGB1 is absent. A variety of polycations, including spermidine and histone H1, promote catenation, and this is most likely due to compaction of the DNA substrate into aggregates with high local DNA concentration favoring the catenated state (39). HMGB1 can promote interlocking of DNA rings by topo IIα only if a crowding agent is present in the course of the catenation reaction. This may reflect the fact that unlike DNA binding of the polyvalent cations, histone H1 or other nuclear proteins (such as nucleolin, DNA-PKcs and Ku70/80) (39,40 and references therein), aggregation of HMGB1–DNA complexes is much less apparent in diluted solutions (14,41,42).
HMGB1 promotes not only topo IIα-mediated catenation of circular DNA, but it can also enhance decatenation of kDNA and relaxation of negatively supercoiled DNA in vitro. The catalytic cycle of topo IIα depends on a coordinate function of several catalytic steps of the enzyme (Figure 10). The first step includes binding of the enzyme to DNA (Step 1 in Figure 10) which has been demonstrated to be significantly enhanced by HMGB1. HMGB1 stimulates catalytic activity of topo IIα by promoting DNA cleavage (Steps 3–4 in Figure 10), without affecting the pattern of specific cleavage sites on DNA (data not shown). Although both ‘pre-strand’ and ‘post-strand passage DNA cleavage’ steps (Steps 3 and 4, respectively in Figure 10) are stimulated by HMGB1, the stimulatory effect by HMGB1 is more prominent at the ‘post-strand passage DNA cleavage’ step of the catalytic cycle of the enzyme. A possible explanation of the HMGB1-mediated stimulation of DNA cleavage by topo IIα is enhanced binding of the enzyme to DNA. The latter may be a consequence of the reported ability of HMGB1 to act as a ‘DNA chaperone’ [e.g. by bending DNA, (6–8)] and/or due to direct interaction of the protein with topo IIα.
Topo IIα (3,23,43), like HMGB1 (7,8,14,44,45), can bind preferentially to supercoiled plasmid (including non B-DNA structures such as Z-DNA and four-way junctions) as compared to the linear B-DNA. Whether the reported preferential binding of HMGB1 to distorted or non B-type DNA structures contributes to recognition of topo IIα-specific DNA-binding sites/structures remains to be established. HMGB1 has also been previously reported to change DNA topology by unwinding the double-helical DNA or introducing negative supercoils in DNA (13,14,20,28). However, the HMGB1-mediated enhancement of decatenation by human topo IIα is similar when assayed using an HMGB1 mutant that does not affect DNA topology. In addition, the activity of E. coli topoisomerase IV, a prokaryotic type-II topoisomerase that has evolved independently from HMG-box proteins, is not influenced by changes in DNA topology induced by HMGB1. Therefore, we favor the hypothesis that the stimulatory effect of HMGB1 on the catalytic activity of the enzyme depends not only on DNA–protein interactions but also on protein–protein interactions. The fact that EMSA assay did not detect any ternary DNA–topo IIα–HMGB1 complex does not necessarily mean the lack of existence of such a complex. It is possible that HMGB1 can ‘deliver’ the topo IIα to its binding/cleavage sites by forming a ternary complex which is transient and unstable. This idea is in agreement with previous reports demonstrating the ability of HMGB1 to enhance the binding a plethora of sequence-specific proteins to their cognate DNA sites without formation of ternary complexes, despite the detection of the corresponding protein–protein interactions in pull-down assays (5,8).
ICRF-193 inhibits Step 6 of the catalytic cycle of topo IIα (Figure 10), the ATP hydrolysis, by trapping the enzyme on the DNA in the closed-clamp form (34). HMGB1 can partially relieve the inhibitory effect of ICRF-193 on decatenation of kDNA by topo IIα. HMGB1 could not significantly stimulate resealing of double-stranded breaks by topo IIα (religation Step 5 in Figure 10) using the temperature-induced religation assay. As the stimulatory effect of HMGB1 on the ATP hydrolysis by topo IIα (and even less on religation of double-stranded breaks) is relatively modest, the enhanced DNA binding of topo IIα and consequently higher DNA cleavage represent the major control steps by which HMGB1 modulates the catalytic activity of the enzyme.
An important question remains whether HMGB1 enhances the activity of topo IIα in vivo, and whether the possible activation involves direct stimulation of the enzymatic activity (as a result of mutual interactions) and/or de novo synthesis of the enzyme. The latter possibility may be indicated by our luciferase reporter gene assays demonstrating a significant (>10-fold) transactivation of the human topo IIα promoter by HMGB1 in Saos-2 cells (Štros et al., unpublished data).
Topo IIα expression is associated with a higher cell proliferation rate, and the level of topo IIα is very low in G1 phase, it increases in S phase, and is maximal in G2/M (1,3,47). Although HMGB1 is expressed throughout the cell cycle with no significant variations (5), the protein is clearly over-expressed in most human tumors, including breast carcinoma, melanoma, gastrointestinal stromal tumors, colon carcinomas and acute myeloblastic leukemia (48 and references therein). Further experiments are needed to clarify whether this over-expression affects the cellular activity and expression of topo IIα, and if so, whether it contributes to the reported enhanced topo IIα activity in tumors (49,50).
ACKNOWLEDGEMENTS
We thank John L. Nitiss (Department of Molecular Pharmacology, St. Jude Children's Research Hospital, Memphis, USA) and Anni H. Andersen (Department of Molecular Biology, University of Aarhus, Arhus, Denmark) for providing yeast strain JEL1Δtop1 and plasmid YEpWOBalphaHT, respectively. Cell culture of Crithidia fasciculata was kindly provided by Paul T. Englund (Haskins Laboratories and Biology Department, Pace University, New York, USA) and Julius Lukeš (Institute of Parasitology, České Budějovice, Czech Republic). ICRF193 was kindly provided by Olivier Hyrien (Laboratoire de Génétique Moléculaire, Ecole Normale Supérieure, Paris, France). We also thank Terezie Keprtová and Jiří Štros for secretarial assistance and preparation of Figure 10, respectively.
This research was supported by grants to M.Š. from the Grant Agency of the Czech Republic (204/05/2031) and the Grant Agency of the Academy of Sciences of the Czech Republic (IAA400040702). F.S. was supported by the Centre National de la Recherche Scientifique (CNRS), and Université Pierre et Marie Curie. M.Š. and F.S. were also supported by the international collaboration program between the Academy of Sciences of the Czech Republic and CNRS. J.Š. was supported by the grant from the Institute of Molecular Genetics, Prague (AVOZ 50520514). Funding to pay the Open Access publication charges for this article was provided by grants to M.Š. from the Grant Agency of the Czech Republic (204/05/2031) and the Grant Agency of the Academy of Sciences of the Czech Republic (IAA400040702).
Conflict of interest statement. None declared.
REFERENCES
- 1.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 2.Osheroff N, Zechiedrich EL, Gale KC. Catalytic function of DNA topoisomerase II. Bioessays. 1991;13:269–273. doi: 10.1002/bies.950130603. [DOI] [PubMed] [Google Scholar]
- 3.Baldwin EL, Osherhoff N. Etoposide, topoisomerase II and cancer. Curr. Med. Chem. Anticancer Agents. 2005;5:363–372. doi: 10.2174/1568011054222364. [DOI] [PubMed] [Google Scholar]
- 4.Denny WA. Emerging DNA toposomerase inhibitors as anticancer drugs. Expert Opin. Emerg. Drugs. 2004;9:105–133. doi: 10.1517/eoed.9.1.105.32948. [DOI] [PubMed] [Google Scholar]
- 5.Agresti A, Bianchi ME. HMGB proteins and gene expression. Curr. Opin. Genet. Dev. 2003;13:170–178. doi: 10.1016/s0959-437x(03)00023-6. [DOI] [PubMed] [Google Scholar]
- 6.Bianchi ME. Significant (re)location: how to use chromatin and/or abundant proteins as messages of life and death. Trends Cell Biol. 2004;14:287–293. doi: 10.1016/j.tcb.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 7.Thomas JO, Travers AA. HMG1 and 2, and related ‘architectural’ DNA-binding proteins. Trends Biochem. Sci. 2001;26:167–174. doi: 10.1016/s0968-0004(01)01801-1. [DOI] [PubMed] [Google Scholar]
- 8.Štros M, Launholt D, Grasser KD. The HMG-box: a versatile protein domain occurring in a wide variety of DNA-binding proteins. Cell. Mol. Life Sci. 2007 doi: 10.1007/s00018-007-7162-3. in press (doi: 10.1007/s00018-007-7160-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paull TT, Haykinson MJ, Johnson RC. The nonspecific DNA-binding and -bending proteins HMG1 and HMG2 promote the assembly of complex nucleoprotein structures. Genes Dev. 1993;7:1521–1534. doi: 10.1101/gad.7.8.1521. [DOI] [PubMed] [Google Scholar]
- 10.Pil PM, Chow CS, Lippard SJ. High-mobility-group 1 protein mediates DNA bending as determined by ring closures. Proc. Natl Acad. Sci. USA. 1993;90:9465–9469. doi: 10.1073/pnas.90.20.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Štros M, Muselíková-Polanská E, Pospíšilová Š, Strauss F. High-affinity binding of tumor-suppressor protein p53 and HMGB1 to hemicatenated DNA loops. Biochemistry. 2004;43:7215–7225. doi: 10.1021/bi049928k. [DOI] [PubMed] [Google Scholar]
- 12.Jaouen S, de Koning L, Gaillard C, Muselíková-Polanská E, Štros M, Strauss F. Determinants of specific binding of HMGB1 protein to hemicatenated DNA loops. J. Mol. Biol. 2005;353:822–837. doi: 10.1016/j.jmb.2005.08.073. [DOI] [PubMed] [Google Scholar]
- 13.Sheflin LG, Spaulding SW. High mobility group protein 1 preferentially conserves torsion in negatively supercoiled DNA. Biochemistry. 1989;28:5658–5664. doi: 10.1021/bi00439a048. [DOI] [PubMed] [Google Scholar]
- 14.Štros M, Štokrová J, Thomas JO. DNA looping by the HMG-box domains of HMG1 and modulation of DNA binding by the acidic C-terminal domain. Nucleic Acids Res. 1994;22:1044–1051. doi: 10.1093/nar/22.6.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calogero S, Grassi F, Aguzzi A, Voigtlander T, Ferrier P, Ferrari S, Bianchi ME. The lack of chromosomal protein Hmg1 does not disrupt cell growth but causes lethal hypoglycaemia in newborn mice. Nat. Genet. 1999;22:276–280. doi: 10.1038/10338. [DOI] [PubMed] [Google Scholar]
- 16.Jensen S, Andersen AH, Kjeldsen E, Biersack H, Olsen EHN, Andersen TB, Westergaard O, Jakobsen BK. Analysis of functional domain organization in DNA topoisomerase II from humans and Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:3866–3877. doi: 10.1128/mcb.16.7.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biersack H, Jensen S, Westergaard O. Rapid purification of DNA topoisomerase II containing a hexahistidine tag by metal ion affinity chromatography. In: Bjornsti M-A, Osheroff N, editors. Methods in Molecular Biology. Totowa, New Jersey: Humana Press; 1999. Vol. 94: Protocols in DNA Topology and Topoisomerases, Part I: DNA Topology and Enzymes, pp. 235–242. [DOI] [PubMed] [Google Scholar]
- 18.Skouboe C, Bjergbaek L, Oestergaard VH, Larsen MK, Knudsen BR, Andersen AH. A human topoisomerase II alpha heterodimer with only one ATP binding site can go through successive catalytic cycles. J. Biol. Chem. 2003;278:5768–5774. doi: 10.1074/jbc.M210332200. [DOI] [PubMed] [Google Scholar]
- 19.Shapiro TA, Klein VA, Englund PT. Isolation of kinetoplast DNA. In: Bjornsti M-A, Osheroff N, editors. Methods in Molecular Biology. Totowa, New Jersey: Humana Press; 1999. Vol. 94: Protocols in DNA Topology and Topoisomerases, Part I: DNA Topology and Enzymes, pp. 61–67. [DOI] [PubMed] [Google Scholar]
- 20.Štros M, Muselíková E. A role of basic residues and the putative intercalating phenylalanine of the HMG-1 box B in DNA supercoiling and binding to four-way DNA junctions. J. Biol. Chem. 2000;275:35699–35707. doi: 10.1074/jbc.M007167200. [DOI] [PubMed] [Google Scholar]
- 21.Štros M. DNA bending by the chromosomal protein HMG1 and its high mobility group box domains. Effect of flanking sequences. J. Biol. Chem. 1998;273:10355–10361. [PubMed] [Google Scholar]
- 22.Štros M, Ozaki T, Bačíková A, Kageyama H, Nakagawara A. HMGB1 and HMGB2 cell-specifically down-regulate the p53- and p73-dependent sequence-specific transactivation from the human Bax gene promoter. J. Biol. Chem. 2002;277:7157–7164. doi: 10.1074/jbc.M110233200. [DOI] [PubMed] [Google Scholar]
- 23.West KL, Austin CA. Human DNA topoisomerase IIβ binds and cleaves four-way junction DNA in vitro. Nucleic Acids Res. 1999;27:984–992. doi: 10.1093/nar/27.4.984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kingma PS, Fortune JM, Osheroff N. Topoisomerase II-catalyzed ATP hydrolysis as monitored by thin-layer chromatography. In: Osheroff N, Bjornsti M-A, editors. Methods in Molecular Biology. Totowa, New Jersey: Humana Press; 2001. Vol. 95: DNA Topoisomerase Protocols, Part II: Enzymology and Drugs, pp. 51–56. [DOI] [PubMed] [Google Scholar]
- 25.Zechiedrich EL, Crisona NJ. Coating DNA with RecA protein to distinguish DNA path by electron microscopy. In: Bjornsti M-A, Osheroff N, editors. Methods in Molecular Biology. Totowa, New Jersey: Humana Press; 1999. Vol. 94: Protocols in DNA Topology and Topoisomerases, Part I: DNA Topology and Enzymes, pp. 99–107. [DOI] [PubMed] [Google Scholar]
- 26.Namork E, Johansen BV. Surface activation of carbon film supports for biological electron microscopy. Ultramicroscopy. 1982;7:321–330. doi: 10.1016/0304-3991(82)90257-1. [DOI] [PubMed] [Google Scholar]
- 27.Štros M, Cherny D, Jovin TM. HMG1 protein stimulates DNA end joining by promoting association of DNA molecules via their ends. Eur. J. Biochem. 2000;267:4088–4097. doi: 10.1046/j.1432-1327.2000.01450.x. [DOI] [PubMed] [Google Scholar]
- 28.Javaherian K, Liu LF, Wang JC. Nonhistone proteins HMG1 and HMG2 change the DNA helical structure. Science. 1978;199:1345–1346. doi: 10.1126/science.628842. [DOI] [PubMed] [Google Scholar]
- 29.Kato J, Suzuki H, Ikeda H. Purification and characterization of DNA topoisomerase IV in Escherichia coli. J. Biol. Chem. 1992;267:25676–25684. [PubMed] [Google Scholar]
- 30.Dintilhac A, Bernues J. HMGB1 interacts with many apparently unrelated proteins by recognizing short amino acid sequences. J. Biol. Chem. 2002;277:7021–7028. doi: 10.1074/jbc.M108417200. [DOI] [PubMed] [Google Scholar]
- 31.Burden DA, Osheroff N. Mechanism of action of eukaryotic topoisomerase II and drugs targeted to the enzyme. Biochem. Biophys. Acta. 1998;1400:139–154. doi: 10.1016/s0167-4781(98)00132-8. [DOI] [PubMed] [Google Scholar]
- 32.Robinson MJ, Osheroff N. Effects of antineoplastic drugs on the post-strand-passage DNA cleavage/religation equilibrium of topoisomerase II. Biochemistry. 1991;30:1807–1813. doi: 10.1021/bi00221a012. [DOI] [PubMed] [Google Scholar]
- 33.Bromberg KD, Osheroff N. DNA cleavage and religation by human topoisomerase IIα at high temperature. Biochemistry. 2001;40:8410–8418. doi: 10.1021/bi010681q. [DOI] [PubMed] [Google Scholar]
- 34.Roca J, Ishida R, Berger JM, Andoh T, Wang JC. Antitumor bisdioxopiperazines inhibit yeast DNA topoisomerase II by trapping the enzyme in the form of a closed protein clamp. Proc. Natl Acad. Sci. USA. 1994;91:1781–1785. doi: 10.1073/pnas.91.5.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ellis RJ. Macromolecular crowding: obvious but underappreciated. Trends Biochem. Sci. 2001;26:597–604. doi: 10.1016/s0968-0004(01)01938-7. [DOI] [PubMed] [Google Scholar]
- 36.Štros M, Reich J. Formation of large nucleoprotein complexes upon binding of the high-mobility-group (HMG) box B-domain of HMG1 protein to supercoiled DNA. Eur. J. Biochem. 1998;251:427–434. doi: 10.1046/j.1432-1327.1998.2510427.x. [DOI] [PubMed] [Google Scholar]
- 37.Nagaki S, Yamamoto M, Yumoto Y, Shirakawa H, Yoshida M, Teraoka H. Non-histone chromosomal proteins HMG1 and 2 enhance ligation reaction of DNA double-strand breaks. Biochem. Biophys. Res. Commun. 1998;246:137–141. doi: 10.1006/bbrc.1998.8589. [DOI] [PubMed] [Google Scholar]
- 38.Corbett AH, Zechiedrich EL, Lloyd RS, Osheroff N. Inhibition of eukaryotic topoisomerase II by ultraviolet-induced cyclobutane pyrimidine dimers. J. Biol. Chem. 1991;266:19666–19671. [PubMed] [Google Scholar]
- 39.Krasnow MA, Cozzarelli NR. Catenation of DNA rings by topoisomerases. Mechanism of control by spermidine. J. Biol. Chem. 1982;257:2687–2693. [PubMed] [Google Scholar]
- 40.Fortune JM, Osheroff N. Osheroff N, Bjornsti M-A. Methods in Molecular Biology. Totowa, New Jersey: Humana Press; 2001. Topoisomerase II-catalyzed relaxation and catenation of plasmid DNA. Vol. 95: DNA Topoisomerase Protocols, Part II: Enzymology and Drugs, pp. 275–281. [DOI] [PubMed] [Google Scholar]
- 41.Polyanichko AM, Chikhirzhina EV, Skvortsov AN, Kostyleva EI, Colson P, Houssier C, Vorob’ev VI. The HMG1 ta(i)le. J. Biomol. Struct. Dyn. 2002;19:1053–1062. doi: 10.1080/07391102.2002.10506808. [DOI] [PubMed] [Google Scholar]
- 42.Takahagi M, Tatsumi K. Aggregative organization enhances the DNA end-joining process that is mediated by DNA-dependent protein kinase. FEBS J. 2006;273:3063–3075. doi: 10.1111/j.1742-4658.2006.05317.x. [DOI] [PubMed] [Google Scholar]
- 43.Bechert T, Diekmann S, Arndt-Jovin DJ. Human 170 kDa and 180 kDa topoisomerases II bind preferentially to curved and left-handed linear DNA. J. Biomol. Struct. Dyn. 1994;12:605–623. doi: 10.1080/07391102.1994.10508762. [DOI] [PubMed] [Google Scholar]
- 44.Sheflin LG, Fucile NW, Spaulding SW. The specific interactions of HMG 1 and 2 with negatively supercoiled DNA are modulated by their acidic C-terminal domains and involve cysteine residues in their HMG 1/2 boxes. Biochemistry. 1993;32:3238–3248. doi: 10.1021/bi00064a005. [DOI] [PubMed] [Google Scholar]
- 45.Štros M. Two mutations of basic residues within the N-terminus of HMG-1 B domain with different effects on DNA supercoiling and binding to bent DNA. Biochemistry. 2001;17:4769–4779. doi: 10.1021/bi002741i. [DOI] [PubMed] [Google Scholar]
- 46.Larsen AK, Escargueil AE, Skladanowski A. Catalytic topoisomerase II inhibitors in cancer therapy. Pharmacol. Ther. 2003;99:167–181. doi: 10.1016/s0163-7258(03)00058-5. [DOI] [PubMed] [Google Scholar]
- 47.Christensen MO, Larsen MK, Barthelmes HU, Hock R, Andersen CL, Kjeldsen E, Knudsen BR, Westergraad O, Boege F, et al. Dynamics of human DNA topoisomerases IIalpha and IIbeta in living cells. J. Cell Biol. 2002;157:31–44. doi: 10.1083/jcb.200112023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Völp K, Brezniceanu M-L, Bösser S, Brabletz T, Kirchner T, Göttel D, Joos S, Zörnig M. Increased expression of high mobility group box 1 (HMGB1) is associated with an elevated level of the antiapoptotic c-IAP2 protein in human colon carcinomas. Gut. 2006;55:234–242. doi: 10.1136/gut.2004.062729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Depowski PL, Rosenthal SI, Brien TP, Stylos S, Johnson RL, Ross JS. Topoisomerase IIalpha expression in breast cancer: correlation with outcome variables. Mod. Pathol. 2000;13:542–547. doi: 10.1038/modpathol.3880094. [DOI] [PubMed] [Google Scholar]
- 50.Watanuki A, Ohwada S, Fukusato T, Makita F, Yamada T, Kikuchi A, Morishita Y. Prognostic significance of DNA topoisomerase IIalpha expression in human hepatocellular carcinoma. Anticancer Res. 2002;22:1113–1119. [PubMed] [Google Scholar]