Abstract
Rhizobia secrete nodulation (Nod) factors, which set in motion the formation of nitrogen-fixing root nodules on legume host plants. Nod factors induce several cellular responses in root hair cells within minutes, but also are essential for the formation of infection threads by which rhizobia enter the root. Based on studies using bacterial mutants, a two-receptor model was proposed, a signaling receptor that induces early responses with low requirements toward Nod factor structure and an entry receptor that controls infection with more stringent demands. Recently, putative Nod factor receptors were shown to be LysM domain receptor kinases. However, mutants in these receptors, in both Lotus japonicus (nfr1 and nfr5) and Medicago truncatula (Medicago; nfp), do not support the two-receptor model because they lack all Nod factor-induced responses. LYK3, the putative Medicago ortholog of NFR1, has only been studied by RNA interference, showing a role in infection thread formation. Medicago hair curling (hcl) mutants are unable to form curled root hairs, a step preceding infection thread formation. We identified the weak hcl-4 allele that is blocked during infection thread growth. We show that HCL encodes LYK3 and, thus, that this receptor, besides infection, also controls root hair curling. By using rhizobial mutants, we also show that HCL controls infection thread formation in a Nod factor structure-dependent manner. Therefore, LYK3 functions as the proposed entry receptor, specifically controlling infection. Finally, we show that LYK3, which regulates a subset of Nod factor-induced genes, is not required for the induction of NODULE INCEPTION.
Legume plants have the unique ability to establish root nodule symbiosis with nitrogen-fixing bacteria of the Rhizobiaceae family. These bacteria—collectively called rhizobia—secrete signal molecules, named nodulation (Nod) factors, which play an important role in the induction of early steps of root nodule formation, including the infection process. All Nod factors are lipochito-oligosaccharides composed of a β-1,4-linked N-acetyl-d-glucosamine backbone of which the nonreducing sugar moiety is substituted with an acyl chain. Depending on the rhizobial species, additional substitutions can be present on the terminal sugar residues of the glucosamine backbone (Perret et al., 2000). These substitutions, as well as the structure of the lipid moiety, determine the biological activity of Nod factors, and specific bacterial Nod genes control the presence and identity of these substitutions. In legumes like pea (Pisum sativum) and Medicago truncatula (Medicago), in particular, the infection process has a highly stringent demand on the structure of the Nod factor. Efficient rhizobial infection requires that specific Nod factor substitutions are present, whereas they are less important for the other Nod factor-induced responses (Ardourel et al., 1994). For example, Nod factors produced by Sinorhizobium meliloti, which is the microsymbiont of most Medicago species, are O-sulfated at the reducing end by NodH, O-acetylated at the nonreducing end by NodL, whereas a specific C16:2 lipid moiety is added by NodF and NodE. This acetate substitution and the specific acyl group are especially required to trigger the formation of infection structures, so-called infection threads, whereas other Nod factor-induced responses, like induction of cell physiological changes (e.g. calcium spiking), transcriptional activation of specific genes, and nodule primordium formation, have less stringent demands to the Nod factor structure (Ardourel et al., 1994; Oldroyd et al., 2001; Wais et al., 2002). Therefore, based on studies with bacterial nod mutants, two types of Nod factor receptors were proposed already more than a decade ago: a signaling receptor that is essential for the induction of early responses and an entry receptor that mediates infection events in the epidermis, which has a more stringent demand on Nod factor structure (Ardourel et al., 1994).
More recently, putative Nod factor receptors were cloned from Medicago and Lotus japonicus (Lotus) and were shown to be LysM domain receptor kinases (Limpens et al., 2003; Madsen et al., 2003; Radutoiu et al., 2003; Arrighi et al., 2006; Mulder et al., 2006). In Lotus, two putative Nod factor receptors have been cloned: LjNFR1 and LjNFR5 (Madsen et al., 2003; Radutoiu et al., 2003). Knockout mutations in either of these genes eliminate almost all Nod factor-inducible responses. Two putative Nod factor receptors were also identified in Medicago, namely, MtLYK3 and MtNFP (Limpens et al., 2003; Arrighi et al., 2006; Mulder et al., 2006). MtNFP is orthologous to LjNFR5 and a knockout mutation in this gene also causes complete loss of Nod factor-inducible responses. The function of MtLYK3 was studied by RNA interference (RNAi) and knockdown of the expression of this gene causes a specific block of infection thread formation, suggesting that MtLYK3 might act as an entry receptor (Limpens et al., 2003). However, based on the syntenic map position, as well as their sequence, MtLYK3 and LjNFR1 are likely orthologous. This might suggest that MtLYK3 has a similar signaling function to LjNFR1 and residual levels of MtLYK3 in the RNAi roots could have masked defects in signaling. Therefore, characterization of allelic series, including knockouts, is essential to determine the role of MtLYK3 in Nod factor perception.
Here, we describe mapping and cloning of the Medicago HAIR CURLING (HCL) gene and show that it encodes MtLYK3. Furthermore, we identified a new, weak allele that renders the control of infection thread formation in a Nod factor structure-dependent manner. These results provide strong support for the two-receptor model in which LYK3 acts as the proposed Nod factor entry receptor.
RESULTS
A Novel hcl Allele with an Infection Thread Phenotype
Upon inoculation with S. meliloti, Medicago root hairs can form a tight curl by which bacteria become entrapped in a cavity. There they form a small colony (microcolony) and subsequently infection thread formation is initiated (Fig. 1A). The latter involves invagination of the plant plasma membrane by which a tubular structure is formed, allowing the rhizobia to enter the root (hair). All currently known Medicago hcl mutants (B56, W1, and AF3 carrying the hcl-1, hcl-2, and hcl-3 alleles, respectively) are affected in root hair curling (Wais et al., 2000; Catoira et al., 2001). S. meliloti induces extensive root hair deformation (e.g. root hairs with one or more outgrowths) in these mutants, as well as continuous curling (in hcl-2 and hcl-3 mutants; Catoira et al., 2001). However, they are unable to induce the formation of tight root hair curls and therefore infection threads are not formed (Fig. 1B).
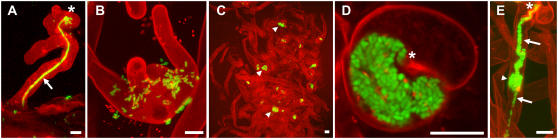
Figure 1.
A, Typical infection by S. meliloti (expressing GFP; green) of a root hair on wild-type Medicago stained with propidium iodide. S. meliloti induce root hair curls, form small microcolonies, and penetrate the root hair via a tubular infection thread. B, Root hair of hcl-3 colonized by S. meliloti inducing multiple outgrowths. C, Part of an hcl-4 root colonized by S. meliloti. Big green dots are microcolonies in root hair curls that, after infection, have developed into a sac-like structure. D, Confocal image (section) of curled root hair in S. meliloti-inoculated hcl-4 plants showing a typical sac-like infection initiated from a microcolony. E, Infection of an hcl-4 root hair that initiated normally from the root hair curl (arrow at bottom); however, after some time, growth becomes anomalous, resulting in a sac-like structure. From this sac-like structure, a new infection thread is initiated, growing further down the shaft of the root hair. Bars in all figures = 10 μm; microcolonies are indicated by asterisks, infection threads by arrows, and sac-like structures by arrowheads.
We identified a novel hcl allele (mutant AC6; hcl-4) in an ethyl methanesulfonate-mutagenized population (Penmetsa and Cook, 2000), which revealed a function of the encoded protein at a later step of infection. In contrast to the previously described hcl mutants, hcl-4 plants do efficiently form root hair curls containing microcolonies of S. meliloti (Fig. 1C). The ability of hcl-4 root hairs to form root hair curls containing a microcolony is not affected as compared to wild-type Medicago. Infection thread formation is initiated, but instead of a tubular structure, a big sac-like infection structure filled with S. meliloti is formed (Fig. 1D). Thus, polar growth of infection threads, as well as maintenance of growth, appears to be markedly affected in hcl-4 plants. However, some infection threads reach the cortex and nodules are formed. These infection threads have disturbed morphology in which sac-like structures alternate with properly grown sections (Fig. 1E). The histology of the nodules formed on the hcl-4 mutant is similar to that of wild-type nodules (data not shown), but the number of nodules is reduced to about 8% of that of wild-type plants (Table I). Crossing hcl-4 with wild-type plants showed that the hcl-4 locus is inherited in recessive Mendelian fashion; 100% of the F1 offspring showed a wild-type nodulation phenotype, whereas the F2 offspring segregated in a 3:1 ratio (61 Nod+:27 Nod−; χ2 = 1.515; P = 0.22).
Table I.
Nodulation phenotypes of Medicago A17 Jemalong and hcl-4
Statistical analysis of the nodulation phenotypes of wild-type and hcl-4 plants inoculated with wild-type and nod mutant bacteria. For each combination, mean number of nodules formed with the confidence at 95% is shown (α = 0.05), as well as the relative efficiency of nodulation compared to wild-type plants. Additionally, the possibility (P) that the nodulation phenotype as a result of inoculation with mutant bacteria occurs by chance is shown; chances are calculated using the t test.
| Medicago | S. meliloti | Mean No. of Nodules | Confidence of Mean | Nodulation Efficiency | P versus Wild Type | P versus hcl-4 |
|---|---|---|---|---|---|---|
| % | ||||||
| Wild type | Wild type | 15.15 | 2.53 | 100.0 | – | |
| Wild type | nodFnodE | 12.20 | 2.28 | 80.5 | 9.82 × 10−2 | |
| Wild type | nodL | 6.45 | 0.83 | 42.6 | 1.57 × 10−6 | |
| hcl-4 | Wild type | 1.24 | 0.44 | 8.2 | – | |
| hcl-4 | nodFnodE | 0.03 | 0.06 | 0.2 | 1.05 × 10−5 | |
| hcl-4 | nodL | 0.00 | 0.00 | 0.0 | 7.01 × 10−6 |
HCL Encodes the Putative Nod Factor Receptor MtLYK3
The hcl-4 mutant shows a similar infection thread phenotype as Medicago roots expressing an MtLYK3 RNAi construct (Limpens et al., 2003). Therefore, we wondered whether HCL might encode MtLYK3. First, we tested whether the HCL map position corresponded to the map position of MtLYK3 using markers DK139L, 10L2, and EH3 (Gualtieri et al., 2002). The HCL locus mapped in between DK139L and EH3, near 10L2, where LYK3 is located as well. Therefore, we sequenced the MtLYK3 gene in the four hcl mutants and identified mutations in all cases (Table II). Complementation using a binary vector containing MtLYK3 resulted in efficient formation of infected nitrogen-fixing root nodules in all four mutants (n ≥ 20 for each mutant), proving that MtLYK3 is encoded by HCL (Supplemental Fig. S1).
Table II.
Summary of hcl mutations
Summary of mutations and their effects in hcl mutant plants. Shown are alternative codes used, which base is mutated, what effect it has, and the EMBL Nucleotide Sequence Database accession number.
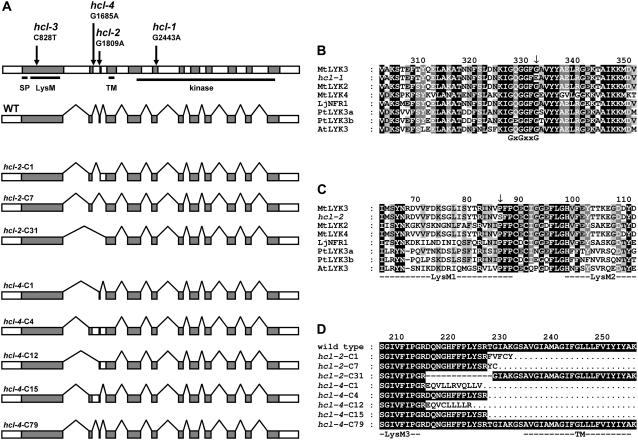
The hcl-1 allele contains a missense mutation converting a conserved Gly of the GxGxxG motif into a Glu (Fig. 2, A and B; Table II). A missense mutation in hcl-3 converts a conserved Pro in LysM domain 1 of LYK3 into a Ser (Fig. 2, A and C; Table II). The mutations in hcl-2 and hcl-4 alter the 3′ and 5′ splice sites of intron 2, respectively (Fig. 2A). To determine the effect of these mutations on splicing, we performed reverse transcription (RT)-PCR on hcl-2 and hcl-4 total RNA. The amplified, partial LYK3 cDNAs of both hcl alleles were cloned and subsequently sequenced (96 clones for each allele). cDNAs from hcl-2 were all mutated due to missplicing; one type of cDNA (approximately 5% of the transcripts) encodes a protein with a 13-amino acid deletion, whereas the two other types of cDNAs encode truncated proteins (Fig. 2D; Table II; accession nos. AM420452–AM420459). Four types of hcl-4 cDNAs also contained mutations resulting in premature stop codons; however, eight cDNA clones (8%) were derived from correctly spliced mRNA that can be translated into wild-type protein (Fig. 2D; Table II). Thus, the ability to form tight curls and the aberrant infection phenotype of hcl-4 is most likely caused by a reduced level of correctly spliced transcripts. It is unlikely that the truncated proteins encoded by the mutated hcl-4 transcripts cause a dominant negative effect because hcl-4 inherits in a recessive manner and the mutant can be complemented by transformation with the wild-type MtLYK3 gene.
Figure 2.
A, LYK3 gene structure. Coding regions are indicated in gray; introns and other noncoding regions in white. Regions encoding the signal peptide (SP), the LysM domains (LysM), the transmembrane helix (TM), and the kinase domain (kinase) are underlined. Arrows indicate the position of the mutations in the different alleles, whereas the text describes the exact position and nature of the nucleotide modification. Directly below a schematic representation of wild-type mRNA, angular lines connecting the coding regions indicate the spliced-out introns. Below, eight different lyk3 mRNAs from the hcl-2 and hcl-4 mutant are schematically represented in similar fashion. Notice that the splice pattern around exons 2 and 3 is markedly altered except in the hcl-4-C79 clone. These nucleotide sequences are deposited in the EMBL nucleotide sequence database with accession numbers AM420452 to AM420459, respectively. B, Alignment of protein kinase subdomain I of LYK3 and homologous proteins. The conserved GxGxxG motif is indicated under the alignment. C, Alignment of the extracellular LysM domains of LYK3 and homologous proteins. LysM domains are underlined and indicated by annotation according to Arrighi et al. (2006). D, Alignment of LYK3 wild-type protein with lyk3 mutant proteins encoded by transcripts isolated from the hcl-2 and hcl-4 mutants. Residues identical to wild-type residues are white letters shaded black; residues dissimilar are bold and black. The putative TM helix and LysM domain 3 (LysM3) are underlined. In all figures, mutations are indicated by arrows, gaps in protein sequences are represented by dashes, and amino acids lacking due to truncation are indicated by dots. The first character of the number above the alignments indicates the position of the amino acid below in the LYK3 amino acids in the immature protein. Unless otherwise noted, shading indicates the degree of conservation; black shading with white letters represents 100% conserved; gray shading with white letters is conserved above 80%; gray shading with black letters represents 60% conserved; no shading and black letters represents amino acids that are conserved less than 60%. The positions where the mutations are present in hcl-1 and hcl-3 show conservation without regarding the mutation itself. PtLYK3a and PtLYK3b were retrieved from the Populus trichocarpa genome database, version 1.1, with the identifiers gw1.II.2929.1 and eugene3.00141039, respectively (http://genome.jgi-psf.org). AtLYK3 was retrieved from the protein database at the National Center for Biotechnology Information (NCBI) Web site database with accession number NP_566689 (http://www.ncbi.nlm.nih.gov).
Involvement of MtLYK3 in Nod Factor-Induced Gene Expression
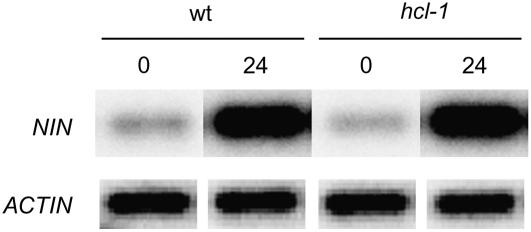
Nod factor-induced gene expression in the hcl-1 mutant has previously been investigated by microarray analysis (Mitra et al., 2004). These studies showed transcriptional changes for 46 genes within 24 h after inoculation with S. meliloti in wild-type plants, whereas in the hcl-1 mutant expression of only 25 of these genes was triggered properly. An interesting gene, whose expression was not studied by Mitra and coworkers, is NODULE INCEPTION (NIN). In Lotus, LjNIN is up-regulated by Nod factor and requires both LjNFR1 and LjNFR5 (Schauser et al., 1999; Radutoiu et al., 2003). Further, the phenotype of Lotus and pea nin knockout mutants resembles that of Medicago hcl mutants—block of infection and extensive root hair deformation (Schauser et al., 1999; Catoira et al., 2001; Borisov et al., 2003). Therefore, we wondered whether the observed phenotype could be related to the down-regulation of expression of MtNIN in hcl mutants. By performing BLAST analysis, we identified the Medicago ortholog of MtNIN on the bacterial artificial chromosome clone Mth2-53m2 (GenBank accession no. CR936325.2) and subsequently a full-size cDNA clone was isolated by RT-PCR. Southern-blot analysis showed that only one copy is present in the Medicago genome (data not shown). We assessed MtNIN expression using semiquantitative RT-PCR on total RNA isolated from wild-type and hcl-1 plants prior to and after inoculation with S. meliloti. One day after inoculation, MtNIN is approximately 10-fold induced in wild-type as well as in hcl-1 plants (Fig. 3), showing that the hcl phenotype or lack of infections is not caused by lack of increased MtNIN expression.
Figure 3.
RT-PCR analyses of MtNIN expression in wild-type and hcl-1 mutant plants. Twenty-four hours after inoculation (24), MtNIN is induced in both wild-type (WT) and hcl-1 mutant (hcl-1) plants compared to uninoculated plants (0). ACTIN expression is used as an internal reference.
MtLYK3 Acts in a Nod Factor Structure-Dependent Fashion
In Medicago species, including M. truncatula (“Medicago” in this article), the structure of the lipid moiety and the acetate at the nonreducing end of the Nod factor are especially important in the induction of the infection process (Ardourel et al., 1994; Catoira et al., 2001; Limpens et al., 2003). The rhizobial Nod genes that are responsible for N-acylation are NodF and NodE, whereas NodL is responsible for O-acetylation. The S. meliloti nodFnodL double mutant is unable to infect Medicago, although this mutant efficiently induces root hair curling, microcolony formation, and the formation of sac-like infection structures (Ardourel et al., 1994; Limpens et al., 2003), a phenotype similar to that of the hcl-4 mutant. Based on the infection phenotype of several S. meliloti nod mutants, a two-receptor model was proposed in which a Nod factor entry receptor is specifically involved in infection and has more stringent demands to Nod factor structure than a Nod factor signaling receptor (Ardourel et al., 1994). LYK3 is an excellent candidate for such an entry receptor and is thus expected to show such stringent demands. Therefore, we tested whether the nodulation efficiency of hcl-4 mutants is affected in a Nod factor structure-dependent manner. We determined nodule numbers on wild-type and hcl-4 plants inoculated with the wild type, nodL, and nodFnodE mutant S. meliloti, respectively. On wild-type Medicago, the S. meliloti nodFnodE mutant forms a similar number of nodules compared to wild-type S. meliloti, whereas the nodL mutant forms a slightly reduced (43%) number of nodules (Table I). This is in agreement with previous reports (Ardourel et al., 1994). Both S. meliloti mutants form microcolonies in root hair curls of hcl-4 as efficiently as wild-type S. meliloti (Supplemental Fig S2). However, the number of nodules formed on hcl-4 roots is more than 40-fold reduced in the case of the S. meliloti nodFnodE mutant, and nodules are completely absent in the case of inoculation with the S. meliloti nodL mutant (Table I). Therefore, these rhizobial nod genes and MtLYK3 show synergistic interaction, indicating that MtLYK3 is involved, directly or indirectly, in recognition of Nod factors during infection. These findings strongly support that LYK3 is an entry receptor with a stringent demand toward Nod factor structure.
DISCUSSION
We showed that the Medicago gene HCL encodes the putative Nod factor receptor MtLYK3 and that this receptor is required for three important steps in the infection process: curling of root hairs and initiation and polar growth of infection threads. Furthermore, it does so in Nod factor structure-dependent fashion and so supports the two-receptor model in which LYK3 acts as the entry receptor (Ardourel et al., 1994).
In addition to its role in root hair curling and infection, LYK3 also appears to have a function in stopping root hair deformation. Previously, it was shown that S. meliloti nodFnodL mutants induce multiple branches on individual root hairs, whereas wild-type S. meliloti does not. Also, multiple branches are formed on root hairs of hcl mutants (see Fig. 1C; Catoira et al., 2001). Because HCL/LYK3 is a putative Nod factor receptor, it is very probable that LYK3 activation by wild-type Nod factors causes down-regulation of continuous root hair branching. The alternative explanation that Nod factors produced by the nodFnodL mutant have gained the ability to hyperstimulate root hair branching has become very unlikely.
The hcl-1, hcl-2, and hcl-3 mutants are all deficient in root hair curling. The mutation in hcl-1 converts the third conserved Gly in the GxGxxG motif of kinase subdomain I into a Glu (Hanks and Hunter, 1995; Fig. 2A). Similar mutations in yeast (Saccharomyces cerevisiae) and Drosophila kinases, changing the third Gly in this motif into a bulky charged amino acid, cause null mutant phenotypes (Melnick et al., 1993; Martin et al. 1996). Mutational analysis of the GxGxxG motif in the insulin receptor kinase also shows that a bulky substitution at this position abolishes protein kinase activity (Odawara et al., 1989). Therefore, it is likely that hcl-1 is a null mutant. Because hcl-1, hcl-2, and hcl-3 have extremely similar phenotypes, which are much more severe than the hcl-4 phenotype, it is likely that hcl-2 and hcl-3 are also strong mutants with minimal LYK3 activity (Catoira et al., 2001). This implies that the Pro at position 87 in LysM domain 1, which is mutated in hcl-3, is essential for functioning of HCL. This Pro is highly conserved in LysM domains of LysM receptor kinases, except for LysM domain 2, where it is equally often a Thr (Arrighi et al., 2006; Mulder et al., 2006; Fig. 2B). This Pro is not likely to be directly involved in binding of Nod factors because its position is not near the predicted peptidoglycan binding sites (Mulder et al., 2006). However, its presence can be required for correct folding or structural integrity and, as a consequence, influences binding indirectly or affects the stability of the protein. We cannot exclude that the different mutated forms of LYK3 still bind Nod factors and still perform part of its function as an inactive kinase. This has been shown for kinase-dead mutant versions of EGFR that can function as a docking protein for other kinases and still mediate signaling (Yamauchi et al., 1997). However, identification of three independent alleles conferring a nearly identical phenotype suggests that hcl-1, hcl-2, and hcl-3 are strong mutant alleles.
We identified and characterized a fourth mutant hcl allele (hcl-4). We showed that hcl-4 is a weak allele because this mutant does efficiently form root hair curls containing rhizobial microcolonies, but infection thread growth is strongly impaired. hcl-4 produces a markedly reduced level of transcripts encoding wild-type MtLYK3. Therefore, it is probable that the MtLYK3 protein level is also reduced. This reduced level still appears to be sufficient to trigger root hair curling. However, this level of expression is too low to support polar growth of infection threads because the vast majority of infection events form sac-like structures that stop growing shortly after they are formed. During root hair curling, an asymmetric endoplasmic microtubule skeleton is formed in the tip of the root hair and this is not formed in hcl knockout mutants (Catoira et al., 2001). When infection thread formation is initiated in the root hair curl, the microtubule skeleton becomes disconnected from the root hair tip and a new microtubule skeleton is formed at the tip of an infection thread. Therefore, it is probable that MtLYK3 mediates Nod factor signaling during root hair curling, as well as during infection thread growth, and controls the configuration and position of the endoplasmic microtubule network.
The two-receptor model proposes that the entry receptor controlling the infection process has a more stringent demand on Nod factor structure than the signaling receptor (Ardourel et al., 1994). This was based on mutations in bacterial Nod genes that interfere specifically with the infection process. For example, the S. meliloti nodFnodL mutant is specifically blocked at the start of infection thread growth in curled root hairs (Ardourel et al., 1994). Because the hcl-4 mutant has a similar phenotype, it is probable that MtLYK3 is most efficiently activated by Nod factors having a C16:2 acyl chain and an acetate substitution at the nonreducing end. A direct interaction of the putative Nod factor receptor MtLYK3 and the nonreducing end of the Nod factor is strongly supported by our studies that show a synergistic genetic interaction of hcl-4 and the nodFnodE or nodL mutant S. meliloti. This synergistic interaction is only detectable during infection thread growth, so this is the step in which the concentration of MtLYK3, in an hcl-4 background, appears to be limiting.
Although MtLYK3 is also essential for root hair curling, a synergistic interaction of hcl-4 and nodFnodE or nodL has not been observed in this early step; both mutants as well as wild-type S. meliloti form microcolonies in tightly curled root hairs of hcl-4 plants. Neither is the S. meliloti nodFnodL double mutant affected in inducing this step in hcl-4 plants (Supplemental Fig S2). We assume that the reduced amount of wild-type LYK3 (in hcl-4), despite less efficient activation by aberrant Nod factors, is still sufficient to induce root hair curling. Thus, root hair curling requires less signaling by MtLYK3 than infection thread formation. This might be comparable to the hypersensitive response that is dependent in a quantitative manner on the level of resistance proteins (Holt et al., 2005).
Previously, it was shown that several early Nod factor-induced responses (e.g. root hair deformation, induction of MtENOD11 expression, Ca2+ spiking) are not affected in hcl mutants (Wais et al., 2000; Catoira et al., 2001). Therefore, MtLYK3 is not required for these early responses but it is essential during root hair curling. In contrast, the putative Nod factor receptor MtNFP is essential for all early responses (Amor et al., 2003; Arrighi et al., 2006). The different role of MtLYK3 in Nod factor signaling is further underlined by the microarray studies of Mitra and coworkers because they showed that NFP is essential for all Nod factor-dependent changes in gene expression induced within 24 h after inoculation, whereas HCL is only essential for about 50% of the induced changes (Mitra et al., 2004). Knowing now that HCL encodes a putative Nod factor receptor, these data strongly support the two-receptor model in which MtNFP acts as a signaling receptor and MtLYK3 acts as an entry receptor.
NIN is a gene that is up-regulated upon Nod factor signaling and, when mutated, causes a hcl-like phenotype in Lotus, pea, and Medicago (Schauser et al., 1999; Borisov et al., 2003; Marsh et al., 2007). Therefore, we wondered whether MtLYK3 controls MtNIN expression. However, MtNIN expression was induced in a wild-type manner in the strong hcl-1 mutant. This shows that MtNIN is not under transcriptional regulation of the entry receptor MtLYK3, which is in agreement with recently published microarray data (Marsh et al., 2007), and therefore the hcl phenotype is not caused by a lack of increased MtNIN expression.
The perception of Nod factors in Lotus, mediated by the NFR receptors, seems somewhat different. The probable ortholog of MtNFP is LjNFR5 and it has a similar loss of Nod factor response phenotype when mutated. However, mutations in LjNFR1, the gene highly homologous to MtLYK3, cause a different effect because they block almost all Nod factor-induced responses (Schauser et al., 1998; Radutoiu et al., 2003). It has been hypothesized that LjNFR1 forms a signaling complex with LjNFR5 in Lotus that controls all Nod factor responses. This is supported by the fact that LjNFR5 (like MtNFP) lacks the activation loop in the kinase domain and, in addition, MtNFP lacks kinase activity in in vitro experiments (Arrighi et al., 2006). Because in Lotus both Nod factor receptors, LjNFR1 and LjNFR5, are essential for all early responses, it seems probable that, in addition to MtNFP, another LysM domain receptor kinase forms a signaling complex with MtNFP, triggering early Nod factor-induced responses in Medicago. An excellent candidate would be MtLYK2, which is also encoded in the NFR1-LYK3 syntenic region, groups together with LYK3 and NFR1 in a phylogenetic tree, and, like MtLYK3, is 77% identical to LjNFR1 (Limpens et al., 2003; Zhu et al., 2006; Supplemental Fig S3; Supplemental Table S1). Genetic screens performed on pea and Medicago might not have identified a receptor functioning like LjNFR1 due to functional redundancy of the different LYKs.
Mutations in MtNFP, LjNFR1, and LjNFR5 did not reveal whether the encoded receptors are also involved in infection processes or are just required at the onset of Nod factor signaling. However, recently it was shown, by means of RNAi, that MtNFP is also involved in infection thread growth (Arrighi et al., 2006). Therefore, we can conclude that both the signaling receptor MtNFP as well as the entry receptor MtLYK3 are required during infection thread growth.
Our data are most consistent with a two-receptor model for Nod factor signaling (see below). However, it cannot be excluded that a single Nod factor receptor complex induces the different responses. For example, the hcl-1 protein with a mutated kinase domain might still bind Nod factors and perform part of its function in early Nod factor signaling as an inactive kinase, whereas regulation of a subset of genes, root hair curling, and infection require LYK3 kinase activity. However, other hcl alleles (hcl-2, hcl-3) that are mutated outside the kinase domain have a very similar phenotype to hcl-1. Therefore, a one-receptor model is highly unlikely.
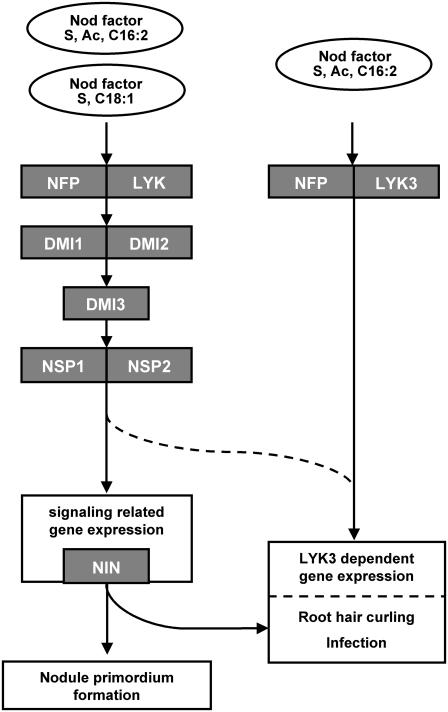
Based on the data reported in this article, in combination with previous reports, we propose a Nod factor perception/signaling model as depicted in Figure 4. Nod factors are perceived by LysM domain receptor kinases. MtNFP acts as a signaling receptor probably together with another LYK protein. In Lotus, this is LjNFR1, whereas in Medicago it has not yet been identified. Activation of this signaling receptor complex is essential for all changes in gene expression (induced within 24 h), nodule primordium formation, and infection. However, activation of a subset of these changes in gene expression, root hair curling, and infection require the additional activation of a second Nod factor receptor, namely, MtLYK3. (Thus, activation of the signaling receptor complex, including MtNFP, is only sufficient to regulate a subset of genes.) MtLYK3 functions as the proposed entry receptor because it is essential for the induction of the infection process and has a more stringent requirement for Nod factor structure than the signaling receptor MtNFP. Knockdown of MtNFP leads to aberrant infection thread formation; therefore, we propose that MtNFP and MtLYK3 together form the entry receptor complex. Up-regulation of the gene encoding the putative transcription factor MtNIN does not require activation of the entry receptor. Hence, activation of both Nod factor signaling complexes is necessary for the infection process because MtNIN, like MtLYK3, is essential for the formation of tight root hair curls and subsequent infection. Whether genes, including MtNIN, that are activated by the signaling receptor cascade are essential for the LYK3-dependent changes in gene expression is not known. Alternatively, it is possible that NSP1 and NSP2, or downstream, yet unidentified, elements of the signaling receptor-activated cascade, are essential for LYK3-dependent gene expression (Fig. 4, dashed lines).
Figure 4.
Model for Nod factor perception in Medicago at the onset of symbiosis. Nod factors produced by wild type (S, Ac, C16:2) and a nodFnodL mutant (S, C18:1) are shown. S. meliloti induce the signaling cascade consisting of NFP, DMI, and the NSP proteins. Most likely another LYK, the ortholog of NFR1, is needed to induce Nod factor signaling. This cascade induces about 50% of the identified Nod factor-induced genes, including NIN. The NFP, DMI, NSP, and NIN proteins are essential for nodule primordium formation, formation of tight root hair curls, and infection, as well. Only Nod factor produced by wild-type S. meliloti is able to activate LYK3 sufficiently (and likely NFP) for LYK3-dependent gene regulation and infection. Whether NIN is needed for regulation of LYK3-dependent gene expression is unknown. As an alternative, it is possible that NSP1 and NSP2 and upstream signaling components are essential for regulation of LYK3-dependent gene expression, indicated by dashed lines. Arrows do not necessarily mean direct interactions.
In conclusion, our data support the model whereby the LysM receptor kinase MtLYK3 functions as the Nod factor entry receptor proposed in the two-receptor model. MtLYK3 acts in a Nod factor structure-dependent fashion, supporting the hypothesis that LysM domain-containing receptors essential to symbiosis interact directly with Nod factor. More specifically, the level of activated MtLYK3, which is Nod factor structure dependent, can strictly control the rhizobial infection process.
MATERIALS AND METHODS
Characterization of hcl-4
Medicago truncatula hcl-4, isolated from an ethyl methanesulfonate-mutagenized population (Penmetsa and Cook, 2000), was crossed with three other hcl mutants to show allelism and with wild-type A17 Jemalong plants to study segregation. Plant growth conditions and rhizobial inoculation for these experiments were as previously described (Catoira et al., 2001). Plants for other experiments were grown under 18 h of 300 μmol m−2 s−1 light and 6-h darkness period, temperature of 22°C, and 75% relative humidity. Nodulation assays with the different rhizobial strains were performed in 15-mL plastic beakers containing perlite (3-mm grain) saturated with liquid Fahraeus medium (Fahraeus, 1957), 20 wild-type plants, and 32 hcl-4 per experimental group for characterization. After 10 to 14 d, plants were flood inoculated by adding 15-mL Fahraeus medium supplemented with 0.01 OD units of Sinorhizobium meliloti 2011 expressing GFP (Limpens et al., 2003); excess of S. meliloti-containing medium is allowed to drain away. After 21 d, roots were analyzed using a fluorescence binocular, a fluorescence microscope, or a confocal laser-scanning microscope. Statistical analysis of data obtained was performed in Excel (Microsoft).
Positional Cloning of the HCL Gene
A segregating population of an hcl-1 × DZA315.16 cross was used for mapping purposes using molecular markers linked to the LYK3 gene: EH3, 10L2, and DK139L, which map 2.6 cM telomeric, 0 cM centromeric (11 kb), and 4 cM centromeric, respectively (Gualtieri et al., 2002). EH3 primers amplify a fragment of 397 bp on hcl-1 genomic DNA and a fragment of 406 bp on DZA315.16 genomic DNA (EH3-F, CCCATGAGCTTCTTACGTACCTTGTT; and EH3-R, TAGGCTGGAGTGATAAGTGATTGGT). 10L2 primers amplify a 325-bp fragment on both hcl-1 and DZA315.16 genomic DNA; the DZA315.16 fragment can be cut using KpnI (10L2-F, ATCCAAAATGGTGTAGTTCTGCTC; and 10L2-R, CTATTGATGATGAAAGTAGGACAGGT). DK139L primers amplify a 352-bp fragment on both hcl-1 and DZA315.16 genomic DNA; the DZA315.16 fragment can be cut using EcoRV; alternatively, TaqI cuts the hcl-1 fragment (DK139L-F, CTAGCAAAACTCAGAAAACCAAGAA; and DK139L-R, CGTTTTAGAGGAAAATATGATGAGCA). The LYK3 gene was sequenced on both strands in all hcl mutants using gene-specific primers. The LYK3 gene was amplified with Phusion DNA polymerase (Finnzymes) using genomic DNA as template (CACCGGACAGATAGCGCAGAGTTG and GCAAAACTAAAATATTCACCCCATTACC). PCR products were cloned into pENTR-TOPO (Invitrogen) and one clone was fully sequenced. Using Gateway recombination technology (Invitrogen), the gene was transferred to the binary vector pKGW-R (Smit et al., 2005). All hcl alleles were complemented with this LYK3 construct using Agrobacterium rhizogenes-mediated transformation (Limpens et al., 2003). Plants were subsequently assayed for nodulation as described above.
Isolation of lyk3 cDNAs
Total RNA was isolated from root material of A17 and the different hcl plants using the RNeasy kit (Qiagen). During isolation, genomic DNA was removed with an on-column DNA treatment (Qiagen). After quantification, 2.5 μg of total RNA was used for RT with Moloney murine leukemia virus (M-MLV) reverse transcriptase according the accompanying protocol (Invitrogen). Synthesized cDNA was used as a template in a PCR reaction with primers in exon 1 and exon 3 using Taq DNA polymerase to amplify partial cDNAs of LYK3 (ACGCAAGTTTGACAACTGTTGAG and CATCTTGAGTTGAAAATGCCCTAG). PCR products were shotgun cloned into pGEMT vector and sequenced using M13 universal primers.
Semiquantitative RT-PCR Analyses of MtNIN Expression
Primers were based on the sequence of bacterial artificial chromosome mth2-53m2 (CR936325) (ATGGAATATGGTGGTGGGATAG and CTAAGGAGGATGGACTGCTGC). A probe was made from the complete MtNIN gene and used to detect MtNIN on 2.5 μg of EcoRI-, HindIII-, and HaeIII-digested A17 Jemalong genomic DNA that was blotted on Hybond-N+ using alkaline transfer (Sambrook and Russell, 2001; GE Healthcare). Hybridization using a 32P-labeled MtNIN probe was carried out in Church buffer without bovine serum albumin (Sambrook and Russell, 2001).
We isolated total RNA from uninoculated and 24-h S. meliloti-inoculated root material from both A17 and hcl-1 plants using the RNeasy plant kit (Qiagen). DNA was removed using on-column DNA digestion with RNase-free DNase (Qiagen). After quantification, 2.5 μg was used for a reverse transcriptase reaction according to the accompanying protocol using M-MLV reverse transcriptase (Invitrogen). We used 0.5% of the total reverse transcriptase reaction per 50-μL PCR reaction performed. As an internal reference, actin expression was determined as described by Smit and coworkers (2005). For determination of MtNIN expression, we used MtNIN-specific primer-bordering introns (GAAGCCATGACTATCCGCGTC and CACCACAACCTGATGCACCAC, GCAGCAGCAGCAGCATGATC and CCATCACCACCATTAGCAGTTGC). Fifteen microliters of the reaction mixture was separated on gel and Southern blotted to Hybond-N+ nylon membrane (GE Healthcare). Products were visualized by hybridization with radioisotope-labeled probes and scanning using a STORM 840 phosphor imager (GE Healthcare). Quantification was performed using the ImageQuant software package and analyzed in Excel (Microsoft).
Light and Fluorescence Microscopy
Samples were observed under a Leica MZFLIII binocular fitted with HQ470/40, HQ525/50, HQ553/30, and HQ620/60 optical filters and a Leica DC300F digital camera (Leica Microsystems). Higher magnification samples were observed on a Nikon Optiphot 2 fitted with both DM510 and DM580 optical filters, and a Nikon Coolpix 990 digital camera (Nikon GmbH).
Laser-Scanning Microscopy
Transgenic roots were analyzed using a Zeiss confocal microscope (Zeiss Axiovert 100 M equipped with a LSM510, an argon laser with a 488-nm laser line, a helium-neon laser with a 543-nm laser line). S. meliloti GFP fluorescence was imaged using the following settings: 488-nm laser → HFT488/543 → sample → HFT488/543 → mirror → NFT545 → BP505-550 → detector. Bright field was imaged using the following setting: 488-nm laser → HFT488/543 → sample → detector. DsRed or propidium iodide fluorescence was imaged using the following settings: 543-nm laser → HFT488/543 → sample → HFT488/543 → mirror → NFT545 → LP560 → detector. The pinhole was adjusted for each channel in such a way that Z resolution is equal to typically 1-μm resolution using a 40× objective (1 Airy unit).
Phylogenetic Analysis of the LYK Gene Family
Protein sequences of LjNFR1b, LjNFR1c, MtLYK2, and MtLYK5 were conceptually translated from mRNAs generated using gene prediction algorithms on genomic sequences (FGENESH 2.5; accession nos. BN001116–BN001119). Manual adjustments were made if the algorithm was unable to correctly identify the gene structure. These sequences were aligned with other LysM receptor kinase proteins using ClustalX 1.64b (MtLYK1 [AAQ73154.1], MtLYK3 [AAQ73159.1], MtLYK4 [AAQ73160.1], MtLYK6 [AAQ73157.1], MtLYK7 [AAQ73158.1], LjNFR1 [CAE02591.1], AtLYK1 [NP_566689.2], AtLYK2 [AAF99862.1], AtLYK3 [NP_175606.2], PtLYK3a [gw1.II.2929.1], PtLYK3b [eugene3.00141039]). (Populus trichocarpa sequences have been retrieved from http://genome.jgi-psf.org, version 1.1.) Settings used were: gap opening 10.00, gap extension 0.05, delay divergent sequences 40%, and a BLOSUM matrix. The unrooted phylogenetic tree was made by bootstrapping the alignment 500 times using different seed numbers. Bootstrapping was not corrected for multiple substitutions, nor were gap positions excluded. Statistics and graphical overview of the alignment were generated using Genedoc 2.5.000.
Nucleotide sequences have been deposited in the EMBL Nucleotide Sequence Database with accession numbers AM420448 to AM420459, and BN001116 to BN001119.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Complementation of hcl.
Supplemental Figure S2. Infection of hcl-4.
Supplemental Figure S3. Phylogeny of LysM receptor kinases.
Supplemental Table S1. Figure legends and comparison of LysM receptor kinases.
Supplementary Material
This work was supported by the Graduate School of Experimental Plant Sciences and the Netherlands Organization for Scientific Research.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Ton Bisseling (ton.bisseling@wur.nl).
The online version of this article contains Web-only data.
References
- Amor BB, Shaw SL, Oldroyd GE, Maillet F, Penmetsa RV, Cook D, Long SR, Denarie J, Gough C (2003) The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J 34 495–506 [DOI] [PubMed] [Google Scholar]
- Ardourel M, Demont N, Debelle F, Maillet F, de Billy F, Prome JC, Denarie J, Truchet G (1994) Rhizobium meliloti lipooligosaccharide nodulation factors: different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell 6 1357–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrighi JF, Barre A, Ben Amor B, Bersoult A, Soriano LC, Mirabella R, de Carvalho-Niebel F, Journet EP, Gherardi M, Huguet T, et al (2006) The Medicago truncatula lysine motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol 142 265–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisov AY, Madsen LH, Tsyganov VE, Umehara Y, Voroshilova VA, Batagov AO, Sandal N, Mortensen A, Schauser L, Ellis N, et al (2003) The Sym35 gene required for root nodule development in pea is an ortholog of Nin from Lotus japonicus. Plant Physiol 131 1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catoira R, Timmers AC, Maillet F, Galera C, Penmetsa RV, Cook D, Denarie J, Gough C (2001) The HCL gene of Medicago truncatula controls Rhizobium-induced root hair curling. Development 128 1507–1518 [DOI] [PubMed] [Google Scholar]
- Fahraeus G (1957) The infection of white clover root hairs by nodule bacteria studied by a simple slide technique. J Gen Microbiol 16 374–381 [DOI] [PubMed] [Google Scholar]
- Gualtieri G, Kulikova O, Limpens E, Kim DJ, Cook DR, Bisseling T, Geurts R (2002) Microsynteny between pea and Medicago truncatula in the SYM2 region. Plant Mol Biol 50 225–235 [DOI] [PubMed] [Google Scholar]
- Hanks SK, Hunter T (1995) Protein kinase 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J 9 576–596 [PubMed] [Google Scholar]
- Holt BF III, Belkhadir Y, Dangl JL (2005) Antagonistic control of disease resistance protein stability in the plant immune system. Science 309 929–932 [DOI] [PubMed] [Google Scholar]
- Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R (2003) LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302 630–633 [DOI] [PubMed] [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N, et al (2003) A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425 637–640 [DOI] [PubMed] [Google Scholar]
- Marsh JF, Rakocevic A, Mitra RM, Brocard L, Sun J, Eschtruth A, Long SR, Schultze M, Ratet P, Oldroyd GED (2007) Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol 144 324–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin H, Castellanos MC, Cenamor R, Sanchez M, Molina M, Nombela C (1996) Molecular and functional characterization of a mutant allele of the mitogen-activated protein-kinase gene SLT2 (MPK1) rescued from yeast autolytic mutants. Curr Genet 29 516–522 [DOI] [PubMed] [Google Scholar]
- Melnick MB, Perkins LA, Lee M, Ambrosio L, Perrimon N (1993) Developmental and molecular characterization of mutations in the Drosophila-raf serine/threonine protein kinase. Development 118 127–138 [DOI] [PubMed] [Google Scholar]
- Mitra RM, Shaw SL, Long SR (2004) Six nonnodulating plant mutants defective for Nod factor-induced transcriptional changes associated with the legume-rhizobia symbiosis. Proc Natl Acad Sci USA 101 10217–10222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder L, Lefebvre B, Cullimore J, Imberty A (2006) LysM domains of Medicago truncatula NFP protein involved in Nod factor perception. Glycosylation state, molecular modeling and docking of chitooligosaccharides and Nod factors. Glycobiology 16 801–809 [DOI] [PubMed] [Google Scholar]
- Odawara M, Kadowaki T, Yamamoto R, Shibasaki Y, Tobe K, Accili D, Bevins C, Mikami Y, Matsuura N, Akanuma Y (1989) Human diabetes associated with a mutation in the tyrosine kinase domain of the insulin receptor. Science 245 66–68 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Mitra RM, Wais RJ, Long SR (2001) Evidence for structurally specific negative feedback in the Nod factor signal transduction pathway. Plant J 28 191–199 [DOI] [PubMed] [Google Scholar]
- Penmetsa RV, Cook DR (2000) Production and characterization of diverse developmental mutants of Medicago truncatula. Plant Physiol 123 1387–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perret X, Staehelin C, Broughton WJ (2000) Molecular basis of symbiotic promiscuity. Microbiol Mol Biol Rev 64 180–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Gronlund M, Sato S, Nakamura Y, Tabata S, Sandal N, et al (2003) Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425 585–592 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell D (2001) Molecular Cloning: A Laboratory Manual, Ed 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schauser L, Handberg K, Sandal N, Stiler J, Thykjær T, Pajuelo E, Nielsen A, Stougaard J (1998) Symbiotic mutants deficient in nodule establishment identified after T-DNA transformation of Lotus japonicus. Mol Gen Genet 259 414–423 [DOI] [PubMed] [Google Scholar]
- Schauser L, Roussis A, Stiller J, Stougaard J (1999) A plant regulator controlling development of symbiotic root nodules. Nature 402 191–195 [DOI] [PubMed] [Google Scholar]
- Smit P, Raedts J, Portyanko V, Debelle F, Gough C, Bisseling T, Geurts R (2005) NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308 1789–1791 [DOI] [PubMed] [Google Scholar]
- Wais RJ, Galera G, Oldroyd G, Catoira R, Penmetsa RV, Cook D, Gough C, Denarie J, Long SR (2000) Genetic analysis of calcium spiking responses in nodulation mutants of Medicago truncatula. Proc Natl Acad Sci USA 97 13407–13412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wais RJ, Keating DH, Long SR (2002) Structure-function analysis of Nod factor-induced root hair calcium spiking in Rhizobium-legume symbiosis. Plant Physiol 129 211–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi T, Ueki K, Tobe K, Tamemoto H, Sekine N, Wada M, Honjo M, Takahashi M, Takahashi T, Hirai H, et al (1997) Tyrosine phosphorylation of the EGF receptor by the kinase Jak2 is induced by growth hormone. Nature 390 91–96 [DOI] [PubMed] [Google Scholar]
- Zhu H, Riely BK, Burns NJ, Ane JM (2006) Tracing nonlegume orthologs of legume genes required for nodulation and arbuscular mycorrhizal symbioses. Genetics 172 2491–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.