Abstract
Gene expression profiling of formalin-fixed and paraffin-embedded (FFPE) specimens, banked from completed clinical trials and routine clinical care, has the potential to yield valuable information implicating and linking genes with clinical parameters. In order to prepare high-quality cDNA from highly fragmented FFPE-RNA, previously precluded from high-throughput analyses, we have designed a novel strategy based on the nucleic acid restoration of incomplete cDNA sequences prior to T7 in vitro transcription (IVT) amplification. We describe this strategy as complementary-template reverse-transcription (CT-RT) because short single-stranded T7-oligo-dT24-VN-DNA sequences, obtained from FFPE-RNA, are used as primers for the RT of complementary RNA templates contained in a sense-RNA library. We validated our assay by determining the correlation between expression profiles of a matched 10-year-old frozen and FFPE breast cancer sample. We show that T7 IVT-amplification of cDNA transcripts restored by CT-RT is a specific and reliable process that allows recovery of transcriptional features undetectable by direct T7 IVT-amplification of FFPE-RNA. Furthermore, CT-RT restored 35–41% of the transcripts from archived breast and cervical specimens when compared to matched frozen tissue; and profiles included tissue-specific transcripts. Our results indicate that CT-RT allows microarray profiling of severely degraded RNA that could not be analyzed by previous methods.
INTRODUCTION
The development of high-density microarrays has enabled the study of thousands of individual transcripts in parallel and helped to identify the distinct transcriptional profiles of tumors (1). As a result, hierarchical clustering of these tumor profiles has proven to be valuable for classification of cancers of the same tissue type. One such example has been the classification of gene expression patterns in primary breast tumors, which led to the identification of four distinct tumor subtypes subsequently linked with different clinical outcomes (2–4). Such studies substantiate the use of molecular taxonomy in clinical medicine for accurate cancer diagnosis and especially for identification or design of suitable therapeutic approaches (5–8).
Retrospective transcriptional profiling of archived tissues, especially those collected from subjects for whom long-term disease outcome is known, represents an attractive objective for identification of novel therapeutic targets. Unfortunately, the specimens that have been collected in surgical pathology have been routinely formalin-fixed and paraffin-embedded (FFPE), a preservation process that has been shown to induce the formation of cross-linkages between proteins and between proteins and nucleic acids (9). This fixation method impedes the extraction of large amounts of RNA and is especially detrimental for single-stranded RNA molecules that appear fragmented and chemically modified (10–14). These characteristics have been major roadblocks for systematic high-throughput analysis of archived tissues. In fact, multi-gene retrospective analyses of FFPE-RNA have been achieved only by in situ hybridization or by relative quantification of target transcripts using real-time RT-PCR (15–19). Although RT-PCR techniques have been enhanced for the study of larger gene sets, this method remains impractical for the analysis of tens of thousands of genes and especially for the identification of novel cancer-related genes, hence the need for developing more appropriate technologies (14,20–22).
One major disadvantage of microarray analysis is the requirement for significant amounts of high-quality RNA, which is essential for increased sensitivity and reproducibility, a standard that cannot be met by fragmented RNA extracted from archived tissues. Although a few commercial kits have been designed to amplify reliably small amounts of starting material, several studies have pointed out that FFPE-RNA is not a good substrate for cDNA synthesis (12,23–26). The RT of fragmented and chemically modified RNA molecules gives rise to a population of small cDNA transcripts that in turn produce short complementary RNA (cRNA) probes by in vitro transcription (IVT). The presence of undersized probes, along with amplified non-specific sequences, contributes to the increase of non-specific signal and thus dilution of specific signal in microarray experiments. There have been a few reports that described the high-throughput transcriptional profiling of FFPE-RNA, but the use of highly fragmented RNA limits studies to a small subset of genes (27,28). Recent studies performed with the Arcturus Paradise Reagents demonstrated that severely degraded RNA could not be analyzed, and showed that high-throughput analysis of clinical samples was limited to only 24% of specimens archived between two and eight years (29,30).
Considering that the fragmentation of RNA and the loss of valuable information increase with archival time, we have developed a reliable technique for microarray analysis of archived specimens that would otherwise be rejected because of the poor quality of the recovered RNA. This strategy has been designed for sequence selection and extension of short single-stranded DNA molecules, reverse-transcribed from FFPE-RNA, by copy of a complementary template (CT). We have termed this process, complementary-template reverse-transcription (CT-RT) because intact complementary sense-RNA templates are annealed to the short single-stranded cDNA primers derived from FFPE tissue, and reverse-transcribed. We investigated the correlation between a matched 10-year-old frozen and 10-year-old FFPE-RNA derived from human breast cancer tissue using the already established T7 IVT-amplification method and compared it to our strategy, by cDNA microarray analysis (31). We demonstrated that the CT-RT process specifically increases the size of the cDNA transcripts utilized in the T7 IVT-amplification reactions. We demonstrated the high reproducibility of this process as well as the increase of specific signal in cDNA microarray experiments by comparison with direct T7 IVT-amplification of FFPE-RNA. We demonstrated that the recovery of single-stranded DNA primers from old FFPE tissue provides access to transcripts that are undetectable by IVT-amplification. Using the same RNA-template library for the restoration of cDNA primers reverse-transcribed either from breast or from cervical archived tissues, we further demonstrated the specificity of CT-RT. Our analysis reveals that highly fragmented poly(A) transcripts recovered from older archived tissues can successfully be restored and lead to the recovery of valuable transcriptional features.
MATERIALS AND METHODS
Reagents
We used universal human reference RNA (UHR; Stratagene), the quality of which was assessed on a Bioanalyzer 2100 expert (Agilent). The linear amplifications were performed using the MessageAmp II aRNA amplification kit from Ambion. The sense RNA template library was generated with the SenseAmp RNA amplification kit from Genisphere (32).
RNA extraction from frozen and FFPE tissues
Matched 10-year-old frozen and 10-year-old FFPE breast cancer samples were obtained from the Montefiore Medical center, Bronx, NY. Matched 3-year-old frozen and FFPE cervical samples were also obtained from the Montefiore Medical center. RNA from 10-year-old frozen tissue was extracted using TRIzol reagent following the manufacturer's instructions (Invitrogen). The 10-year-old breast cancer and the 3-year-old cervical FFPE tissues were macro-dissected and underwent the same RNA extraction procedure. Areas of interest were identified on H&E slides. Macro-dissection consisted of removing two to three punches of 1 mm from the FFPE tissue, which yielded 5 μg of RNA for the 10-year-old tissue, and between 5 and 20 μg of RNA for the 3-year-old tissue. The FFPE tissue was de-paraffinized using 500 μl of Hemo-De at room temperature on an agitator, three times (10). The tissue was washed with 1 ml of 100% RNase-free ethanol three times followed by three washes with 1 ml of 95% RNase-free ethanol on ice, for 8 min each time (11). The tissue was then washed with 1 ml of 1× PBS DEPC treated and incubated in 200 μl of RNase-Free 1× PBS and 6.5 μl of RNase-Out (Invitrogen) for 90 min on ice, for rehydration. Prior to proteinase K digestion, each tissue was homogenized in a 7 ml Wheaton homogenizer using 2.010 ml digesting buffer (50 mM Tris–HCl pH 7.5, 75 mM NaCl, 5 mM CaCl2 and 0.1% SDS). The homogenized tissue was separated in 15 tubes of 134 μl, to which was added 1 μl of RNase out (Invitrogen). A volume of 15 μl of proteinase K at 30 mg/ml was added to each tube (Roche Diagnostics). The digestion was carried out at 59°C for 1 h, upon agitation every 5 min. After 1 h, digests were gathered in two tubes and spun down at 12 000 r.p.m. for 1 min. The pellets were kept on ice, and 1 ml of butanol-1 was added to the supernatants. The tubes were vortexed and spun at 10 000 r.p.m. for 1 min. The butanol extractions were repeated to achieve partial removal of aqueous phase and obtain a final volume of 100 μl. This solution was used to re-suspend the tissue pellets, which led to a final volume of 150 μl. The solution was homogenized in 1 ml of TRIzol (Invitrogen) following the manufacturer's instructions. The RNA present in the supernatant was precipitated with 1 μl of 0.1 mg/ml of linear acrylamide and 3 M sodium acetate and 600 μl of isopropanol. The tubes were incubated for 12 h at −20°C and then spun at 14 000 r.p.m. for 30 min at 4°C. The precipitated RNA was washed with 200 μl of 70% RNase-free ethanol, dried and re-suspended in RNase-free water (Promega). The RNA was quantified on a Nanodrop ND-100 spectrophotometer and analyzed on Bioanalyzer using the Agilent-2100 software.
First strand cDNA synthesis and purification for the CT-RT process
The synthesis of single-stranded DNA primers from FFPE-RNA was achieved by using 5 μg of RNA in a 20 μl reaction. For the RT of the FFPE-RNA, we used Arrayscript (Ambion), the same enzyme as the one provided in the MessageAmp II RNA kit (Ambion). Each reaction contained 5 μg of RNA, 2 μl of 10× arrayscript buffer, 1 μl of RNase inhibitor mix, 4 μl dNTP mix 2.5 mM each (Ambion), 1 μl (100 ng) of T7-Oligo dT(24)-VN(5′-GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGGdT(24) (A/C/G)(A/C/G/T)-3′) and 1 μl of arrayscript reverse-transcriptase. The reaction proceeded at 42°C for 2 h. The 20 μl reaction was brought to 400 μl with RNase-free water (Sigma) for purification on a microcon YM-50 (Millipore), as instructed by the manufacturer. The purification was performed by spinning the YM-50 at 500g for 12 min to cut-off double-stranded cDNA/RNA fragments under 100-bp and single-stranded T7-oligo dT(24) -VN. The filter was washed three times using 400 μl of RNase-free water (Sigma). A volume of 88 μl was recovered and incubated with 10 μl of 10× RNase H buffer and 2 μl of RNase H 10 U/μl (Ambion) for 30 min at 37°C. The solution was transferred to a boiling water bath for 3 min and transferred on ice for 5 min. The single-stranded cDNA was purified using a MinElute purification column, following manufacturer's instruction (Qiagen). The single-stranded cDNA was recovered in 10 μl and quantified on a Nanodrop NO-100 Spectrophometer.
T7 in vitro amplifications and CT-RT amplifications
The direct amplification of RNA extracted from matched 10-year-old frozen and FFPE breast cancer tissues and from the 3-year-old frozen cervical tissue were performed with the MessageAmpII aRNA kit from Ambion, using 5 μg of total RNA for each reaction, as instructed by manufacturer. The T7 IVT-amplifications proceeded for 14 h at 37°C. For the restorations, the CT-RT process was performed using the single-stranded DNA primers obtained from 5 μg of FFPE-RNA from the 10-year-old breast cancer and the 3-year-old cervical samples. In order to prevent the T7-oligo dT(24) sequences of the purified cDNA primers from priming poly(A) sequences of random templates in the Sense-RNA template library, 1 μl of the Non-Sense Knock-out oligonucleotide (NSK), 5′-(A/C/T/G)(C/T/G)dA(24) CCGCCTCCCTATAGTGAGTCGTATTACAATTCACTGGCC-3′ (0.5 μg/μl was added to the 9 μl single-stranded cDNA solution. The mix was incubated for 10 min at 70°C, 10 min from 70 to 37°C and at 37°C for 10 min for hybridization. The solution was then speed-vacuum-dried to obtain a final volume of 1 μl. The sense-RNA template library was prepared using 2.5 μg of fresh UHR RNA (Stratagene) and amplified using the SenseAmp RNA Amplification Kit from Genisphere, following the manufacturer's instructions. We made one minor change to the procedure, by adding DNase-I prior to the sense-RNA purification in order to remove any trace of DNA. The integrity of the UHR-RNA was checked on a Bioanalyzer (Agilent). The sense-RNA was quantified using the Nanodrop ND-100 and stored in 10 μg/5 μl samples at −80°C. For the CT-RT reaction, we annealed 8.8 μl of sense-RNA and 1 μl of purified cDNA/NSK1 primers in presence of 1.2 μl of 10× first strand buffer and one drop of RNase-free mineral oil (Sigma). The solution was incubated in a 0.5 μl microfuge PCR tube in a Perkin–Elmer Cetus DNA thermal cycler with a thermocycle file as follows: 70°C for 10 min, 70–42°C in 90 min. The first strand cDNA synthesis was prepared by adding 1.2 μl of RNase-free water, 0.8 μl of 10× first strand buffer, 4 μl of dNTP mix, 1 μl of ribonuclease inhibitor and 1 μl of arrayscript reverse-transcriptase from the MessageAmp II aRNA kit (Ambion) and added at the end of the cycle, at 42°C and incubated for 2 h at 42°C. At the end of the cycle, the tube was transferred on ice. The second strand cDNA synthesis mix was prepared (63 μl of RNase-free water, 10 μl of 10× second strand cDNA buffer, 4 μl of dNTPs, 2 μl of DNA polymerase and 1 μl of RNase-H) and added under the mineral oil to the 20 μl solution, following the manufacturer's instructions (Ambion). The tubes were incubated for 2 h at 16°C, the cDNA was purified and the aRNA synthesized following the IVT instructions for 14 h (Ambion). The aRNA was quantified on a Nanodrop spectrophotometer and analyzed on a Bioanalyzer.
PCR analyses of double-stranded cDNA
Double-stranded cDNA obtained from 5 μg of UHR RNA, 10-year-old frozen RNA, 10-year-old FFPE-RNA and 10-year-old restored FFPE-cDNA was used for the PCR reactions. Half the volume of the cDNA recovered from MessageAmpII cDNA purifying columns was used to prepare a master mix for each set of PCR reactions (nine reactions). Three sets of four primers (Invitrogen), containing three forward and one reverse-primer, were synthesized for the three corresponding genes, human Cyclin D1 (Ccnd1; GenBank accession number: 053056), human tumor protein 53 (p53; GenBank accession number: 000546) and human tyrosine-kinase type-receptor (HER-2/neu/ERBB2; GenBank accession number: 004448). For Ccnd1, from 5′ to 3′ end, forward-primer 1; 5′-GTGATGGGGCAAGGGCACAAGTC-3′, Primer 2; 5′-CGGCTGGGTCTGTG-CATTTCTGG-3′, primer 3; 5′-CCCAGCACCAACATGTAACCGGC-3′ and reverse-primer 5′-TGGGGTTTTACCAGTTTTATTTC-3′. For p53, from 5′ to 3′ end, forward-primer 1; 5′-GCTGGTCTCAAACTCCTGGGCTC-3′, primer 2; 5′-GTGGAGCTGGAA-GGGTCAACATC-3′, primer 3; 5′-CCCACCCTTCCCCTCCTTCTCCC-3′ and reverse-primer; 5′-GCAGCAAAGTTTTATTGTAAAATAAG-3′. For Her-2, from 5′ to 3′ end, forward-primer 1; 5′-GCGACCCATTCAGAGACTGTCCC-3′, primer 2; 5′-GTGTCAG-TATCCAGGCTTTGTAC-3′, primer 3; 5′-GGGGAGAATGGGTGTTGTATGGG-3′ and reverse-primer; 5′-TGCAAATGGACAAAGTGGGTGTGGAG-3′. Each forward-primer was paired with the corresponding reverse-primer for each gene (Ccnd1, p53 and Her-2). All PCR reactions were performed using the Platinum Taq DNA polymerase high-fidelity kit, in 50 μl reactions (Invitrogen). Each reaction contained 1 μl of cDNA, 2 μl of forward- and reverse-primer (1 μg/μl), 2 μl of 50 mM MgCl2, 5 μl of 10× high fidelity buffer (600 mM Tris–SO4 pH 8.9, 180 mM ammonium sulfate), 5 μl of 0.2 mM of dNTP (2.5 mM), 33 μl of distilled water and a drop of mineral oil. Negative controls were performed using 1 μl of sterile distilled water instead of cDNA. Platinum Taq high-fidelity polymerase (2 μl of 0.5 U/μl) was added after cDNA denaturation for 5 min at 95°C, reactions were performed in a Perkin–Elmer Cetus DNA thermal cycler for 30 cycles (95°C for 1 min, 50°C for 1 min 30 s and 68°C for 2 min, ending with a final extension step at 68°C for 10 min). The PCR amplicons were visualized using a UV light box after electrophoresis on a 1.5% agarose gel containing 0.5 μg/μl ethidium bromide. The gels were photographed using a Fluorchem Imager (Alpha Innotech Corporation, CA, USA).
Cy3/Cy5 labeling of cRNA and microarray hybridization
The cRNA produced from each amplification was used to make fluorescent probes by RT. For each microarray experiment, 5 μg of cRNA synthesized from UHR RNA (reference) and 5 μg of cRNA from our samples were labeled. The aRNA was incubated in the presence of 2.67 μl of random primers (3 μg/μl) from Invitrogen in a final volume of 19 μl at 70°C for 10 min, spun down and put on ice for 5 min. Labeling reactions were performed by adding 8 μl of 5× first strand buffer, 4 μl of 0.1 M DTT, 4 μl of dNTP labeling mix (2.5 mM of each), 4 μl of 25 nM Cy3-labeled deoxyuridine triphosphate (Cy3-dUTP) for cRNA amplified from UHR (reference) or 4 μl of 25 nM Cy5-labeled deoxyuridine triphosphate (Cy5-dUTP; Amersham Pharmacia Biotech, NJ, USA) for cRNA from our samples, 1 μl of RNase-out at 40 U/μl (Invitrogen), 1.5 μl of Superscript II reverse-transcriptase 200 U/μl (Invitrogen) and incubated at 42°C for 1 h. After 1 h of incubation, 1.5 μl of Superscript II reverse-transcriptase was added for another 60 min at 42°C. The 40 μl reactions containing the labeled cDNAs were incubated in the presence of 44 μl of RNase-free water, 10 μl of 10× RNase-One buffer and 2 μl of RNase-One 10 U/μl (Promega) for 35 min at 37°C for removal of cRNA templates. The RNase-One was then inactivated by transferring the tubes at 95°C for 3 min and kept on ice. The 100 μl of Cy3-labeled UHR cDNA and the 100 μl of Cy5-labeled sample cDNA were combined in a tube containing 20 μl sodium acetate 3 M (PH5.1), 2 μl of 0.1 μg/μl linear acrylamide (Ambion) and 500 μl of 100% ethanol and precipitated at 14 000 r.p.m. for 30 min. The probes were washed with 200 μl of 70% RNase-free ethanol and air dried before being re-suspended in 16.5 μl of RNase-free water for 28 000 (28k) features microarrays and 6.5 μl of RNase-free water for 9000 (9k) features microarrays. The microarray slides were incubated at 50°C with 60 or 15 μl of pre-hybridization buffer for 28k or 9k microarrays, respectively. The 16.5 μl probes were incubated with 40.5 μl of hybridization buffer and 3 μl of human block solution for 28k microarrays and the 6.5 μl probes with 12.5 μl hybridization buffer and 1 μl of human block for 9k microarrays. The pre-hybridization and hybridization buffers were supplied by the Albert Einstein College of Medicine (AECOM) microarray facility, and human block solution prepared as described at: http://microarray1k.aecom.yu.edu. After 1 h the slides were washed in distillated water and dried. The 60 μl labeled probe solution was added onto the 28k microarray slide and covered with a 22 × 60 premium cover glass, and the 20 μl of labeled probes added onto the 9k microarray slides and covered with a 22 × 22 Premium Cover Glass (Fisher). The microarrays were placed in a sealed chamber in a water bath at 50°C overnight, in the dark. The slides were washed the following day as described in Belbin et al. (33). Dry slides were scanned using the GenePix 4000A microarray scanner (Axon Instruments, Foster City, CA, USA). The UHR Cy3 (Green) and the FFPE-RNA Cy5 (Red) images were acquired and gridded with GenePix Pro 6.0 Software to generate signal intensities.
cDNA microarray experiments and statistical analysis
Arrays used for the studies were designed and printed at the cDNA Microarray Facility, Albert Einstein College of Medicine (AECOM), Bronx, NY. The 28K arrays (28 704 spots) represent 14 725 human cDNA clones of the distinct genes with sequence annotation and gene ontology information, 8376 expressed sequence tags (ESTs) with incomplete sequence annotation, 5603 ESTs without any available annotation as of the date of the study and 192 bacterial sequences for quality control of the arrays. The 9K arrays contained 9216 spots (http://microarray1k.aecom.yu.edu/). The background-subtracted signals were normalized with a global LOWESS algorithm using in-house Perl scripts and the R statistical package. Two-parameter normalization was done between different arrays when necessary. Mean and SD of the signal intensity were calculated for each gene in every group of the repeated experiments. Missing and low intensity signals (<1000) were excluded from the correlation analysis.
RESULTS
Method design
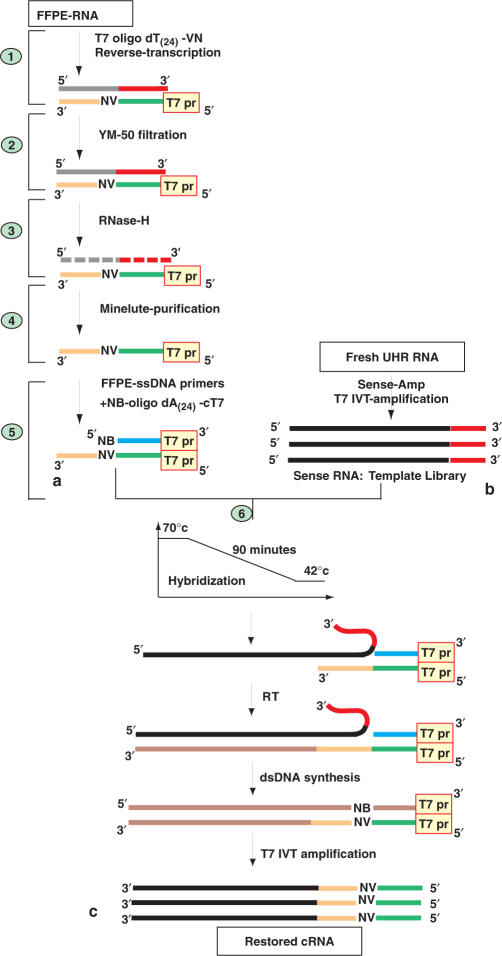
The fragmented messenger RNA extracted from FFPE tissue is primed with a T7-oligo dT(24) primer and reverse-transcribed. The RNA/cDNA duplex is filtered to allow the removal of single-stranded T7-oligo dT(24) primers that have not been used up in the reaction, providing double-stranded fragments longer than 100 nt (Figure 1a). The cDNA that has been synthesized and purified is single-stranded by RNA degradation with RNase-H. A blocking primer, complementary to the T7-oligo dT(24) sequence of the purified cDNA, is added to prevent random annealing of the oligo-dT(24) to the poly(A) tail of sense-RNA templates. The sense-RNA template library is obtained by IVT of a T7 promoter incorporated into the 3′ end of the first strand cDNA, which provides RNA with the same orientation as messenger RNA (Figure 1b) (30). Optimal annealing conditions between single-stranded DNA regions and sense-RNA templates are obtained in 90 min through a temperature gradient from 70 to 42°C (Figure 1c). The RT of the hybridized sense-RNA templates allows for the extension and thus restoration of incomplete DNA sequences. The process of CT-RT is followed by double-stranded DNA synthesis, T7 IVT-amplification and cDNA microarray analysis of the cRNA (see Supplementary Data for the details of the method).
Figure 1.
Complementary-template reverse-transcription (CT-RT) of single-stranded DNA primers reverse-transcribed from FFPE-RNA. (a) RNA extracted from FFPE tissue is reverse-transcribed, the mRNA/DNA duplex is filtered on an YM-50 column and the DNA is single-stranded with RNase-H and column purified. The 5′-NB-Oligo-dA(24)-cT7-3′ (complementary to the T7 promoter) is annealed to the FFPE-cDNA primers. (b) Total RNA from universal human reference (UHR, Stratagene) is amplified using the Sense-Amp cRNA amplification kit from Genisphere to provide RNA with the same orientation as messenger RNA (32). (c) Single-stranded DNA primers are hybridized to their sense-RNA template between 70 and 42°C for 90 min. The hybridized products are reverse-transcribed by a process described as CT-RT. The restored FFPE-cDNAs are doubled stranded and transcribed in vitro using T7 polymerase. (See Supplementary Data for technical description of points 1 through 6.)
CT-RT of a 10-year-old archived breast cancer sample
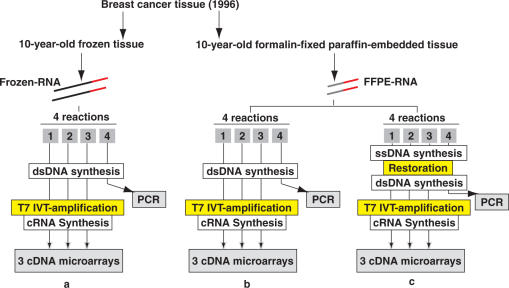
We tested our strategy on matched 10-year-old frozen and 10-year-old FFPE samples obtained from the same breast cancer tissue. After extracting RNA from the frozen tissue, we performed one round of T7 IVT-amplification with 5 μg of total RNA in four individual reactions (Figure 2a). One of the reactions was stopped after purification of double-stranded DNA for PCR experiments, while the remaining three reactions underwent T7 IVT-amplification. Using the FFPE tissue, we extracted RNA from the same region as from the frozen tissue and obtained a 260/280 ratio of 1.90. We tested the direct T7 IVT-amplification of FFPE-RNA by preparing four reactions using each 5 μg of RNA (Figure 2b). One of the reactions was stopped after purification of double-stranded DNA (dsDNA) and used for PCR experiments, while the three remaining reactions underwent T7 IVT-amplification. In order to evaluate the restoration strategy, we prepared four reactions containing 5 μg of FFPE-RNA each and performed RTs with the reverse-transcriptase provided in the MessageAmpII amplification kit from Ambion (Figure 2c). After purification, for each reaction we obtained a set of T7-dT (24)-single-stranded DNA primers that we hybridized with 10 μg of sense-RNA template library. The sense-RNA template library contained templates with sizes ranging between 250 and 1000 nt, peaking at 500 nt as observed on an Agilent bioanalyzer 2100 expert (Figure 3a). The size of these transcripts was shorter than the expected size of transcripts obtain by IVT amplification of fresh RNA. The CT-RT was carried out by adding the reagents provided in the MessageAmpII aRNA kit (Ambion). The second strand DNA synthesis was carried out. One reaction was stopped after cDNA synthesis for PCR experiments, while the other three underwent T7 IVT-amplification.
Figure 2.
Experimental procedure utilized for the analysis of 10-year-old matched frozen and formalin-fixed paraffin embedded breast cancer samples. (a) Five micrograms of RNA extracted from the 10-year-old frozen portion of the sample, is reverse-transcribed and the cDNA is double stranded (dsDNA), in four individual reactions. The dsDNA of three reactions undergoes IVT-amplification (MessageAmpII, Ambion), which gives rise to complementary RNA (cRNA) for cDNA microarray analyses. The dsDNA of one reaction is used for PCR experiments. (b) Five micrograms of RNA extracted from the 10-year-old FFPE portion of the sample underwent the exact same process. (c) Single-stranded DNA (ssDNA) obtained by RT of 5 μg of FFPE-RNA is purified and hybridized to the sense-RNA template library. The restored ssDNA is double stranded and purified. Three of the CT-RT reactions undergo IVT-amplification, while the dsDNA of one reaction is used for PCR experiments.
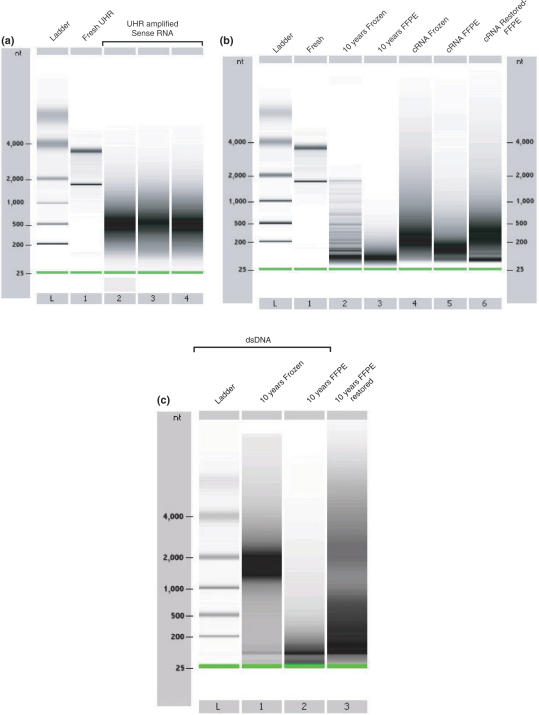
Figure 3.
Size distribution of mRNA, cRNA and dsDNA on Agilent 2100 Bioanalyzer 6000 Nanochips. (a) Universal Human Reference (UHR) RNA and sense-RNA template library after IVT-amplification. Lane L displays the ladder (25, 200, 500, 1000, 2000 and 4000 nt). Lane 1 contains fresh UHR RNA. Lanes 2, 3 and 4 display three individual IVT-amplifications of sense-RNA using total RNA displayed in lane 1. (b) Size distribution of fresh, frozen, FFPE-RNA and amplified cRNA. Lane L displays the same ladder as observed in (a). Lane 1 contains fresh human breast RNA. Lane 2 contains total RNA from the 10-year-old frozen human breast cancer tissue. Lane 3 contains total RNA from the matched 10-year-old FFPE human breast cancer tissue. Lane 4 contains cRNA obtained by IVT-amplifications of 10-year-old frozen RNA (lane 2). Lanes 5 contains cRNA obtained by direct IVT-amplification of the 10-year-old FFPE-RNA (lane 3). Lanes 6 contains amplified cRNA obtained by CT-RT and IVT-amplification of the same 10-year-old FFPE-RNA. (c) Size distribution of double-stranded DNA on a Bioanalyzer 2100 Agilent nanochip. Lane L displays the ladder. Lane 1 displays dsDNA obtained from 10-year-old frozen RNA. Lane 2 displays dsDNA obtained from 10-year-old FFPE-RNA. Lane 3 shows dsDNA obtained after CT-RT and double-strand DNA synthesis of the same 10-year-old FFPE-RNA. (See Supplementary Data for fragmented RNA profiles)
The CT-RT process provides access to larger cDNA and cRNA transcripts
In order to investigate the benefit of the CT-RT process over the single RT of short FFPE-RNA, we compared the size distribution of cRNA and cDNA products for each of our T7 IVT-amplifications on a bioanalyzer 2100 Agilent. The 10-year-old frozen RNA displayed a degradation pattern characterized by the absence of 28s ribosomal RNA and low abundance of 18s RNA, with a ladder-like pattern (Figure 3b, lane 2). The T7 IVT-amplification of this frozen RNA gave rise to products ranging between 50 and 1000 nt, peaking at 200 nt (lane 4). Frozen RNA has been shown to provide shorter cRNA in previous studies (32). The three amplifications generated 105, 88 and 88.5 μg of cRNA. The RNA extracted from the 10-year-old FFPE-RNA provided us with much smaller products, as most fragments were <200 nt (lane 3). The T7 IVT-amplification of this FFPE-RNA gave rise to products ranging between 50 and 250 nt, which peaked at 125 nt, in each of the triplicate reactions (lane 5). Although the RNA appeared largely degraded, the amplification reactions yielded 44, 40.4 and 32.9 μg of cRNA, thereby providing sufficient amounts for microarray analyses. We then analyzed the restored cRNA obtained by CT-RT reactions, performed on the same 10-year-old FFPE-RNA, and observed a large increase in the size of the products. The cRNA products ranged between 50 and 850 nt with a peak at 300 nt (lane 6). The CT-RT process produced lower amounts of cRNA in three reactions, with 28.2, 27.8 and 27.3 μg. Considering that the CT-RT process is based on the increase of available DNA sequences by a secondary RT of a longer complementary sense-RNA template, we compared the dsDNA obtained in our three different experiments on a Bioanalyzer 2100 Agilent (Figure 3c). The dsDNA generated from 10-year-old frozen RNA appeared the largest with sizes ranging between 1000 and 2000 nt (lane 1). The dsDNA obtained by RT of FFPE-RNA appeared the smallest with sizes no larger than 200 nt (lane 2), whereas dsDNA obtained by CT-RT of sense-RNA templates exhibited products as large as 750 nt (lane 3). Taken together, these results indicate that frozen RNA provides a good template for IVT-amplification. Degraded FFPE-RNA, however, provides much smaller templates for IVT-amplification. The CT-RT of cDNA obtained from FFPE-RNA appears to provide lengthened transcripts.
cRNA transcripts obtained by CT-RT contain more gene-specific information
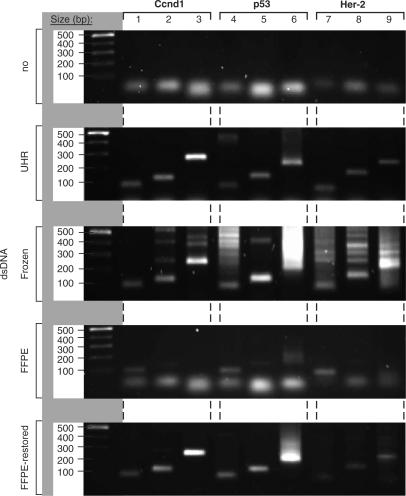
In order to verify that the increase in cRNA size, for the restored material, was due to the RT of CTs, we designed a PCR experiment to test for the presence of gene and size-specific dsDNA molecules. We chose three genes of significance for breast cancer, the human Cyclin D1 (Ccnd1), human tumor protein 53 (p53) and human tyrosine-kinase type-receptor (HER-2/neu) (34). For each gene, we designed three sense oligonucleotides increasingly spaced from the 3′end of the transcripts. Then, by combining each of these different sense primers with the anti-sense primer, in individual PCR reactions, we determined the amount of sequence available from the 3′ end of dsDNA for each gene (see Supplementary Data for details on the PCR). We first tested our PCR reactions in the absence of a known source of dsDNA, which did not generate any amplicons (Figure 4, panel 1). Using dsDNA, obtained by RT and dsDNA synthesis of fresh UHR RNA with the MessageAmp II amplification kit from Ambion, we validated the presence of each of the three genes, and demonstrated that their RNA templates were larger than 250 nt by obtaining the largest PCR products for each of the three genes (panel 2, lanes 3, 6 and 9). We performed these PCR reactions with dsDNA obtained from RNA extracted from 10-year-old frozen tissue and detected each of the three different PCR products for each of the three genes, thus demonstrating the presence of RNA transcripts larger than 250 nt for each gene (Figure 4, panel 3). Although the PCR experiments were performed with the same conditions as with dsDNA obtained from fresh UHR-RNA, we detected an increased number of non-specific products. These results suggested that fragmented frozen RNA may provide partial templates for annealing of the short PCR primers. We then performed PCR reactions with dsDNA obtained from RNA extracted from 10-year-old FFPE tissue, and verified the presence of the three genes by detecting the smallest PCR amplicons (Figure 4, panel 4, lanes 1, 4 and 7). The detection of these products revealed that short dsDNA templates were present, for all three genes, but did not exceed at 150 bp, thus demonstrating the absence of dsDNA of 251 bp for Ccnd1, 214 bp for p53 and 225 bp for Her-2 (Figure 4, panel 4). These PCR results corroborated the bioanalyzer analyses, which suggested that RNA from 10-year-old FFPE tissue contained transcripts around 100 nt and no larger than 165 nt. When we performed these PCR reactions with dsDNA obtained by RT of the FFPE-RNA and restored by the CT-RT process, we were able to detect much larger products for each of the three genes (Figure 4, panel 5). We then cloned and sequenced the largest PCR amplicons of each of the three genes, and verified that these sequences were 100% specific to each of the three genes (Figure 4, panel 5, lanes 3, 6 and 9). Taken together, these results demonstrated that the RT of older FFPE-RNA provides short but specific DNA transcripts that effectively match the ones detected in frozen RNA. When these cDNA transcripts were used for the CT-RT of cRNA templates, a physical restoration of gene-specific sequences was detected. The T7 IVT-amplification of larger cDNA templates produces larger cRNA transcripts.
Figure 4.
The size of double-stranded DNA molecules is determined by PCR experiments. Size distribution of the PCR products on a 1% agarose gel. The first lane of each gel displays the ladder (500, 400, 300, 200 and 100 bp from top to bottom of gel). Lanes 1–9 show the PCR products from three primer pairs for Ccnd1, three primer pairs for p53 and three primer pairs for Her-2. Panel 1 shows PCR experiments performed without DNA, panel 2 with dsDNA generated from UHR RNA, panel 3 with dsDNA obtained from 10-year-old frozen RNA, panel 4 with dsDNA obtained from 10-year-old FFPE-RNA and panel 5 with dsDNA restored by CT-RT (See Supplementary Data for PCR details).
The CT-RT process provides reliable cRNA for transcriptional profiling
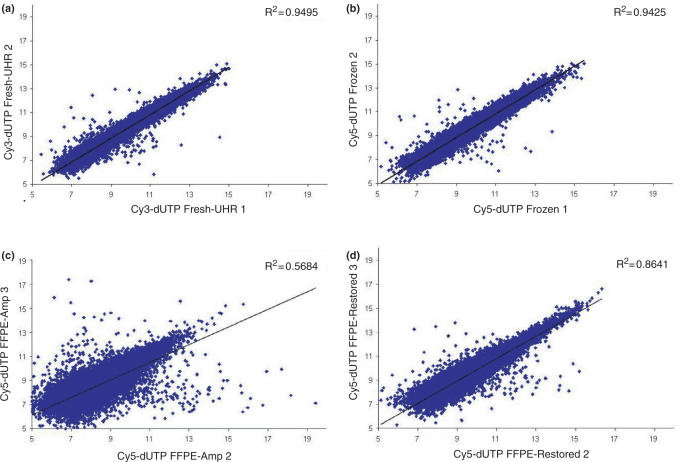
In order to evaluate the robustness of the CT-RT process we analyzed the cRNA, obtained after subsequent IVT-amplification, on cDNA microarrays (Figure 5). For quality control, each cDNA microarray was hybridized with both Cy5-labeled material (the experiment) and Cy3-labeled UHR cRNA (the reference). We first investigated the robustness of the IVT-amplification process. We examined the correlation in gene expression between technical repeats using cRNA obtained from fresh UHR RNA (Cy3) on cDNA microarrays (Figure 5a). The coefficient of determination (R2 = 0.9495), which was high, demonstrated the strength of the linear relationship between repeats and thus the reliability of the IVT-amplification. We performed the same measurements with cRNA amplified from partially degraded RNA extracted from 10-year-old frozen tissue, and obtained similarly high coefficient of determination (Figure 5b, R2 = 0.9425). These results demonstrated that the MessageAmpII aRNA kit from Ambion provided high performance IVT-amplifications even with partially degraded RNA. We then evaluated the quality of the cRNA amplified directly from RNA extracted from 10-year-old FFPE tissue. The coefficient of determination was much lower (R2 between 0.5684 and 0.6201), reflecting that the correlation between technical repeats might be impeded by the degradation and/or the chemical modification of FFPE-RNA (Figure 5c). We then evaluated the robustness of the CT-RT process, when performed with RNA extracted from the 10-year-old FFPE tissue, by cDNA microarray analysis of technical repeats (Figure 5d). The coefficient of determination was significantly higher (R2 between 0.8621 and 0.8495) than the ones obtained by direct IVT-amplification of FFPE-RNA. Together, these results demonstrated that single-stranded DNA, obtained from highly degraded and chemically modified and older FFPE-RNA, provides a highly reliable material for CT-RT reactions and IVT-amplifications.
Figure 5.
Scatter plots comparing the log2 ratios of experimental repeats. (a) Graph displaying the correlation between IVT-amplifications of fresh universal human reference RNA (Figure 3a, lane 1) in the green channel (Cy3), used as the reference. The coefficient of determination is displayed in the top right corner, R2 = 0.9495. (b) Graph displaying the correlation between IVT-amplifications from 10-year-old frozen RNA (Figure 3b, cRNA lanes 4 and 5) in the red channel (Cy5). The coefficient of determination between both experiments is displayed in the top right corner, R2 = 0.9425. (c) This scatter plot shows the correlation between IVT-amplification from 10-year-old FFPE-RNA (Figure 3b, cRNA lanes 8 and 9) in the red channel (Cy5). The R2 is displayed in the top right corner, and is 0.5684. (d) This scatter plot displays the correlation between restorations using the same 10-year-old FFPE-RNA (Figure 3b, lanes 11 and 12) in the red channel (Cy5). The coefficient of determination between restoration 2 and restoration 3 is displayed in the top right corner, R2 = 0.8641.
CT-RT improves the correlation between FFPE and frozen RNA expression profiles
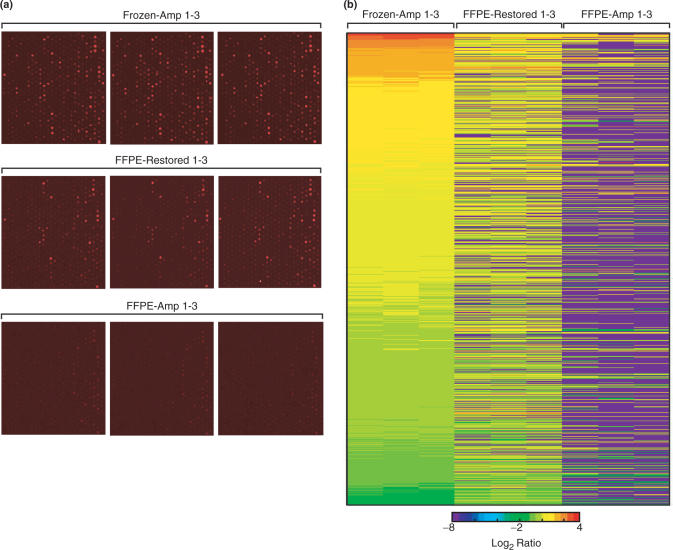
We then sought to evaluate the amount of information that could be retrieved from RNA extracted from the 10-year-old FFPE tissue by comparison with transcriptional information contained in messenger RNA from the 10-year-old frozen tissue, using either direct IVT-amplification or the CT-RT process followed by IVT-amplification. Using the microarray data, we determined that the number of observable spots with a signal higher than 1000 in each the red (Cy5) and the green (Cy3) channels was almost four times higher with the CT-RT process (4583) than by direct IVT-amplification of FFPE-RNA (1306; Table 1). Our data also revealed that the number of spots, displaying <20% of variability with the signal of matching spots from frozen material, was five times higher by the CT-RT process (2475) than by direct IVT-amplification (490) of the 10-year-old FFPE-RNA (Table 1). These results indicated that the CT-RT process allowed for a more reliable and a significantly higher recovery of features from the 10-year-old FFPE tissue. These features could not be detected by microarray analysis of material obtained by direct IVT-amplification of RNA from 10-year-old FFPE tissue. We chose one sample grid from our different microarray experiments to display the signal intensity generated by Cy5-labeled probes obtained by RT of the different cRNAs (Figure 6a). We determined that the cRNA obtained by direct IVT-amplification of RNA from 10-year-old FFPE tissue generated the lowest intensity (FFPE-Amp 1–3), where cRNA obtained by CT-RT and IVT-amplification (FFPE-Restored 1–3) provided access to transcripts with matching signal intensities to the transcripts detected in 10-year-old frozen tissue (Frozen 1–3). In order to compare the amount of signal available on the arrays, we chose to analyze genes with expression ratios ranging between 0.5 and 2 in frozen tissue. We selected a set of genes detected in both frozen RNA and in UHR RNA, and generated a heat map for 1044 genes with expression ratios between 0.5 and 2 (Figure 6b, Frozen-Amp 1–3). The UHR RNA was utilized to generate sense-RNA library that provided the templates used by CT-RT in restoration experiments. We observed that the restoration by CT-RT (FFPE-Restored 1–3) provided access to a much larger set of genes with expression ratios overlapping the ones from frozen material, than direct IVT-amplification of FFPE-RNA (FFPE-Amp 1–3). We determined that the total number of transcripts recovered from 10-year-old FFPE tissue with ratios matching the ones of transcripts from frozen tissue was three times higher after restoration (3562) than after direct IVT-amplification (1218, Table 2). Furthermore, the coefficient of determination was four times higher (R2 = 0.38) than after IVT-amplification of FFPE-RNA (R2 = 0.10), despite the increased number of features. When we selected features with expression ratios ranging between 0.5 and 2, we detected after restoration (2395) more than three times the number of genes after IVT-amplification (785). For these genes, the CT-RT process provided a coefficient of determination 25 times higher (R2 = 0.50) than the one obtained by IVT-amplification (R2 = 0.02). Together, these results demonstrated that the restoration of FFPE material provides access to a larger set of transcripts, which display higher intensities possibly due to their elongation during the process of CT-RT. Our results demonstrate that the transcripts detected in restored experiments correlate better with the ones detected in frozen RNA than those detected by direct IVT-amplification of FFPE-RNA.
Table 1.
Transcripts detected after IVT-amplification of 10-year-old frozen, IVT-amplification of 10-year-old FFPE-RNA and after restoration by CT-RT followed by IVT-amplification of 10-year-old FFPE-RNA
| Frozen | FFPE- Amplified | FFPE- Restored | |
|---|---|---|---|
| Number of good spots | 25 791 | 22 838 | 22 997 |
| Number of spots with red (Cy5) and green (Cy3) intensities ⩾1000 | 4535 | 1306 | 4583 |
| Number of measurements with variability ⩽20% | 2878 | 490 | 2475 |
Figure 6.
Signal intensity and heat-map analysis of the correlation between the log2 ratios measured by cDNA microarrays. (a) Signal intensity of one sample grid in the red channel (Cy5) across all microarrays. Top three panels display the grids obtained from three repeats using cRNA from 10-year-old frozen RNA (Frozen-Amp 1–3). Three mid-panels show the signal of three repeats using cRNA obtained by restoration and IVT-amplification of RNA from 10-year-old FFPE tissue (FFPE-Restored 1–3). Three bottom panels display the signal of three repeats using cRNA obtained by direct IVT-amplification of RNA from 10-year-old FFPE tissue. (b) Heat map displaying the log2 of expression ratios ranging between 0.5 and 2 for 1044 genes detected in frozen tissue on a 28 032 features cDNA microarray and represented in the UHR library. From left to right are displayed the ratios obtained by IVT-amplification of RNA from 10-year-old frozen tissue (Frozen-Amp 1–3), restoration and IVT-amplification of RNA from 10-year-old FFPE tissue (FFPE-Restored 1–3) and direct IVT-amplification of RNA from 10-year-old FFPE tissue (FFPE-Amp 1–3). Each column represents an individual hybridization and each line a different feature. Red and blue represent up-regulated and down-regulated genes, respectively.
Table 2.
Correlation between transcriptional profiles obtained from 10-year-old matched frozen and FFPE tissues
| Frozen versus FFPE-Amplified | Frozen versus FFPE-Restored | |||
|---|---|---|---|---|
| Number of genes | R2 (Cy5) | Number of genes | R2 (Cy5) | |
| Correlation between spots with intensities ⩾1000 | 1218 | 0.10 | 3568 | 0.38 |
| Correlation between spots with ratios between 0.5 and 2 | 785 | 0.02 | 2395 | 0.50 |
Material obtained by IVT-amplification of RNA from 10-year-old frozen is compared to material recovered by IVT-amplification of RNA from 10-year-old FFPE tissues. Material obtained by IVT-amplification of RNA from 10-year-old frozen is compared to material recovered by CT-RT and IVT-amplification of RNA from 10-year-old FFPE tissues.
CT-RT of single-stranded DNA primers provides tissue-specific information
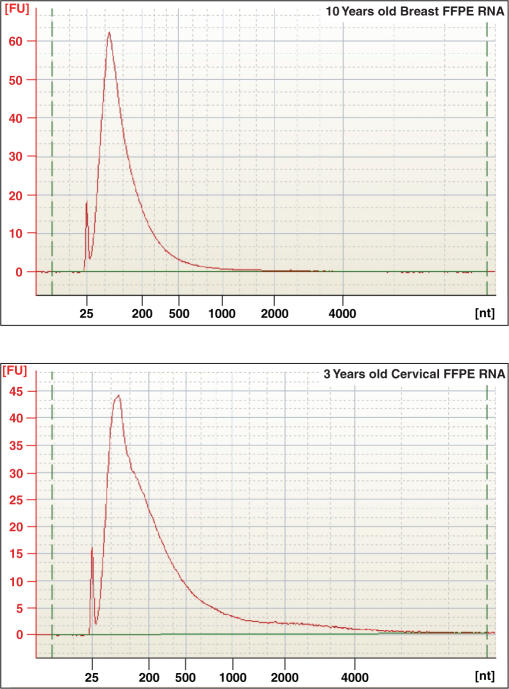
We then assessed the reproducibility and the reliability of the CT-RT process for distinguishing archived clinical samples that provide severely degraded RNA. We chose to compare a 3-year-old matched frozen and formalin-fixed normal cervical sample to the 10-year-old matched frozen and formalin-fixed breast cancer sample (Figure 7). For these experiments, total RNA isolated from the 3-year-old cervical frozen and formalin-fixed tissues, as well as the cRNA obtained by IVT-amplification of frozen RNA or by CT-RT and IVT-ampliication of FFPE-RNA were analyzed on the Agilent 2100 bioanalyzer (Supplementary Data). In order to compare the cRNA derived from IVT-amplifications of 10-year-old breast cancer and the 3-year-old cervical frozen tissues, we performed a dye-flip experiment to minimize labeling bias. By overlapping the genes detected by Cy3 and by Cy5 dyes, for each sample, we identified the expressed genes in each tissue (Table 3). This dye-flip experiment was then performed with cRNA obtained by CT-RT and IVT-amplification of 10-year-old breast cancer and 3-year-old cervical FFPE RNAs. Comparison to the transcripts detected in frozen samples revealed that 41.3% of the breast transcripts and 35.7% of the cervical transcripts were restored (Table 3). The overlap between transcripts detected in frozen and restored FFPE materials was calculated to be ∼89% with ∼10% false positives (Table 3). We analyzed the transcriptional profiles of the two tissues for the presence of tissue-specific features. By comparing the microarray profiles of frozen breast cancer and cervical tissues, we identified 905 breast-specific genes and 1013 cervix-specific genes (Table 4). The comparison between restored transcriptional profiles from the breast cancer and cervical samples identified 42 breast-specific and 109 cervix-specific transcripts. Comparison of tissue-specific transcripts revealed that 3.4% of the breast-specific and 6.8% of the cervix-specific transcripts were restored. It is important to note that the sense-RNA library (UHR), which was used for the restoration of our FFPE samples, was obtained by pooling 10 different tissues and primarily provided high concentrations of transcripts common to all the tissues giving a selection bias for common transcripts. Thus, transcripts specific to one tissue were diluted by a 1/10 ratio and therefore might have been too low in abundance, which could explain the relatively higher yield for all transcripts (35–41%) versus the lower yield for tissue-specific transcripts (3–7%). Altogether, our experiments demonstrated that single-stranded DNA primers representing the 3′ region of mRNA, adjacent to the poly(A) tail, contain sufficient information for retrieval of tissue-specific transcripts by -CT-RT.
Figure 7.
Electropherogram of total RNA recovered from FFPE samples utilized in CT-RT experiments. Total RNA extracted from 10-year-old breast cancer and 3-year-old cervical FFPE tissues were loaded on a Agilent 2100 Bioanalyzer 6000 Nanochip. Electropherograms were obtained with the 2100 expert software from Agilent, version B.02.03.SI307. 18S and 28S ribosomal RNA peaks are not detected in either FFPE sample, indicative of severe degradation.
Table 3.
Transcript detection by CT-RT of 10-year-old breast cancer and 3-year-old cervical FFPE samples
| Breast tissues | Cervical tissues | |
|---|---|---|
| Number of genes detected in Frozen, intensities ⩾1000 | 2639 | 3273 |
| Number of genes detected in FFPE-Restored, intensities ⩾1000 | 1222 | 1299 |
| Number of genes restored detected in Frozen | 1091 | 1167 |
| % of genes in restored detected in Frozen | 89.3% | 89.8% |
| Number of genes restored not detected in Frozen | 131 | 132 |
| % of genes in restored not detected in Frozen | 10.7% | 10.2% |
| % of overall recovery | 41.3% | 35.7% |
Table 4.
Detection of tissue-specific transcripts after CT-RT in 10-year-old breast cancer and 3-year-old cervical FFPE samples
| Breast tissue | Cervical tissue | |
|---|---|---|
| Tissue-specific genes detected in Frozen, intensities ⩾1000 | 905 | 1013 |
| Tissue-specific genes detected in FFPE-Restored, intensities ⩾1000 | 42 | 109 |
| Number of tissue-specific genes restored and detected in Frozen | 31 | 69 |
| % of tissue-specific genes restored and detected in Frozen | 3.4% | 6.8% |
DISCUSSION
The successful analysis of RNA extracted from archived samples has been achieved in studies that targeted a limited number of genes. One such remarkable example is the resolution of the crystal structure of a major surface antigen of the extinct 1918 ‘Spanish’ influenza virus, which killed over 20 million people worldwide. This was determined after reassembly of the hemagglutinin gene from viral RNA fragments retrieved from 1918 formalin-fixed lung tissues (35,36). Although RNA from FFPE tissue is fragmented and chemically modified, RT-PCR experiments have successfully demonstrated the presence of valuable stretches of information spanning over a 100 nt (14,20,21,37,38). Taking advantage of the presence of these sequences, we devised a novel strategy for the molecular restoration of valuable portions of information lost through fixation and extended storage. This novel technology has the potential to provide a tool for the retrospective high-throughput analysis of older archived samples and therefore the recovery of valuable transcriptional information.
The transcriptional profiling of moderately degraded RNA, submitted to multiple rounds of IVT-amplifications, has been shown to provide reasonable results (39). Similar studies using IVT-amplifications based on the random priming of degraded RNA indicated also that microarray analyses might also be feasible with FFPE-RNA (25,40,41). It has been shown, however, that the extent of fragmentation, which significantly increases with archive storage time, may become a detriment to the efficient detection of transcripts by microarray analysis (14). Our results strongly corroborated these findings, as RNA recovered from a 10-year-old archived tissue appeared largely degraded, with fragments peaking at 165 nt and smaller than 200 nt. The microarray analysis of cRNA obtained by IVT-amplification of 10-year-old FFPE-RNA revealed that the correlation between experimental repeats was much lower than repeats performed on 10-year-old frozen RNA (23). Although the amplifications provided sufficient amounts of cRNA, our experiments revealed that the short size of the products might have contributed to the loss of signal as well as the generation of non-specific signal. Our findings suggest that the process of T7 driven IVT-amplification may have primarily benefited short transcripts, non-specific messages or other species of RNA carrying poly(A) stretches, thereby severely decreasing the correlation between expression profiles of experimental repeats as well as the correlation with frozen material. It is important to note that T7 RNA polymerase has been shown to be abortive and less efficient with linear cDNA transcripts containing the T7 promoter, a process that might also contribute to the shortening of cRNA transcripts during amplification of small cDNA molecules (42, 43, 44).
The purpose of our study was to improve the quality and the specificity of microarray analyses performed with cRNA obtained from old FFPE material that contains severely fragmented RNA and that is routinely rejected from high-throughput analyses (29,30). RNA fragmentation and/or degradation are processes encountered with older FFPE tissues or samples that have been improperly fixed, and have remained major roadblocks for reliable microarray analyses (29,30). In fact, when Penland et al. used the state-of-the-art Paradise Reagent System from Arcturus on 2 to 8-year-old FFPE tissues, 76% of the archived clinical samples were rejected from microarray profiling due to poor RNA quality (30). Therefore, we designed the process of CT-RT to strengthen transcript selection prior to IVT-amplification and in turn strengthen signal detection in microarray experiments, when using RNA of the poorest quality. This process was designed to investigate the small single-stranded DNA primers synthesized from highly fragmented messenger RNA, by hybridization and RT of sense cRNA templates. This strategy was built around the already established and widely used T7 IVT-amplification method in order to minimize variables in our experiments. Using PCR experiments, we showed that this process increased the 5′ region of cDNA molecules by at least 100 nt in length. Although the CT-RT process required the preliminary purification of DNA primers on a filter and a column, our results indicated that these steps contributed to the improvement of T7 IVT-amplification of older FFPE material. Our experiments also showed that the RT of CTs was a highly reproducible mechanism as we obtained coefficient of determination between 0.84 and 0.86, for material as old as 10 years. When we correlated the transcriptional profiles from frozen and FFPE tissues, our method allowed 35–41% of transcript recovery for gene ratios ranging between 0.5 and 2, where the direct IVT-amplification of FFPE-RNA only provided 2% of recovery. Our results support the hypothesis that short transcripts recovered from archival material might be a major source of non-specific signal as well as a contributing factor to the loss of signal. By contrast, transcript restoration by CT-RT dramatically improves transcript selection and in turn the correlation between transcriptional profiles of matched frozen and FFPE material, and this process has the advantage to provide T7 RNA polymerase with longer cDNA molecules for IVT.
The extension of short single-stranded DNA primers by RT of complementary sense-RNA templates, represented in excess in a library, is a reaction that gives access to genes that are still represented in highly fragmented FFPE-RNA but that cannot be detected by direct IVT-amplification. CT-RT is a multiplex process that can be performed in a single reaction and may lead to the recovery of thousands of biologically significant transcripts from valuable archived clinical samples. Sense-RNA templates are selected by the DNA primers, synthesized from the archived RNA, during the annealing reaction. These short DNA primers, although representative of the 3′ region of mRNA transcripts, adjacent to the poly(A) tail, demonstrated to contain specific and sufficient information to recognize CTs contained in a sense-RNA library (45,46). Therefore, the restoration of single-stranded DNA primers, synthesized from highly degraded RNA, is a reaction that has the potential to lead to the retrospective identification of transcriptional biomarkers that have remained in the archived tissues as fragmented sequences.
The focus of our work was to establish a reliable method for the high-throughput analysis of severely degraded RNA typically recovered in 76% of archived clinical samples (30). Our results demonstrated that CT-RT provided access to biologically relevant genes, which could discriminate between two different tissues. It is important to note that the use of a generic library, where transcripts from 10 different tissues have been gathered, might not provide optimal tissue-specific sense-RNA concentration for the annealing of single-stranded DNA transcripts that are recovered in low abundance in a single tissue. Our experiments suggested that transcript recovery by CT-RT might depend upon the presence of the CT in the sense-RNA library as much as its representation in the single-stranded DNA primer pool. Therefore to improve the process of CT-RT as well as its efficiency, we propose that the selection of specific libraries containing a complete array of tissue, disease and/or tumor-specific transcripts might provide access to more informative and more complete transcriptional profiles when using highly fragmented FFPE-RNA from a specific tissue. The CT-RT process might also be improved by increasing the amount of recovered single-stranded DNA primers. The use of magnetic micro-beads may provide better mRNA recovery and thus increase the amount of primers for application of the CT-RT process to smaller FFPE-RNA fractions. (47). It should be noted that the CT-RT process may bias alternative splicing profiles of transcripts containing identical 3′ regions, towards that of the reference template, but the overall cumulative expression level of the splicing variants in the group should remain intact. In summary, this novel technique provides the basis for a new molecular approach to the microarray analysis of severely degraded RNA, recovered from most archived clinical samples.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors wish to thank Dr Thomas Harris, Dr Kenny Ye, Leslie Adrien and Antonio Nakouzi for their technical assistance. Funding to pay the Open Access publication charges for this article was provided by the Albert Einstein College of Medicine, New York.
Conflict of interest statement. None declared.
REFERENCES
- 1.Mohr S, Leikauf GD, Keith G, Rihn BH. Microarrays as cancer keys: an array of possibilities. J. Clin. Oncol. 2002;20:3165–3175. doi: 10.1200/JCO.2002.12.073. [DOI] [PubMed] [Google Scholar]
- 2.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 3.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl Acad. Sci. USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc. Natl Acad. Sci. USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Golub TR. Genome-wide views of cancer. N. Engl. J. Med. 2001;344:601–602. doi: 10.1056/NEJM200102223440809. [DOI] [PubMed] [Google Scholar]
- 6.Abramovitz M, Leyland-Jones BR. A systems approach to clinical oncology: focus on breast cancer. Proteome Sci. 2006;4:5. doi: 10.1186/1477-5956-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robison JE, Perreard L, Bernard PS. State of the science: molecular classifications of breast cancer for clinical diagnostics. Clin. Biochem. 2000;37:572–578. doi: 10.1016/j.clinbiochem.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 8.Dietel M, Sers C. Personalized medicine and development of targeted therapies: the upcoming challenge for diagnostic molecular pathology. A review. Virchows Arch. 2006;448:744–755. doi: 10.1007/s00428-006-0189-2. [DOI] [PubMed] [Google Scholar]
- 9.Werner M, Chott A, Fabiano A, Battifora H. Effect of formalin tissue fixation and processing on immunohistochemistry. Am. J. Surg. Pathol. 2000;24:1016–1019. doi: 10.1097/00000478-200007000-00014. [DOI] [PubMed] [Google Scholar]
- 10.Krafft AE, Duncan BW, Bijwaard KE, Taubenberger JK, Lichy JH. Optimization of the isolation and amplification of RNA from formalin-fixed, paraffin-embedded tissue: the armed forces institute of pathology experience and literature review. Mol. Diagn. 1997;2:217–230. doi: 10.1054/MODI00200217. [DOI] [PubMed] [Google Scholar]
- 11.Stanta G, Bonin S, Perin R. RNA extraction from formalin-fixed and paraffin-embedded tissues. Methods Mol. Biol. 1998;86:23–26. doi: 10.1385/0-89603-494-1:23. [DOI] [PubMed] [Google Scholar]
- 12.Masuda N, Ohnishi T, Kawamoto S, Monden M, Okubo K. Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucleic Acids Res. 1999;27:4436–4443. doi: 10.1093/nar/27.22.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coombs NJ, Gough AC, Primrose JN. Optimisation of DNA and RNA extraction from archival formalin-fixed tissue. Nucleic Acids Res. 1999;27:e12. doi: 10.1093/nar/27.16.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cronin M, Pho M, Dutta D, Stephans JC, Shak S, Kiefer MC, Esteban JM, Baker JB. Measurement of gene expression in archival paraffin-embedded tissues: development and performance of a 92-gene reverse transcriptase-polymerase chain reaction assay. Am. J. Pathol. 2004;164:35–42. doi: 10.1016/S0002-9440(10)63093-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lehmann U, Kreipe H. Real-time PCR analysis of DNA and RNA extracted from formalin-fixed and paraffin-embedded biopsies. Methods. 2001;25:409–418. doi: 10.1006/meth.2001.1263. [DOI] [PubMed] [Google Scholar]
- 16.Lewis F, Maughan NJ, Smith V, Hillan K, Quirke P. Unlocking the archive—gene expression in paraffin-embedded tissue. J. Pathol. 2001;195:66–71. doi: 10.1002/1096-9896(200109)195:1<66::AID-PATH921>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 17.Relf BL, Machaalani R, Waters KA. Retrieval of mRNA from paraffin-embedded human infant brain tissue for non-radioactive in situ hybridization using oligonucleotides. J. Neurosci. Methods. 2002;115:129–136. doi: 10.1016/s0165-0270(02)00003-1. [DOI] [PubMed] [Google Scholar]
- 18.Capodieci P, Donovan M, Buchinsky H, Jeffers Y, Cordon-Cardo C, Gerald W, Edelson J, Shenoy SM, Singer RH. Gene expression profiling in single cells within tissue. Nat. Methods. 2005;2:663–665. doi: 10.1038/NMETH786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paik S, Kim CY, Song YK, Kim WS. Technology insight: application of molecular techniques to formalin-fixed paraffin-embedded tissues from breast cancer. Nat. Clin. Pract. Oncol. 2005;2:246–254. doi: 10.1038/ncponc0171. [DOI] [PubMed] [Google Scholar]
- 20.Bibikova M, Talantov D, Chudin E, Yeakley JM, Chen J, Doucet D, Wickham E, Atkins D, Barker D, et al. Quantitative gene expression profiling in formalin-fixed, paraffin-embedded tissues using universal bead arrays. Am. J. Pathol. 2004;165:1799–1807. doi: 10.1016/S0002-9440(10)63435-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bibikova M, Yeakley JM, Chudin E, Chen J, Wickham E, Wang-Rodriguez J, Fan JB. Gene expression profiles in formalin-fixed, paraffin-embedded tissues obtained with a novel assay for microarray analysis. Clin. Chem. 2004;50:2384–2386. doi: 10.1373/clinchem.2004.037432. [DOI] [PubMed] [Google Scholar]
- 22.Ma XJ, Patel R, Wang X, Salunga J, Desai R, Tuggle JT, Wang W, Chu S, Stecker K, et al. Molecular classification of human cancers using a 92 real-time quantitative polymerase chain reaction assay. Arch. Pathol. Lab. Med. 2006;130:465–473. doi: 10.5858/2006-130-465-MCOHCU. [DOI] [PubMed] [Google Scholar]
- 23.Karsten SL, Van Deerlin VM, Sabatti C, Gill LH, Geschwind DH. An evaluation of tyramide signal amplification and archived fixed and frozen tissue in microarray gene expression analysis. Nucleic Acids Res. 2002;30:e4. doi: 10.1093/nar/30.2.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klur S, Toy K, Williams MP, Certa U. Evaluation of procedures for amplification of small-size samples for hybridization on microarrays. Genomics. 2004;83:508–517. doi: 10.1016/j.ygeno.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 25.Xiang CC, Chen M, Ma L, Phan QN, Inman JM, Kozhich OA, Brownstein MJ. A new strategy to amplify degraded RNA from small tissue samples for microarray studies. Nucleic Acids Res. 2003;31:e53. doi: 10.1093/nar/gng053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Hu L, Hamilton SR, Coombes KR, Zhang W. RNA amplification strategies for cDNA microarray experiments. Biotechniques. 2003;34:394–400. doi: 10.2144/03342mt04. [DOI] [PubMed] [Google Scholar]
- 27.Onken MD, Worley LA, Ehlers JP, Harbour JW. Gene expression profiling in uveal melanoma reveals two molecular classes and predicts metastatic death. Cancer Res. 2004;64:7205–7209. doi: 10.1158/0008-5472.CAN-04-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chung CH, Parker JS, Ely K, Carter J, Yi Y, Murphy BA, Ang KK, El-Naggar AK, Zanation AM, et al. Gene expression profiles identify epithelial-to-mesenchymal transition and activation of nuclear factor-(kappa) B signaling as characteristics of a high-risk head and neck squamous cell carcinoma. Cancer Res. 2006;66:8210–8218. doi: 10.1158/0008-5472.CAN-06-1213. [DOI] [PubMed] [Google Scholar]
- 29.Coudry AR, Meireles RS, Cooper HS, Carpino A, Wang X, Engstrom PF, Clapper ML. Successful application of microarray technology to microdissected formalin-fixed, paraffin-embedded tissue. J. Mol. Diagn. 2007;9:70–79. doi: 10.2353/jmoldx.2007.060004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Penland SK, Keku TO, Torrice C, He X, Krishnamurthy J, Hoadley KA, Woosley JT, Thomas NE, Perou CM, et al. RNA expression analysis of formalin-fixed paraffin-embedded tumors. Lab. Invest. 2007:e1–e9. doi: 10.1038/labinvest.3700529. [DOI] [PubMed] [Google Scholar]
- 31.Van Gelder RN, von Zastrow ME, Yool A, Dement WC, Barchas JD, Eberwine JH. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc. Natl Acad. Sci. USA. 1990;87:1663–1667. doi: 10.1073/pnas.87.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goff LA, Bowers J, Schwalm J, Howerton K, Getts RC, Hart RP. Evaluation of sense-strand mRNA amplification by comparative quantitative PCR. BMC Genomics. 2004;5:76. doi: 10.1186/1471-2164-5-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Belbin TJ, Bhuvanesh S, Barber I, Socci N, Wenig B, Smith R, Prystowsky MB, Childs G. Molecular classification of head and neck squamous cell carcinoma using cDNA microarrays. Cancer Res. 2002;62:1184–1190. [PubMed] [Google Scholar]
- 34.Lebeau A, Unholzer A, Amann G, Kronawitter M, Bauerfeind I, Sendelhofert A, Iff A, Lohrs U. EGFR, HER-2/neu, cyclin D1, p21 and p53 in correlation to cell proliferation and steroid hormone receptor status in ductal carcinoma in-situ of the breast. Breast Cancer Res. Treat. 2003;79:187–198. doi: 10.1023/a:1023958324448. [DOI] [PubMed] [Google Scholar]
- 35.Reid AH, Taubenberger JK, Fanning TG. The 1918 spanish influenza: integrating history and Biology. Microbes Infect. 2001;3:81. doi: 10.1016/s1286-4579(00)01351-4. [DOI] [PubMed] [Google Scholar]
- 36.Stevens J, Corper AL, Basler CF, Taubenberger JK, Palese P, Wilson IA. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science. 2004;303:1866–1870. doi: 10.1126/science.1093373. [DOI] [PubMed] [Google Scholar]
- 37.Abrahamsen HN, Steiniche T, Nexo E, Hamilton-Dutoi SJ, Sorensen BS. Towards quantitative mRNA analysis in paraffin-embedded tissues using real-time reverse transcriptase-polymerase chain reaction: a methodological study on lymph nodes from melanoma patients. J. Mol. Diagn. 2003;5:34–41. doi: 10.1016/S1525-1578(10)60449-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Antonov J, Goldstein DR, Oberli A, Baltzer A, Pirotta M, Fleischmann A, Altermatt HJ, Jaggi R. Reliable expression measurements from degraded RNA by quantitative real-time PCR depend on short amplicons and a proper normalization. Lab. Invest. 2005;85:1040–1050. doi: 10.1038/labinvest.3700303. [DOI] [PubMed] [Google Scholar]
- 39.Schoor O, Weinschenk T, Hennenlotter J, Corvin S, Stenzl A, Rammensee H-G, Stefanovic S. Moderate degradation does not preclude microarray analysis of small amounts of RNA. Biotechniques. 2003;35:1192–1201. doi: 10.2144/03356rr01. [DOI] [PubMed] [Google Scholar]
- 40.Tomlins SA, Mehra R, Rhodes DR, Shah RB, Rubin MA, Bruening E, Makarov V, Chinnaiyan AM. Whole transcriptome amplification for gene expression profiling and development of molecular archives. Neoplasia. 2006;8:153–162. doi: 10.1593/neo.05754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scicchitano MS, Dalmas MA, Bertiaux MA, Anderson SM, Turner LR, Thomas RA, Mirable R, Boyce RW. Preliminary comparison of quantity, quality, and microarray performance of RNA extracted from formalin-fixed, paraffin-embedded, and unfixed frozen tissue samples. J. Histochem. Cytochem. 2006;54:1229–1237. doi: 10.1369/jhc.6A6999.2006. [DOI] [PubMed] [Google Scholar]
- 42.Diaz GA, Rong M, McAllister WT, Durbin RK. The stability of abortively cycling T7 RNA polymerase complexes depends upon template conformation. Biochemistry. 1996;35:10837–10843. doi: 10.1021/bi960488+. [DOI] [PubMed] [Google Scholar]
- 43.Lopez PJ, Guillerez J, Sousa R, Dreyfus M. The low processivity of T7 RNA polymerase over the initially transcribed sequence can limit productive initiation in vivo. J. Mol. Biol. 1997;269:41–51. doi: 10.1006/jmbi.1997.1039. [DOI] [PubMed] [Google Scholar]
- 44.Gong P, Martin CT. Mechanism of instability in abortive cycling by T7 RNA polymerase. J. Biol. Chem. 2006;281:23533–23544. doi: 10.1074/jbc.M604023200. [DOI] [PubMed] [Google Scholar]
- 45.Edwalds-Gilbert G, Veraldi KL, Milcarek C. Alternative poly(A) site selection in complex transcription units: means to an end. Nucleic Acids Res. 1997;25:2547–2561. doi: 10.1093/nar/25.13.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hughes TA. Regulation of gene expression by alternative untranslated regions. Trends Genet. 2006;22:119–122. doi: 10.1016/j.tig.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 47.De Andres B, del Pozo V, Gallardo S, de Arruda-Chaves E, Cardaba B, Martin-Orozco E, Posada M, Palomino P, Lahoz C. Improved method for mRNA extraction from paraffin-embedded tissues. Biotechniques. 1995;18:42–44. [PubMed] [Google Scholar]