Abstract
Plants have evolved complicated regulatory systems to control immune responses. Both positive and negative signaling pathways interplay to coordinate development of a resistance response with the appropriate amplitude and duration. AtNUDT7, a Nudix domain-containing protein in Arabidopsis (Arabidopsis thaliana) that hydrolyzes nucleotide derivatives, was found to be a negative regulator of the basal defense response, and its loss-of-function mutation results in enhanced resistance to infection by Pseudomonas syringae. The nudt7 mutation does not cause a strong constitutive disease resistance phenotype, but it leads to a heightened defense response, including accelerated activation of defense-related genes that can be triggered by pathogenic and nonpathogenic microorganisms. The nudt7 mutation enhances two distinct defense response pathways: one independent of and the other dependent on NPR1 and salicylic acid accumulation. In vitro enzymatic assays revealed that ADP-ribose and NADH are preferred substrates of NUDT7, and the hydrolysis activity of NUDT7 is essential for its biological function and is sensitive to inhibition by Ca2+. Further analyses indicate that ADP-ribose is not likely the physiological substrate of NUDT7. However, the nudt7 mutation leads to perturbation of cellular redox homeostasis and a higher level of NADH in pathogen-challenged leaves. The study suggests that the alteration in cellular antioxidant status caused by the nudt7 mutation primes the cells for the amplified defense response and NUDT7 functions to modulate the defense response to prevent excessive stimulation.
Recognition of a potentially pathogenic microorganism by plant cells triggers coordinated disease resistance responses, leading to the deployment of appropriate defense mechanisms. A strong form of disease resistance is mediated by race-specific gene-for-gene interaction in which the host Resistance (R) protein directly or indirectly recognizes the cognate Avirulence (Avr) protein delivered into the host by a specific pathogen strain (Dangl and Jones, 2001). Such an interaction activates a hypersensitive response culminating in rapid cell death of the infected site, leading to a halt of pathogen growth. Several signaling molecules/second messengers, such as reactive oxygen/nitrogen species, salicylic acid (SA), jasmonic acid, and ethylene, play important roles in relaying, amplifying, and integrating the initial pathogen signal through complex signaling networks to activate defense mechanisms (Dangl and Jones, 2001; Hammond-Kosack and Parker, 2003). Phospholipid derivatives such as phosphoinositols and nucleotide derivatives such as cyclic GMP and cyclic ADP-Rib (cADPR) are also involved in the defense signaling pathways (Durner et al., 1998; Laxalt and Munnik, 2002).
In the absence of the R-Avr interactions, host cells are still capable of activating the basal defense response (Glazebrook et al., 1997; Belkhadir et al., 2004). The basal defense response can be rapidly induced upon recognition of molecular signatures common to a whole class of microorganisms (whether pathogenic or not), termed pathogen-associated molecular patterns/microbe-associated molecular patterns (MAMPs) or general elicitors (Gomez-Gomez and Boller, 2002; Eulgem, 2005). The basal defense response, together with constitutive physical and chemical barriers, successfully prevents most infections from becoming established. Pathogens have evolved Avr proteins as virulence effectors to suppress the basal defense response, whereas R proteins have evolved to detect action of effectors and to amplify the basal defense response (Jones and Dangl, 2006). Because activation of the induced defense response will likely cause deleterious effects on normal cell functioning, delicate regulation of the immune response by both positive and negative pathways is essential for an organism's functioning (Alexander and Hilton, 2004). However, regulatory systems that modulate the defense response in plants remain largely unknown.
Nudix box-containing proteins generally hydrolyze a nucleoside diphosphate linked to some other moiety X, and the proteins containing the domain were termed Nudix proteins (Bessman et al., 1996). Substrates of Nudix hydrolases include dinucleoside polyphosphates, nucleotide sugars, phosphoinositol derivatives, and capped mRNA. Some of these molecules play regulatory roles, whereas others are coenzymes and mutagenic or toxic components. It has been suggested that many Nudix hydrolases function as cellular surveillance enzymes to maintain physiological homeostasis by sensing and modulating the levels of their substrates (Xu et al., 2001). The first plant Nudix protein was isolated from Lupinus angustifolius as a diadenosine tetraphosphate (Ap4A) hydrolase (Maksel et al., 1998). The Arabidopsis (Arabidopsis thaliana) genome encodes 24 members of the Nudix family (named AtNUDT; Ogawa et al., 2005). One of the Nudix proteins, At1g68760, was found to remove pyrophosphate from dihydroneopterin triphosphate, a step in the folate synthesis pathway (Klaus et al., 2005). In vitro enzymatic analyses on substrate specificity have been carried out for nine of the AtNUDTs (abbreviated as NUDTs here) that are predicted to be cytosolic (Ogawa et al., 2005). Biological functions of plant Nudix proteins are little known. NUDT7 was recently identified as a multiple stress-induced gene, and its knockout mutant was found to show constitutive resistance against Pseudomonas syringae (Jambunathan and Mahalingam, 2006). Furthermore, the NUDT7 gene was identified as one of the pathogen-responsive genes whose induction is dependent on EDS1 and PAD4 (Bartsch et al., 2006). The authors also showed that the nudt7 mutant exhibited enhanced resistance to Hyaloperonospora parasitica and the enhanced disease resistance phenotype requires functional EDS1. These results indicate that NUDT7 plays an important role as a repressor in the disease resistance pathways. However, the mechanism by which NUDT7 regulates these pathways is little understood. Interestingly, Nudix proteins have been identified as putative TTSS effectors from phytopathogenic bacteria (Mukaihara et al., 2004; Tamura et al., 2005; Koebnik et al., 2006), suggesting that pathogens may deliver Nudix protein to promote pathogenesis presumably by modulating the levels of nucleotide derivatives in host cells.
In this report, we describe the identification and characterization of NUDT7 as a negative regulator of the basal defense response. Its loss-of-function mutation leads to acceleration and amplification of the basal defense response. Two distinct pathways, one dependent on and the other independent of NPR1 and SA, are modulated by NUDT7 to prevent an excessive defense response. Our study indicates that alteration in redox homeostasis caused by the nudt7 mutation may lead to the excessive defense response that can be triggered not only by pathogens but also by other inciting agents.
RESULTS
Identification of nudt7 as an Enhanced Disease Resistance Mutant
The basal defense response can be quickly triggered upon pathogen invasion through recognition of MAMPs by host cells. It is plausible that many of the genes rapidly induced following pathogen infection may play important roles in the basal defense response. Based on gene expression profiles from our Arabidopsis Genechip microarray experiment (data not shown), we selected some early pathogen-responsive genes (induced within 1.5 h of infection by the bacterial pathogen P. syringae) and obtained their T-DNA insertion mutagenized lines for further analyses to determine their roles in the basal defense response. Among the T-DNA insertion lines, the nudt7 mutant was found to have an enhanced disease resistance phenotype against the virulent P. syringae pv. tomato (Pst) strain DC3000. The nudt7 mutant line (Salk_046441) carries a T-DNA insertion at the locus At4g12720, which encodes a protein containing a Nudix box (see below). The gene was previously named AtNUDT7 (abbreviated here as NUDT7), one of 24 putative Arabidopsis genes encoding Nudix proteins, and was reported to possess a hydrolase activity in an in vitro enzymatic assay (Ogawa et al., 2005). Growth of Pst in nudt7 was 3- to 9-fold lower than in wild-type plants in several independent experiments using the in planta bacterial growth assay. Figure 1A shows the results of one of the experiments. In addition, growth of the avirulent strain Pst avrRpm1was also reduced in the mutant by severalfold (Fig. 1B). Heterozygous nudt7/+ plants were indistinguishable from wild-type plants in resistance to Pst (data not shown), indicating that the nudt7 mutation is recessive.
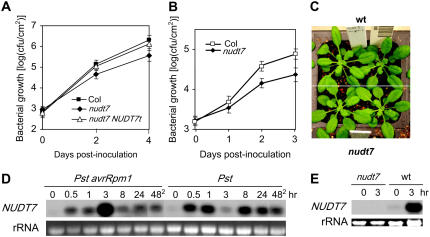
Figure 1.
The nudt7 mutant exhibits enhanced resistance to P. syringae. A, In planta growth of Pst in wild type (Columbia [Col]), nudt7, and the mutant complemented with the wild-type genomic clone (nudt7 NUDT7t). B, In planta growth of Pst avrRpm1 in wild-type and nudt7 plants. Each data point in A and B represents the average of three replicates ± sd. C, Five-week-old wild-type and mutant plants showing no obvious morphological difference. D, A northern-blotting result showing expression profiles of NUDT7 in wild-type plants in response to inoculation with Pst avrRpm1 and Pst. The lanes labeled with 482 contain RNA samples from uninfected leaves harvested 48 h after four other leaves of the same plants had been inoculated. E, An RNA blot showing that NUDT7 transcript was not detectable in the mutant. RNA was harvested from wild-type and mutant leaves 0 and 3 h postinoculation with Pst avrRpm1. wt, Wild type. [See online article for color version of this figure.]
The morphological phenotype of the nudt7 plants differs under different growth environments. When the plants were grown in our communal growth rooms where pathogens (mainly Erysiphe cichoracearum) and insects (mainly aphids) were frequently present and pesticides were routinely used, the nudt7 plants grew much smaller than the wild-type plants (data not shown). However, in isolated growth chambers where no obvious damage by pathogens or insects was observed and no pesticide or other chemical (except fertilizer) was applied, the nudt7 plants were often morphologically indistinguishable from the wild-type plants (Fig. 1C) and were slightly smaller than the wild-type plants in some batches. All experimental data presented here were obtained using plants growing in the isolated growth chamber on soil unless otherwise specifically indicated in the text. In two recent reports, the same nudt7 mutant was described as having a stunting growth phenotype (Bartsch et al., 2006; Jambunathan and Mahalingam, 2006). Our results indicated that the growth stunting is likely indirectly caused by the nudt7 mutation due to the mutant's super-sensitivity to some stimuli in growth environments. Indeed, excessive oxidative stresses could cause growth stunting of the mutant (see below).
NUDT7 consists of eight exons and the mutant has a T-DNA insertion in its first exon at 117 bp 3′ to the initial codon. A NUDT7 genomic clone (NUDT7t) including the 2-kb promoter region was obtained by PCR from genomic DNA of wild-type plants. When transformed into the nudt7 mutant, it was able to complement the enhanced disease resistance phenotype of the mutant (Fig. 1A), confirming that the mutation is indeed caused by the insertion at the NUDT7 gene.
NUDT7 was expressed at a low level in the uninfected wild-type leaves (Fig. 1D). Its expression was significantly induced in response to infection by both avirulent Pst avrRpm1 and virulent Pst strains. Strong induction of NUDT7 could be detected within 0.5 h of the inoculation. In the Pst-inoculated leaves, its transcript level dropped at 3 h postinoculation following the initial increase and then increased again at the later time points (Fig. 1D). This expression profile was repeatedly observed in multiple experiments. NUDT7 was also induced systemically by both the virulent Pst and avirulent Pst avrRpm1 strains (Fig. 1D). NUDT7 transcripts were not detectable in the mutant (Fig. 1E), indicating that the insertion likely results in a null allele.
The nudt7 Mutation Results in Hyperactivation of the Defense Response
To understand the mechanism by which the nudt7 mutation results in enhanced disease resistance, we used RNA-blotting analysis to reveal differences in activation of the defense response between the mutant and wild-type plants by examining pathogen-induced expression patterns of several pathogen-induced genes, including PATHOGENESIS-RELATED (PR) genes (PR1, PR2) and AVRRPT2-INDUCED GENE1 (AIG1; Reuber and Ausubel, 1996). The mutant did not accumulate high levels of PR1, PR2, or AIG1 in the absence of pathogen infection; however, induction of these genes was accelerated and amplified in the mutant following pathogen inoculation (Fig. 2, A–D; data not shown). In wild-type plants, transcripts of these genes were detectable only after several hours postinoculation, whereas in the mutant plants induction of these genes became apparent at 1 to 2 h postinoculation. Hyperactivation of the pathogen-responsive genes in the mutant can be induced not only by the avirulent Pst strain (Fig. 2A) but also by the virulent Pst (Fig. 2, B and D), as well as by the Pst mutant hrcC and a nonhost pathogen P. syringae pv. phaseolicola (Psp; Fig. 2, C and D). Pst hrcC carries a deletion in the hrcC gene that encodes for a component of the type III secretion system (Boch et al., 2002) and is defective in delivering type III virulence effectors into host cells and therefore is unable to grow or cause disease in plant tissues. The RNA-blotting analysis also revealed that the nudt7 mutant carrying the wild-type NUDT7 transgene no longer showed hyperactivation of the defense-related genes (Fig. 2E), again demonstrating that the nudt7 mutation is responsible for the hyperresponsiveness phenotype.
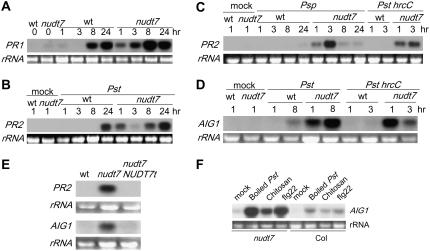
Figure 2.
The nudt7 mutation leads to hyperactivation of defense-related genes. A, Expression patterns of PR1 in wild-type and mutant leaves following inoculation with Pst avrRpm1. B, Expression patterns of PR2 induced by Pst. “Mock” represents an RNA sample from water-inoculated leaves. C, Accumulation of PR2 transcripts in wild-type and the mutant after inoculation with the nonhost pathogens Psp and Pst hrcC. D, Hyperactivation of AIG1 in the mutant in response to Pst or Pst hrcC. E, The mutant plants carrying the genomic clone of NUDT7 no longer exhibited hyperactivation of PR2 (top) or AIG1 (bottom) by Pst avrRpm1. The leaves were collected 1.5 h postinoculation. F, Induction of AIG1 in the nudt7 and wild-type (Col) plants 1.5 h postinfiltration with boiled Pst, chitosan, and flg22. wt, Wild type.
The above results indicate that the hyperresponsiveness of the nudt7 mutant is likely triggered upon the initial recognition of the microorganisms, presumably through detection of MAMPs. Indeed, infiltration of the mutant leaves with boiled Pst and individual MAMPs such as chitosan (100 μg/mL; a main cell wall component of fungi) and flg22 (5 μm; the 22-amino acid peptide from bacterial flagellin that triggers the innate immune response) resulted in quick and strong induction of AIG1 and PR2 in the mutant (Fig. 2F; data not shown).
The nudt7 Mutation Does Not Affect Hypersensitive Cell Death
The nudt7 mutant does not develop spontaneous lesions visible by the naked eye. When examined under microscopes after trypan blue staining, the leaves of uninfected mutant plants were found to display very small lesions, most of which appear to consist of a single cell (Fig. 3A). The estimated number of the microlesions in expanded leaves from the mutant was approximately 150 per leaf, whereas leaves from a wild-type plant generally displayed fewer than 20 stained spots per leaf (Fig. 3B). Diaminobenzidine (DAB) staining (to detect hydrogen peroxide [H2O2] in situ) revealed no DAB-positive spots in the uninfected leaves of the mutant or wild-type plants (data not shown), indicating that the microcell death of the mutant is not associated with a high level of H2O2 production. Similarly, we did not detect significant difference between wild-type and mutant leaves in levels of H2O2 (Fig. 3G), as determined by a quantitative H2O2 measurement method.
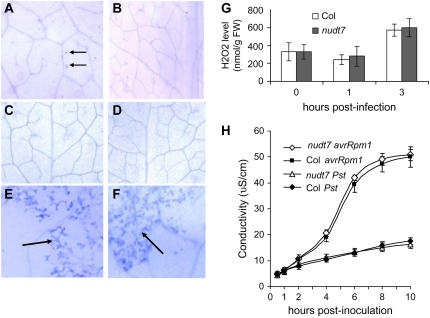
Figure 3.
The nudt7 mutation does not enhance the hypersensitive cell death. A, Spontaneous microlesion formation in an uninfected mutant leaf observed after trypan blue staining. B, A portion of a leaf from a wild-type plant grown in the same environment as the mutant plant shown in A. C and D, Trypan blue-stained leaves from a mutant plant (C) and a wild-type plant (D) grown in a petri dish with sterile conditions. E and F, Hypersensitive cell death in wild-type (E) and nudt7 (F) leaves induced by Pst avrRpm1. The leaves were taken 8 h postinoculation, and massive cell death (blue staining) was observed in the inoculated sites (arrows) of the both leaves. G, H2O2 levels in mutant and wild-type (Col) leaves. H, Electrolyte leakage following inoculation of wild-type (Col) and nudt7 leaves with Pst and Pst avrRpm1. Values are the averages of four replicates ± sd.
The microlesions associated with the mutant plants may be a consequence of disruption of the host cells' normal function due to their hyperresponsiveness to microorganisms and/or other stimuli in the environment. Examination of green healthy leaves from plants grown in sterile environments in culture tubes revealed no significant difference in microlesions between the mutant and wild-type plants (Fig. 3, C and D): Both the mutant and wild-type leaves had fewer than 10 trypan blue-positive spots per leaf.
To determine whether the nudt7 mutation enhances hypersensitive cell death triggered by avirulent pathogens, we inoculated leaves with Pst avrRpm1 at a concentration of approximately 107 cfu/mL and monitored appearance of dead cells by trypan blue staining. No obvious difference was found between the mutant and wild-type plants in the timing of hypersensitive cell death. In both wild-type and mutant leaves, dead cells were observed after approximately 6 h postinoculation (Fig. 3, E and F). Tissue collapse, the visible symptom of infected leaves, appeared approximately 10 h postinoculation for both mutant and wild-type plants (data not shown). The virulent Pst strain was not able to induce cell death in either wild-type or mutant leaves when the leaves were observed by trypan blue staining in 5- to 36-h intervals after the inoculation (data not shown). We further monitored electrolyte leakage to provide a quantitative measurement of cell death in leaf discs infiltrated with Pst avrRpm1 and Pst (107 cfu/mL). Compared with the Pst-inoculated leaf discs, Pst avrRpm1 caused significant iron leakage, which began after approximately 2 h postinoculation (Fig. 3H). However, there is no significant difference in electrolyte leakage between the infected leaf discs of the wild-type and nudt7 mutant plants. Besides, no difference in hypersensitive cell death was seen between the mutant and wild-type plants when infection was carried out with a low concentration of the bacteria (2 × 105 cfu/mL; data not shown).
The Hyperactivation of the Defense Response in the nudt7 Mutant Is Not Primed by an Elevated Level of SA
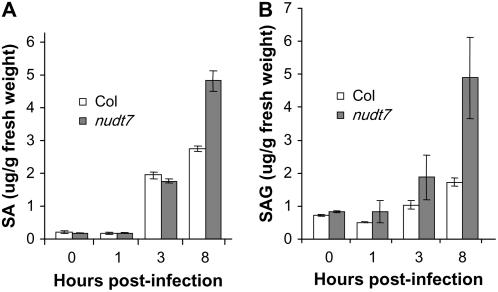
Many enhanced disease resistance mutants constitutively accumulate high levels of SA. To investigate whether the hyperresponsiveness phenotype of nudt7 is caused by higher levels of SA before and after inoculation, we measured SA and its glucoside conjugate (SAG, the storage form of SA). As shown in Figure 4, the mutant accumulated significantly higher levels of SA and SAG 8 h postinoculation with Pst. However, within 3 h postinoculation, although the mutant had slightly higher levels of SAG, there was little difference in the levels of SA between the mutant and wild-type plants. The results indicate that the rapid activation of the defense-related genes in the mutant is unlikely due to an elevated level of SA, although the high level of SA at the later stages likely contributes to the enhanced disease resistance phenotype of the mutant.
Figure 4.
Levels of SA and SAG in uninoculated and Pst-inoculated leaves of nudt7 and wild-type (Col) plants. Values are the averages of three replicates ± sd.
The nudt7 Mutation Enhances Two Distinct Defense Response Pathways
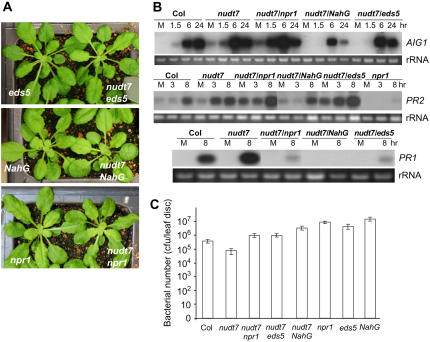
The nudt7 mutant was crossed with previously identified disease resistance mutants, npr1 and eds5, and with the NahG plant to generate double mutants. NPR1 is a key component required for disease resistance (Cao et al., 1994), the eds5 mutant is defective in the SA biosynthetic pathway (Nawrath et al., 2002), and the NahG plants overexpress a bacterial hydroxylase that converts SA to catechol (Delaney et al., 1994). Catechol could also cause susceptibility to nonhost pathogens (van Wees and Glazebrook, 2003). These double mutants are slightly smaller than the single mutants (Fig. 5A). Analyses of expression profiles of the defense-related genes using northern blotting (Fig. 5B) revealed that like the nudt7 single mutant, both the nudt7 npr1 and the nudt7 eds5 double mutants exhibited pathogen-induced hyperactivation of PR2 and AIG1 genes, although eds5 nudt7 showed slightly attenuated AIG1 induction. On the other hand, the npr1 and eds5 mutations not only blocked hyperactivation of PR1 associated with the nudt7 mutation but also suppressed normal induction of the PR1 gene by the pathogen infection. In the nudt7 NahG plants, induction of both AIG1 and PR2 was suppressed and PR1 expression was abolished (Fig. 5B). Further analyses of the disease resistance phenotypes of the double mutants using the in planta bacterial growth assays (Fig. 5C) showed that nudt7 npr1 and nudt7 eds5 double mutants were slightly more susceptible than the wild-type plants but were more resistant than the eds5 and npr1 single mutants. The nudt7 NahG plants supported significantly more bacterial growth than the wild-type plants, although they are still more resistant than the NahG plants. These results indicate that NUDT7 modulates two distinct branches of the defense response pathways: one independent of NPR1 and SA and the other dependent on both of the two components.
Figure 5.
NUDT7 modulates two distinct branches of the defense response. A, Morphological phenotypes of the single and double mutants. B, Northern-blotting results revealing effects of npr1, eds5, and NahG on pathogen-induced expression of the defense-related genes associated in the nudt7 mutant. Leaves were inoculated with Pst avrRpm1. “M” represents mock samples harvested 1.5 h after treatment with water. C, Effects of npr1, eds5, and NahG on disease resistance of the nudt7 mutant. Shown is the result of an in planta bacterial growth assay. Bacterial numbers were counted 3 d postinoculation with Pst. Values are the averages of three replicates ± sd. [See online article for color version of this figure.]
ADPR and NADH are Preferred in Vitro Substrates of NUDT7
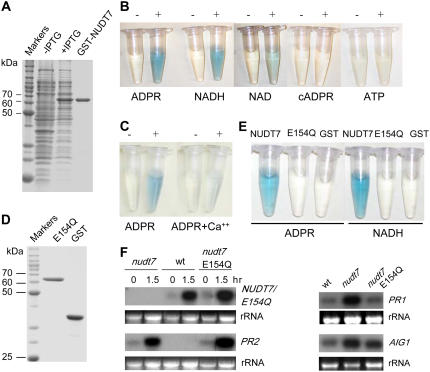
NUDT7 encodes a 32-kD polypeptide that contains the conserved sequence motif called the Nudix box that is involved in hydrolyzing a nucleoside diphosphate linked to some other moiety X (Bessman et al., 1996). The Nudix motif consists of the highly conserved consensus sequence GX5EX7REUXEEXGU, where X can be any amino acid and U is an aliphatic hydrophobic acid. A general method for the activity assay of Nudix hydrolases (Xu et al., 2004) was used to determine enzymatic activity of NUDT7 expressed in and purified from Escherichia coli as a fusion protein to glutathione S-transferase (GST; Fig. 6A). In this assay, hydrolysis of a substrate (nucleoside diphosphate-X) will generate phosphatase-sensitive products that, in the presence of intestinal phosphatase, will release inorganic phosphate that can then be quantified colorimetrically. NUDT7 was found to be active on several known Nudix hydrolase substrates, such as ADPR, NADH, NAD, NADPH, NADP, and diadenosine tetraphosphate (Ap4A), but showed essentially no activity on cADPR, GDP-Man, UDP-Glc, ATP, or GTP (Fig. 6B; Table I). Besides, NUDT7 was not able to hydrolyze ADPR in poly- or mono-ADP-ribosylated polypeptides (data not shown). Among these nucleotide analogs, ADPR and NADH are NUDT7's preferred substrates. NUDT7's hydrolase activity was inhibited in the presence of Ca2+ (Fig. 6C; Table I). Later, we used E. coli-expressed NUDT7 without the GST tag for the activity assays on these nucleotide analogs and observed similar results (data not shown).
Figure 6.
NUDT7 is a nucleoside diphosphate hydrolase. A, SDS-PAGE analyses of recombinant NUDT7 purified from E. coli. The first two lanes are soluble protein extracts of the E. coli cultures without or with induction of the recombinant protein by IPTG. The protein bands were visualized with Coomassie staining. B, Hydrolase activities of NUDT7 against several nucleotides. The intensity of the blue color is directly related to the NUDT7 activity. The reaction was carried out for each substrate in the presence (+) or absence (−) of the purified NUDT7 protein. C, NUDT7 is sensitive to inhibition by Ca2+. D, SDS-PAGE analysis of purified E154Q. E, E154Q is inactive on the nucleotide derivatives. F, E154Q is unable to complement the nudt7 mutant phenotype. One and half hours postinoculation with Pst avrRpm1, the nudt7 plants carrying the NUDT7p∷E154Q construct show normal accumulation of E154Q transcripts; however, they still show hyperactivation of PR1 (3 h postinoculation), PR2, and AIG1 (8 h postinoculation).
Table I.
Hydrolysis activities of NUDT7 on nucleotide derivatives
| Substrate | Activitya |
|---|---|
| ADPR | 43.3 ± 5.5 |
| NADH | 43.7 ± 7.7 |
| NAD | 12.9 ± 2.6 |
| NADPH | 17.0 |
| NADP | 4.8 |
| FAD | 19.1 ± 1.1 |
| Ap4A | 14.6 ± 1.6 |
| NAADP | 6.9 |
| cADPR | 2.3 |
| GDP-Man | 4.0 |
| UDP-Glc | 0.3 |
| ATP | 2.5 |
| GTP | 0.1 |
| ADPR + Ca2+ | 6.8 ± 0.5 |
The rates of hydrolysis are presented as μmol−1 mg protein−1 30 min (±sd).
We generated a mutant form of NUDT7 (NUDT7 E154Q) in which the conserved Glu residue E154 in the Nudix box is mutagenized to Gln. E154 likely binds to Mg2+ to carry out the nucleophilic attack and therefore is considered essential for its hydrolase activity (Mildvan et al., 2005). E. coli-expressed E154Q was found inactive against the nucleotide derivatives such as ADPR and NADH (Fig. 6, D and E). The E154Q mutant, whose expression is under the control of the NUDT7 promoter, was then transformed into the nudt7 mutant plants and was found unable to complement the nudt7 mutant phenotype. Like nudt7, the nudt7 E154Q plants still showed hyperactivation of PR1, PR2, and AIG1 (Fig. 6F) and enhanced resistance to Pst (data not shown), demonstrating that NUDT7's hydrolase activity is required for its normal biological function although it is possible that the E154Q mutation may cause another unknown effect on the protein function in addition to abolishing its hydrolase activity.
The nudt7 Mutation Affects Redox Homeostasis
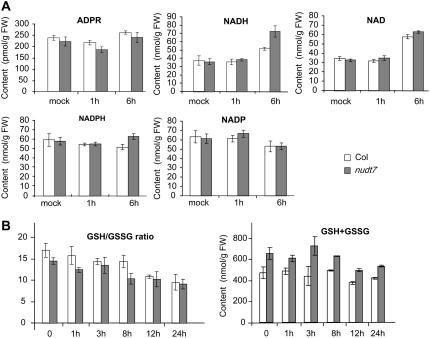
The in vitro hydrolase activity assays described above raise the possibility that ADPR could be the physiological substrate of NUDT7. We reasoned that if NUDT7 functions to modulate the ADPR level, the nudt7 mutation is expected to lead to an increase in the level of ADPR. To test the working hypothesis, we determined levels of free ADPR. Free ADPR levels in Arabidopsis leaves were found to be too low to be detected in our initial nucleotide profiling analyses using regular HPLC (data not shown). We then applied a HPLC technique for detection of ethano-ADPR, a fluorescence analog of ADPR for measuring ADPR (see “Materials and Methods”). Our results showed that the concentration of ADPR in Arabidopsis leaves is around 200 pmol/g fresh weight (Fig. 7A), which will give a rough estimate of about 20 to 40 μm in cellular compartments (such as the cytosol) according to an estimation method assuming that amount of ADPR is inappreciable in the vacuole (Noctor, 2006). We did not detect significant difference in the ADPR levels between the mutant and wild-type leaves or between pathogen-infected and uninfected leaves. These results argued against ADPR being a physiological substrate of NUDT7. This notion is further supported by the result of our enzyme kinetic assay that showed that Km for the hydrolysis of ADPR by NUDT7 is 0.602 ± 0.056 mm, apparently too high for APDR to be an in vivo substrate.
Figure 7.
The nudt7 mutation affects cellular redox homeostasis. Levels of ADPR, NAD, NADH, NADP, NADPH (A), and glutathione (B) in pathogen-challenged (infiltrated with 2 × 107 cfu/mL Pst avrRpm1) and control plants were determined. The bottom left panel shows the ratios of GSH to GSSG. Values are the averages of three replicates ± se.
The result from the in vitro enzymatic assays also suggested that NUDT7 could be a redox modulator since it hydrolyzes NADH and NADPH. We determined levels of NAD(P) and NAD(P)H in wild-type and nudt7 leaves before and after infection with P. syringae (Fig. 7A). Concentration of each of these nucleotide analogs in leaves ranged from 30 to 80 nmol/g tissue (roughly several mm to 20 mm in cellular compartments excluding the vacuole based on the estimation method mentioned earlier). Km for the hydrolysis reaction of NADH by NUDT7 was found to be 0.1281 ± 0.0155 mm. As shown in Figure 7A, the levels of NADPH and NADP did not change much before and after pathogen incubation. However, slightly more NADPH was detected in the nudt7 mutant at 6 h postinfection (the difference is not statistically significant). The levels of both NADH and NAD significantly increased in the mutant and wild-type leaves at 6 h postinfection. At that time point, the concentration of NADH in the mutant leaves was significantly higher than that in the wild-type leaves.
To get more information on overall cellular redox states, we further determined levels of GSH and GSSG, an important antioxidant buffer system involved in scavenging attacking radicals as well as in regenerating oxidized biomolecules. In both wild-type and mutant leaves, GSH to GSSG ratios decreased gradually following the pathogen infection, reflecting an increasing oxidative stress (Fig. 7B). However, the ratios in the mutant were consistently lower than those in the wild type in both uninfected and infected leaves. However, the mutant leaves contained a consistently higher level of total glutathione. These results indicate that the nudt7 mutation leads to an increase in oxidation of cellular components that in turn promotes cells to increase production of total glutathione and NAD(P)H to counteract the perturbation of cellular redox balance. To determine whether the escalated oxidative state associated with the mutant makes it more sensitive to extra oxidative stresses, we examined tolerance of plants to application of paraquat (PQ), a reactive oxygen species-generating chemical. As shown in Figure 8, the mutant plants were more sensitive to the externally applied oxidative stress. Leaves of the mutant plants, especially older leaves, became wilted within 24 h after spraying with 10 μm PQ (Fig. 8A) and showed clear necrotic lesions several days later (Fig. 8B), while the effect on the wild-type plants was very mild. The nudt7 eds5 (Fig. 8A), nudt7 npr1 (Fig. 8B), and nudt7 NahG (data not shown) plants were as sensitive as the nudt7 single mutant to the PQ treatments. Multiple applications of a lower concentration of PQ starting at a younger seedling stage caused striking growth stunting to the mutant plants (Fig. 8C).
Figure 8.
The nudt7 mutant is hypersensitive to PQ. A, One day after spraying 4-week-old plants with 10 μm PQ, some leaves of nudt7 and nudt7 eds5 plants became wilted, whereas the wild-type plant (Col) showed a much milder symptom. B, Five days after the PQ treatment, some leaves of nudt7 and nudt7 npr1 plants showed the necrotic symptom. C, PQ application causes pronounced growth stunting to the nudt7 mutant. The plants were sprayed with 5 μm PQ every 4 d starting at the 3-week-old seedling stage. The photo was taken 4 d after the third application of PQ.
DISCUSSION
Basal immunity is apparently controlled by complex pathways. Many important genes that control the basal defense response have likely eluded identification through classical forward genetic approaches because of the complexity of the pathways and functional redundancy. We have taken the reverse/functional genomic approach to identify components involved in basal immunity. Loss of function of NUDT7 was found to enhance resistance against P. syringae, indicating that it acts as a negative regulator of disease resistance. The mutant does not display constitutive accumulation of high levels of PR gene transcripts in absence of pathogen infection; however, the mutation potentiates a remarkably stronger defense response that can be triggered by both pathogenic and nonpathogenic bacterial strains.
In the two previous reports by other groups characterizing the same mutant line, the mutant plants were found to have a stunting growth phenotype and showed constitutive accumulation of very high levels of PR gene transcripts and SA (Bartsch et al., 2006; Jambunathan and Mahalingam, 2006). Our study reveals that the nudt7 mutation does not directly cause such a strong constitutive disease resistance phenotype. Instead, the mutation causes the plants to be “primed” for an accelerated defense response upon recognition of an inciting agent. Besides pathogen infection, other inciting agents can also trigger activation of the abnormally regulated defense response in the mutant. These stimuli may include nonpathogenic microorganisms in growth environments or some chemicals.
Innate immunity is considered a double-edged sword. Delicate regulation of the innate immune response is not only important for host defense against invading pathogens but also essential for cohabitation with nonpathogenic microorganisms in the environment (Kobayashi and Flavell, 2004). NUDT7 likely acts as a component in a negative feedback loop of the basal defense response to prevent unnecessary cell stimulation. The analyses of the nudt7 mutation coupled with the other disease resistance mutants indicate that NUDT7 modulates two defense response pathways. The nudt7 mutation enhances one branch of the pathway that leads to activation of a subset of pathogen-induced genes, including PR1. This branch of the pathway is dependent on both NPR1 and SA. The mutation also enhances another branch of the defense response that activates expression of another subset of pathogen-induced genes, including PR2 and AIG1, which is largely independent of NPR1 or SA. It has been reported that neither NPR1 nor SA was required for flagellin-induced innate immunity (Zipfel et al., 2004). Because the nudt7 mutant also exhibited hyperactivation of the defense-related genes triggered by the MAMPs, including flagellin and chitosan, NUDT7 likely functions as a modulator of the innate immune response.
NUDT7 may play its role by sensing and modulating the levels of its substrate(s) to fine-tune the defense response. Identification of ADPR and NADH as its preferred in vitro substrates raises the possibility that these nucleotide analogs may be physiological substrates of NUDT7 and act as regulatory molecules in plant disease resistance signaling. ADPR and NADH have previously been reported to be preferred in vitro substrates of NUDT7 (Ogawa et al., 2005; Olejnik and Kraszewska, 2005; Jambunathan and Mahalingam, 2006). However, our results suggest that ADPR is unlikely the physiological substrate of NUDT7 since the mutant does not accumulate a higher level of ADPR. Neither did we detect a significant change of the level of ADPR in response to pathogen infection. Although Jambunathan and Mahalingam (2006) reported that untreated nudt7 plants accumulated over 2-fold more NADH than wild-type plants, we did not find any significant difference in the levels of NADH in untreated mutant and wild-type leaves. Besides, the levels of NADH in wild-type plants determined by Jambunathan and Mahalingam (2006) are about several 100-fold higher than those from our results and from other reports (Noctor, 2006). It is not clear whether the difference is caused by different growth conditions. However, we did find that the NADH level in mutant leaves at 6 h postinfection was approximately 40% higher than that in wild-type plants. Although the results suggest that NADH could be a biologically important substrate of NUDT7, we could not rule out the possibility that the higher level of NADH in the mutant may be indirectly caused by the mutation. The nudt7 mutation leads to an escalated oxidative stress, as indicated by having a lower GSH to GSSG ratio than wild-type plants in both infected and untreated leaves, which could cause cells to produce more NADH as an attempt to maintain the antioxidative power of glutathione. Another indication that NADH may not be the physiological substrate is that we did not find any significant difference in the levels of NADH between uninfected leaves of wild-type and mutant plants. In addition, NUDT7 overexpression lines did not differ from wild-type plants in the NADH levels, and neither have we observed any difference in disease resistance or defense-related gene induction between wild type and the overexpression lines (data not shown).
A more direct effect caused by the nudt7 mutation was found to be the perturbation of cellular redox homeostasis. Unlike the levels of NADH or SA, which were not different between the untreated wild-type and mutant plants, the ratios of GSH to GSSG were consistently lower in the mutant, suggesting that more GSH molecules were used to regenerate oxidized biomolecules. Although the precise mechanism by which the nudt7 mutation causes the change in the glutathione status remains to be determined, the perturbation in redox balance may “prime” cells to hyperrespond to inciting agents such as pathogen infection, resulting in an excessive defense response and enhanced disease resistance. It is believed that antioxidant status sets the threshold for general defense responses provoked by biotic or other stresses (Foyer and Noctor, 2005).
It is worth mentioning that the primary biochemical function of a Nudix protein may not always be hydrolysis of its substrate. Instead, a Nudix box may be involved in conformational change of a Nudix-containing protein through its binding to a nucleotide analog. In animal immunocytes, the Ca2+-permeable cation channel LTRP2 mediates Ca2+ influx to trigger the immune responses (Perraud et al., 2001; Hara et al., 2002). LTRP2 contains a Nudix box at its C-terminal portion. ADPR regulates the channel's activity by binding to the Nudix box, which also acts as an intrinsic deactivation mechanism for LTRP2 through its ability to break down ADPR. Interestingly, a Nudix protein from cauliflower (Brassica oleracea) was found to be an inhibitor of Gln synthetase and nitrate reductase, and the inhibition is dependent on NADH but relieved by ATP and AMP (Moorhead et al., 2003). The mechanism of the inhibition and its biological significance are not known. Recently, it was shown that Arabidopsis DCP2, a Nudix protein, interacts with DCP1 and VCS to form an mRNA decapping complex (Xu et al., 2006). It would be interesting to explore whether NUDT7 regulates activity of other proteins through protein-protein interaction mediated by binding of a nucleotide analog to its Nudix domain.
The Arabidopsis genome encodes over 20 members of the Nudix family. Their biological functions and their modes of action remain largely unknown. It is plausible that biological substrates of some AtNUDT proteins are nucleotide analogs. A variety of nucleotide derivatives, including ADPR, cyclic GMP, cADPR, Ap4A, and cytokinins, play regulatory roles in many biological processes. Nudix hydrolases may be involved in biological processes by sensing and modulating levels of nucleotide analogs. The identification of the Nudix proteins as putative virulence effectors of phytopathogens (Mukaihara et al., 2004; Tamura et al., 2005; Koebnik et al., 2006) suggests that pathogens may use Nudix proteins to mimic a host's cellular functions. IalA, a Nudix protein encoded by one of two invasion-associated loci of Bartonella bacilliformis, the only bacterium that infects human red blood cells, is also a virulence factor essential for infectivity (Mitchell and Minnick, 1995; Conyers and Bessman, 1999). When expressed in E. coli, IalA can markedly increase E. coli's capacity to invade red blood cells (Mitchell and Minnick, 1995). Definitive understanding of how Nudix proteins such as NUDT7 are involved in modulating the host defense response will provide novel and important insights into the molecular mechanisms of disease resistance in plants.
MATERIALS AND METHODS
Plant Growth
Plants were grown at 22°C with 50% humidity and under short-day conditions (9/15 h photoperiod at a light intensity of 125 mol m−2 s−1).
PCR Analysis of the nudt7 Mutant Allele
The T-DNA-flanking genomic fragment was amplified by PCR from Salk_046441 using the gene-specific primer 4g12720p5 (5′-CAACGAGATGAGTCCAATAAACAAA-3′) and the T-DNA left-border primer LBb1 (AGTTGCAGCAAGCGGTCCACGC). The wild-type fragment of this region was amplified using the primer pair 4g12720p5 and 4g12720p3 (CTAAACAATACGCTGACACCCTTCA).
Nucleic Acid Analysis
Arabidopsis (Arabidopsis thaliana) genomic DNA was isolated from leaves using a modified CTAB procedure (Saghai-Maroof et al., 1984). Total RNA was isolated from leaf tissues using the Trizol (Promega) method for northern analysis or the on-column DNase digestion RNA preparation columns (Sigma) for Q-PCR analysis. Nucleic acid electrophoresis, transferring onto nylon membranes, 32P labeling of DNA probes (27-9240-01; Amersham Biosciences), and hybridization were performed according to the standard protocols (Sambrook et al., 1989).
Plant Pathogens and Inoculation
Pseudomonas syringae was grown as described (Cameron et al., 1994). Psp was provided by Dr. Jian-Min Zhou, and all other strains were from Dr. Barbara Kunkel. To prepare inoculated leaf tissues for RNA preparation, leaves were infiltrated with 1 × 107 cfu/mL bacterial suspension (in water) with a syringe. For the in planta bacterial growth assays, 3 × 104 cfu/mL (for virulent strains) or 1 × 105 cfu/mL (for avirulent strains) bacterial suspension was infiltrated into leaves. Eight leaf discs were collected per replicate, and each data point represents three replicates. All bacterial growth assays were repeated, and only results that were observed consistently are shown.
Treatments with MAMPs
To prepare boiled Pst and Escherichia coli, the bacterial suspensions (107 cfu/mL in double-distilled water [ddH2O]) were boiled for 10 min and cooled before being infiltrated into leaves. flg22 (QRLSTGSRINSAKDDAAGLQIA) was synthesized by GenScript. Chitosan (52368; Sigma) and lipopolysaccharide (62326; Sigma) were purchased from Sigma. Induction of the defense response was conducted by pressure infiltration of the MAMP solutions into Arabidopsis leaves using 1-mL syringes (approximately 20 μL/leaf).
Complementation of the nudt7 Mutant
A 4.2-kb fragment of NUDT7 genomic DNA (NUDT7t), which includes a 2-kb promoter region and a 0.4-kb 3′ untranslated region, was amplified from Col-0 by using the primer pair 4g12720p5Kpn (GGATCCGGAGCTAAGCATCTGAATCAG) and 4g12720p3Sal (GTCGACCAGTGTAGTAAATGGTCAAGAGAC). The PCR product was cloned into pCR-Blunt II-TOPO (Invitrogen), subcloned into the binary vector pPZP221 using the restriction sites KpnI and SalI, and then transformed into the mutant plants.
Measurement of SA and SAG
SA and SAG were extracted and analyzed as described previously (Uknes et al., 1993). Reverse-phase HPLC on a C-18 column (ODS-AM analytical column from Waters) was used in the analysis.
DAB Staining, Trypan Blue Staining, H2O2 Measurement, and Electrolyte Leakage Analysis
DAB staining and trypan blue staining were carried out as described previously (Alvarez et al., 1998). Determination of H2O2 levels was carried out using the modified chromogenic peroxidase-coupled assay (Veljovic-Jovanovic et al., 2002). The electrolyte leakage assay was performed according to the previously described method (Torres et al., 2002). Briefly, leaves were infiltrated with bacterial suspensions at 107 cfu/mL in ddH2O. Leaf discs (0.25 cm2 each) were collected from the inoculated areas 15 min postinoculation and washed four times with ddH2O for about 15 min. Six leaf discs were placed in a test tube with 4 mL of ddH2O. Each treatment was replicated three times. Conductivity was measured over time using an ORKTON pH/conductivity meter (510 series).
NUDT7 Expression in E. coli and Activity Assays
NUDT7 cDNA was cloned through reverse transcription-PCR from an RNA sample extracted from leaves inoculated with P. syringae. After reverse transcription with oligo(dT) as a primer, NUDT7 cDNA was amplified using the primer set TAGGATCCGGTACTAGAGCTCAGCAG and CAGTCGACTCAGAGAGAAGCAGAGGC. The 0.8-kb PCR product was cloned into pCR-Blunt II-TOPO, and the cDNA insert was then dropped by digesting with BamHI/HindIII and ligated into pET41a (Novagen) to generate a fusion protein with GST at its N terminus. The fusion construct was transformed into DL21(DE3). After the bacterial culture grew to OD600 = 0.6 to approximately 0.8, 1 mm IPTG was added to the culture, which was then grown at 30 for additional 4 h. Purification of GST-NUDT7 was preformed using a glutathione-agarose affinity resin (G4510; Sigma) according to the manufacturer's instructions.
A general method for the activity assay of Nudix hydrolases was used to determine enzymatic activity of NUDT7 (Xu et al., 2004). The assays were carried out in a 50-μL hydrolysis mixture of 50 mm Tris-HCl, pH 8.5, 2 μg of GST-NUDT7, 5 mm MgCl2, 1 mm dithiothreitol, 2 mm substrate, and 4 units of alkaline phosphatase. After incubation for 30 min at 37°C, the reaction was stopped by addition of 150 μL of 1 n HCl, followed by addition of 100 μL of water and 700 μL of a mixture that was freshly made by mixing 0.42% molybdate·4H2O with 10% ascorbic acid (6:1). The reaction tubes were put in a 45°C water bath for 20 min for color development and then cooled down to room temperature. The solutions were measured using a spectrophotometer at A820. The reaction without GST-NUDT7 was used as a blank control for each substrate. As for nucleoside triphosphate substrates ATP and GTP, alkaline phosphatase was replaced by inorganic pyrophosphatase and the substrates were removed by charcoal before color development. CaCl2 was added to the hydrolysis mixture to a concentration of 5 mm when investigating effects of Ca2+ on NUDT7 activity. All the substrates were purchased from Sigma.
For enzyme kinetic assays, the reaction was stopped by adding 1 n HCl at different time points. Concentrations of the substrates used in the kinetic assay were 0.5 mm, 2 mm, 5 mm, 10 mm, and 20 mm. The kinetic assay for each substrate concentration was repeated at least three times. Km and Vmax were calculated using the Lineweaver-Burk plot based on the Michaelis-Menton equation.
Site-Directed Mutagenesis
The NUDT7 genomic clone used for the complementation experiment and the NUDT7 cDNA clone used for expression in E. coli were mutagenized using the QuikChange XL site-directed mutagenesis kit (Stratagene) according to the manufacturer's instruction manual. E154Q mutation was generated by using the primer pair E154Qf (GATATATGGACTGGAGTAGCTAGGCAAGTGGAAGAAGAAACTGG) and E154Qr (CCAGTTTCTTCTTCCACTTGCCTAGCTACTCCAGTCCATATATC). The underlined nucleotides indicate the position of mutagenesis. The mutagenized genomic clone was then subcloned into the binary vector pPZP221 and transformed into nudt7 mutant plants. The mutagenized cDNA clone was subcloned into the E. coli expression vector. Expression and purification of NUDT7 E154Q followed the same protocol as that for wild-type NUDT7.
Determination of Glutathione, ADPR, NAD(P), and NAD(P)H
Glutathione levels were determined according to a previously described method (Griffith, 1980). Plant tissues were extracted and deproteinated with metaphosphoric acid. Total glutathione was determined based on a recycling enzymatic reaction in which GSH was oxidized by DTNB to form the 412-nm chromophore TNB and another product, GS-TNB, which can be reduced to GSH again by glutathione reductase in the presence of NADPH. The rate of TNB formation was monitored at 412 nm and the glutathione concentration was evaluated by comparison with a standard calibration curve. For determination of GSSG, 2-vinylpyridine was used to mask GSH.
For ADPR determination, extraction, purification, and HPLC separation of cellular ADP-Rib was performed essentially as described previously (Gasser and Guse, 2005). Extracted nucleotide samples from 100 mg of plant tissues were injected into the Beckman's Gold HPLC system (Beckman) with a YMC-ODS column (250 mm × 4.6 mm, particle size 5 μm) protected by a guard column (17 mm × 4.6 mm; both columns were from Waters). Fractions (400 μL) corresponding to the retention time of ADP-Rib (22.8–23.6 min) were collected. Fluorescence labeling and the HPLC detection for ethano-ADPR were carried out as described (Bobalova et al., 2002). The HPLC system and columns were the same as above but equipped with a fluorescence detector (model FP150; JASCO). The fluorescence detector was set to record signals at an excitation wavelength of 230 nm and emission wavelength of 410 nm.
The pyrimidine nucleotides were measured by the cycling assay as described previously (Matsumura and Miyachi, 1980). NADH and NADPH were extracted with 0.1 m NaOH, while NAD and NADP were extracted with 0.1 m HCl. The assays involve the phenazine ethosulfate-catalyzed reduction of thiazolyl blue tetrazolium bromide (MTT) in the presence of ethanol and alcohol dehydrogenase (for NAD and NADH) or Glc-6-P and G6PDH (for NADP and NADPH). The rate of reduction of MTT was monitored at 570 nm.
Treatments of Plants with PQ
PQ (methyl viologen; 856177; Sigma) was dissolved in water and sprayed onto plants using a hand spray bottle in solution containing 0.02% Silwet-77 as a detergent.
Acknowledgments
We thank Barbara Kunkel for all Pst strains, including Pst hrcC; Jian-min Zhou for the Psp strain; Xinnian Dong for the npr1 mutant seeds; and Christine Ehret for editing the manuscript.
This work was supported by the National Institutes of Health (grant no. GM076420 to Y.X.), by the Natural Science Foundation of China (grant no. 30670178 to X.G.), and by the Shanghai Pujiang Program (grant no. 06PJ14005 to X.G.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Yiji Xia (yxia@danforthcenter.org).
Some figures in this article are displayed in color online but in black and white in the print edition.
Open Access articles can be viewed online without a subscription.
References
- Alexander WS, Hilton DJ (2004) The role of suppressors of cytokine signaling (SOCS) proteins in regulation of the immune response. Annu Rev Immunol 22 503–529 [DOI] [PubMed] [Google Scholar]
- Alvarez ME, Pennell RI, Meijer PJ, Ishikawa A, Dixon RA, Lamb C (1998) Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 92 773–784 [DOI] [PubMed] [Google Scholar]
- Bartsch M, Gobbato E, Bednarek P, Debey S, Schultze JL, Bautor J, Parker JE (2006) Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell 18 1038–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir Y, Subramaniam R, Dangl JL (2004) Plant disease resistance protein signaling: NBS-LRR proteins and their partners. Curr Opin Plant Biol 7 391–399 [DOI] [PubMed] [Google Scholar]
- Bessman MJ, Frick DN, O'Handley SF (1996) The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J Biol Chem 271 25059–25062 [DOI] [PubMed] [Google Scholar]
- Bobalova J, Bobal P, Mutafova-Yambolieva VN (2002) High-performance liquid chromatographic technique for detection of a fluorescent analogue of ADP-ribose in isolated blood vessel preparations. Anal Biochem 305 269–276 [DOI] [PubMed] [Google Scholar]
- Boch J, Joardar V, Gao L, Robertson TL, Lim M, Kunkel BN (2002) Identification of Pseudomonas syringae pv. tomato genes induced during infection of Arabidopsis thaliana. Mol Microbiol 44 73–88 [DOI] [PubMed] [Google Scholar]
- Cameron RK, Dixon RA, Lamb CJ (1994) Biologically induced systemic acquired resistance in Arabidopsis thaliana. Plant J 5 715–725 [Google Scholar]
- Cao H, Bowling SA, Gordon AS, Dong X (1994) Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6 1583–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conyers GB, Bessman MJ (1999) The gene, ialA, associated with the invasion of human erythrocytes by Bartonella bacilliformis, designates a nudix hydrolase active on dinucleoside 5′-polyphosphates. J Biol Chem 274 1203–1206 [DOI] [PubMed] [Google Scholar]
- Dangl JL, Jones JD (2001) Plant pathogens and integrated defence responses to infection. Nature 411 826–833 [DOI] [PubMed] [Google Scholar]
- Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E, et al (1994) A central role of salicylic acid in plant disease resistance. Science 266 1247–1250 [DOI] [PubMed] [Google Scholar]
- Durner J, Wendehenne D, Klessig DF (1998) Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc Natl Acad Sci USA 95 10328–10333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T (2005) Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci 10 71–78 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G (2005) Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell 17 1866–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser A, Guse AH (2005) Determination of intracellular concentrations of the TRPM2 agonist ADP-ribose by reversed-phase HPLC. J Chromatogr B Analyt Technol Biomed Life Sci 821 181–187 [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Rogers EE, Ausubel FM (1997) Use of Arabidopsis for genetic dissection of plant defense responses. Annu Rev Genet 31 547–569 [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez L, Boller T (2002) Flagellin perception: a paradigm for innate immunity. Trends Plant Sci 7 251–256 [DOI] [PubMed] [Google Scholar]
- Griffith OW (1980) Determination of glutathione and glutathione disulfide using glutathione reductase and 2-vinylpyridine. Anal Biochem 106 207–212 [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack KE, Parker JE (2003) Deciphering plant-pathogen communication: fresh perspectives for molecular resistance breeding. Curr Opin Biotechnol 14 177–193 [DOI] [PubMed] [Google Scholar]
- Hara Y, Wakamori M, Ishii M, Maeno E, Nishida M, Yoshida T, Yamada H, Shimizu S, Mori E, Kudoh J, et al (2002) LTRPC2 Ca2+-permeable channel activated by changes in redox status confers susceptibility to cell death. Mol Cell 9 163–173 [DOI] [PubMed] [Google Scholar]
- Jambunathan N, Mahalingam R (2006) Analysis of Arabidopsis growth factor gene 1 (GFG1) encoding a nudix hydrolase during oxidative signaling. Planta 224 1–11 [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444 323–329 [DOI] [PubMed] [Google Scholar]
- Klaus SM, Wegkamp A, Sybesma W, Hugenholtz J, Gregory JF III, Hanson AD (2005) A nudix enzyme removes pyrophosphate from dihydroneopterin triphosphate in the folate synthesis pathway of bacteria and plants. J Biol Chem 280 5274–5280 [DOI] [PubMed] [Google Scholar]
- Kobayashi KS, Flavell RA (2004) Shielding the double-edged sword: negative regulation of the innate immune system. J Leukoc Biol 75 428–433 [DOI] [PubMed] [Google Scholar]
- Koebnik R, Kruger A, Thieme F, Urban A, Bonas U (2006) Specific binding of the Xanthomonas campestris pv. vesicatoria AraC-type transcriptional activator HrpX to plant-inducible promoter boxes. J Bacteriol 188 7652–7660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxalt AM, Munnik T (2002) Phospholipid signalling in plant defence. Curr Opin Plant Biol 5 332–338 [DOI] [PubMed] [Google Scholar]
- Maksel D, Guranowski A, Ilgoutz SC, Moir A, Blackburn MG, Gayler KR (1998) Cloning and expression of diadenosine 5′,5‴-P1,P4-tetraphosphate hydrolase from Lupinus angustifolius L. Biochem J 329 313–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura H, Miyachi S (1980) Cycling assay for nicotinamide adenine dinucleotides. Methods Enzymol 69 465–470 [Google Scholar]
- Mildvan AS, Xia Z, Azurmendi HF, Saraswat V, Legler PM, Massiah MA, Gabelli SB, Bianchet MA, Kang LW, Amzel LM (2005) Structures and mechanisms of Nudix hydrolases. Arch Biochem Biophys 433 129–143 [DOI] [PubMed] [Google Scholar]
- Mitchell S, Minnick M (1995) Characterization of a two-gene locus from Bartonella bacilliformis associated with the ability to invade human erythrocytes. Infect Immun 63 1552–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorhead GB, Meek SE, Douglas P, Bridges D, Smith CS, Morrice N, MacKintosh C (2003) Purification of a plant nucleotide pyrophosphatase as a protein that interferes with nitrate reductase and glutamine synthetase assays. Eur J Biochem 270 1356–1362 [DOI] [PubMed] [Google Scholar]
- Mukaihara T, Tamura N, Murata Y, Iwabuchi M (2004) Genetic screening of Hrp type III-related pathogenicity genes controlled by the HrpB transcriptional activator in Ralstonia solanacearum. Mol Microbiol 54 863–875 [DOI] [PubMed] [Google Scholar]
- Nawrath C, Heck S, Parinthawong N, Metraux JP (2002) EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 14 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G (2006) Metabolic signalling in defence and stress: the central roles of soluble redox couples. Plant Cell Environ 29 409–425 [DOI] [PubMed] [Google Scholar]
- Ogawa T, Ueda Y, Yoshimura K, Shigeoka S (2005) Comprehensive analysis of cytosolic Nudix hydrolases in Arabidopsis thaliana. J Biol Chem 280 25277–25283 [DOI] [PubMed] [Google Scholar]
- Olejnik K, Kraszewska E (2005) Cloning and characterization of an Arabidopsis thaliana Nudix hydrolase homologous to the mammalian GFG protein. Biochim Biophys Acta 1752 133–141 [DOI] [PubMed] [Google Scholar]
- Perraud AL, Fleig A, Dunn CA, Bagley LA, Launay P, Schmitz C, Stokes AJ, Zhu Q, Bessman MJ, Penner R, et al (2001) ADP-ribose gating of the calcium-permeable LTRPC2 channel revealed by Nudix motif homology. Nature 411 595–599; comment Cahalan MD (2001) 411: 542–543 [DOI] [PubMed] [Google Scholar]
- Reuber TL, Ausubel FM (1996) Isolation of Arabidopsis genes that differentiate between resistance responses mediated by the RPS2 and RPM1 disease resistance genes. Plant Cell 8 241–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW (1984) Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci USA 81 8014–8018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
- Tamura N, Murata Y, Mukaihara T (2005) Isolation of Ralstonia solanacearum hrpB constitutive mutants and secretion analysis of hrpB-regulated gene products that share homology with known type III effectors and enzymes. Microbiology 151 2873–2884 [DOI] [PubMed] [Google Scholar]
- Torres MA, Dangl JL, Jones JD (2002) Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc Natl Acad Sci USA 99 517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uknes S, Winter AM, Delaney T, Vernooij B, Morse A, Friedrich L, Nye G, Potter S, Ward ER, Ryals J (1993) Biological induction of systemic acquired resistance in Arabidopsis. Mol Plant Microbe Interact 6 682–689 [Google Scholar]
- van Wees SCM, Glazebrook J (2003) Loss of non-host resistance of Arabidopsis NahG to Pseudomonas syringae pv. phaseolicola is due to degradation products of salicylic acid. Plant J 33 733–742 [DOI] [PubMed] [Google Scholar]
- Veljovic-Jovanovic S, Noctor G, Foyer CH (2002) Are leaf hydrogen peroxide concentrations commonly overestimated? The potential influence of artefactual interference by tissue phenolics and ascorbate. Plant Physiol Biochem 40 501–507 [Google Scholar]
- Xu J, Yang JY, Niu QW, Chua NH (2006) Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell 18 3386–3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Dunn CA, Jones CR, D'Souza G, Bessman MJ (2004) The 26 Nudix hydrolases of Bacillus cereus, a close relative of Bacillus anthracis. J Biol Chem 279 24861–24865 [DOI] [PubMed] [Google Scholar]
- Xu W, Shen J, Dunn CA, Desai S, Bessman MJ (2001) The Nudix hydrolases of Deinococcus radiodurans. Mol Microbiol 39 286–290 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JDG, Felix G, Boller T (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428 764–767 [DOI] [PubMed] [Google Scholar]