Abstract
Cryptochromes mediate blue light-dependent photomorphogenic responses, such as inhibition of hypocotyl elongation. To investigate the underlying mechanism, we analyzed a genetic suppressor, scc7-D (suppressors of cry1cry2), which suppressed the long-hypocotyl phenotype of the cry1cry2 (cryptochrome1/cryptochrome2) mutant in a light-dependent but wavelength-independent manner. scc7-D is a gain-of-expression allele of the GA2ox8 gene encoding a gibberellin (GA)-inactivating enzyme, GA 2-oxidase. Although scc7-D is hypersensitive to light, transgenic seedlings expressing GA2ox at a level higher than scc7-D showed a constitutive photomorphogenic phenotype, confirming a general role of GA2ox and GA in the suppression of hypocotyl elongation. Prompted by this result, we investigated blue light regulation of mRNA expression of the GA metabolic and catabolic genes. We demonstrated that cryptochromes are required for the blue light regulation of GA2ox1, GA20ox1, and GA3ox1 expression in transient induction, continuous illumination, and photoperiodic conditions. The kinetics of cryptochrome induction of GA2ox1 expression and cryptochrome suppression of GA20ox1 or GA3ox1 expression correlate with the cryptochrome-dependent transient reduction of GA4 in etiolated wild-type seedlings exposed to blue light. Therefore we propose that in deetiolating seedlings, cryptochromes mediate blue light regulation of GA catabolic/metabolic genes, which affect GA levels and hypocotyl elongation. Surprisingly, no significant change in the GA4 content was detected in the whole shoot samples of the wild-type or cry1cry2 seedlings grown in the dark or continuous blue light, suggesting that cryptochromes may also regulate GA responsiveness and/or trigger cell- or tissue-specific changes of the level of bioactive GAs.
Cryptochromes are blue light receptors that regulate various photomorphogenic responses in plants, including deetiolation and photoperiodic control of floral initiation (Cashmore, 2003; Lin and Shalitin, 2003). Arabidopsis (Arabidopsis thaliana) CRYPTOCHROME1 (CRY1) and CRY2 mediate blue light inhibition of hypocotyl elongation, although CRY1 plays a more prominent role in this response (Koornneef et al., 1980; Ahmad and Cashmore, 1993; Guo et al., 1998; Koornneef et al., 1998; Lin et al., 1998). Cryptochromes and phytochromes regulate many overlapping physiological and developmental responses, and they may do so by regulating similar genes (Fankhauser and Chory, 1997; Quail, 2002; Lin and Shalitin, 2003; Sullivan and Deng, 2003). Many hormones, including auxin, GA, brassinosteroids, ethylene, and cytokinin, are known to be involved in hypocotyl growth (Vandenbussche et al., 2005). In etiolated pea (Pisum sativum) seedlings exposed to light, the levels of indole-3-acetic acid, GA, and abscisic acid were found to change to various extents. A reduction of GA1 (the major bioactive GA in pea) was the first detected and most dramatically changed (Symons and Reid, 2003). Results of genome-wide expression profiling also indicate that many photoreceptor-regulated genes encode enzymes involved in the biosynthesis and catabolism of phytohormones, including GA (Ma et al., 2001; Folta et al., 2003; Ohgishi et al., 2004; Zimmermann et al., 2004; Jiao et al., 2005; X. Yu, D. Shalitin, and C. Lin, unpublished data). Those enzymes are often encoded by multiple-member gene families, but few of those gene families have been characterized in detail sufficient to assess the blue light effects on hormonal homeostasis and photomorphogenesis.
GAs are tetracyclic diterpenoid hormones that promote growth, such as hypocotyl elongation (Olszewski et al., 2002; Sun and Gubler, 2004). Only a few of the presently known 126 different GAs have been shown to be physiologically active, including GA1, GA3, GA4, and GA7 (Hedden and Phillips, 2000). GA4 is believed to be the major bioactive GA in Arabidopsis. Key enzymes involved in the metabolism and catabolism of bioactive GAs include ent-kaurene synthase, P450 monooxygenases, and dioxygenases. Two dioxygenases, GA 20-oxidase (GA20ox) and GA 3β-hydroxygenase (GA3ox), catalyze the last few steps in the synthesis of bioactive GAs (Hedden and Phillips, 2000; Reid et al., 2004). Another dioxygenase, GA 2-oxidase (GA2ox), catalyzes catabolism and inactivation of bioactive GAs or their precursors (Lester et al., 1999; Thomas et al., 1999; Hedden and Phillips, 2000; Schomburg et al., 2003). GA signal transduction involves DELLA proteins; it is believed that GA promotes degradation of DELLA proteins to release their suppression on the GA signaling pathway (Peng and Harberd, 1997; Silverstone et al., 1997; Peng et al., 1999; Olszewski et al., 2002; Sun and Gubler, 2004).
GA is well known for its involvement in phytochrome-regulated hypocotyl elongation (Kamiya and Garcia-Martinez, 1999; Hedden and Phillips, 2000; Garcia-Martinez and Gil, 2001; Halliday and Fankhauser, 2003; Vandenbussche et al., 2005). Phytochromes affect GA levels by regulating expression of the GA2ox and GA3ox genes (Reid et al., 2002). Phytochromes may also regulate GA responsiveness (Reed et al., 1996; Cao et al., 2005; Foo et al., 2006). It has been recently shown that phyA and phyB mediate light stabilization of the DELLA proteins, which may, at least partially, result from the phytochrome-dependent regulation of GA homeostasis (Achard et al., 2007). In comparison to the phytochrome-regulated responses, the relationship between cryptochromes and GA in the blue light responses is less clear in Arabidopsis. It has been found in pea that cry1 and phyA redundantly regulate GA2ox and GA3ox expression and GA signaling (Symons and Reid, 2003; Foo et al., 2006). Arabidopsis cry1 has been reported to regulate hypocotyl elongation via its effect on GA and auxin homeostasis or signaling (Folta et al., 2003).
Prompted by a study of the genetic suppressor of cry1cry2 that corresponds to the Arabidopsis GA2ox8 gene, we investigated the relationship between GA homeostasis and cryptochrome-mediated deetiolation. We showed that increased expression of a GA2ox gene caused hypersensitive or constitutive photomorphogenesis, depending on the relative levels of GA2ox overexpression. We further demonstrated that cryptochromes are required for the blue light induction of GA2ox1 expression and blue light suppression of GA20ox1 and GA3ox1 expression. Although all those observations are consistent with a simple hypothesis that cryptochromes may inhibit accumulation of bioactive GAs to suppress hypocotyl growth, our analyses of the GA4 content in whole shoot samples indicate the involvement of a more complex mode of regulation in the cryptochrome-mediated photomorphogenic responses.
RESULTS
Overexpression of GA2ox8 Suppresses the Long Hypocotyl Phenotype of the cry1cry2 Mutant
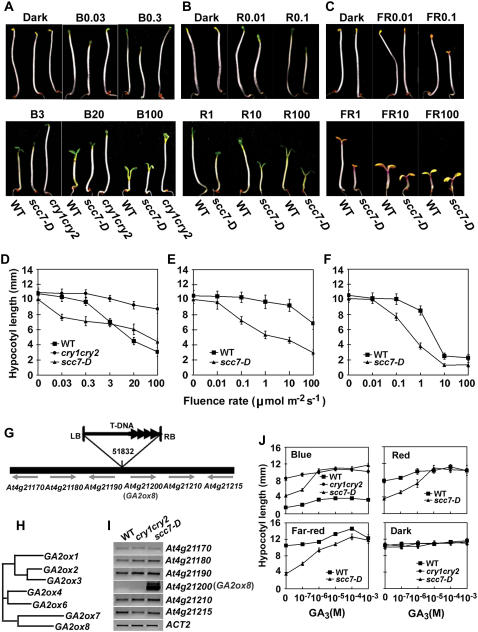
To investigate the genetic mechanisms underlying cryptochrome-mediated light responses in plants, we prepared an activation-tagging population (see “Materials and Methods”) in the cry1cry2 double mutant, which exhibits a long hypocotyl phenotype when grown in blue light (Guo et al., 1998; Mockler et al., 1999). scc7-D was one of the dominant mutants identified in this screen. The scc7-D mutant suppressed the long-hypocotyl phenotype of the cry1cry2 parent when grown in blue light, but it showed a normally elongated hypocotyl when grown in the dark (Fig. 1, A–C). However, scc7-D is hypersensitive to not only blue light, but also red and far-red (FR) lights (Fig. 1, D–F), suggesting that the corresponding gene is associated with light inhibition of cell elongation mediated by both cryptochromes and phytochromes.
Figure 1.
Characterization of the scc7-D mutant (A–F). Phenotypic analyses of scc7-D. Six-day-old seedlings of scc7-D, cry1cry2 mutant (cry1cry2), and wild type (WT) grown in dark or continuous blue (A and D), red (B and E), and FR (C and F) light with indicated fluence rates. Hypocotyl lengths (D–F) and representative seedlings grown under indicated light conditions are shown (A–C). The symbols of B0.03, B0.3, B3, B20, and B100 indicate that seedlings were grown under continuous blue light with fluence rates of 0.03, 0.3, 3, 20, and 100 μmol m−2 s−1, respectively. Similarly, R0.01 or FR0.01 symbolize red light or FR light, respectively, with fluence rates (μmol m−2 s−1) indicated by the numbers. G to J, Molecular characterization of scc7-D. G, Diagram depicting the scc7-D locus and the T-DNA insertion. H, A putative phylogenetic relationship of members of the GA2ox gene family as predicted using ClustalW (http://www.ebi.ac.uk/clustalw/). I, mRNA expression of genes flanking the T-DNA insert of scc7-D. J, Hypocotyl lengths of scc7-D seedlings grown in the dark or in continuous lights of different wavelength (100 μmol m−2 s−1 blue or red light, and 1 μmol m−2 s−1 FR light) in the presence of different concentrations of GA3.
The scc7-D locus contains a T-DNA inserted in an intergenic region, at 4,293 bp upstream from the start codon of GA2ox8 (Fig. 1, G–H), and GA2ox8 seems the only one overexpressed among the six T-DNA flanking genes tested (Fig. 1I). The light-hypersensitive phenotype of scc7-D can be rescued by exogenous GA3 (Fig. 1J). In addition, scc7-D also showed a dwarf and late-flowering phenotype (data not shown). Similar dominant alleles of the GA2ox8 gene have been reported previously, and it was shown that GA2ox8 encodes a GA2ox that catalyzes 2β-hydroxylation of C20-GAs (Schomburg et al., 2003). Importantly, plants overexpressing GA2ox8 contain reduced amounts of bioactive GAs and precursors, while loss-of-function ga2ox7 and ga2ox8 mutants exhibited a long hypocotyl phenotype (Schomburg et al., 2003). We concluded that the exaggerated light response in the scc7-D allele was also caused by the increased GA2ox8 expression and reduced accumulation of bioactive GAs.
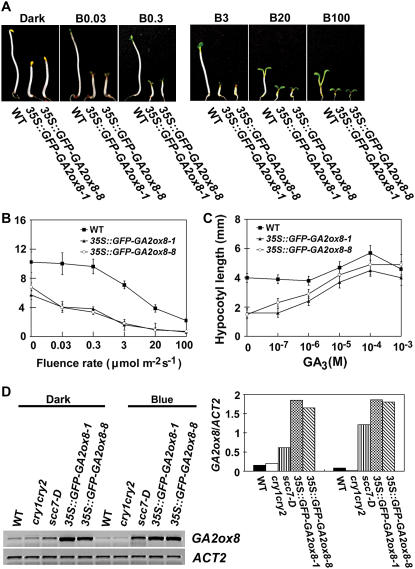
The level of GA2ox8 mRNA in scc7-D was probably high enough to cause exaggerated hypocotyl inhibition in blue light via phytochromes, some of which can act as blue light receptors, but not high enough to suppress hypocotyl elongation in the absence of light. Indeed, transgenic plants overexpressing the GFP-GA2ox8 fusion protein under the control of a strong 35S promoter exhibited a GA-rescuable constitutive photomorphogenic phenotype (Fig. 2, A–C). Similar to scc7-D, the 35S∷GFP-GA2ox8 seedlings are also hypersensitive to light (Fig. 2, A and B). But contrary to scc7-D, the etiolated 35S∷GFP-GA2ox8 transgenic seedlings grown in the dark also showed short hypocotyls, as well as unhooked and partially opened cotyledons (Fig. 2, A and B). The phenotype of 35S∷GFP-GA2ox8 was rescued by exogenous GA3 (Fig. 2C; data not shown). The levels of the 35S∷GFP-GA2ox8 mRNA were markedly higher in transgenic 35S∷GFP-GA2ox8 seedlings than that of the GA2ox8 mRNA in scc7-D or wild-type seedlings (Fig. 2D), which explains why 35S∷GFP-GA2ox8 lines are constitutively photomorphogenic regardless of light whereas scc7-D seedlings showed exaggerated photomorphogenic response only in light. Our observation that overexpression of a GA2ox enzyme, which catalyzes inactivation of GAs, caused constitutive photomorphogenesis is consistent with a previous report that the ga1 mutant impaired in the ent-copalyl diphosphate synthase gene showed constitutive photomorphogenesis (Sun et al., 1992; Alabadi et al., 2004). Both observations suggest that photomorphogenic development is at least partially regulated by the levels of bioactive GA.
Figure 2.
The 35S∷GFP-GA2ox8 transgenic plants are constitutively photomorphogenic (A–C). GA-rescuable constitutive photomorphogenic phenotype of two independent transgenic lines overexpressing the GFP-GA2ox8 protein. D, A markedly higher level of GFP-GA2ox mRNA in the 35S∷GFP-GA2ox8 transgenic plants than the level of GA2ox8 mRNA in scc7-D. Seedlings were grown in the dark or continuous blue light for 6 d.
Association between GA Homeostasis and Blue Light Inhibition of Hypocotyl Elongation
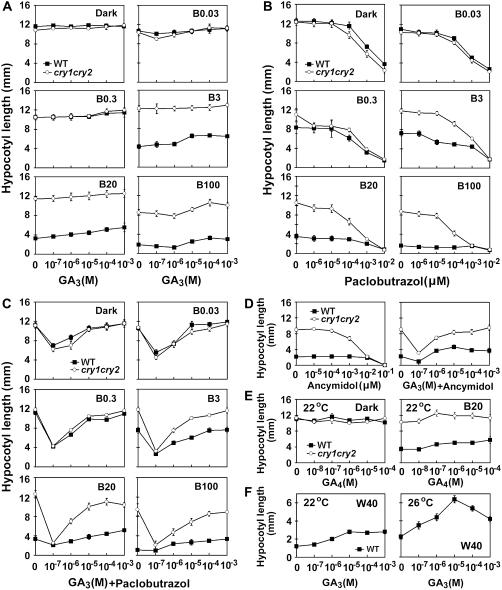
The study of scc7-D prompted us to further explore a possible association between cryptochrome function and GA homeostasis. The wild-type and the cry1cry2 mutants (grown at 22°C) responded only weakly to bioactive GA3 or GA4, regardless of light conditions (Fig. 3). Even under the highest fluence rate of blue light tested, whereby the wild-type seedlings normally exhibited minimum hypocotyl elongation, high concentrations of GA3 or GA4 (0.1–1 mm) failed to elicit a large increase in hypocotyl growth or to phenocopy the cry1cry2 mutant in the wild-type seedlings (Fig. 3, A and E). It is noteworthy that although various hormones have been shown to antagonize light inhibition of hypocotyl growth when applied exogenously, such treatments never completely reverse the inhibitory effect of light on growth (Vandenbussche et al., 2005). At a higher temperature (26°C), the wild-type Arabidopsis seedlings responded relatively more strongly to exogenous GAs as reported previously (Fig. 3F; Collett et al., 2000). A thermoperiodic regulation of PsGA2ox expression has been reported to control thermoperiodic fluctuation of GA1 content and stem elongation of pea stems (Stavang et al., 2005). Whether our observation represents a temperature-sensitive GA responsiveness or that it may be related to the high temperature promotion of auxin-mediated hypocotyl elongation (Gray et al., 1998) remains to be further investigated.
Figure 3.
Effects of GAs or GA biosynthesis inhibitors on blue light inhibition of hypocotyl elongation. Seedlings were grown on Murashige and Skoog medium containing different concentrations of GA3 (A), GA4 (E), GA inhibitors paclobutrazol (B) or ancymidol (D), and GA inhibitors plus GA3 at different concentrations (C and D). The GA inhibitor used in C or D is paclobutrazol (0.02 μm) or ancymidol (0.1 μm), respectively. Hypocotyl lengths of 6-d-old seedlings grown under blue light of different fluence rates are shown. The symbols of B0.03, B0.3, B3, B20, and B100 indicate that seedlings were grown under continuous blue light with fluence rates of 0.03, 0.3, 3, 20, and 100 μmol m−2 s−1, respectively. Hypocotyl responses of seedlings grown on Murashige and Skoog medium containing different concentrations of GA3 under white light (W40, 40 μmol m−2 s−1) at two different temperatures are shown in F.
We next examined how the cry1cry2 mutant responded to GA biosynthesis inhibitors. The GA biosynthesis inhibitors, paclobutrazol (Fig. 3B) or ancymidol (Fig. 3D), can both rescue the long-hypocotyl phenotype of the cry1cry2 mutant, which is consistent with the observation of the cry1 mutant (Folta et al., 2003). These results suggest that GA is likely involved in the development of the long hypocotyl of cry1cry2, and that the level of bioactive GA is important for cryptochrome function. It is interesting that, when GA synthesis was inhibited, the cry1cry2 mutant seedlings grown in blue light with relatively high fluence rates exhibited greater hypocotyl elongation in response to GA3 than the wild-type seedlings (Fig. 3C; B3, B20, B100). These results indicate that cryptochromes may also be required to suppress GA response under blue light.
Cryptochromes Mediate Blue Light-Activated mRNA Expression of GA2ox Genes, Especially GA2ox1, under Dark-to-Light Transfer, Continuous Illumination, and Photoperiodic Conditions
The Arabidopsis genome encodes up to eight GA2ox-related sequences, referred to as GA2ox1 to GA2ox8 (see “Materials and Methods”). However, the mRNA expression of only six members (GA2ox1, GA2ox2, GA2ox4, GA2ox6, GA2ox7, and GA2ox8) was readily detected in our study. GA2ox5 seems to be a pseudogene (Hedden et al., 2001; Schomburg et al., 2003), whereas GA2ox3 is normally expressed at a low level in the absence of auxin induction (Frigerio et al., 2006). Results of various DNA microarray studies concerning the blue light effect on GA2ox expression were not always consistent. For example, the expression of GA2ox1 shows an 8-fold increase in response to blue light in the wild-type seedlings in one study (https://www.genevestigator.ethz.ch), but a 2.4-fold increase of GA2ox1 transcript was reported in the cry1 mutant in another study (Folta et al., 2003). We decided to systematically examine the blue light and cryptochrome effects on the expression of each member of the GA2ox gene family upon transfer from dark to blue light and under photoperiodic and continuous light conditions.
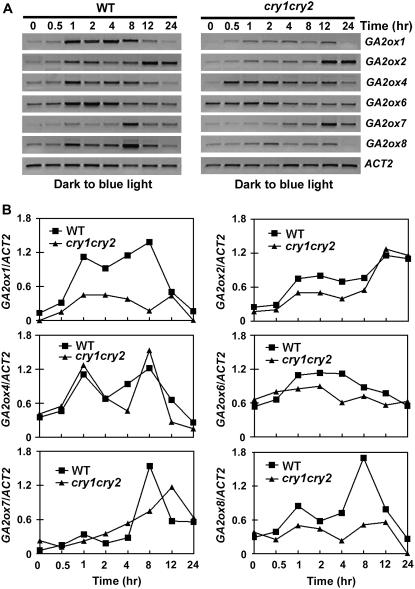
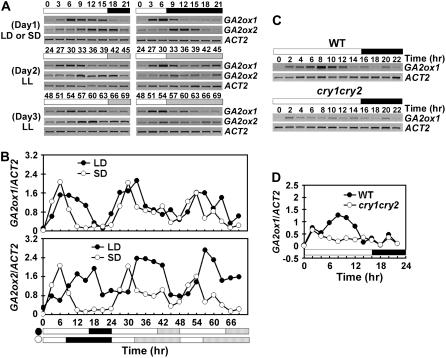
We first analyzed blue light regulation of GA2ox expression in response to inductive blue light treatment. In this experiment, 6-d-old etiolated seedlings were exposed to blue light (100 μmol m−2 s−1) for 0.5 to 24 h, and the level of mRNA expression of the GA2ox genes was examined at various time points. In wild-type seedlings, all six GA2ox genes tested showed various degrees of blue light induction of mRNA expression (Fig. 4). For example, the expression of GA2ox1 and GA2ox8 increased about 2- to 3-fold within 60 min of blue light treatment, or about 6- to 8-fold within 8 h of blue light treatment, respectively (Fig. 4B). The blue light-induced mRNA expression is transient for most GA2ox genes tested. The expression generally increased immediately after blue light treatment, peaked within 12 h, and returned to the dark level within 24 h of blue light treatment. Four of the GA2ox genes tested (GA2ox1, GA2ox2, GA2ox6, and GA2ox8) showed reduced blue light induction in the cry1 or cry1cry2 mutant (Fig. 4; data not shown). The blue light induction of GA2ox1 expression was partially impaired in the cry1 mutant (data not shown), but it was almost completely abolished in the cry1cry2 mutant (Fig. 4). Our results are consistent with a previous DNA microarray study by Thomas Kretsch and colleagues (http://www.uni-tuebingen.de/plantphys/AFGN/atgenex.htm) but opposite to the result of another DNA microarray study (Folta et al., 2003). It is particularly interesting that GA2ox1 is much more responsive to cryptochrome-dependent blue light induction than other GA2ox genes, because GA2ox1 is one of the GA2ox genes that are not induced by auxin (Frigerio et al., 2006). The observation that GA2ox1 is more responsive to blue light than other auxin-inducible GA2ox genes indicates that GA2ox1 may be associated with the auxin-independent growth response reported previously (Collett et al., 2000).
Figure 4.
Blue light-induced change of mRNA expression of the GA2ox genes. Six-day-old etiolated wild-type or cry1cry2 mutant seedlings were exposed to 100 μmol m−2 s−1 blue light, and samples were collected at the time points indicated for RNA analyses. Levels of mRNA expression are shown as the RT-PCR gel images (A) and the relative signal intensities (B).
The transient nature of the blue light induction of GA2ox expression observed in the 24 h inductive blue light treatment (Fig. 4) may be due to the diurnal or circadian rhythmic expression of the GA2ox genes. To test this possibility, seedlings were entrained in long-day (16-h light/8-h dark) and short-day (8-h light/16-h dark) photoperiods for 10 d and then transferred to continuous white light for 2 d. Samples were collected every 3 h for 1 d before transfer and 2 d after transfer to the free-running condition, and the expression of each GA2ox gene was analyzed. The mRNA expression of all six GA2ox genes exhibited either a diurnal rhythm or circadian rhythm (Fig. 5; data not shown). Among them, GA2ox1 and GA2ox2 showed the most robust circadian rhythms that were sustained in seedlings transferred from the long-day or short-day photoperiod to continuous white light for at least 2 d (Fig. 5, A and B; data not shown). The phase of the circadian rhythm of GA2ox2 was dramatically different in plants grown in long-day and short-day photoperiods (Fig. 5B). It has been shown recently that GA4 is the active GA regulating floral initiation (Eriksson et al., 2006). Whether the photoperiod-dependent phase change of GA2ox2 expression affects photoperiodic flowering in Arabidopsis remains to be further investigated (Blazquez and Weigel, 1999; Yu et al., 2006; Zhao et al., 2007). The high-amplitude rhythmic expression of GA2ox1 was all but abolished in the cry1cry2 mutant grown in long-day photoperiod (Fig. 5, C and D). The similar pattern of changes of GA2ox1 expression in the cry1cry2 mutant shown in Figures 4B and 5D indicate that the transient nature of the blue light induction of GA2ox1 expression is at least partially attributed to its rhythmic expression. It is also noteworthy that although GA2ox1 expression is regulated by both cryptochrome and phytochrome, cryptochrome alone had significant effect on the GA2ox1 expression in seedlings grown in white light under the photoperiodic condition (Fig. 5D).
Figure 5.
Cryptochromes control the circadian rhythm of GA2ox1 mRNA expression. mRNA expression of GA2ox1 and GA2ox2 genes in seedlings grown under long day (LD) or short day (SD) for 10 d and then transferred to continuous white light were examined using RT-PCR. Samples were collected every 3 h for 1 d in photoperiod and 2 d in continuous white light (A and B), or every 2 h for 1 d in long-day photoperiod (C and D). The white/black bars indicate light/dark phases, and the dashed bars indicate subjective night phase under continuous light. The time (hour) of light on of the first day of sample collection is set as zero.
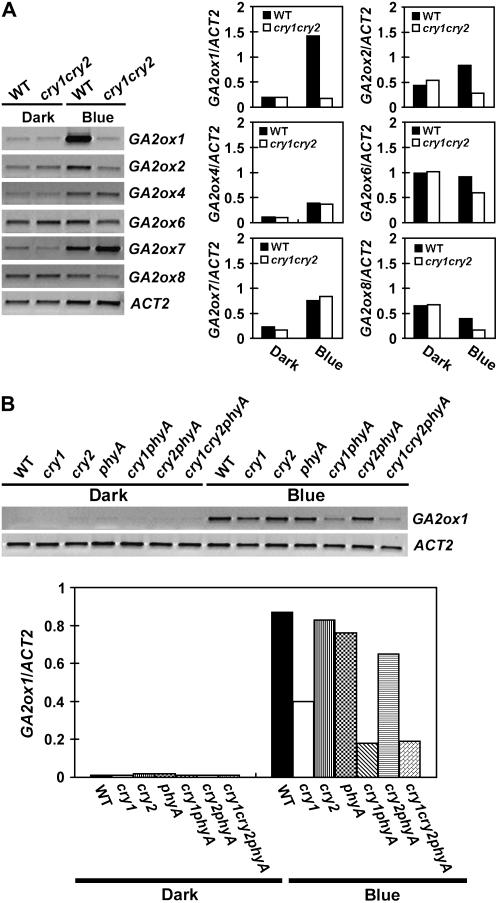
We next compared the steady-state levels of GA2ox expression in seedlings grown in continuous blue light or dark (Fig. 6). Four GA2ox genes (GA2ox1, GA2ox2, GA2ox4, GA2ox7) tested showed higher steady-state levels of mRNA in seedlings grown in continuous blue light than in etiolated seedlings (Fig. 6A). Under continuous blue light, the expression of GA2ox1 showed a more pronounced blue light response than other GA2ox genes; the mRNA expression of GA2ox1 was at least 7-fold higher in light-grown wild-type seedlings than that in etiolated wild-type seedlings (Fig. 6A). A more pronounced blue light effect on the GA2ox1 expression is consistent with that observed under other conditions (Figs. 4 and 5) and that found in continuous white light (Achard et al., 2007). Although our study was initially prompted by the observation that overexpression of GA2ox8 suppressed cry1cry2 phenotype in the scc7-D mutant (Fig. 1), the level of GA2ox8 mRNA expression was not dramatically affected by continuous blue light or cryptochromes (Figs. 2D and 6B). The blue light-induced GA2ox1 expression in seedlings grown under continuous blue light was moderately impaired in the cry1 mutant (Fig. 6B), but it was markedly reduced in the cry1cry2 (Fig. 6A), cry1phyA (Fig. 6B), and cry1cry2phyA mutants (Fig. 6B). These results demonstrate that CRY1 acts redundantly with CRY2 or phyA to mediate blue light induction of GA2ox1 expression. We concluded that cryptochromes are the major blue light receptors required for the transient blue light induction of GA2ox1 expression in etiolated seedlings exposed to blue light, for the circadian rhythmic GA2ox1 expression in seedlings grown in long-day photoperiods under white light, and for the high level steady-state mRNA expression of GA2ox1 in seedlings grown in continuous blue light.
Figure 6.
Comparison of the level of GA2ox mRNAs in seedlings grown in dark or continuous blue light. Six-day-old seedlings were grown in dark or continuous blue light (20 μmol m−2 s−1), the levels of mRNA expression are shown as the RT-PCR gel images (left) and the relative signal intensities (right). The mRNA expression of GA2ox genes in the wild-type or cry1cry2 mutant are shown in A. An independent experiment shows the levels of GA2ox1 mRNA expression in 6-day-old wild type and the indicated photoreceptor mutants grown in the dark or continuous blue light (B).
Cryptochromes Mediate Blue Light Suppression of GA20ox1 and GA3ox1 mRNA Expression
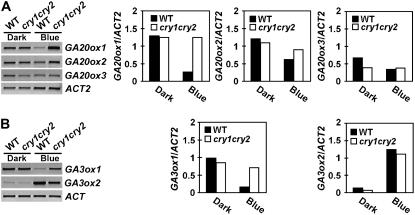
In addition to the GA2ox enzymes that catalyze catabolism or inactivation of bioactive GAs such as GA4, two other dioxygenases, GA20ox and GA3ox, that catalyze synthesis of bioactive GAs are also critical to GA homeostasis (Hedden and Phillips, 2000; Reid et al., 2004). Therefore, we next examined blue light and cryptochrome effects on the expression of GA20ox and GA3ox genes. Figure 7 shows that expression of the three GA20ox genes and one of the two GA3ox genes tested were lower (to differing extents) in seedlings grown in continuous blue light than in etiolated seedlings (Fig. 7A). The blue light suppression of GA20ox1 and GA3ox1 expression are dependent on cryptochromes, because the sustained blue light suppression of both genes was largely abolished in the cry1cry2 mutant (Fig. 7). Therefore, GA20ox1 and GA3ox1 are also major targets of cryptochromes. Similar to what was observed for the GA2ox genes (Fig. 4), but in the opposite direction, the expression of most GA20ox and GA3ox genes tested (except GA3ox2) showed a transient decrease in etiolated seedlings exposed to blue light (Supplemental Fig. S1). The blue light suppression of GA20ox1 and GA3ox1 expression was moderately affected in the cry1cry2 mutant upon transfer from dark to blue light, although their expression was more significantly affected in continuous blue light (Fig. 7; Supplemental Fig. S1). In contrast to all the other GA biosynthesis genes tested, GA3ox2 expression exhibited cryptochrome-dependent blue light induction upon transfer from dark to blue light and under continuous light conditions (Fig. 7; Supplemental Fig. S1). In contrast to all the other GA biosynthesis genes tested, GA3ox2 expression exhibited blue light induction upon transfer from dark to blue light and under continuous light conditions, and it was impaired in the cry1cry2 mutant only under the inductive condition (Fig. 7; Supplemental Fig. S1). It is known that phyB mediates red light activation of GA3ox2 expression in germinating seeds to facilitate germination (Yamaguchi et al., 1998). The blue light activation of GA3ox2 expression may also be associated with light stimulation of cotyledon expansion rather than light inhibition of hypocotyl elongation, but this response appears redundantly regulated by phytochromes and cryptochromes. We might predict that overall, the cryptochrome-dependent blue light suppression of the expression of GA biosynthesis genes such as GA20ox1 and GA3ox1, and cryptochrome-dependent blue light activation of the expression of GA catabolic genes such as GA2ox1, may cause a reduction in bioactive GA levels. However, the discrepancy regarding GA3ox2 expression (discussed above) and the relatively small effect of blue light on the expression of the GA20ox2, GA20ox3, GA2ox2, GA2ox6, and GA2ox8 genes make such predictions very uncertain. Direct measurement of endogenous bioactive GAs is clearly required to accurately assess the effect of blue light on GA level.
Figure 7.
Expression of GA20ox and GA3ox genes in seedlings grown in the dark or continuous blue light. Six-day-old wild-type or cry1cry2 mutant seedlings grown in dark or continuous blue light (20 μmol m−2 s−1) were analyzed for the expression of the GA20ox genes (A) or the GA3ox genes (B). Levels of mRNA expression are shown as the RT-PCR gel images (left) or the relative signal intensities (right).
Cryptochromes Mediate Transient Blue Light Suppression of GA4 Accumulation
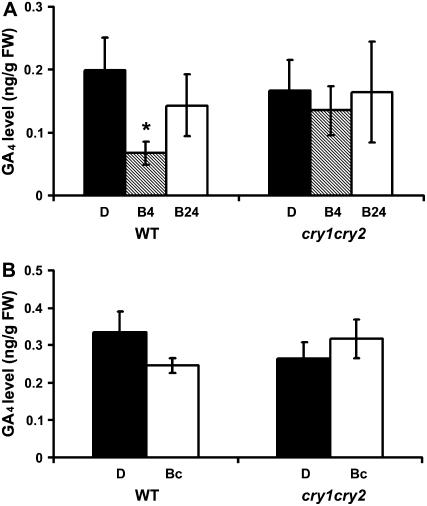
To test whether cryptochrome-mediated blue light regulation of gene expression changes correlate with changes in the level of bioactive GA4, we examined GA4 levels in etiolated wild-type and cry1cry2 mutant seedlings exposed to blue light. As expected, the level of GA4 showed a statistically significant decline in the wild-type seedlings exposed to blue light for 4 h (Fig. 8A). The blue light-induced decrease of GA4 was dependent on cryptochromes, because no statistically significant reduction of GA4 was detected in the cry1cry2 mutant (Fig. 8A). After a blue light treatment for 24 h, the level of GA4 in the etiolated wild-type seedlings exposed to blue light became indistinguishable from that in the etiolated seedlings (Fig. 8A), suggesting the cryptochrome-dependent blue light inhibition of GA4 accumulation was transient or rhythmic. The transient decrease of GA4 upon transfer from dark to blue light appears to correlate with the transient (or rhythmic) increase of GA2ox1 expression (Figs. 4 and 5) and transient (or rhythmic) decrease of GA20ox1 and GA3ox1 expression (Supplemental Figs. S1 and S2). A transient decline of bioactive GA accumulation in etiolated seedlings exposed to blue light has also been reported previously in pea (Foo et al., 2006). In this case the major bioactive GA in pea, GA1, showed a significant but transient decrease in etiolated seedlings exposed to blue light. In contrast to Arabidopsis, wherein cryptochromes mediate the transient blue light suppression of GA4, cry1 and phyA redundantly control the transient blue light suppression of GA1 in pea (Foo et al., 2006).
Figure 8.
Cryptochromes mediate transient blue light suppression of GA4. The GA4 content of 6-d-old etiolated wild-type (WT) and cry1cry2 mutant seedlings exposed to blue light (60 μmol m−2 s−1) for 4 or 24 h are shown in A. The GA4 content of 5-d-old wild-type (WT) and cry1cry2 mutant seedlings grown in continuous dark or continuous blue light are shown in B. Asterisk indicates a significant difference (P < 0.05, two-way ANOVA). D, Dark; B4 and B24, etiolated seedlings exposed to blue light for 4 and 24 h, respectively; Bc, seedlings grown in continuous blue light. GA4 in whole seedling samples except roots was separated by HPLC and analyzed by gas chromatography-MS.
Finally, we compared GA4 levels in etiolated wild-type and cry1cry2 seedlings with those grown under continuous blue light. Interestingly, we detected no significant difference in GA4 level in those samples (Fig. 8B). However, because whole-shoot samples were used in our GA analyses, we cannot exclude the possibility that a localized or cell-specific change of GA4 content may occur in seedlings grown in continuous blue light.
DISCUSSION
In this study, we showed that increased expression of a GA2ox gene genetically suppressed the cry1cry2 mutant (Figs. 1 and 2). Although this observation by itself may have alternative interpretations, our follow-up photophysiological (Fig. 3) and gene expression studies (Figs. 4–5) demonstrate that cryptochromes are indeed positive regulators of GA2ox genes, especially GA2ox1. We showed that cryptochromes are required for the transient induction of GA2ox1 expression in etiolated seedlings exposed to blue light, for the sustained elevation of GA2ox1 expression in seedlings grown in continuous blue light, and for maintaining a high amplitude of the circadian rhythm of GA2ox1 expression in seedlings grown in long-day photoperiods. Consistent with the cryptochrome-mediated blue light stimulation of the expression of the GA catabolic gene GA2ox1, we also demonstrated that cryptochromes mediate blue light suppression of the expression of GA biosynthesis genes GA20ox1 and GA3ox1 (Fig. 7). We concluded that cryptochromes are positive regulators of GA2ox1 but negative regulators of GA20ox1 and GA3ox1. These cryptochrome-regulated gene expression changes may result in a blue light-dependent reduction of bioactive GAs. Given the well-established role of GA as a growth promoter, we propose that cryptochrome-regulated change in GA homeostasis is an important mechanism underlying blue light inhibition of hypocotyl elongation.
Consistent with this hypothesis, our analyses of GA4 content showed a cryptochrome-dependent transient reduction of GA4 in etiolated wild-type seedlings exposed to blue light (Fig. 8A). This result correlates with the transitory/rhythmic expression patterns of many GA metabolism/catabolism genes (Figs. 4 and 5; Supplemental Figs. S1 and S2). However, in contrast to the transient reduction in GA levels in deetiolating seedlings exposed to blue light, we did not detect a significant reduction of GA4 in the wild-type seedlings grown in continuous blue light (Fig. 8B) or white light (G.M. Symons and J.B. Reid, unpublished data). Neither did we detect a significant effect of the cry1cry2 mutation on the GA4 level in seedlings grown in continuous blue light (Fig. 8B). These results impose a significant challenge to the hypothesis that cryptochromes inhibits hypocotyl elongation solely by reducing GA4 levels. Similar observations have been previously reported in pea (O'Neill et al., 2000; Symons and Reid, 2003; Foo et al., 2006). In comparison to etiolated seedlings, the level of bioactive GA also showed a transient reduction in pea seedlings transferred from dark to blue light but not in seedlings grown under prolonged illumination (Reid et al., 2002; Foo et al., 2006). It was hypothesized that phyA and cry1 regulate not only GA homeostasis but also GA responsiveness to affect shoot elongation (Reid et al., 2002; Foo et al., 2006). Therefore, it is possible that light regulation of GA responsiveness or GA signal transduction may account for the inhibition of hypocotyl elongation in Arabidopsis plants grown under continuous blue light.
However, we cannot escape the question why continuous blue light caused markedly changed mRNA expression of the GA2ox1, GA20ox1, and GA3ox1 genes (Figs. 4–7) without a significant change in the level of GA4 (Fig. 8B). It would be interesting to examine whether those mRNA changes actually resulted in corresponding changes in the protein levels of the respective key enzymes in GA homeostasis. Alternatively, a localized change of GA4 levels in response to continuous blue light may provide another possible explanation of our puzzling observations. Specifically, a blue light-dependent reduction of GA4 may be limited to specific organs or cells. For example, light inhibits hypocotyl elongation but stimulates cotyledon expansion, suggesting that light may trigger a decrease or increase of GA4 in hypocotyls or cotyledons, respectively. Moreover, hypocotyl elongation is accomplished mostly by a limited number of cells in the elongation zone (Vandenbussche et al., 2005). Therefore, it is conceivable that light may suppress GA4 accumulation only in cells located in the elongation zone, but not in cells located in other regions of the same hypocotyl. Such localized changes of GA4 homeostasis may not be readily discernable when the whole shoot samples were examined as in this study. The localized change of hormone homeostasis is not uncommon in plants. For example, differential distribution of GA1 was previously reported in pea seedlings (O'Neill et al., 2000). Developmental regulation by cell-specific auxin and cytokinin biosynthesis have also been reported recently in Arabidopsis and rice (Oryza sativa), respectively (Cheng et al., 2006; Kurakawa et al., 2007). A cell-specific change of GA levels in response to blue light may be regulated by a posttranslational mechanism specific to the respective cells, because the blue light-dependent changes of mRNA expression of GA metabolism/catabolism genes were readily detectable in the whole shoot samples (Figs. 4–7). Whether cryptochromes differentially regulate changes of GA4 levels in specific cells and what molecular mechanism(s) may be responsible for such changes remain to be further investigated.
MATERIALS AND METHODS
Plant Materials
Arabidopsis (Arabidopsis thaliana) mutants cry1, cry2, cry1cry2, phyA, and cry1cry2phyA used in this study are in the Columbia background as described previously (Mockler et al., 2003).
To prepare an activation-tagging population, the cry1cry2 plants were transformed using the binary vector pSKI015 and Agrobacterium strain GV3101 as described (Weigel et al., 2000). The T1 seeds were harvested in pools and each pool contains seeds harvested from approximately 50 T0 plants. T1 seeds were germinated in white light and grown on compound soil, submerged in the herbicide Basta (approximately 0.006% ammonium glufosinate), and approximately 250,000 Basta-resistant T1 individuals were obtained. The herbicide-resistant individuals that showed hypocotyl length shorter than that of cry1cry2 parent were selected as putative scc-D mutants, and they were separated from the rest of the mutant populations. Those scc-D lines that showed short hypocotyls in the T2 generation grown in continuous blue light were subject to further genetic analysis. Genomic sequence flanking the T-DNA insert was identified using plasmid rescue (Weigel et al., 2000) or the Tail-PCR method (Liu et al., 1995). Hypocotyl lengths were measured manually for 6-d-old or 7-d-old seedlings (Lin et al., 1998). The sample size is larger than 20 seedlings, and the sds are calculated.
To investigate the response of hypocotyl elongation to exogenous GA3, paclobutrazol, or ancymidol, seeds were surface sterilized for 30 s in 70% ethanol, placed in 0.1% Hgcl2 for 8 min, and rinsed five times with sterile, distilled water. About 100 seeds were placed in Murashige and Skoog agar growth medium. All hormone and inhibitor stocks were dissolved in 70% (V/V) ethanol at a concentration 500 times greater than the final concentration used. GA3 (Shanghai Solvent), GA4 (Sigma), and/or GA biosynthesis inhibitors ancymidol (Sigma) or paclobutrazol (J&K Chemical Ltd), were added into the Murashige and Skoog medium to the final concentrations indicated in the respective figures. Seeds were placed in the dark at 4°C for 4 d, exposed to white light for 12 h to enhance germination before transferring to temperature-controlled growth chambers, and grown under continuous blue, red, or FR light or in the dark at 22°C unless it is indicated otherwise (i.e. 26°C).
For studies of light- or clock-regulated gene expression, about 300 sterile seeds were sown on Murashige and Skoog agar medium, cold treated at 4°C for 4 d, exposed to white light for 12 h, and grown in the dark for 6 d before transfer to various light treatments. Alternatively, seedlings were grown under white light (20 ± 3 μmol m−2 s−1) with long-day (16-h light/8-h dark) or short-day (8-h light/16-h dark) photoperiod for 10 d, and some petri dishes were transferred to continuous white light for 2 d. At the end of treatment, petri dishes were dipped in liquid nitrogen, and tissues (mostly shoot devoid of roots) were harvested by gentle scraping and stored at −80°C for RNA extraction.
Light Sources
In addition to light sources reported previously (Shalitin et al., 2002; Yu et al., 2007): LED-B (peak: 470 nm, half band width: 30 nm), LED-R (peak: 660 nm, half band width: 20 nm), and LED-FR (peak: 740 nm, half band width: 25 nm) were also used. Fluence rates of white, red, and blue light were measured using a Li-250 quantum photometer (LI-COR). The approximate fluence rates of FR light were estimated by plotting the relative fluence rates measured with a Li-250 quantum photometer to a near-linear standard curve of fluence rates measured with a spectroradiometer (T. Mockler and C. Lin, unpublished data).
GA Analysis
Whole shoot (all tissues except root) samples were used to analyze the GA content. For inductive light experiments, seeds were sown thickly on pots filled with potting mix, covered with a fine mesh, and placed in weak fluorescent light at 4°C for 4 d. Plants were transferred to dark at 22°C for 6 d and then transferred to 60 μmol m−2 s−1 blue light at 22°C using light source described (Platten et al., 2005).
For GA measurement, 3 to 4 g whole shoot samples were harvested, placed in ice-cold 80% methanol, homogenized, and the extract was filtered as described (Symons and Reid, 2003). Samples were concentrated to approximately 1 mL and loaded onto a preconditioned Sep-pak C18 cartridge in 0.4% acetic acid. GAs were eluted in 80% methanol in 0.4% acetic acid, dried, and fractionated using the reverse-phase C18 HPLC system (Jager et al., 2005). HPLC fractions corresponding to retention times of the relevant GAs were pooled, dried, methylated, and further purified by solvent partitioning as described (Jones et al., 2005). A total of 10 μL dry pyridine and 40 μL N,O-bis(trimethylsilyl) trifluoroacetamide were added to each vial and samples were heated at 80°C for 20 min. The samples were then dried before the addition of 15 μL N,O-bis(trimethylsilyl) trifluoroacetamide, incubated at 80°C for 15 min, dried under nitrogen gas, resuspended in 20 μL of chloroform, and analyzed by gas chromatography/tandem mass spectrometry (MS/MS) on a Varian 1200 triple quadrupole mass spectrometer (Jones et al., 2005). Injections of 1.5 μL were made in splitless mode at an injection temperature of 260°C onto a Varian VF5-ms column (30 m × 0.25 mm × 0.25 micron). The oven temperature was held at 50°C for 2 min, ramped to 230°C at 30°C per min, increased to 270°C at 5°C per min, and held at 270°C for 3 min. The transfer line temperature was 290°C, and the ion source was held at 220°C. The electron multiplier gain was 1,950 V. Carrier gas was helium at 1.2 mL per min. The MS was operated in selected reaction monitoring mode, with a Q1 peak width of 1.5 mass-to-charge ratio (m/z) units, and a Q3 peak width of 2.0 m/z units. The collision energy was −12 V, with the collision gas being argon at 1 mTorr. Prior to analyses instrument tuning was manually optimized in MS/MS mode rather than using the autotune feature. Based on preliminary full scan MS/MS experiments of standards, for endogenous GA4 (methyl ester TMS ether) the selected precursor ion (Q1) was m/z 284 and the selected product ion (Q3) was m/z 224. For 2H2 GA4 (methyl ester TMS ether) the ions were 286 (Q1) and 226 (Q3). The amount of endogenous GA4 was calculated from the peak areas. The internal standards ([2H2]GA4, [2H2]GA34, and [2H2]GA9) were supplied by Professor L.N. Mander (Research School of Chemistry, Australian National University, Canberra).
Although the transient reduction in GA4 level may result from reduced conversion of the precursor GA9 to GA4 and/or increased inactivation of GA4 to the 2β-hydroxylated GA34, we detected no significant changes in the GA9 or GA34 levels. These discrepancies may be due to the fact that GA9 level (less than 0.0025 ng g−1) was below our detection limit, whereas GA34 levels were approximately 10-fold higher than that of GA4, resulting in technical difficulties to detect relatively small or transient changes in the GA34 level under our experimental conditions. To compare GA content in etiolated seedlings with that grown in continuous blue light, 5-d-old whole shoot samples were used.
RNA Analyses
The accession numbers of genes discussed in this report are: GA2ox1 (At1g78440), GA2ox2 (At1g30040), GA2ox3 (At2g34555, mRNA not detectable), GA2ox4 (At1g47990), GA2ox5 (pseudogene), GA2ox6 (At1g02400), GA2ox7 (At1g50960), GA2ox8 (At4g21200), GA20ox1 (At4g25420), GA20ox2 (At5g51580), GA20ox3 (At5g07200), GA3ox1 (At1g15550), and GA3ox2 (At1g80340).
Total RNA was isolated using Puprep RNAeasy mini kit (Ambiogen Life Tech Ltd). DNA-free RNA was obtained by RQ1 DNase I treatment according to the manufacturer's instructions (Promega). The amount of mRNA was analyzed using semiquantitative reverse transcription (RT)-PCR as decribed (Mockler et al., 2003). cDNA was prepared from 2 μg of total RNA by using Moloney murine leukemia virus reverse transcriptase according to the manufacturer's instructions (Promega). The cDNA was generally diluted 10-fold, and 1 μL of diluted cDNA was used in a 20 μL PCR reaction. DNA sequences of the PCR primers used in this study are the following: ACT2F (5′-CACTGTGCCAATCTACGAGGGT-3′), ACT2R (5′-CACAAACGAGGGCTGGAACAAG-3′); GA2ox1F (5′-CACTATCCACCATGTCCTCTTA-3′), GA2ox1R (5′-CAGACCAAGTAAACTCCTCGTA-3′); GA2ox2F (5′-AGAGGCGGAGAAGATGGTGAA-3′), GA2ox2R (5′-GACAAGGCATGGCAATGGTGC-3′); GA2ox4F (5′-CCGATCAATTCTTTGGTGAAG-3′), GA2ox4R (5′-AATGTTTGGTACAACCGTGGC-3′), GA2ox6F (5′-ATGATTACATACGCACGGTTAG-3′), GA2ox6R (5′-ACATACGTGGCTTCTTTGCTG-3′); GA2ox7F (5′-GGGAAACAAGTGAACGTGAGT-3′), GA2ox7R (5′-GAGAAACCTGGACAAGCCTAC-3′); GA2ox8F (5′-CGGAATCAGAGGCATTAGC-3′), GA2ox8R (5′-CCACCTTTGGGTTCGTCAT-3′); A20ox1F (5′-CAGCCATTTGGGAAGGTGTATC-3′), GA20ox1R (5′-CAAGCAGCTCTTGTATCTATCGT-3′); GA20ox2F (5′-TCAATATTGGTGACACTTTCAT-3′), GA20ox2R (5′-GATGGGATTGTTGTTGGTAATA-3′); GA20ox3F (5′-AAAATGGGCGATGGATACGAAG-3′), GA20ox3R (5′-CGAAAGCGTGAGGGTTAGGAG-3′), GA3ox1F (5′-CCGAAGGTTTCACCATCACTG-3′), GA3ox1R (5′-GAGGCGATTCAACGGGACTAAC-3′); GA3ox2F (5′-CCAGCCACCACCTCAAATAC-3′), GA3ox2R (5′-GTGAAGCACGCTCGGGAAGA-3′).
PCR was generally performed with a 5 min denaturation at 95°C followed by 24 to 35 cycles with each cycle composed of 95°C for 30 s, 55 to 60°C for 30 s, and 72°C for 30 s. PCR products were analyzed using 1.5% agarose gel eletrophoresis. RT-PCR reactions for each experiment were repeated at least three times, and the representative gel images were shown. The expression level of the ACTIN2 gene was used as the internal control to normalize and calculate relative expression levels of genes tested using ImageJ (http://rsb.info.nih.gov/ij/).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At1g78440, At1g30040, At2g34555, At1g47990, At1g02400, At1g50960, At4g21200, At4g25420, At5g51580, At5g07200, At1g15550, and At1g80340.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Blue light-reduced mRNA expression of members of the GA20ox and GA3ox genes.
Supplemental Figure S2. The circadian rhythm of the expression of GA20ox genes.
Supplementary Material
Acknowledgments
The authors thank Detlef Weigel for providing the activation-tagging vectors, Professor L.N. Mander for the GA standards, and John Klejnot for critical reading of the manuscript.
This work was supported by the National Institutes of Health (grant no. GM56265 to C.L.), Changjiang scholarship (to C.L.), and 985 higher education enhancement fund to Hunan University. J.L. and K.B. were partially supported by UC MEXUS-CONACYT fellowship from the University of California and the BOYSCAST award from India, respectively.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) are: Xuanming Liu (xml05@126.com) and Chentao Lin (clin@mcdb.ucla.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Achard P, Liao L, Jiang C, Desnos T, Bartlett J, Fu X, Harberd NP (2007) DELLAs contribute to plant photomorphogenesis. Plant Physiol 143 1163–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad M, Cashmore AR (1993) HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366 162–166 [DOI] [PubMed] [Google Scholar]
- Alabadi D, Gil J, Blazquez MA, Garcia-Martinez JL (2004) Gibberellins repress photomorphogenesis in darkness. Plant Physiol 134 1050–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazquez MA, Weigel D (1999) Independent regulation of flowering by phytochrome B and gibberellins in Arabidopsis. Plant Physiol 120 1025–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Hussain A, Cheng H, Peng J (2005) Loss of function of four DELLA genes leads to light- and gibberellin-independent seed germination in Arabidopsis. Planta 223 105–113 [DOI] [PubMed] [Google Scholar]
- Cashmore AR (2003) Cryptochromes: enabling plants and animals to determine circadian time. Cell 114 537–543 [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20 1790–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collett CE, Harberd NP, Leyser O (2000) Hormonal interactions in the control of Arabidopsis hypocotyl elongation. Plant Physiol 124 553–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson S, Bohlenius H, Moritz T, Nilsson O (2006) GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell 18 2172–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Chory J (1997) Light control of plant development. Annu Rev Cell Dev Biol 13 203–229 [DOI] [PubMed] [Google Scholar]
- Folta KM, Pontin MA, Karlin-Neumann G, Bottini R, Spalding EP (2003) Genomic and physiological studies of early cryptochrome 1 action demonstrate roles for auxin and gibberellin in the control of hypocotyl growth by blue light. Plant J 36 203–214 [DOI] [PubMed] [Google Scholar]
- Foo EJ, Platten D, Weller JL, Reid JB (2006) PhyA and cry1 act redundantly to regulate gibberellin levels during de-etiolation in blue light. Physiol Plant 127 149–156 [Google Scholar]
- Frigerio M, Alabadi D, Perez-Gomez J, Garcia-Carcel L, Phillips AL, Hedden P, Blazquez MA (2006) Transcriptional regulation of gibberellin metabolism genes by auxin signaling in Arabidopsis. Plant Physiol 142 553–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Martinez JL, Gil J (2001) Light regulation of gibberellin biosynthesis and mode of action. J Plant Growth Regul 20 354–368 [DOI] [PubMed] [Google Scholar]
- Gray WM, Ostin A, Sandberg G, Romano CP, Estelle M (1998) High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci USA 95 7197–7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Yang H, Mockler TC, Lin C (1998) Regulation of flowering time by Arabidopsis photoreceptors. Science 279 1360–1363 [DOI] [PubMed] [Google Scholar]
- Halliday KJ, Fankhauser C (2003) Phytochrome-hormonal signalling networks. New Phytol 157 449–463 [DOI] [PubMed] [Google Scholar]
- Hedden P, Phillips AL (2000) Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci 5 523–530 [DOI] [PubMed] [Google Scholar]
- Hedden P, Phillips AL, Rojas MC, Carrera E, Tudzynski B (2001) Gibberellin biosynthesis in plants and fungi: a case of convergent evolution? J Plant Growth Regul 20 319–331 [DOI] [PubMed] [Google Scholar]
- Jager CE, Symons GM, Ross JJ, Smith JJ, Reid JB (2005) The brassinosteroid growth response in pea is not mediated by changes in gibberellin content. Planta 221 141–148 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Ma L, Strickland E, Deng XW (2005) Conservation and divergence of light-regulated genome expression patterns during seedling development in rice and Arabidopsis. Plant Cell 17 3239–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SE, Demeo JS, Davies NW, Noonan SE, Ross JJ (2005) Stems of the Arabidopsis pin1-1 mutant are not deficient in free indole-3-acetic acid. Planta 222 530–534 [DOI] [PubMed] [Google Scholar]
- Kamiya Y, Garcia-Martinez JL (1999) Regulation of gibberellin biosynthesis by light. Curr Opin Plant Biol 2 398–403 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Peeters AJM, Soppe W (1998) Genetic control of flowering time in Arabidopsis. Annu Rev Plant Physiol Plant Mol Biol 49 345–370 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Rolff E, Spruit CJP (1980) Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh. Z Pflanzenphysiol 100 147–160 [Google Scholar]
- Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Sakakibara H, Kyozuka J (2007) Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445 652–655 [DOI] [PubMed] [Google Scholar]
- Lester DR, Ross JJ, Smith JJ, Elliott RC, Reid JB (1999) Gibberellin 2-oxidation and the SLN gene of Pisum sativum. Plant J 19 65–73 [DOI] [PubMed] [Google Scholar]
- Lin C, Shalitin D (2003) Cryptochrome structure and signal transduction. Annu Rev Plant Biol 54 469–496 [DOI] [PubMed] [Google Scholar]
- Lin C, Yang H, Guo H, Mockler T, Chen J, Cashmore AR (1998) Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc Natl Acad Sci USA 95 2686–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8 457–463 [DOI] [PubMed] [Google Scholar]
- Ma L, Li J, Qu L, Hager J, Chen Z, Zhao H, Deng XW (2001) Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13 2589–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler T, Yang H, Yu X, Parikh D, Cheng YC, Dolan S, Lin C (2003) Regulation of photoperiodic flowering by Arabidopsis photoreceptors. Proc Natl Acad Sci USA 100 2140–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler TC, Guo H, Yang H, Duong H, Lin C (1999) Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126 2073–2082 [DOI] [PubMed] [Google Scholar]
- O'Neill DP, Ross JJ, Reid JB (2000) Changes in gibberellin A(1) levels and response during de-etiolation of pea seedlings. Plant Physiol 124 805–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgishi M, Saji K, Okada K, Sakai T (2004) Functional analysis of each blue light receptor, cry1, cry2, phot1, and phot2, by using combinatorial multiple mutants in Arabidopsis. Proc Natl Acad Sci USA 101 2223–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski N, Sun TP, Gubler F (2002) Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell (Suppl) 14 S61–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Harberd NP (1997) Gibberellin deficiency and response mutations suppress the stem elongation phenotype of phytochrome-deficient mutants of Arabidopsis. Plant Physiol 113 1051–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Richards DE, Moritz T, Cano-Delgado A, Harberd NP (1999) Extragenic suppressors of the Arabidopsis gai mutation alter the dose-response relationship of diverse gibberellin. responses. Plant Physiol 119 1199–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platten JD, Foo E, Elliott RC, Hecht V, Reid JB, Weller JL (2005) Cryptochrome 1 contributes to blue-light sensing in pea. Plant Physiol 139 1472–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail PH (2002) Phytochrome photosensory signalling networks. Nat Rev Mol Cell Biol 3 85–93 [DOI] [PubMed] [Google Scholar]
- Reed JW, Foster KR, Morgan PW, Chory J (1996) Phytochrome B affects responsiveness to gibberellins in Arabidopsis. Plant Physiol 112 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid JB, Botwright NA, Smith JJ, O'Neill DP, Kerckhoffs LH (2002) Control of gibberellin levels and gene expression during de-etiolation in pea. Plant Physiol 128 734–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid JB, Symons GM, Ross JJ (2004) Regulation of gibberellin and brassinosteriod biosynthesis by genetic, environmental and hormonal factors. In PJ Davies, ed, Plant Hormones: Biosynthesis, Signal Transduction, Action. Kluwer Academic Publishers, New York, pp 179–203
- Schomburg FM, Bizzell CM, Lee DJ, Zeevaart JA, Amasino RM (2003) Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell 15 151–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalitin D, Yang H, Mockler TC, Maymon M, Guo H, Whitelam GC, Lin C (2002) Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature 417 763–767 [DOI] [PubMed] [Google Scholar]
- Silverstone AL, Mak PY, Martinez EC, Sun TP (1997) The new RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana. Genetics 146 1087–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavang JA, Lindgard B, Erntsen A, Lid SE, Moe R, Olsen JE (2005) Thermoperiodic stem elongation involves transcriptional regulation of gibberellin deactivation in pea. Plant Physiol 138 2344–2353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JA, Deng XW (2003) From seed to seed: the role of photoreceptors in Arabidopsis development. Dev Biol 260 289–297 [DOI] [PubMed] [Google Scholar]
- Sun T, Goodman HM, Ausubel FM (1992) Cloning the Arabidopsis GA1 locus by genomic subtraction. Plant Cell 4 119–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TP, Gubler F (2004) Molecular mechanism of gibberellin signaling in plants. Annu Rev Plant Biol 55 197–223 [DOI] [PubMed] [Google Scholar]
- Symons GM, Reid JB (2003) Hormone levels and response during de-etiolation in pea. Planta 216 422–431 [DOI] [PubMed] [Google Scholar]
- Thomas SG, Phillips AL, Hedden P (1999) Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc Natl Acad Sci USA 96 4698–4703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F, Verbelen JP, Van Der Straeten D (2005) Of light and length: regulation of hypocotyl growth in Arabidopsis. Bioessays 27 275–284 [DOI] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blazquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrandiz C, Kardailsky I, Malancharuvil EJ, Neff MM, et al (2000) Activation tagging in Arabidopsis. Plant Physiol 122 1003–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Smith MW, Brown RG, Kamiya Y, Sun T (1998) Phytochrome regulation and differential expression of gibberellin 3β-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell 10 2115–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Klejnot J, Lin C (2006) Florigen: one found, more to follow? J Integr Plant Biol 48 617–621 [Google Scholar]
- Yu X, Shalitin D, Liu X, Maymon M, Klejnot J, Yang H, Lopez J, Zhao X, Bendehakkalu KT, Lin C (2007) De-repression of the NC80 motif is critical for the photoactivation of Arabidopsis CRY2. Proc Natl Acad Sci USA 104 7289–7294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XY, Yu XH, Liu XM, Lin C (2007) Light regulation of gibberellins metabolism in seedling development. J Integr Plant Biol 49 21 [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.