Abstract
Despite the great potential of RNAi, ectopic expression of shRNA or siRNAs holds the inherent risk of competition for critical RNAi components, thus altering the regulatory functions of some cellular microRNAs. In addition, specific siRNA sequences can potentially hinder incorporation of other siRNAs when used in a combinatorial approach. We show that both synthetic siRNAs and expressed shRNAs compete against each other and with the endogenous microRNAs for transport and for incorporation into the RNA induced silencing complex (RISC). The same siRNA sequences do not display competition when expressed from a microRNA backbone. We also show that TAR RNA binding protein (TRBP) is one of the sensors for selection and incorporation of the guide sequence of interfering RNAs. These findings reveal that combinatorial siRNA approaches can be problematic and have important implications for the methodology of expression and use of therapeutic interfering RNAs.
INTRODUCTION
The phenomenon of RNA interference was first observed almost a decade ago (1) and seems to have evolved as a defense mechanism against foreign double-stranded RNA. It is triggered by short RNA duplexes (∼21–23 nt in length), which are processed from longer double-stranded RNA transcripts (2,3). Two major classes of small double-stranded RNA molecules have thus far been identified: small interfering RNAs (siRNAs) and microRNAs (miRNAs). SiRNAs are fully complementary double-stranded molecules while miRNAs originate as duplexes that have bulges. Both classes of double-stranded RNAs assemble in the RNA induced silencing complex (RISC), which contains key proteins for the processing and functioning of double-stranded RNAs in RNAi. A member of the Argonaute family, Ago-2, has been identified as the catalytic core of this complex (4–8).
Since their discovery, the use of siRNAs has quickly widened to diverse areas of research and as potential therapeutic agents (9). High-throughput analysis based on the use of siRNA libraries is also revolutionizing the field of functional genomics (9–11). However, RNAi is an important component of endogenous cellular processes (12) involved in post-transcriptional regulation of endogenous gene expression (13,14) as well as antiviral protection (15–18). Interfering with the endogenous functions of RNAi could lead to severe toxicity as recently demonstrated (19).
The key component for the cellular export of shRNAs and microRNAs is the nuclear karyopherin Exportin-5 (20–23). In the presence of Ran-GTP, Exportin-5 binds to both shRNAs and microRNAs transporting them from the nucleus to the cytoplasm. Exportin-5 is a saturable transport pathway so the excessive production of small interfering RNAs could result in a decrease of cellular miRNA function (19,24). This export function is not required for the activity of synthetic siRNAs (20). In this work, we show that the competition between shRNAs with cellular miRNAs is a general phenomenon that also takes place with synthetic siRNA sequences. The ability to compete varies with the sequence and does not depend solely on the saturation of Exportin-5. We show that siRNAs, which do not depend on Exportin-5 for their transport to the cytoplasm, retain their ability to compete against shRNAs and microRNAs. Ectopic expression of Exportin-5 only partially relieves the competition between shRNAs and endogenous miRNAs or exogenous siRNAs. Ectopically expressed shRNAs and synthetic siRNAs, but not ectopically expressed microRNAs are able to interfere with each other and with the endogenous microRNA pathway through their ability to be incorporated into RISC. However, the relative strength of competition can be equalized for all the tested siRNA sequences by reducing the cellular concentration of the HIV-1 TAR RNA binding protein (TRBP). Thus, our results suggest that TRBP is a sensor in the loading of the RNA guide sequence into RISC.
MATERIALS AND METHODS
ShRNAs and MicroRNAs constructions
U6shRNA-expressing constructs were synthesized via a PCR-based reaction that includes primers encoding the shRNA with a 3′ region complementary to the U6 promoter and a primer complementary to the 5′ end of the U6 promoter as described previously (25). All PCR reactions were carried out as follows: 1 min at 94°C, 1 min at 55°C and 1 min at 72°C for 30 cycles. The resulting PCR cassettes were cloned directly into the pCR2.1 plasmid (TA cloning vector, Invitrogen). The shRNA constructs used for the competition experiments are targeted against sequences present in the HIV-rev (shSII and shSI) HIV tat (shSI, shTAT), HIV vif (shVif) and the EnvPb1 retrovirus envelope (shL) genes. The shRNA Luc (Luc) is targeted to a sequence present in the coding region of the firefly luciferase gene. All shRNAs are expressed by transcription from the U6 promoter. The synthetic siRNAs (siH3, siH6 and siH1) are targeted against sequences present in the coding region of the hnRNPH gene. Refer to the Supplementary Information for specific sequences for the shRNAs and siRNAs. All other oligonucleotide sequences are available upon request.
The microRNA constructs were similarly constructed using overlapping primers containing the wild type or a variant mir30 sequence. The mir30 guide sequence was replaced with 21 nt complementary to a site present in the HIV-rev transcript (SII) by including the corresponding nucleotide mutations in the PCR primers. The resulting PCR products were cloned into pcDNA3.1 under the control of the CMV promoter.
A cassette containing the U1 promoter, the firefly luciferase coding sequence, and the SV40 polyadenylation signal was digested with EcoRI and DraII and cloned into the Bluescript vector (Stratagene) to create the firefly luciferase target (FLT).
The Renilla luciferase target (RLT) was generated by cloning a 21-nt HIV-rev sequence (SII) into the XhoI site located in the 3′-untranslated region (3′-UTR) of the humanized Renilla luciferase gene of plasmid psiCHECK-2™ (Promega).
Cell lines and transfection conditions
Cells were grown in DMEM (Irvine Scientific, Santa Ana, CA, USA) supplemented with 10% fetal calf serum (Irvine Scientific), 1 mM l-glutamine. To generate the HCT116-GFPmiR21 cell line, HCT116 cells were transfected (using Lipofectamine 2000, Invitrogen) with SapI linearized pcDNA4/GFPmir21 plasmid DNA (4) (generous gift from Thomas Tuschl) and cells with stable plasmid integrations were selected by applying 200 μg Zeocin per ml media for 10 days. Single colonies were picked and screened for expression of EGFP upon transfection with a 2′O-methyl RNA complementary to miR21 relative to a control 2′O-methyl RNA complementary to miR33.
For the competition experiments, 293 cells were transfected with the shRNA expression constructs and the corresponding targets in 24-well plates using Lipofectamine Plus™ reagent (Life Technologies, GibcoBRL) as described by the manufacturer. One hundred nanogram of the FLT plasmid was co-transfected with 12.5, 25, 50 or 100 ng of the shRNA-expressing constructs and 0.2 ng of the Renilla luciferase plasmid which was included to normalize for transfection efficiency for each reaction. The synthetic siRNAs were transfected to achieve 10 nanomolar concentrations in all co-transfection experiments. However, 100 nanomolar concentrations of each siRNA were used for transfection in the HCT116-GFPmiR21 cell line. One hundred nanomolar concentrations of the 2′O-Me oligonucleotides were also used for transfection in this cell line.
One hundred nanogram of the microRNA constructs were co-transfected with 20, 50 or 100 ng of RTL and yielded the same relative results in each case. Here, 0.5 ng of firefly luciferase were used to normalize for transfection efficiency in each sample. However, 800 ng of both the shRNAs and the microRNAs were used for the transfections in the HCT116-GFPmiR21 cell line.
For the overexpression of Exportin-5, 100 ng of an Exportin-5 expression plasmid (kindly provided by Thomas Tuschl) was co-transfected with 100 ng of the FLT plasmid and 50 ng of each shRNA construct. Ten (or) twenty nanomolar of each synthetic siRNA were also used for this experiments and yielded analogous results.
The TRBP down-regulation was achieved by transfecting 40 nM of anti-TRBP siRNAs (26) (siTRBP-A siTRBP-B and siTRBP1; sequences are available upon request) in a 100 mm plate of 293 cells. Cells were transfected in suspension at ∼50% confluency using the siQuest transfection reagent (Mirus, Madison, WI, USA) as suggested by the manufacturer. The following day cells reached ∼70% confluency and were transfected a second time using the siQuest transfection reagent. Twenty-four hours after the second transfection, cells were lifted, seeded in a 24-well plate, and transfected with the various constructs as described in the Results section. Cells were lysed 24 h after this final transfection and processed for luciferase assays.
Each experimental sample was normalized against its own target control, which included the corresponding target co-transfected with one or more irrelevant controls. For example, down-regulation of the FLT by the U6 anti-luciferase shRNA (Luc) was calculated by comparing the luciferase down-regulation obtained with the Luc-specific shRNA to the luciferase units obtained when the target was co-transfected with an irrelevant shRNA. When specific shRNAs were co-transfected with microRNAs, the luciferase units were compared to those obtained when the target was co-transfected with irrelevant shRNA and the microRNA controls. Similarly the impairment of Luc activity by the synthetic oligonucleotides was determined by comparing the luciferase units with those obtained when the luciferase target was transfected with only the irrelevant synthetic siRNA. It is important to normalize the experimental samples to their specific controls since si, sh or microRNA constructs co-transfected with a target could non-specifically alter the results by increasing or decreasing the expression of that target. All the experiments were normalized for transfection efficiency, carried out in duplicate, and repeated a minimum of three times. Different amounts of Bluescript plasmid (Stratagene) were added to all the transfections to obtain the same total amount of DNA transfected in each experiment, which corresponded to 400 ng per reaction (1 μl Lipofectamine-2000) except for the transfections in the HCT116-GFPmiR21 cell line in which the total amount of transfected DNA was 800 ng (2 μl Lipofectamine-2000; no Bluescript was added). The cells were analyzed for luciferase or EGFP expression 48 h following transfections.
Luciferase assays
A total of 293 cells were seeded in 24-well plates and transfected the next day at 80% confluency. Twenty-four or 48 h post-transfection, the medium was removed and cells were washed once with DPBS (Dulbecco's Phosphate Buffer Saline, Cellgro). One hundred microliter of 1× Passive Lysis Buffer (Promega) was then added to each well. Cells were lysed at room temperature with gentle shaking for 20 min. Ten microliter of cell lysates were assayed for dual luciferase activity according to the manufacturer's instructions (Dual Luciferase Reporter System, Promega). Changes in the levels of the FLT were calculated after normalization against the Renilla luciferase units. When the Renilla luciferase was used as the target, changes in expression were calculated relative to firefly luciferase (transfection control). The luminescence was determined using a Veritas luminometer (Turner BioSystems, Inc. Sunnyvale, CA, USA). Results are presented in relative light units and the SD is indicated with error bars as shown in the graph data.
Northern analyses
Total RNA was isolated using RNA STAT-60 (TEL-TEST B Inc., Friendswood, TX, USA) according to the manufacturer's instructions. Fifteen microgram of total RNA were fractionated in 8 M–6% PAGE, and transferred onto a Hybond-N+ membrane (Amersham Pharmacia Biotech). 32P-radiolabeled 21-mer probes complementary to the siRNA-antisense sequences were used for the hybridization reactions, which were performed for 16 h at 37°C. A 21-mer DNA oligonucleotide was electrophoresed alongside the RNA samples and used as a size marker and hybridization control (not shown). For TRBP detection, 30 μg RNA was fractionated on a 1% MOPS agarose gel, transferred on a Hybond-N+ membrane and hybridized with a 32P labeled probe complementary to TRBP overnight at 37°C.
Immunodepletion assay
Confluent HEK293 cells in a 10 cm dish were harvested with 1× PBS after trypsinizing and resuspended in 0.5 ml of Buffer D (20 mM HEPES, pH 7.9, 0.2 mM EDTA, 0.5 mM DTT, 50 mM KCl, 10% Glycerol and 0.2 mM PMSF). The cells were sonicated for 15 s in ice water, and the supernatant was collected following 5 min centrifugation.
The immunodepletion was carried out by incubating 100 μl of the extract with 30 μl of anti-TRBP antibody (27) or 30 μl (2 μg/ml) of anti-β-tubulin (Abcam Inc., Cambridge, MA, USA) antibody. Following overnight incubation at 4°C, 30 μl of protein A agarose beads (Upstate USA, Inc. Chicago, IL, USA) were added and incubated for 1.5 h at 4°C with gentle agitation. Following the removal of the beads by a brief centrifugation, 3 μl of the supernatant were resolved in a 10% SDS-PAGE for western blot analyses, and 10 μl were used for the electrophoretic mobility gel shift assay. The pellet beads were boiled with 25 μl of 2× SDS loading buffer and 10 μl were resolved alongside the supernatant sample for western blot analyses. After transferring proteins overnight, the PVDF membranes were blocked with 5% milk in 1× PBS for 2 h at room temperature and washed with 1× PBST for 10 min three times. Primary and secondary antibody incubations were carried out each for 1.5 h in 5% milk in 1× PBST at room temperature followed by washing with 1× PBST for 10 min three times. The membranes were rinsed with 1× PBS for 5 min before drying. The secondary antibody labeled with a 700 channel dye, anti-rabbit-680 nm, was used to detect rabbit polyclonal anti-TRBP Ab672 and β-tubulin.
Gel shift assays
For each reaction, 10 μl of the TRBP or β-tubulin immunodepleted cell extract was incubated with 10 fmol of 5′ end-labeled siRNA for 30 min at room temperature. The samples were mixed with 2× native gel loading dye and resolved in a 5% non-denaturing polyacrylamide gel for 3 h at 200 V at 4°C. The HEK 293 total cell extract and the tubulin immunodepleted cell extract were used as controls.
P24 assays
The human T cell line CEM was maintained in RPMI 1640 medium supplemented with 10% FBS. A total of 1 × 106 transduced CEM T cells were infected with HIV-1 strain IIIB at an MOI of 0.01. After overnight incubation, cells were washed three times with Hanks’ balanced salts solution and cultured in medium with R10 (RPMI 1640 plus 10% FBS). At designated time points, culture supernatants were collected for p24 analyses (HIV-1 p24 antigen EIA kit, Beckman Coulter Inc.).
RESULTS
Competition for RNAi components by co-transfected ShRNAs
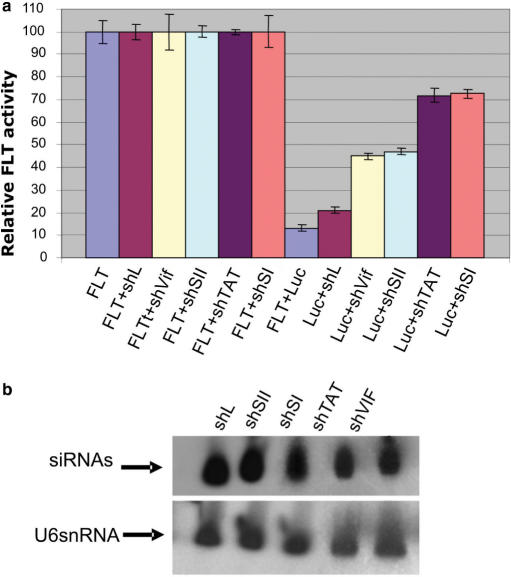
We have been interested in investigating the possibility that in the presence of multiple interfering RNAs, the saturation of RNAi components could diminish the activity of a particular sh or siRNA and generate misleading results. Most importantly, the degree of saturation could be such as to interfere with the endogeneous microRNA pathway, thus disrupting potentially critical regulatory functions in the cell. To test this possibility, we selected a series of shRNAs (shL, shVif, shSII, shTAT, shSI) and co-transfected each with a FLT and an anti-firefly-luciferase shRNA (Luc), which routinely generates ∼90% down-regulation of the target (Figure 1a). Each shRNA was also co-transfected alone with the luciferase target (constructs 2–6, Figure 1a) to monitor any non-specific reduction or increase of luciferase expression. The results clearly show that there is a direct competition among non-related shRNAs (Figure 1a). The degree of competition varies with the shRNA used in the co-transfection. The down-regulation of Luc was reduced by co-transfected, non-specific shRNAs from 10 to 60%, yielding in two cases only 30% down-regulation of the luciferase target (Figure 1, a Luc+shSI and Luc+shTAT) without any targeting of RNAi components and/or presumably other cellular genes. Northern analyses of total RNA from cells transfected with these shRNA constructs did not show a correlation between the degree of competition and their intracellular expression levels (Figure 1, compare a and b). Thus, competition must be dictated by the ability of each shRNA to be processed into siRNA and incorporated into RISC. Many parameters have been analyzed in the literature that can improve shRNA designs and possibly their incorporation into RISC (28). However, none of the popular rules for the optimal shRNA design can explain the different levels of competition exerted by each shRNA with the Luc shRNA construct (Supplementary Data).
Figure 1.
Irrelevant shRNA sequences compete with a luciferase-directed shRNA and reduce its ability to down-regulate the target. (a) FLT is a U1 expressed firefly luciferase gene, shL is targeted against the EnvPb1 retrovirus envelope gene, shVif is targeted against the Vif gene, shSII against sequences present in the HIV-Rev, shTAT is designed against the HIV Tat gene and shSI is directed against both TAT and Rev genes. The shRNA Luc is targeted to a sequence present in the coding region of the firefly luciferase gene. The irrelevant shRNAs were transfected with the luciferase target to monitor non-specific effects (constructs 2–6). FLT (construct 1) indicates the luciferase target transfected alone. The Bluescript vector was used to adjust the total amount of DNA transfected to be the same in each sample. Fifty nanogram of the firefly luciferase target and 50 ng of the shRNA constructs were used for these transfection reactions. The luciferase units were calculated by comparing each experimental sample to its own control. A Renilla luciferase plasmid was co-transfected in each sample as transfection control and used to normalize the firefly luciferase units. (b) Northern analysis of total RNA from 293 cells transfected with the relevant shRNAs as indicated on the top of the gel. Probes complementary to the guide sequence corresponding to each shRNA were used to detect the processed siRNA products (siRNA). A probe complementary to the endogenous U6 small nuclear RNA (U6snRNA) was used as internal control for the amount and integrity of the RNA loaded in each lane.
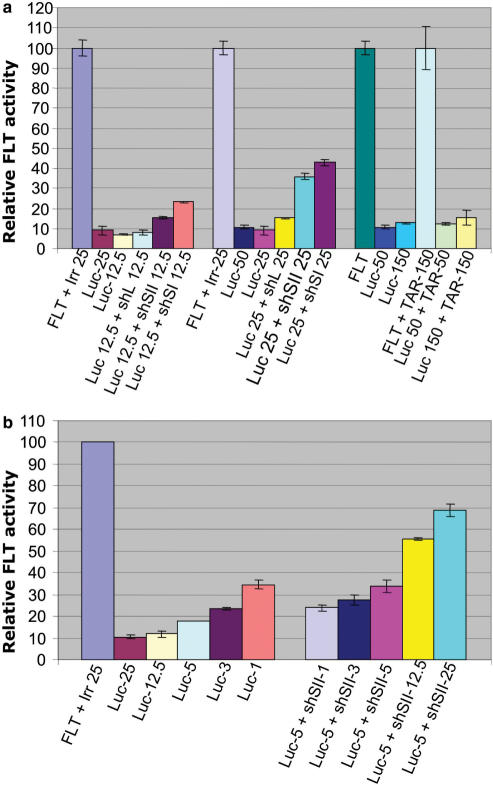
Although the relative intracellular expression of each shRNA could not by itself explain the degree of competition for each of the shRNAs, the total amount of shRNA present in the cell can render Exportin 5 rate limiting and affect the overall shRNA activity. To test this possibility, we co-transfected low concentrations (either 12.5 or 50 ng) of three irrelevant shRNA expression plasmids (shL, shSII and shSI) with equal amounts of the Luc shRNA construct and its luciferase target. As expected, the higher amount of shRNAs present in the cell increased the competition while keeping the relative strength of each shRNA as a competitor unchanged (Figure 2a). The Luc shRNA showed the same ability to down-regulate the target when 12.5, 25 or 50 total ng were transfected with the luciferase target (Figure 2a, Luc-12.5, Luc-25, Luc-50). A double-stranded RNA stem-loop expressing construct (a mutated HIV TAR element that retains its structure) was also co-transfected with the Luc shRNA and the luciferase target to show that the competition is specific to shRNAs. The U6 TAR element is highly expressed in cells (not shown) yielding levels of cellular transcripts that are comparable to any of the shRNAs tested in these experiments. As shown in Figure 2a, the TAR element did not significantly affect the efficacy of the Luc shRNA even when transfected at a 1:3 ratio (50 ng of Luc and 150 ng TAR). To test if the competition persists when the shRNAs are present in a linear range we performed a dose response curve using the Luc shRNA and the shSII as a competitor (Figure 2b). We used 1, 3, 5, 12.5 and 25 ng of shLuc in a luciferase down-regulation assay to select the lowest amount of shRNA that still achieved optimal target down-regulation (Figure 2b). Based on these results, we co-tranfected 5 ng of shLuc with increasing amounts of the shSII competitor. The results show that even 1 ng of added competitor is able to reduce the activity of shLuc (Luc-5 + shSII-1, Figure 2b).
Figure 2.
Different shRNA sequences compete with different strengths against the Luc shRNA. (a) Different concentrations of irrelevant shRNA-expressing plasmids (shL, shSII, shSI) were co-transfected with the firefly luciferase target (FLT) and the specific anti-luciferase shRNA (Luc). The amount used in each transfection is indicated in ng (12.5, 25, 50 or 150). The luciferase units for each sample were calculated as described in Figure 1 and in the Materials and Methods section. Each irrelevant shRNA was transfected independently with the luciferase target to monitor non-specific effects. Each Luc co-transfection experiment is normalized to its corresponding target-irrelevant control and to the Renilla luciferase transfection control. However, for convenience only one bar is shown to represent the target for each group and it is indicated by FLT+Irr (Irrelevant). Bluescript was used to adjust the total amount of DNA transfected in each sample. TAR indicates the co-transfection of a U6 construct expressing a mutated HIV-1 TAR structure (see text). (b) Non-saturating amounts of shRNAs can still compete for incorporation into RISC. A titration of U6 shLuc (Luc) was used to establish a concentration in the linear range for activity (left side of the chart). Five nanogram of U6sh Luc were then selected to be co-transfected with increasing amounts of a shRNA competitor (shSII) (right side of the chart). The numbers 1–25 indicate the amount in nanograms used in the reactions.
SiRNAs compete for incorporation into RISC
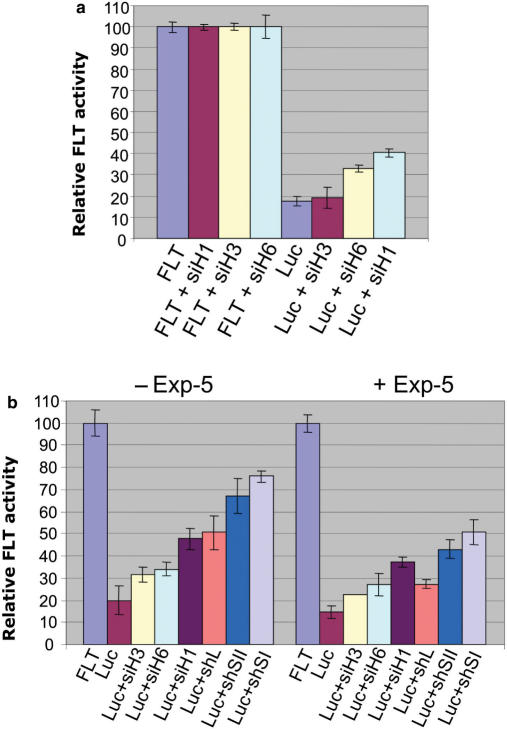
The observation that the shRNA competition increases with the total amount of transfected shRNA could be explained by the increasing saturation of Exportin 5, although the different relative strengths of each shRNA to compete against the shLuc when used in equal amounts suggest that they are more likely due to a preferential incorporation into RISC. Moreover, the shRNA can also compete when used in non-saturating conditions (Figure 2b). However, to bypass Exportin 5 we repeated the co-transfection experiment using synthetic siRNAs (siH3, siH6 and siH1; Figure 3a). The luciferase target was transfected alone or with each synthetic siRNA (Figure 3a). The results of this experiment show that the synthetic siRNAs were able to affect the down-regulation of the luciferase target when co-transfected with the Luc construct. Similar to what was shown with the shRNAs, the siRNAs competed to differing degrees with the Luc shRNA. Since synthetic siRNAs do not need to be transported from the nucleus to the cytoplasm for their function and their activity is not affected by Exportin-5 (20), the observed competition must be at the level of incorporation into RISC. Although a recent report shows that synthetic siRNAs can potentially shuttle from the cytoplasm back into the nucleus using Exportin-5 (29), this should not be a major competition pathway for synthetic siRNAs. Therefore, these results show that sh and siRNAs compete at the level of incorporation into RISC but may also compete for the Exportin 5 pathway to some extent.
Figure 3.
(a) Synthetic siRNAs can reduce the ability of a shRNA to down-regulate its target. A 10 nM concentration of each synthetic siRNA (siH3, siH6 and siH1) was co-transfected with the firefly luciferase target (FLT) independently or together with the anti-firefly shRNA (Luc). The firefly luciferase units were normalized against Renilla luciferase, which was used as transfection control. (b) Over-expression of Exportin-5 decreases shRNA competition but does not eliminate competition among interfering RNAs. Twenty nanomolar of each synthetic siRNA (siH3, siH6, siH1) or 50 ng of each shRNA construct (shL, shSII, shSI) were co-transfected with the luciferase target and the Luc shRNA with or without 100 ng of the Exportin-5-expressing plasmid (Exp-5) as indicated in the figure (+Exp-5, -Exp-5). Firefly luciferase units were calculated as previously described.
Sh and siRNAs compete for Exportin 5
To characterize any contribution made by the saturation of Exportin-5 on the competition detected among interfering RNAs, we included a plasmid-expressing Exportin-5 in the co-transfection experiments. It has previously been shown that co-transfecting Exportin-5 relieves the saturation of transport and improves shRNA cleavage of its target (19,24). As expected, our results show that the overall activity of shLuc is improved, but competition among the different sequences still persists (Figure 3b). It should be noted that for siRNAs, the effect could be indirect, and based only on more efficient export of shLuc.
Micro RNAs do not compete for RNAi components
Our results show that when different exogenously supplied interfering RNAs are introduced into cells, competition for export and incorporation into RISC can decrease or completely block the activity of some RNAi triggers. These findings suggest that transfected siRNAs and shRNAs can also compete with endogenous microRNAs. To test this hypothesis, we inserted the siRNA directed against the site II HIV-rev-1 sequence (SII) into the backbone of the mir-30 microRNA. We made two different versions of this microRNA, one containing the entire wild-type backbone (Mir-B, and the other with some additional mutations designed to increase the stability of the microRNA structure (Mir-CG). Both microRNAs are processed (Figure 4c and data not shown) and are effective in down-regulating a RLT which contains the corresponding complementary target sequence within its 3′ UTR (Mir-B, Mir-CG; Figure 4a). When these microRNAs were co-transfected with an irrelevant shRNA-expressing construct (shSI), their inhibitory activity was dramatically reduced (Mir-B+shSI, Mir-CG+shSI; Figure 4a). Conversely, the microRNA constructs did not exert significant inhibition on the activity of shRNAs as shown by co-transfecting Mir-B and Mir-CG with a U6 shRNA targeting the firefly luciferase gene (compare Luc to Luc+Mir-B, Luc+Mir-CG; Figure 4b). Importantly, the guide sequence expressed by the microRNA becomes a good competitor when it is expressed in the form of a shRNA (shSII, Figure 1a).
Figure 4.
ShRNAs reduce the activity of microRNA constructs while microRNAs are not effective competitors. (a) A target site for the Mir-B and Mir-CG is inserted in the 3′ UTR of the Renilla luciferase gene, which is used as reporter construct in this experiment (RLT). Co-transfection of an irrelevant shRNA (shSI) nearly abolished the activity of the microRNAs (compare Mir-B and Mir-CG with Mir-B+shSI, Mir-CG+shSI) while an irrelevant microRNA (Mir-Irr) had no effect on Mir-B and Mir-CG (Mir-B+Mir-Irr and Mir-CG+Mir-Irr). The Renilla luciferase units were normalized against the firefly luciferase which was co-transfected with the experimental samples and used as an internal transfection control. (b) An irrelevant shRNA (shSI) strongly reduces the efficacy of Luc (Luc versus Luc+shSI), but an irrelevant microRNA (Mir-B or Mir-CG) did not have a significant effect on the activity of the specific shRNA (compare Luc versus Luc+Mir-B and Luc+Mir-CG). The units of the firefly reporter target were normalized against the Renilla luciferase which was used as internal transfection control. (c) Left; northern analysis of total RNA from 293 cells transfected with shSII or mirCG. A 4-fold excess of mirCG over shSII was used for the transfection. The probe is complementary to the mature SII which is the sequence expressed from both constructs. A probe complementary to the endogenous U6snRNA (U6) was used as control. Right; shSII or mirCG were used as competitors against the shLuc construct. Five hundred nanogram or 5 ng of mirCG and shSII respectively were used to compete against 5 ng of shLuc. (d) Synthetic si and shRNAs, but not expressed microRNAs, can compete with the endogenous microRNA pathway. A stable clonal cell line with an integrated EGFP gene that includes the target site for the endogenous mir21 at its 3′ end (HCT116-GFPmiR21) was mock transfected (Lip-2000) or transfected with: (i) a 2′O-Me-oligonucleotide designed to bind and block mir21 activity (Mir-21–2′O-Me), (ii) an irrelevant 2′O-Me-oligonucleotide to test for non-specific reactivation of EGFP (IRR-2′O-Me), (iii) a microRNA construct expressing the SII guide sequence (Mir-CG), (iv) an shRNA construct expressing the SII guide sequence (shSII) and (v) a synthetic siRNA (siH1). EGFP reactivation indicates interference with the activity of the endogenous mir21.
Finally, when Mir-B and Mir-CG were co-transfected with an irrelevant microRNA-expressing construct (Mir-Irr), there was no significant change in their ability to down-regulate their target (Mir-B+Mir-Irr, Mir-CG+Mir-Irr; Figure 4a), indicating that perhaps the safest method for down-regulation of cellular targets is to express the siRNA of choice within a microRNA backbone.
To test if the differences among sh and microRNAs in their ability to compete resides primarily with their levels of expression, we repeated the competition experiment using concentrations that would generate comparable amounts of mature sequences as determined by Northern blot analysis (Figure 4c). A 4-fold excess of mirCG over shSII was used for the northern blot. Based on the differences in the processed SII sequence, which is expressed from both constructs, a 100-fold excess of mirCG over shSII was used in a competition assay (Figure 4c). The data show that a siRNA sequence is not an effective competitor when expressed from a microRNA and yet it is very proficient in down-regulating its target even when transfected at lower concentrations (Figure 4a).
Si and shRNAs can compete with endogenous microRNAs
To confirm that si and shRNAs are able to interfere with the microRNA regulatory activities in an intracellular environment, we constructed a stable clonal cell line with an integrated EGFP gene that includes the target site for mir21 at its 3′ end (HCT116-GFPmiR21). In this cell line, the EGFP gene is silenced by the endogenous expression of the mir21 microRNA. Any inhibition with the processing, transport and/or incorporation of mir21 would cause a reactivation of EGFP expression. It has previously been shown that 2′O-Me-oligonucleotides (Antagomirs) can block microRNA activity when designed to bind the microRNAs’ guide sequences (30,31). As expected, transfection of an Antagomir fully complementary to the mir21 guide sequence in the HCT116-GFPmiR21 cells reactivated EGFP fluorescence by blocking the activity of the endogenous mir21 (Mir-21–2′ O-Me, Figure 4d), while an irrelevant Antagomir used as control did not have any effect on the level of fluorescence (IRR-2′O-Me, Figure 4d). Both the non-specific sh and siRNAs were able to increase EGFP fluorescence, demonstrating that these constructs can interfere with the endogenous microRNA pathway (shSII, siH1, Figure 4d). However, when the same siRNA guide sequence was expressed from a microRNA backbone, it did not affect the mir21 activity, as shown by the lack of EGFP fluorescence (Mir-CG, Figure 4d). Two additional microRNA constructs were transfected in HCT116-GFPmiR21 with the same experimental outcome (data not shown).
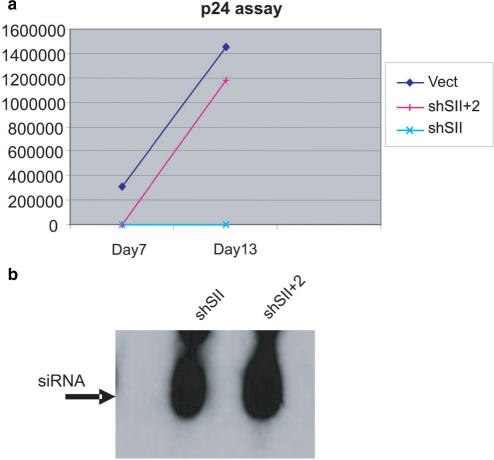
To confirm that competition can alter the efficacy of shRNAs when used in a combinatorial fashion, we tested the anti HIV inhibitory activity of shSII alone (shSII) or in combination with two other shRNAs (shSII+2). CEM cells were stably transduced with a single shRNA or with three different shRNAs each independently expressed from a separate U6 promoter and then infected with HIV IIIB. The results from these experiments show that shSII results in potent inhibition of HIV replication whereas when combined with two additional shRNAs provide protection for the first seven days, but viral replication breaks through after one week (Figure 5a). The loss of inhibitory activity by the triple construct was not due to loss of expression, since the levels of siRNAs were comparable for the single shRNA and triple constructs (Figure 5b). The most plausible explanation for the viral breakthrough in the triple construct expressing cells is competition among the shRNAs/siRNAs for export and incorporation into RISC, allowing the virus to escape RNAi surveillance and replicate.
Figure 5.
(a) p24 HIV challenge assay shows escape from RNAi when multiple shRNAs are simultaneously expressed. Stable CEM cells expressing shSII (shSII) are protected from HIV infection. This protection is lost when two other shRNAs are expressed simultaneously. Cells transduced with the vector backbone (Vect) are used as control. (b) Northern analysis shows comparable expression of shSII in transduced cells when expressed alone (shSII) or in combination with two other shRNAs (shSII + 2). A probe complementary to the mature SII RNA sequence was used to detect the processed siRNA. Probes for the other two shRNAs also revealed robust expression (data not shown).
TRBP contributes to the preferential loading of different sequences into RISC
Our experiments show that each siRNA sequence competes to a different extent with other siRNAs and shRNAs. Incubation of 32P end-labeled siRNAs in HEK293 cytoplasmic extracts results in the formation of a gel-shifted complex using native polyacrylamide gel electrophoresis (Figure 6a, lane 2). The amount of binding in this assay is directly proportional to the intracellular efficacy of the siRNAs analyzed (Sakurai et al., unpublished data). We speculated that TRBP, a double-stranded RNA-binding protein that has been shown to be part of the minimal RISC (26,32), could contribute to the loading of siRNA guide sequences into RISC and that it would be part of this complex. Depletion of TRBP from the cellular extracts (anti-TRBP) resulted in a marked reduction of siRNA complex formation (Figure 6a, lanes 4), while depletion of β-tubulin (anti-β-tubulin), which was used as control, had no effect (Figure 6a, compare lanes 2 and 3). Identical results were obtained when depletion for β-actin was used as control (data not shown). Western analyses of the extracts showed that depletion of both TRBP and β-tubulin were nearly complete (Figure 6b). However, the small amount of residual gel-shifted complex may reflect incomplete removal of TRBP or could be due to binding of the siRNAs by other components of RISC. We repeated our competition experiment using cells in which TRBP levels were reduced by siRNA-mediated targeting of the TRBP2 mRNA (see Materials and Methods section). TRBP levels were reduced by ∼80% as determined by western (Figure 6c) and northern (data not shown) analyses. Depletion of TRBP resulted in more equivalent competition by each shRNA sequence (shL, shVif, shSII, shTAT, shSI) or siRNA sequence (siH3, siH6, siH1), but competition with shLuc still persisted (Figure 6d).
Figure 6.
(a) Gel shift assay shows loss of complex formation when TRBP depleted cellular extracts are mixed with labeled siRNAs. A 32P end-labeled anti-EGFP siRNA (without cellular extracts in lane 1) was mixed with: lane 2, total cellular extract; lane 3, extracts depleted of β-tubulin; lane 4 extracts depleted of TRBP. (b) Western detection of TRBP and β-tubulin after immunodepletion. Extracts were treated with TRBP or β-tubulin antibodies and precipitated as described in the Materials and Methods section. S, supernatant; P, pellet. The supernatant fractions show to be substantially depleted of TRBP or β-tubulin and were subsequently used in the gel shift assay for detection of complex formation (a). (c) Western analyses of lysates from cells treated with (+) or without (−) TRBP siRNA as described in Materials and Methods section. Lysates from cells transfected with a TRBP-expressing plasmid (TRBP) were used for size determination. Detection of β-tubulin was used as an internal control. (d) TRBP down-regulation eliminates the differential competition among interfering RNAs. Twenty nanomolar of each synthetic siRNA (siH3, siH6, siH1) or 50 ng of each shRNA construct (shL, shVif, shSII, shTAT, shSI) were co-transfected with the firefly luciferase target (FLT) and the Luc shRNA in cells that were pre-treated for 5 days with a siRNAs targeted against TRBP. Firefly luciferase units were calculated as previously described.
DISCUSSION
We have shown competition between siRNAs and shRNAs when they are co-transfected into cultured cells. This competition is partially dependent upon saturation of Exportin-5 by the shRNAs and is largely sequence independent. The activity of a luciferase-targeted si or shRNA can be compromised by the ability of another siRNA to be preferentially loaded into RISC.
The competition among shRNAs raises some important issues. Our data show that both siRNAs and shRNAs can compete with the endogenous microRNA pathway (Figure 4d) thus posing the potential for perturbation of endogenous cellular miRNA processes. In a recent publication, Grimm et al. (19) reported that some shRNA sequences were able to block the activity of the liver-specific miR122 and additionally caused severe liver toxicity and death of some of the treated mice. The authors attributed this adverse effect to saturation of Exportin-5, postulating that despite being expressed from the same AAV vector backbone, some sequences were expressed at higher levels and were more toxic. Our results demonstrate that competition with endogenous microRNAs can be a general phenomenon and that Exportin-5 is only one of the components involved, as shown by the inability to fully restore the U6sh-luciferase activity when Exportin-5 was ectopically expressed (Figure 3b). Moreover, synthetic siRNAs, which do not rely on Exportin-5 for their transport, can still affect the activity of both shRNAs (Figure 3a) and microRNAs (Figure 4d). These results indicate that the ability of a specific sequence to compete is also determined by its ability to be loaded into RISC. Interestingly, the relative competitive strength of the synthetic siRNAs with the Luc shRNA correlated with their efficacy to down-regulate their cognate targets, which in turn is directly proportional to the complex formation in cell extracts (Sakurai et al., unpublished data). These results are consistent with a previously published study (33) in which competition was detected between heavily modified and unmodified synthetic siRNAs. The competition was found to correlate with the potency of the siRNAs and it was attributed to a possible saturation of the export system or competition for other RISC components.
The affinity of each sequence for one or more RNAi component(s) may determine the efficiency of loading of that sequence into RISC, thereby potentiating the competition with other sh/siRNAs and miRNAs. TRBP is a double-stranded RNA-binding protein that has been shown to associate with both Dicer and Ago-2 (26,32). It has been proposed that TRBP couples the initiation and executions steps of RNAi (26) and that it identifies the guide strand prior for passage on to Ago-2 (34). Our data reveal that cellular down-regulation of TRBP results in loss of differential competition by the various sh/siRNAs, although competition per se still persists (Figure 6d), perhaps via a bypass mechanism for incorporation into the Ago2-binding domain. Our results support the role of TRBP as a sensor for the RISC loading or Ago-2 handoff (26). However, it is possible that TRBP does not act alone and loading may include other RISC proteins.
In our experimental system, sequences expressed from a microRNA backbone are poor competitors relative to shRNA or siRNAs of the same sequences (Figure 4). Two different sequences were expressed from three different microRNA backbones and none of them was able to compete (Figure 4 and data not shown) despite the fact that these sequences were still effective in RNAi. The overall level of shRNA expression does not seem to be the primary reason for this lack of competition (Figure 4c), which is probably determined by the overall kinetics of incorporation. Since miRNAs are being shunted through the endogenous miRNA-processing pathway, this may slow their entry into RISC relative to highly expressed shRNAs and transfected siRNAs.
Si and sh-RNAs are routinely used to down-regulate cellular genes, siRNA libraries have been widely employed to screen for gene function (9–11), and siRNAs are already in human clinical trials for wet adult macular degeneration, respiratory syncitial virus (SRV) infection and acute kidney failure. The potential for competition of the clinically applied siRNAs with endogenous miRNAs is a concern as is the competition among combinations of si/shRNAs. Due to the rapid emergence of resistance mutations in viruses such as HCV and HIV, the use of combinations of sh/siRNAs represents a potential mechanism for circumventing this problem. If one of the siRNAs is more efficiently loaded into RISC, it could compromise the effectiveness of one or more of the other siRNAs. For example, our data clearly show that an effective shRNA targeted against the HIV-1 rev gene, when combined with two other hairpins, no longer provides stable, long-term inhibition of HIV replication (Figure 5). This result is consistent with previous finding by Nishitsuji et al. (35), which showed a negative effect on shRNA antiviral activities when used in a combinatorial fashion. However, based upon our findings, concern about obstructing the endogenous RNAi machinery can potentially be overcome by modifying the siRNA expression system to produce a microRNA, or by combining si and shRNAs that do not compete with one another. By finding siRNA/target combinations that work at the lowest possible concentrations, it should be possible to mitigate the potential for competition with endogenous miRNAs or among combinations of applied si/shRNAs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by NIH AI29329, AI42552 and HL07470 to J.J.R. We are grateful to Dr M. Amarzguioui for helpful discussion during this work. Funding to pay the Open Access publication charges for this article was provided by the National Institutes of Health, USA.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 3.Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- 4.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates rNA cleavage targeted by miRNAs and siRNAs. Molecular Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 5.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 6.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 7.Rand TA, Ginalski K, Grishin NV, Wang X. Biochemical identification of Argonaute 2 as the sole protein required for RNA-induced silencing complex activity. Proc. Natl Acad. Sci. USA. 2004;101:14385–14389. doi: 10.1073/pnas.0405913101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 9.Hannon GJ, Rossi JJ. Unlocking the potential of the human genome with RNAinterference. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- 10.Moffat J, Sabatini DM. Building mammalian signaling pathways with RNAi screens. Nat. Rev. Mol. Cell. Biol. 2006;7:177–187. doi: 10.1038/nrm1860. [DOI] [PubMed] [Google Scholar]
- 11.Chatterjee-Kishore M. from genome to phenome – RNAi library screening and hit characterization using signaling pathway analysis. Curr. Opin. Drug Discov. Devel. 2006;9:231–239. [PubMed] [Google Scholar]
- 12.Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 13.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 14.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 15.Gitlin L, Karelsky S, Andino R. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature. 2002;418:430–434. doi: 10.1038/nature00873. [DOI] [PubMed] [Google Scholar]
- 16.Gitlin L, Andino R. Nucleic acid-based immune system: the antiviral potential of mammalian RNA silencing. J. Virol. 2003;77:7159–7165. doi: 10.1128/JVI.77.13.7159-7165.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vargason JM, Szittya G, Burgyan J, Tanaka Hall TM. Size selective recognition of siRNA by an RNA silencing suppressor. Cell. 2003;115:799–811. doi: 10.1016/s0092-8674(03)00984-x. [DOI] [PubMed] [Google Scholar]
- 18.Cullen BR. Viruses and microRNAs. Nat. Genet. 2006;38:S25–30. doi: 10.1038/ng1793. [DOI] [PubMed] [Google Scholar]
- 19.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 20.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 22.Bohnsack MT, Czaplinski K, Gorlich D. Exportin-5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim VN. MicroRNA precursors in motion: exportin-5 mediates their nuclear export. Trends Cell Biol. 2004;14:156–159. doi: 10.1016/j.tcb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 24.Yi R, Doehle BP, Qin Y, Macara IG, Cullen BR. Overexpression of exportin-5 enhances RNA interference mediate by short hairpin RNAs and microRNAs. RNA. 2005;11:220–226. doi: 10.1261/rna.7233305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castanotto D, Scherer L. Targeting cellular genes with PCR cassettes expressing short interfering RNAs. Methods Enzymol. 2005;392:173–185. doi: 10.1016/S0076-6879(04)92010-1. [DOI] [PubMed] [Google Scholar]
- 26.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daviet L, Erard M, Dorin D, Duarte M, Vaquero C, Gatignol A. Analysis of a binding defference between the two dsRNA-binding domains in TRBP reveals the modular function of a KR-helix motif. Eur. J. Biochem. 2000;267:2419–2431. doi: 10.1046/j.1432-1327.2000.01256.x. [DOI] [PubMed] [Google Scholar]
- 28.Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 29.Ohrt T, Merkle D, Birkenfeld K, Echeverri CJ, Schwille P. In situ fluorescence analysis demonstrates active siRNA exclusion from the nucleus by Exportin 5. Nucleic Acids Res. 2006;34:1369–1380. doi: 10.1093/nar/gkl001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hutvágner G, Simard MJ, Mello CC, Zamore PD. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:e98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meister G, Landthaler M, Dorsett Y, Tuschl T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA. 2004;10:544–550. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koller E, Propp S, Murray H, Lima W, Bhat B, Prakash TP, Allerson CR, Swayze EE, Marcusson EG, et al. Competition for RISC binding predicts in vitro potency of siRNA. N ucleic Acids Res. 2006;34:4467–4476. doi: 10.1093/nar/gkl589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples MicroRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 35.Nishitsuji H, Kohara M, Kannagi M, Masuda T. Effective suppression of human immunodeficiency virus type 1 through a combination of short- or long-hairpin RNAs targeting essential sequences for retroviral integration. J. Virol. 2006;80:7658–66. doi: 10.1128/JVI.00078-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.