Abstract
Glucan phosphorylating enzymes are required for normal mobilization of starch in leaves of Arabidopsis (Arabidopsis thaliana) and potato (Solanum tuberosum), but mechanisms underlying this dependency are unknown. Using two different activity assays, we aimed to identify starch degrading enzymes from Arabidopsis, whose activity is affected by glucan phosphorylation. Breakdown of granular starch by a protein fraction purified from leaf extracts increased approximately 2-fold if the granules were simultaneously phosphorylated by recombinant potato glucan, water dikinase (GWD). Using matrix-assisted laser-desorption ionization mass spectrometry several putative starch-related enzymes were identified in this fraction, among them β-AMYLASE1 (BAM1; At3g23920) and ISOAMYLASE3 (ISA3; At4g09020). Experiments using purified recombinant enzymes showed that BAM1 activity with granules similarly increased under conditions of simultaneous starch phosphorylation. Purified recombinant potato ISA3 (StISA3) did not attack the granular starch significantly with or without glucan phosphorylation. However, starch breakdown by a mixture of BAM1 and StISA3 was 2 times higher than that by BAM1 alone and was further enhanced in the presence of GWD and ATP. Similar to BAM1, maltose release from granular starch by purified recombinant BAM3 (At4g17090), another plastid-localized β-amylase isoform, increased 2- to 3-fold if the granules were simultaneously phosphorylated by GWD. BAM activity in turn strongly stimulated the GWD-catalyzed phosphorylation. The interdependence between the activities of GWD and BAMs offers an explanation for the severe starch excess phenotype of GWD-deficient mutants.
Starch consists of the two Glc polymers, amylose and amylopectin, and is deposited as semicrystalline granules inside plastids. The structure of amylopectin, which normally accounts for 70% or more of the dry weight of starch, is responsible for the semicrystalline nature of starch. In amylopectin α-1,4-linked glucan chains are connected by α-1,6 branches. The chain length distribution and the arrangement of the branch points in amylopectin lead to the formation of ordered arrays of densely packed double helices in the semicrystalline zones of the starch granule. The essentially linear amylose is probably present in the so-called amorphous zones of the granule that also contain amylopectin in a less ordered structure (Smith, 2001).
Enzymes required for starch synthesis are ADP-Glc pyrophosphorylase, starch synthases, branching enzymes, and also distinct isoforms of debranching enzymes (Tetlow et al., 2004; Zeeman et al., 2007). The breakdown of the starch particle is less well understood. α-Amylase, which cleaves α-1,4 bonds within the polyglucan, plays an important role in the degradation of cereal endosperm starch (Smith et al., 2005). However, this enzyme is not essential for starch breakdown in Arabidopsis (Arabidopsis thaliana) leaves (Yu et al., 2005). In contrast, Arabidopsis plants in which the plastidial β-amylase isoform β-AMYLASE3 (BAM3; BMY8, At4g17090) is repressed by means of RNAi show a starch excess phenotype in their leaves (Kaplan and Guy, 2005). The same phenotype was observed in potato (Solanum tuberosum) antisense plants with reduced expression of the plastidial β-amylase PCT-BMY1, the putative ortholog of BAM3 (Scheidig et al., 2002). β-Amylases are exoamylases that release maltose from the nonreducing ends of glucans or dextrins by cleavage of α-1,4 linkages. α-1,6 linkages are hydrolyzed by debranching enzymes. Most higher plants contain four different debranching enzymes: three isoforms of isoamylase and one limit dextrinase (Lloyd et al., 2005). It has been shown that the debranching enzyme ISOAMYLASE3 (ISA3) is required for normal rates of starch breakdown in leaves from Arabidopsis (Wattebled et al., 2005; Delatte et al., 2006) and potato (Hussain, 2002). ISA3 from either potato (Hussain et al., 2003) or Arabidopsis (Delatte et al., 2006) displays high activity with β-limit dextrins (glucans that are produced as a result of β-amylase activity during starch breakdown).
In addition to enzymes that cleave α-1,4 or α-1,6 linkages, starch phosphorylating enzymes are also required for normal starch mobilization in leaves. Phosphate monoesterified to the C6 or the C3 position of glucosyl residues is a minor constituent of most starches (Blennow et al., 2002). Glucan, water dikinase (GWD; formerly designated as R1 or SEX1) specifically phosphorylates the C6 position (Ritte et al., 2006). The formation of the less frequent C3-phosphate esters is catalyzed by phosphoglucan, water dikinase (PWD; Baunsgaard et al., 2005; Kötting et al., 2005; Ritte et al., 2006). Activity of PWD strictly relies on a preceding starch phosphorylation by GWD (Kötting et al., 2005). The catalytic mechanism of both dikinases includes autophosphorylation of the enzyme. The β-P of ATP is first transferred to a conserved His residue and then to the (phospho) glucan (Ritte et al., 2002; Mikkelsen et al., 2004; Baunsgaard et al., 2005; Kötting et al., 2005). Transgenic or mutant plants in which the activity of GWD (Lorberth et al., 1998; Yu et al., 2001) or PWD (Baunsgaard et al., 2005; Kötting et al., 2005) is reduced, display increased starch levels in their leaves, with the phenotype of the GWD-deficient plants, such as the Arabidopsis mutant sex1-3, being more severe. Only traces of C6- and C3-P esters could be detected in starch of the sex1-3 mutant that has no detectable GWD (Ritte et al., 2006).
The interaction of GWD and PWD with starch particles is affected by the physiological state of the cells. Both dikinases were recovered in the fraction of starch-associated proteins if the granules were prepared from starch degrading (darkened) leaves, whereas the proportion of granule-bound GWD or PWD was negligible during starch synthesis in the light (Ritte et al., 2000; Kötting et al., 2005). However, even in darkness the granule-associated fraction of GWD and PWD represents only a rather small proportion of the total and no light/dark-dependent differences in the amounts of the respective dikinases in the soluble fractions could be observed (Ritte et al., 2000; Kötting et al., 2005). It was proposed that potato GWD is regulated by redox modification and that the starch-bound fraction is in an inactive, oxidized form, whereas the stroma-soluble fraction is reduced and active (Mikkelsen et al., 2005). The fact that only a proportion of GWD (a rather tiny one, see above) appeared to be inactivated at night was attributed to the midpoint redox potential of GWD, which is more positive than that of any (other) known redox-regulated enzyme (Mikkelsen et al., 2005). However, subsequent investigations clearly indicate that the starch-bound GWD is active (G. Ritte, unpublished data). It is, therefore, unlikely, that the proposed redox modification is involved in the regulation of GWD activity in leaves during the diurnal cycle. This view is supported by the analysis of starch phosphorylation in vivo (Ritte et al., 2004). Labeling studies using photoautotrophic cultures of Chlamydomonas reinhardtii showed that the phosphorylation rate increased when starch was mobilized in darkness. Furthermore, in potato leaves the phosphorylation level of the granule surface was considerably higher during starch breakdown than during starch synthesis. The phosphate residues incorporated into starch during its mobilization were subjected to a significant turnover, which was not observed during starch synthesis-associated glucan phosphorylation (Ritte et al., 2004).
Why are the glucan phosphorylating enzymes required for normal starch mobilization? It was suggested that phosphate in starch affects the hydrophilicity and structure of starch and thereby renders the rather hydrophobic starch particle accessible for enzymatic attack (Yu et al., 2001; Ritte et al., 2002). However, a starch degrading enzyme whose activity relies on (or is increased by) glucan phosphorylation has not yet been reported. Using recombinant potato GWD (StGWD) and granules of the Arabidopsis sex1-3 mutant as tools we studied if breakdown of the native starch particles by proteins extracted from Arabidopsis leaves is stimulated by glucan phosphorylation. Purification of the affected activities and the analysis of purified recombinant proteins provided evidence for an interdependence between the activities of GWD and plastidial β-amylases.
RESULTS
Purification of Starch Degrading Arabidopsis Enzymes Whose Activities Increase if the Starch Is Simultaneously Phosphorylated
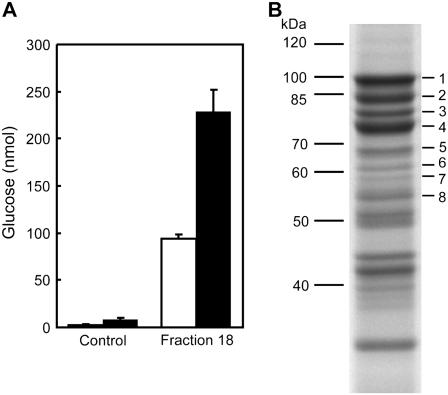
To identify starch degrading enzymes, whose activity is affected by glucan phosphorylation, we analyzed the ability of proteins extracted from Arabidopsis wild-type leaves to degrade sex1-3 starch granules with and without simultaneous starch phosphorylation by StGWD (nonradioactive test, see “Materials and Methods”). However, the nonradioactive test is time consuming and was deemed impractical to analyze a large number of fractions during a protein purification. For this purpose we used a second assay (radioactive test). In the radioactive test, sex1-3 starch granules that had been radiolabeled in vitro using StGWD and 33P-ATP, served as substrate. The radiolabeled granules were incubated with the different protein fractions. At the end of the incubation period the starch granules were sedimented by centrifugation and the radioactivity in the supernatant was quantified. In principle enzyme activities that release phosphoglucans, phospho-Glc, or orthophosphate from starch could be detected using the radioactive test. Since the release of phosphoglucans from starch could also be catalyzed by enzymes that do not discriminate between phosphorylated and nonphosphorylated substrates, the (combined) active fractions identified using the radioactive test were then also analyzed using the nonradioactive test (see above). Only those fractions that showed higher in vitro starch breakdown under conditions of simultaneous glucan phosphorylation were used for further purification. The breakdown of granular starch by the active protein fraction obtained after four purification steps increased more than 2-fold in the presence of StGWD and ATP (Fig. 1A). This increase was not observed if either ATP (Fig. 1A) or StGWD (data not shown) were lacking. This indicates that glucan phosphorylation is required for the elevated starch breakdown. SDS-PAGE and matrix-assisted laser-desorption ionization (MALDI) time-of-flight mass spectrometry (MS) analyses revealed that the protein fraction contained several known starch-related enzymes (Fig. 1B). Among these were starch branching enzyme SBE3 (At2g36390, Dumez et al., 2006), isoamylase ISA3 (At4g09020, Wattebled et al., 2005; Delatte et al., 2006), disproportionating enzyme DPE1 (At5g64860, Critchley et al., 2001), and β-amylase BAM1 (At3g23920, Sparla et al., 2006). Another interesting protein in this fraction was the putative phosphatase At3g01510 (Fig. 1B) that is closely related to the SEX4 phosphatase At3g52180 (Kerk et al., 2006; Niittylä et al., 2006; Sokolov et al., 2006). It was reported recently that animal and plant phosphatases homologous to At3g01510 can dephosphorylate amylopectin (Worby et al., 2006; see also “Discussion”). Thus, the phosphatase At3g01510 could be involved in the release of radiolabel from 33P-labeled starch in the radioactive test. The analysis of the products of the radioactive test using high performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) revealed that label was predominantly released from starch as phosphooligosaccharides. Orthophosphate accounted for approximately 20% of the total radioactivity released (data not shown). It is not known if the orthophosphate originated directly from granular starch or if it was released from phosphodextrins derived therefrom. The subcellular localization of the phosphatase At3g01510 has not yet been established and it is, therefore, unclear whether or not starch can serve as a substrate of this enzyme in vivo.
Figure 1.
Breakdown of granular starch by Arabidopsis proteins is stimulated by simultaneous glucan phosphorylation. A, Starch degrading activity of the protein fraction obtained from Arabidopsis leaves using four purification steps. Sex1-3 starch granules (2.5 mg) were incubated with 60 μL fraction 18 (MonoQ) or 60 μL buffer (control) with (black bars) or without (white bars) 0.5 mm ATP in the presence of 1.6 μg recombinant potato GWD. Final volume of the assay: 120 μL. Following incubation at 25°C for 90 min the starch was sedimented by centrifugation. Starch breakdown products present in the supernatant were hydrolyzed with acid and subsequently Glc was quantified. The slightly increased Glc content in the ATP-containing control was not consistently observed in independent experiments. B, SDS-PAGE of fraction 18 and protein identification using MALDI-MS. Proteins were separated by SDS-PAGE (10% acrylamide in the separation gel). Following staining with Coomassie Blue bands were excized, digested with trypsin, and the peptides were analyzed by MALDI-MS and database search. The following proteins were identified: 1, 2, 3 = SBE3 (At2g36390); 4 = ISA3 (At4g09020) + SBE3; 5 = putative Phosphatase (At3g01510); 6 = unknown protein (At3g55760); 7 = BAM1 (At3g23920); 8 = DPE1 (At5g64860). Proteins ≤55 kD were also analyzed but were not related to carbohydrate metabolism.
To narrow down which enzymes are affected by glucan phosphorylation we used homozygous insertion mutants defective in either SBE3 (SALK_048089, be3-1; Dumez et al., 2006), ISA3 (GABI_KAT_280G10, Atisa3-2; Delatte et al., 2006), BAM1 (SALK_039895; Kaplan and Guy, 2005), or DPE1 (dpe1-1; Critchley et al., 2001). First we isolated leaf proteins that precipitated if the solution was brought to 45% saturation with ammonium sulfate (first step of the purification procedure) and analyzed the release of radioactivity from 33P-labeled granules (radioactive test). Compared to wild type, activities were reduced in extracts from isa3, bam1, and sbe3 plants but not in those from dpe1 plants (data not shown). Leaf extracts from isa3, bam1, and sbe3 were then analyzed in more detail and were subjected to a two-step purification procedure (ammonium sulfate precipitation and Q-Sepharose fast-flow anion-exchange chromatography). In addition to the radioactive test starch breakdown with and without simultaneous glucan phosphorylation (nonradioactive test) was analyzed. In the protein fractions prepared from isa3 or bam1 plants activities were reduced in both tests by approximately 50%, whereas in extracts from sbe3 plants only the activity in the radioactive test was reduced (Supplemental Fig. S1). This indicates that SBE3 contributed to the release of phosphodextrins from starch but its activity was not stimulated in the presence of StGWD and ATP.
GWD Activity Stimulates β-Amylolytic Attack on Granular Starch
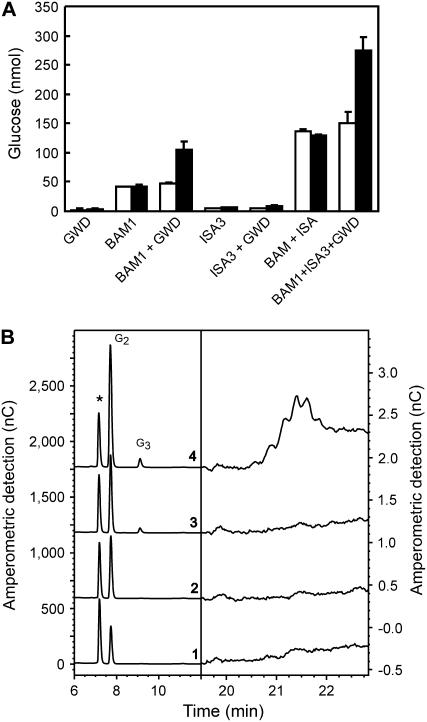
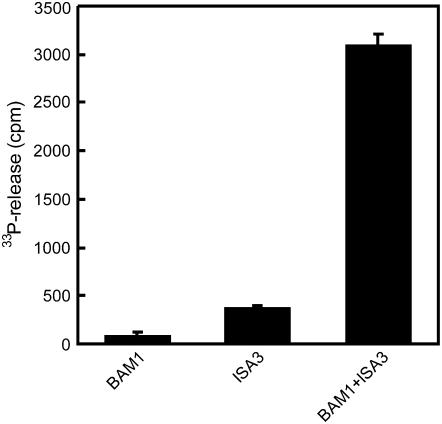
To further investigate if the activities of ISA3 and BAM1 are stimulated by glucan phosphorylation these enzymes were heterologously expressed in Escherichia coli and purified. Arabidopsis BAM1 was expressed as a GST fusion protein. Instead of Arabidopsis ISA3 we used recombinant potato ISA3 (StISA3, Hussain et al., 2003). The sequences of the ISA3 proteins from Arabidopsis and potato are highly similar, implying that their activities are closely related (Hussain et al., 2003). ISA3 is required for normal leaf starch breakdown in both Arabidopsis and potato (Hussain, 2002; Wattebled et al., 2005; Delatte et al., 2006). The purities of typical preparations of the different recombinant proteins as analyzed by SDS-PAGE are depicted in Supplemental Figure S2. Breakdown of granular starch by the GST-BAM1 fusion protein increased 2-fold in the presence of StGWD and ATP. No increase was observed if either StGWD or ATP were omitted (Fig. 2A). Significant starch breakdown by StISA3 alone or in combination with GWD (+ATP) could not be detected. However, starch degradation by a mixture of BAM1 and StISA3 was more than 2-fold higher than that of BAM1 alone and further doubled in the presence of ATP and GWD (Fig. 2A). Analysis of the starch breakdown products using HPAEC-PAD indicated that maltose is the exclusive product of BAM1 (Fig. 2B). In samples containing BAM1 and StISA3 maltotriose appeared as an additional product. Under conditions of simultaneous glucan phosphorylation the amounts of the respective products increased (Fig. 2B). The sample derived from starch incubated with BAM1, StISA3, StGWD, and ATP in addition contained low amounts of late eluting compounds (20–23 min), which according to our previous studies (Ritte et al., 2004) likely represent phosphooligosaccharides. Using starch that had been prelabeled with 33P we confirmed that BAM1 is not able to release phosphorylated products from starch (Fig. 3). Little radioactivity was released from 33P starch by StISA3 alone. However, the release of labeled products considerably increased if BAM1 and StISA3 acted simultaneously on starch (Fig. 3). As shown above (Fig. 2) and in previous studies (Hussain et al., 2003; Delatte et al., 2006), the debranching activity of ISA3 strongly increased if the glucan chains were shortened by β-amylase. Apparently, this also holds true for phosphorylated glucan chains.
Figure 2.
Glucan phosphorylation by GWD stimulates degradation of granular starch by BAM1. Sex1-3 starch granules were incubated in buffer with (black bars) or without (white bars) 0.25 mm ATP for 90 min. The following amounts of recombinant enzymes were added: BAM1 (7.5 μg, GST-fusion protein), StGWD (4.5 μg), StISA3 (0.45 μg). A, The starch degradation products released into the soluble phase were hydrolyzed with acid and Glc was quantified. The Glc present in control samples that lacked any recombinant protein was subtracted from all other samples. B, HPAEC-PAD analyses of the starch degradation products without acid hydrolysis. All samples contained ATP and the following recombinant proteins: GST-BAM1 (1), GST-BAM1 and StGWD (2), GST-BAM1 and StISA3 (3), GST-BAM1, StISA3, and StGWD (4). The peak labeled with an asterisk (*) comprises HEPES. Maltose (G2) or maltotriose (G3) were not detectable in samples containing either no recombinant protein or only StISA3 and/or StGWD (data not shown).
Figure 3.
Release of radioactive products from 33P-labeled starch by BAM1 and/or StISA3. 33P-labeled starch granules (equivalent to 50,000 cpm) were incubated with 4 μg GST-BAM1, 0.75 μg StISA3, or a mixture of both for 60 min. The release of label into the soluble phase was quantified. In a control lacking any recombinant protein 230 cpm were present in the supernatant. This value was subtracted from all other samples. The experiment was repeated under similar conditions with essentially the same result.
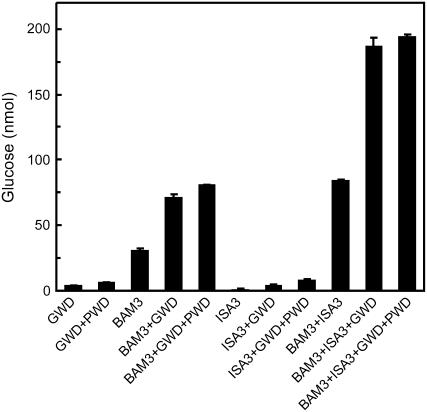
There is evidence that β-amylases and ISA3 are involved in the initial attack at the starch granule surface in vivo (Zeeman et al., 2007). Arabidopsis plants lacking ISA3 show a starch excess phenotype (Wattebled et al., 2005; Delatte et al., 2006), but those lacking BAM1 do not (Kaplan and Guy, 2005). However, elevated starch levels were reported for Arabidopsis plants with reduced expression of BAM3 (At4g17090; Kaplan and Guy, 2005) and this has been confirmed with a null mutant of BAM3 (S.M. Smith, unpublished data). To determine whether the activity of BAM3 is also affected by glucan phosphorylation the BAM3 cDNA was cloned, a vector was constructed allowing the expression of a GST-BAM3 fusion protein in E. coli, and the protein was purified (Supplemental Fig. S2). As shown in Figure 4 breakdown of sex1-3 starch granules by BAM3, alone or in combination with StISA3, increased 2- to 3-fold upon simultaneous phosphorylation of glucosyl residues by StGWD. If the second starch phosphorylating enzyme PWD was also present, the further increase in starch breakdown by BAM3 was only slight (≤15%; Fig. 4). Similar results were obtained using BAM1 (data not shown). No significant effect of PWD on glucan release by a mixture of BAM3, StISA3, and StGWD was observed (Fig. 4). As shown for BAM1 (Fig. 3), BAM3 cannot release phosphorylated products from starch and HPAEC-PAD analysis of the products revealed that maltose is the exclusive product of this enzyme (data not shown).
Figure 4.
Glucan phosphorylation by StGWD stimulates degradation of granular starch by BAM3. Sex1-3 starch granules were incubated in buffer with 0.25 mm ATP for 45 min. The following amounts of recombinant enzymes were added: GST-BAM3 (2 μg), StGWD (3 μg), AtPWD (3 μg), and StISA3 (0.45 μg). The starch degradation products released into the soluble phase were hydrolyzed with acid and Glc was quanitified. The Glc present in control samples that lacked any recombinant protein was subtracted from all other samples. Increased starch breakdown in the presence of StGWD was ATP dependent and was also observed if BAM3 devoid of the GST tag was used instead of GST-BAM3 (data not shown).
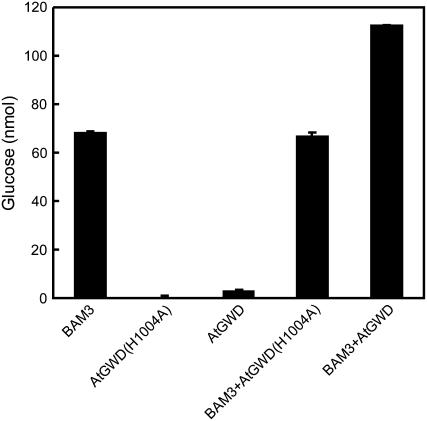
Release of maltose from starch granules by BAM3 also significantly increased in the presence of recombinant AtGWD (instead of StGWD) and ATP (Fig. 5). No stimulation of BAM3-catalyzed starch breakdown was observed, if the wild-type AtGWD was replaced by the mutant AtGWD(H1004A), in which the conserved His residue within the phosphohistidine domain (Yu et al., 2001) was replaced by Ala (Fig. 5). As has been shown before using the equivalent potato GWD mutant (Mikkelsen et al., 2004), the Arabidopsis mutant GWD was no longer capable of autophosphorylation and, consequently, glucan phosphorylation (data not shown). The binding of AtGWD(H1004A) to sex1-3 starch granules in vitro, however, was not reduced compared to the AtGWD wild-type protein (data not shown).
Figure 5.
An inactive Arabidopsis mutant GWD cannot stimulate β-amylolysis of granular starch. Sex1-3 starch granules were incubated in buffer containing 0.25 mm ATP for 60 min. The following amounts of recombinant enzymes were added: GST-BAM3 (8 μg), AtGWD (3 μg), and AtGWD(H1004A) (3 μg).
The failure of the inactive GWD mutant to accelerate BAM-catalyzed breakdown of native starch suggests strongly that phosphorylation of the glucan chains enhances the capability of BAM to attack the granular starch. Although β-amylolytic degradation of the phosphorylated chain is blocked downstream of the phosphorylation site, the introduction of a charged group could open up the densely packed amylopectin structure and render neighboring unphosphorylated chains accessible to BAM. This view is supported by the fact that the BAM3-catalyzed breakdown of solubilized sex1 starch or solubilized potato tuber starch, whose crystalline structure has already been destroyed, was not increased if these substrates were simultaneously phosphorylated by StGWD (data not shown).
β-Amylolytic Attack Leads to Increased Phosphorylation of Granular Starch by GWD
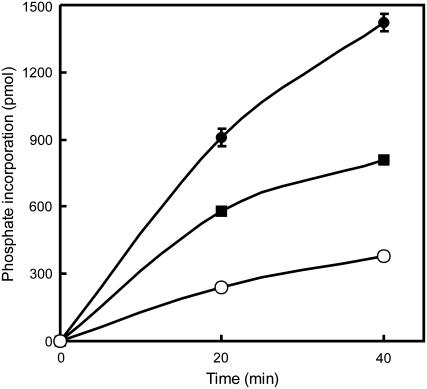
Interestingly, GWD activity not only stimulates maltose release from starch by β-amylases but the latter process, in turn, significantly accelerates starch phosphorylation by GWD. Phosphate incorporation into sex1-3 starch granules increased up to 4-fold if the granules were simultaneously attacked by BAM3 (Fig. 6) or by BAM1 (data not shown). Pretreating the starch with BAM3 also caused enhanced starch phosphorylation but the effect was less than with simultaneous β-amylolytic attack (Fig. 6).
Figure 6.
β-Amylolytic attack leads to increased phosphorylation of granular starch by GWD. sex1-3 starch granules (4 mg) were phosphorylated with 0.5 μg StGWD and 50 μm ATP containing 1 μCi [β-33P]ATP in a final volume of 0.4 mL in the presence (black circles) or absence (white circles) of 1 μg GST-BAM3 for the times indicated. In another sample 5 mg starch was pretreated with 1 μg GST-BAM3 for 40 min. GST-BAM3 was then removed by washing the starch in 2% SDS and water. Subsequently, 4 mg starch was phosphorylated with GWD only (squares).
We also tested if pretreatment of granules with GWD and ATP affects the subsequent β-amylolysis. Sex1-3 starch was prephosphorylated with GWD. GWD was then removed or, alternatively, any further glucan phosphorylation was stopped by adding EDTA in excess. Subsequently, BAM3 was added to the granule suspension and maltose release was quantified. Breakdown of the prephosphorylated granules was not or only slightly (≤15%) increased compared to nonphosphorylated control samples (data not shown). Thus, the interplay of both enzymes is needed for increased starch breakdown. The analysis of in vitro degradation of Arabidopsis wild-type leaf starch granules also indicates that simultaneous activity of GWD and BAM is required for effective starch mobilization. The wild-type granules already contained phosphate, which was incorporated during their biosynthesis. Nonetheless, breakdown of these granules by recombinant BAM1 approximately doubled in the presence of StGWD and ATP (data not shown) as observed with the phosphate-free sex1-3 granules.
DISCUSSION
Here we show that the in vitro breakdown of semicrystalline starch particles by β-amylases increases significantly if they act together with GWD. This effect was demonstrated for the Arabidopsis enzymes BAM1 and BAM3 and also for PCT-BMY1 (Scheidig et al., 2002), the potato ortholog of BAM3 (Supplemental Fig. S3A). In planta all three enzymes are located in plastids (Lao et al., 1999; Scheidig et al., 2002; Sparla et al., 2006). In contrast, the activity of a commercially available β-amylase from barley (Hordeum vulgare; Megazyme) was hardly affected by a simultaneous phosphorylation of the granules by GWD (Supplemental Fig. S3B). BAM3 and also its potato ortholog PCT-BMY1 are required for normal leaf starch degradation at night in Arabidopsis (Kaplan and Guy, 2005; S.M. Smith, unpublished data) and potato (Scheidig et al., 2002), respectively. Thus, our findings offer an explanation for the starch excess phenotype of the GWD-deficient Arabidopsis sex1 mutants and potato GWD-antisense plants (Lorberth et al., 1998; Yu et al., 2001). BAM3- as well as GWD-deficient Arabidopsis mutants are also impaired in cold induced starch breakdown in Arabidopsis leaves (Kaplan and Guy, 2005; Yano et al., 2005), which indicates that these enzymes also cooperate during cold shock. BAM1 is not essential for leaf starch degradation at night in Arabidopsis since mutants lacking this enzyme show wild-type-like starch levels in their leaves (Kaplan and Guy, 2005). BAM1 is strongly expressed during heat shock (Kaplan and Guy, 2004), osmotic, and salt stress and is probably the only β-amylase that is expressed in nonphotosynthetic tissues of Arabidopsis (Sparla et al., 2006). Thus, this enzyme might play a role in leaf starch breakdown under specific stress conditions and in starch mobilization in nonphotosynthetic tissues. The starch excess phenotype of the GWD-deficient Arabidopsis mutants is not restricted to leaves. Obviously, GWD is involved in granule degradation in all Arabidopsis organs (Caspar et al., 1991).
Interestingly, GWD activity not only stimulates β-amylolysis, but the latter process in turn causes considerably increased phosphate incorporation into starch particles by GWD. In a previous study we had shown that starch granules extracted from potato leaves that were harvested at night, were superior in vitro substrates for recombinant StGWD than granules prepared from leaves harvested during the day (Ritte et al., 2004). This also indicates that phosphorylation sites are formed or become accessible for GWD during starch degradation.
What kind of glucans within the granule may serve as substrates for BAM and GWD, respectively? The Glc polymers in native starch have different levels of molecular organization. Crystalline regions with a high level of molecular order alternate with amorphous zones with lower, but so far poorly resolved molecular organization. In vitro BAM1 and BAM3 display approximately 100-fold higher activity with solubilized starch compared to granules, whereas the activities of GWD with particulate and solubilized starch have an equal order of magnitude (data not shown). Starch is solubilized (gelatinized) by heating in an excess of water (or alkaline solutions). Gelatinization is an order-disorder phase transition and is associated with uncoiling and dissociation of double helices (Hoover, 2001). However, upon cooling some reorganization of the glucans can occur (Miles et al., 1985). The strong preference for solubilized starch indicates that BAM1 and BAM3 act on less ordered structures, for example single helices or random coil chains. In contrast, GWD can likely (also) act on substrates with a higher degree of molecular order, for example double helices. Since GWD stimulates the activity of plastidic β-amylases we propose that GWD activity causes a decrease in the molecular order of glucans in starch. This could be accomplished if GWD not only catalyzes glucan phosphorylation but also the (partial) unwinding of the glucan chains.
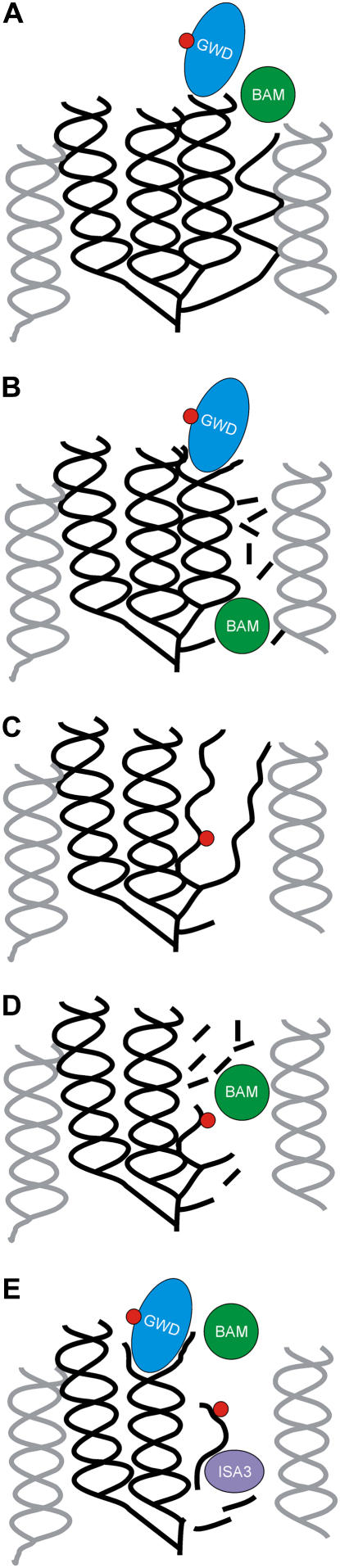
A model of the possible interplay of BAM, GWD, and ISA3 during breakdown of starch granules is depicted in Figure 7. BAM could degrade single helix chains within amylopectin and thereby render neighboring double helices accessible for GWD attack (Fig. 7, A and B). GWD activity may cause uncoiling of the double helix and phosphorylation of one chain (Fig. 7, B and C). Double helices are stabilized by hydrogen bonding between oxygen atoms of C6 and C2 atoms of glucosyl residues of the different chains (Imberty et al., 1988). Phosphorylation at the C6 position, therefore, likely hinders reassociation of the double helical structure. Given that GWD activity leads to (partial) unwinding of the double helix BAM could then degrade the single chains. Maltose release by BAM stops at (or one to two glucosyl residues ahead of) the phosphorylated glucosyl residue (Takeda and Hizukuri, 1981) and prior to the α-1,6 branch (Fig. 7D). Experiments in which sex1-3 granules were pretreated with recombinant BAM3 and then digested with StISA3 showed that BAM3 degrades chains until stubs of three to four glucosyl residues remain upstream of the α-1,6 linkages. These stubs were then removed by the recombinant potato isoamylase (Supplemental Fig. S4). ISA3 of Arabidopsis likely displays a similar substrate preference since amylopectin prepared from isa3 mutants is strongly enriched in chains of degree of polymerization 3 and (to a lesser extent) degree of polymerization 4 (Delatte et al., 2006). The purified recombinant StISA3 can also release phosphodextrins from starch if it acts concomitantly with BAM (Fig. 3). This likely also holds true for ISA3 from Arabidopsis (Supplemental Fig. S1B). Thus, ISA3 removes the unphosphorylated and phosphorylated leftovers of BAM. Due to the degradation of the glucan chains by BAM and ISA3 new space becomes available in the amylopectin molecule. The next double helix can be attacked by GWD and a new cycle of phosphorylation and degradation starts (Fig. 7E). The interplay between the different enzymes could explain why a clear effect of GWD-catalyzed starch phosphorylation on β-amylolysis was observed only when GWD and BAM acted simultaneously.
Figure 7.
Hypothetical interplay of GWD, BAM, and ISA3 during breakdown of starch granules. A, Densely packed amylopectin (double) helices. Six glucosyl residues are required for a full helix turn. Chains as depicted here have a degree of polymerization of approximately 15. The exact lengths were arbitrarily chosen but they are in a realistic order of magnitude for chains within the crystalline lamellae of starch (Smith, 2001). Gray double helices represent neighboring clusters of glucan chains. B, A single chain is degraded by BAM. The space thereby provided allows GWD attack on a neighboring double helix. C, GWD unwinds a double helix and phosphorylates one strand. The phosphate residue (red dot) stabilizes the open chain conformation. D, BAM attacks the single chains but cannot go beyond the phosphorylated residue and the α-1,6 linkages. E, ISA3 releases the remaining stubs and also phosphodextrins. GWD can attack another double helix and a new cycle of phosphorylation and degradation starts.
An alternative explanation for the absence of a significant increase of BAM activity with prephosphorylated starch would be that the glucan phosphorylation itself is not sufficient (or not required at all) for increased BAM activity but rather a protein-protein interaction between BAM and GWD is important. Binding of GWD to starch is strongly enhanced during granule breakdown (Ritte et al., 2000; Kötting et al., 2005) and, therefore, GWD might accelerate starch breakdown by targeting other proteins to starch. The mutated Arabidopsis GWD AtGWD(H1004A), which is incapable of auto and glucan phosphorylation but not affected in starch binding, however, could not stimulate BAM (Fig. 5). This does not exclude the possibility that BAM and GWD only interact if the latter is autophosphorylated. However, immunoprecipitation experiments in the presence or absence of ATP did not provide any evidence for a physical interaction between these proteins (data not shown). Furthermore, AtGWD used as bait in yeast (Saccharomyces cerevisiae) two-hybrid experiments fails to detect any BAM proteins as prey (D. Villadsen and S.M. Smith, unpublished data). Although it is still possible that protein-protein interactions are involved, it is more likely that the phosphate incorporation itself is important for the mobilization of semicrystalline starch. Phosphorylation of starch strongly increases if GWD and BAM act together (Fig. 6) in vitro and we have shown before that glucan phosphorylation in chloroplasts is also considerably accelerated during starch granule breakdown in vivo (Ritte et al., 2004).
Starch breakdown in vivo is probably more complex than depicted in Figure 7. At least in vitro, the release of phosphodextrins from starch by StISA3 was rather low and other enzymes (e.g. limit dextrinase and the plastidic α-amylase) may contribute to the release of phosphodextrins from starch in vivo. The further catabolism of the phosphooligosaccharides is unknown. Possibly phosphodextrins are first dephosphorylated and then degraded by BAMs and DPE1. It remains to be determined if the phosphatases At3g01510 (see Fig. 1) or SEX4 (DSP4, At3g52180; Kerk et al., 2006; Niittylä et al., 2006; Sokolov et al., 2006) are involved in the dephosphorylation of phosphooligosaccharides or starch. These enzymes have been classified as dual-specificity phosphatases that are able to dephosphorylate phospho-Ser/-Thr as well as phosphotyrosine residues (Fordham-Skelton et al., 2002; Kerk et al., 2006). According to Worby et al. (2006) the animal phosphatase laforin, an enzyme homologous to At3g01510 and SEX4, dephosphorylates amylopectin in vitro, and in the discussion of their results the authors mention that this also holds true for SEX4. SEX4 can bind to starch and sex4 mutants display a starch excess phenotype (Kerk et al., 2006; Niittylä et al., 2006; Sokolov et al., 2006). However, the ratio of glucosyl-6-P to glucosyl residues in starch of the sex4-3 mutant (SALK_102567, Niittylä et al., 2006) was not significantly different from wild-type starch (data not shown). More detailed studies are needed to reveal if SEX4 is involved in dephosphorylation of starch or phosphodextrins in vivo.
In addition to GWD, PWD is also required for normal starch breakdown in leaves of Arabidopsis, although the phenotype of the pwd mutants is less severe than that of the GWD-deficient sex1 mutants. In Arabidopsis both dikinases are coexpressed (Smith et al., 2004; Fettke et al., 2007) and both enzymes bind to leaf starch granules during their degradation (Kötting et al., 2005). The phosphorylation of the C3 position strictly depends on a preceding C6 phosphorylation by GWD. Possibly PWD phosphorylates chains presented by GWD activity (see Fig. 7C) and this may further stabilize an open conformation and prepare these chains for degradation. However, we observed only a small further increase of BAM3-catalyzed starch breakdown in samples containing PWD in addition to GWD (Fig. 4). This could be due to the fact that the phosphate residues themselves are barriers for BAM and that enzymes involved in dephosphorylation in vivo (see above) were possibly not included in the in vitro assays. Alternatively, different degrading enzymes might be affected by C6 or C3 phosphorylation, respectively.
Starch synthesis and degradation have evolved from the procaryotic ADP-Glc-dependent metabolism of glycogen (or semiamylopectin; Nakamura et al., 2005) in the cyanobacterial ancestor of plastids. The interplay between BAMs and ISA3 in transitory starch breakdown is reminiscent to the interaction of glycogen phosphorylase and the isoamylase-type debranching enzyme (GlcX) in E. coli. Glycogen phosphorylase catalyzes the phosphorolytic degradation of the external glycogen chains until maltotetraose stubs remain upstream of the α-1,6 linkages (Alonso-Casajús et al., 2006), which are then removed by GlcX (Dauvillée et al., 2005). The specificity of GlcX for phosphorylase-limit chains of glycogen molecules ensures that its debranching activity does not interfere with glycogen synthesis (Ball and Morell, 2003). Likewise, the interference of ISA3 activity with starch synthesis is probably prevented by its high specificity for β-limit dextrin-like structures in amylopectin. However, there are also fundamental differences in the degradation of the water-soluble glycogen and the semicrystalline starch particle. So far sequences related to the starch phosphorylating dikinases have only been detected in organisms that accumulate semicrystalline storage polysaccharides but not in glycogen synthesizing organisms like E. coli or yeast (Coppin et al., 2005). In plastids β-amylases seem to replace phosphorylase and degrade the external glucan chains in a phosphorylation-dependent manner. This view is supported by the fact that the plastidial starch phosphorylase is dispensable for starch breakdown in leaves of Arabidopsis plants grown under standard conditions (Zeeman et al., 2004). Furthermore, it has been reported that plastidial starch phosphorylase from spinach (Spinacia oleracea) is incapable of degrading native starch granules when acting on its own (Steup et al., 1983). However, as shown here, strong synergistic effects between the various enzymes have to be taken into consideration. Thus, it cannot be excluded that plastidic phosphorylase attacks starch granules if it acts together with GWD and/or other enzymes and thereby constitutes an additional degradative path in leaves of wild-type plants. Likewise, it remains to be tested whether the activities of other starch-related enzymes, such as DPE1, plastidial α-amylase, and limit dextrinase are affected by glucan phosphorylation.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were cultivated in a growth cabinet under controlled conditions (14 h light/10 h dark, 22°C/17°C, 70% relative humidity, and approximately 100 μmol quanta m−2 s−1). Rosette leaves from plants in the early flowering stage were harvested at the end of the light period.
In Vitro Phosphorylation of Starch Using Recombinant GWD and [β-33P]ATP
Starch granules were extracted from Arabidopsis leaves as described (Kötting et al., 2005). Unless otherwise stated, dried starch granules of the GWD-deficient Arabidopsis mutants sex1-3 (Yu et al., 2001) or sex1-8 (Ritte et al., 2006) were resuspended in 50 mm HEPES-KOH, pH 7.5, 1 mm EDTA, 6 mm MgCl2, 50 μm ATP, and were radioactively phosphorylated using 0.4 μg recombinant potato (Solanum tuberosum) GWD (StGWD; Ritte et al., 2002) and 0.25 μCi [β-33P]ATP (Hartmann Analytic) per milligram starch. Samples were agitated on a rotating wheel at room temperature overnight and the reaction was terminated by adding SDS to a final concentration of 2%. The starch granules were washed three times each, first with 2% (w/v) SDS, then with 2 mm ATP, and finally with water. Incorporated label was calculated by subjecting aliquots to scintillation counting.
Purification of Arabidopsis Proteins That Are Affected by Glucan Phosphorylation
Approximately 500 g of Arabidopsis wild-type (ecotype Columbia) leaves were harvested and immediately frozen in liquid nitrogen. Following grinding in a mortar two volumes of ice-cold buffer A (20 mm Tris-HCl, pH 7.8, 1 mm EDTA, 2 mm dithioerythritol [DTE], 2 mm benzamidine, 2 mm ɛ-aminocaproic acid, 0.5 mm phenylmethylsulfonylfluoride) were added. All following steps were carried out at 4°C. The leaf material was further homogenized in a Waring blender, passed through a nylon net (100 μm mesh width), and centrifuged for 20 min (20,000g) to yield the crude extract. This solution was made up to 45% ammonium sulfate and precipitated proteins were collected by centrifugation, resolved in buffer A, and desalted using PD-10 columns (GE Healthcare).
Q-Sepharose Fast-Flow Anion-Exchange Chromatography
Thirty milliliters of desalted ammonium sulfate precipitate were diluted 2-fold with buffer A and loaded onto a Q-Sepharose Fast-Flow (GE Healthcare) anion-exchange column (1.6 × 12.4 cm, 25 mL bed volume) that had been equilibrated with buffer A. After 60 mL of washing with buffer A, bound proteins were eluted using a linear 350-mL gradient of 0 to 0.75 m NaCl in buffer A at a flow rate of 2 mL min−1. Fractions containing the desired activity were pooled and concentrated using ultrafiltration (Amicon Diaflo PM30, Millipore).
Affinity Chromatography Using Amylose Resin
The concentrated sample from Q-Sepharose chromatography was buffer exchanged into buffer B (buffer A supplemented with 3 mm CaCl2, 200 mm NaCl, and 500 mm citrate) using PD-10 columns. Approximately 130 mg of protein were applied to a column (1.6 × 5.0 cm) containing 10 mL amylose resin (New England Biolabs) that had been equilibrated with buffer B. Unbound proteins were washed off the column until a stable UV baseline was reached. Active fractions were recovered by elution with buffer C (buffer A containing 3 mm CaCl2 and 50 mg/mL Maltodextrin [Sigma, 419672]), pooled, and concentrated using an Amicon Ultra-4 centrifugal filter-unit concentrator (molecular weight cutoff 10,000, Millipore). The buffer of the concentrate was exchanged by passage through a NAP-5 column (GE Healthcare) equilibrated with buffer A.
Anion-Exchange Chromatography on Mono-Q
To remove the excess maltooligosaccharides the protein fraction obtained using the amylose resin was subjected to Mono-Q 5/50 GL (GE Healthcare) anion-exchange chromatography. The column was washed with 10 mL buffer A and eluted with consecutive steps of 37.5, 300, and 750 mm NaCl in buffer A. The active fractions (300 mm NaCl step) were frozen in liquid nitrogen and stored at −80°C.
Radioactive Test: Release of 33P from Prelabeled Starch Granules
One hundred microliters of (diluted) sample was mixed with 50 μL of 33P-labeled starch (equivalent to 0.0125 μCi) suspended in 20 mm Tris-HCl, pH 7.8, 1 mm EDTA, 15 mm MgCl2, and 15 mm CaCl2. Unless otherwise stated, samples were agitated in a thermomixer for 45 min at 25°C. Reactions were terminated by adding 50 μL of a 10% (w/v) SDS solution and starch granules were pelleted by centrifugation for 5 min at 16,000g. A total of 150 μL of supernatant was mixed with 3 mL scintillation fluid and radioactivity was counted.
Nonradioactive Test: In Vitro Starch Degradation with and without Glucan Phosphorylation
Starch granules (2.5 mg, suspended in water) were mixed with the protein sample (proteins extracted from Arabidopsis leaves or recombinant proteins) and ATP and/or GWD (or PWD) as indicated. Total volume of the assays was 105 μL with final concentrations of 30 mm HEPES-KOH, pH 7.5, 5 mm MgCl2, 5 mm CaCl2, if not otherwise indicated. The buffer was supplemented with 1 mg mL−1 bovine serum albumin in experiments in which the activities of recombinant proteins were analyzed. Samples were agitated in a thermomixer at 25°C for the times indicated. Reactions were stopped by centrifugation for 1 min at 20,000g. The supernatant was centrifuged once more (5 min) to remove residual starch granules. For total hydrolysis of the released glucans, equal volumes of supernatant and 2 n HCl were mixed and incubated at 100°C for 2 h. Samples were neutralized with 2 n NaOH and Glc was quantified (Stitt et al., 1989).
Recombinant Proteins
Recombinant GWD of potato (StGWD) and PWD of Arabidopsis (AtPWD) were purified as described (Ritte et al., 2002; Kötting et al., 2005).
AtGWD, Wild Type, and Mutant H1004A
The Arabidopsis GWD (At1g10760) cDNA (Yano et al., 2005) was a generous gift from Professor I. Nishida (Saitama University, Japan). Using the cDNA as template the AtGWD sequence lacking those nucleotides encoding the putative transit peptide (1–225 bp, corresponding to amino acids 1–75) was amplified by PCR with primers containing NdeI and SalI restriction sites (forward primer: 5′-CATATGGTCCTTGCCATGGATCCTCAG-3′; reverse primer: 5′-GTCGACCACTTGTGGTCGTGTCTGGAC-3′, restriction sites underlined) using the Expand High Fidelity PCR system (Roche). The PCR product was subcloned into a pGEM-T Easy cloning vector (Promega). For expression and purification of C-terminally His-tagged AtGWD, a NdeI/SalI-AtGWD fragment (4.1 kb) was then cloned into the expression vector pET23b (Novagen). Sequencing of the resulting plasmid (pETAtGWD) confirmed the accuracy of the AtGWD gene in the pET vector. A His-to-Ala mutation at residue 1004 of AtGWD was introduced using the QuickChange site-directed mutagenesis kit (Stratagene). The plasmid pETAtGWD was used as a template and the following primers were designed to produce the site-specific mutation including a BtsI restriction site, fw. 5′-CATGCCGGATGTACTATCTGCAGTGTCTGTTCGAGCAAGAAATG-3′ and rev. 5′-CATTTCTTGCTCGAACAGACACTGCAGATAGTACATCCGGCATG-3′ (exchanged nucleotides in bold, BtsI recognition site underlined). The construct was confirmed by DNA sequencing.
For heterologous expression of wild-type or mutant AtGWD Escherichia coli BL21(DE3) cells (Novagen) were transformed with the respective recombinant plasmids and cells were grown in terrific broth (TB) medium containing 100 μg/mL ampicillin to an OD600 of approximately 0.7 before 1 mm isopropylthio-β-galactoside was added. Cells were harvested 2 h after induction, washed in 50 mm Tris-HCl, pH 7.5, and frozen at −80°C until use. Cells were resuspended in extraction buffer (20 mm NaH2PO4, 500 mm NaCl, 20 mm imidazole, 2.5 mm DTE, 1% [v/v] protease inhibitor cocktail III [Calbiochem, 539134], pH 7.4) and were lysed by sonication. Cell debris was removed by centrifugation and the supernatant was applied to a His-Trap-HP column (GE Healthcare). The His-tagged AtGWD was eluted by gradually increasing the imidazole concentration of the extraction buffer (lacking DTE and protease inhibitors) from 50 to 500 mm. Fractions with adequate amount and purity of the desired protein were combined and then concentrated and buffer exchanged into 50 mm HEPES-KOH, pH 7.5, 1 mm EDTA, 1 mm DTE using an Amicon Ultra-4 centrifugal filter-unit concentrator (MWCO 10,000, Millipore). The activities of independent AtGWD preparations using soluble potato starch as a substrate (Ritte et al., 2003) ranged from 20 to 40 nmol (mg protein)−1 min−1 and were comparable to typical activities of recombinant StGWD. The mutant AtGWD(H1004A) was inactive.
StISA3
StISA3 fused to a S-Tag (Hussain et al., 2003) was expressed in E. coli BL21(DE3) cells under conditions as described for the heterologous expression of AtGWD. StISA3 devoid of the S-Tag was purified using the S-Tag Thrombin Purification kit (Novagen) according to the manufacturer's instructions. The protein was buffer exchanged into 50 mm HEPES-KOH, pH 7.5, 1 mm EDTA, and 1 mm DTE. Aliquots were frozen in liquid nitrogen and stored at −80°C until use. Activity was analyzed with appropriate dilutions of the purified enzyme using β-limit dextrin as substrate (Hussain et al., 2003). Activities of independent preparations ranged from 30 to 40 μmol reducing sugar (mg protein)−1 min−1.
AtBAM1
To produce and amplify cDNA coding for BAM1 (BMY7, TR-BAMY, At3g23920) without the N-terminal 38 amino acids (amino acids 1–41 are the predicted transit peptide), a high fidelity tag Titan one tube reverse transcription-PCR system (Roche) was employed using gene-specific primers containing EcoRI and NotI restriction sites (forward primer: 5′-CCGGAATTCTGACACCTAAAGCAATGAA-3′; reverse primer: 5′-ACGTGCGGCCGCCACAATCTATCTCTCTA-3′; restriction sites are underlined) and total RNA extracted from Arabidopsis leaves exposed to 40°C for 1 h. The cDNA was gel purified using Wizard PCR Preps DNA purification system (Promega). EcoRI and NotI cut fragments were inserted in the vector pGEX-4T-2 (GE-Healthcare) such that GST is fused in frame at the N terminus of BAM1. E. coli strain Rosetta gami (DE3; Novagen) was transformed with the recombinant plasmid. Cells were incubated in TB medium containing 50 μg/mL ampicillin, 34 μg/mL chloramphenicol, 15 μg/mL kanamycin, 12.5 μg/mL tetracyclin at 30°C. Expression of the GST-BAM1 fusion protein was initiated by adding 1 mm isopropylthio-β-galactoside at an OD600 of approximately 0.7. Two hours later cells were harvested, washed in 50 mm Tris-HCl, pH 7.5, and frozen at −80°C until use. Cells were extracted in 10 mm Na2HPO4, 1.8 mm KH2PO4, 140 mm NaCl, 2.7 mm KCl, pH 7.3 (phosphate-buffered saline [PBS]), supplemented with 5 mm dithiothreitol (DTT). The extract was clarified by centrifugation and passed through a 0.45 μm filter. GST-tagged fusion protein was subsequently purified from the bacterial lysate using a GSTrapFF column (GE Healthcare). The sample was applied to a column that had been equilibrated in PBS plus 5 mm DTT using a peristaltic pump at a flow rate of 0.25 mL min−1. After binding, the matrix was washed with 20 volumes of PBS/DTT. Proteins were eluted with 10 mm reduced glutathione in 50 mm Tris-HCl, pH 8.0, then concentrated and buffer exchanged into 50 mm HEPES-KOH, pH 7.5, 1 mm EDTA, 2 mm DTT. Activity of the purified GST-BAM1 using soluble potato starch as a substrate was approximately 400 μmol reducing sugar (mg protein)−1 min−1.
AtBAM3
A cDNA encoding BAM3 (CT-BMY, BMY8; At4g17090) was a generous gift of Dr Jychian Chen (Academica Sinica, Taiwan). PCR primers were designed to amplify the coding region of the predicted mature protein starting at amino acid residue 86, based on ChloroP (http://www.cbs.dtu.dk/services/ChloroP/) and Lao et al. (1999). The primers used for the amplification were Bam3F and Bam3R with sequences of 5′-CATAGGGATCCGTTCCGGTGTTCGTCATGTTA-3′ and 5′-GCTCGGGATCCTTACACTAAAGCAGCCTCCT-3′, respectively, and included BamHI restriction enzyme sites (underlined) for subsequent cloning. The amplified fragment was gel purified, digested with BamHI, and cloned into pGEX-2T (GE Healthcare). E. coli strain BL21Codon-Plus (DE3)-RIL (Stratagene) was transformed with the recombinant plasmid. Cells were grown in TB medium supplemented with 100 μg/mL ampicillin and 34 μg/mL chloramphenicol. Expression and purification of the recombinant GST-BAM3 fusion protein was performed as described for AtBAM1. Activities of independent GST-BAM3 preparations with soluble potato starch ranged from 300 to 600 μmol reducing sugar (mg protein)−1 min−1.
Removal of the GST tag from purified BAM1 or BAM3 fusion protein was achieved by proteolytic cleavage. GST-tagged BAM was incubated with 10 units of biotinylated thrombin (Novagen) for 2.5 h at room temperature according to the manufacturer's instructions. The protease was subsequently removed from the solution by binding to streptavidin agarose (Novagen). Untagged BAM3 protein was separated from the free tag and uncleaved fusion protein by passage through a GSTrapFF column (GE Healthcare). Purity of the final cleavage product was verified by SDS-PAGE.
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Analytical Methods
MALDI-MS and HPAEC-PAD analyses were performed as described in Kötting et al. (2005). Activities of the GST-BAM1 and GST-BAM3 preparations were analyzed in 30 mm HEPES-KOH, pH 7.5, 5 mm MgCl2, 5 mm CaCl2, and 1 mg mL−1 bovine serum albumin using soluble potato starch as a substrate (6 mg mL−1) at 25°C. The amount of reducing sugar produced was determined according to Waffenschmidt and Jaenicke (1987).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Analysis of protein fractions purified from wild-type, bam1, isa3, and sbe3 plants.
Supplemental Figure S2. SDS-PAGE analysis of purified recombinant proteins.
Supplemental Figure S3. Breakdown of starch granules by recombinant potato PCT-BMY and a commercial barley β-amylase.
Supplemental Figure S4. StISA3 releases maltotriose and maltotetraose from starch following granule pretreatment with BAM3.
Supplementary Material
Acknowledgments
The authors are grateful to Professor Cathie Martin (John Innes Centre, Norwich, UK) for valuable discussions and critically reading the manuscript. We thank Kerstin Pusch (University of Potsdam, Germany) for excellent technical assistance and Dr. Oliver Kötting (ETH Zurich) for generation of the PWD expression vector. We are grateful to Professor Samuel Zeeman (ETH Zurich) and Dr. Thierry Delatte (ETH Zurich) for providing seeds of the homozygous mutant Atisa3-2 (GABI_KAT_280G10). We thank Dr. Susan Blauth (University of Redlands, CA), Hannah Dunstan (University of Edinburgh), and Professor Alison Smith (John Innes Centre, Norwich, UK) for provision of seeds of the homozygous mutants be3-1 (SALK_048089), bam1 (SALK_039895), and dpe1-1, respectively. The PCT-BMY1 expression vector was a kind gift of Professor Jens Kossmann (University of Stellenbosch, South Africa), Dr. James Lloyd (University of Stellenbosch, South Africa), and Dr. Andreas Scheidig (DIREVO Biotech AG, Cologne, Germany). We thank the Salk Institute and the Nottingham Arabidopsis Stock Center for provision of T-DNA insertion lines.
This work was supported by the Deutsche Forschungsgemeinschaft (grant nos. SFB 429 TP–B2 to M.S. and TP–B7 to G.R.). S.M.S. acknowledges receipt of an Australian Research Council Federation Fellowship and Discovery Grant (grant no. DP0666434) to support this research.
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Gerhard Ritte (ritte@uni-potsdam.de).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso-Casajús N, Dauvillée D, Viale AM, Munoz J, Baroja-Fernández E, Morán-Zorzano MT, Eydallin G, Ball S, Pozueta-Romero J (2006) Glycogen phosphorylase, the product of the glgP gene, catalyzes glycogen breakdown by removing glucose units from the nonreducing ends in Escherichia coli. J Bacteriol 188 5266–5272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball SG, Morell MK (2003) From bacterial glycogen to starch: understanding the biogenisis of the starch granule. Annu Rev Plant Biol 54 207–233 [DOI] [PubMed] [Google Scholar]
- Baunsgaard L, Lütken H, Mikkelsen R, Glaring MA, Pham TT, Blennow A (2005) A novel isoform of glucan, water dikinase phosphorylates pre-phosphorylated α-glucans and is involved in starch degradation in Arabidopsis. Plant J 41 595–695 [DOI] [PubMed] [Google Scholar]
- Blennow A, Engelsen SB, Nielsen TH, Baunsgaard L, Mikkelsen R (2002) Starch phosphorylation: a new front line in starch research. Trends Plant Sci 7 445–450 [DOI] [PubMed] [Google Scholar]
- Caspar T, Lin TP, Kakefuda G, Benbow L, Preiss J, Somerville C (1991) Mutants of Arabidopsis with altered regulation of starch degradation. Plant Physiol 95 1181–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppin A, Varre J-S, Lienard L, Dauvillee D, Guerardel Y, Soyer-Gobillard M-O, Buleon A, Ball S, Tomavo S (2005) Evolution of plant-like storage polysaccharides in the protozoan parasite Toxoplasma gondii argues for a red alga ancestry. J Mol Evol 60 257–267 [DOI] [PubMed] [Google Scholar]
- Critchley JH, Zeeman SC, Takaha T, Smith AM, Smith SM (2001) A critical role of disproportionating enzyme in starch breakdown is revealed by a knock-out mutation in Arabidopsis. Plant J 26 89–100 [DOI] [PubMed] [Google Scholar]
- Dauvillée D, Kinderf IS, Li Z, Kosar-Hashemi B, Samuel MS, Rampling L, Ball S, Morell MK (2005) Role of the Escherichia coli glgX gene in glycogen metabolism. J Bacteriol 187 1465–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delatte T, Umhang M, Trevisan M, Eicke S, Thorneycroft D, Smith SM, Zeeman SC (2006) Evidence for distinct mechanisms of starch granule breakdown in plants. J Biol Chem 281 12050–12059 [DOI] [PubMed] [Google Scholar]
- Dumez S, Wattebled F, Dauvillee D, Delvalle D, Planchot V, Ball SG, D'Hulst C (2006) Mutants of Arabidopsis lacking starch branching enzyme II substitute plastidial starch synthesis by cytoplasmic maltose accumulation. Plant Cell 18 2694–2709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fettke J, Eckermann N, Kötting O, Ritte G, Steup M (2007) Novel starch-related enzymes and carbohydrates. Cell Mol Biol 52 OL883–OL904 [PubMed] [Google Scholar]
- Fordham-Skelton AP, Chilley P, Lumbreras V, Reignoux S, Fenton TR, Dahm CC, Montserrat P, Gatehouse JA (2002) A novel higher plant protein tyrosine phosphatase interacts with SNF1-related protein kinases via a KIS (kinase interaction sequence) domain. Plant J 29 705–715 [DOI] [PubMed] [Google Scholar]
- Hoover R (2001) Composition, molecular structure, and physicochemical properties of tuber and root starches: a review. Carbohydr Polymer 45 253–267 [Google Scholar]
- Hussain H, Mant A, Seale R, Zeeman S, Hincliffe E, Edwards A, Hylton C, Bornemann S, Smith AM, Martin C, et al (2003) Three isoforms of isoamylase contribute different catalytic properties for debranching of potato glucans. Plant Cell 15 133–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain MH (2002) Analysis of debranching enzymes from pea and potato. PhD thesis. University of East Anglia, Norwich, UK
- Imberty A, Chanzy H, Perez S, Buleon A, Tran V (1988) The double-helical nature of the crystalline part of A-starch. J Mol Biol 201 365–378 [DOI] [PubMed] [Google Scholar]
- Kaplan F, Guy CL (2004) β-Amylase induction and the protective role of maltose during temperature shock. Plant Physiol 135 1674–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F, Guy CL (2005) RNA interference of Arabidopsis beta-amylase8 prevents maltose accumulation upon cold shock and increases sensitivity of PSII photochemical efficiency to freezing stress. Plant J 44 730–743 [DOI] [PubMed] [Google Scholar]
- Kerk D, Conley TR, Rodriguez FA, Tran HT, Nimick M, Muench DG, Moorhead GBG (2006) A chloroplast-localized dual-specificity protein phosphatase in Arabidopsis contains a phylogenetically dispersed and ancient carbohydrate-binding domain, which binds the polysaccharide starch. Plant J 46 400–413 [DOI] [PubMed] [Google Scholar]
- Kötting O, Pusch K, Tiessen A, Geigenberger P, Steup M, Ritte G (2005) Identification of a novel enzyme required for starch metabolism in Arabidopsis leaves: the phosphoglucan, water dikinase (PWD). Plant Physiol 137 242–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao NT, Schoneveld O, Mould RM, Hibberd J, Gray JC, Kavanagh TA (1999) An Arabidopsis gene encoding a chloroplast-targeted β-amylase. Plant J 20 519–527 [DOI] [PubMed] [Google Scholar]
- Lloyd JR, Kossmann J, Ritte G (2005) Leaf starch degradation comes out of the shadows. Trends Plant Sci 10 130–137 [DOI] [PubMed] [Google Scholar]
- Lorberth R, Ritte G, Willmitzer L, Kossmann J (1998) Inhibition of a starch-granule-bound protein leads to modified starch and repression of cold sweetening. Nat Biotechnol 16 473–477 [DOI] [PubMed] [Google Scholar]
- Mikkelsen R, Baunsgaard L, Blennow A (2004) Functional characterization of α-glucan, water dikinase, the starch phosphorylating enzyme. Biochem J 377 525–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen R, Mutenda KE, Mant A, Schürmann P, Blennow A (2005) α-Glucan, water dikinase (GWD): a plastidic enzyme with redox-regulated and coordinated catalytic activity and binding affinity. Proc Natl Acad Sci USA 102 1785–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles M, Morris VJ, Orford PD, Ring S (1985) The roles of amylose and amylopectin in the gelation and retrogradation of starch. Carbohydr Res 135 271–281 [Google Scholar]
- Nakamura Y, Takahashi JI, Sakurai A, Inaba Y, Suzuki E, Satoko N, Fujiwara S, Tsuzuki M, Miyashita H, Ikemoto H, et al (2005) Some cyanobacteria synthesize semi-amylopectin type α-polyglucans instead of glycogen. Plant Cell Physiol 46 539–545 [DOI] [PubMed] [Google Scholar]
- Niittylä T, Comparot-Moss S, Lue WL, Messerli G, Trevisan M, Seymour MDJ, Gatehouse JA, Villadsen D, Smith SM, Chen J, et al (2006) Similar protein phosphatases control starch metabolism in plants and glycogen metabolism in mammals. J Biol Chem 281 11815–11818 [DOI] [PubMed] [Google Scholar]
- Ritte G, Heydenreich M, Mahlow S, Haebel S, Kötting O, Steup M (2006) Phosphorylation of C6- and C3-positions of glucosyl residues in starch is catalysed by distinct dikinases. FEBS Lett 580 4872–4876 [DOI] [PubMed] [Google Scholar]
- Ritte G, Lloyd JR, Eckermann N, Rottmann A, Kossmann J, Steup M (2002) The starch-related R1 protein is an α-glucan, water dikinase. Proc Natl Acad Sci USA 99 7166–7171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritte G, Lorberth R, Steup M (2000) Reversible binding of the starch-related R1 protein to the surface of transitory starch granules. Plant J 21 387–391 [DOI] [PubMed] [Google Scholar]
- Ritte G, Scharf A, Eckermann N, Haebel S, Steup M (2004) Phosphorylation of transitory starch is increased during degradation. Plant Physiol 135 2068–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritte G, Steup M, Kossmann J, Lloyd JR (2003) Determination of the starch-phosphorylating enzyme activity in plant extracts. Planta 216 798–801 [DOI] [PubMed] [Google Scholar]
- Scheidig A, Fröhlich A, Schulze S, Lloyd JR, Kossmann J (2002) Downregulation of a chloroplast-targeted β-amylase leads to a starch-excess phenotype in leaves. Plant J 30 581–591 [DOI] [PubMed] [Google Scholar]
- Smith AM (2001) The biosynthesis of starch granules. Biomacromolecules 2 335–341 [DOI] [PubMed] [Google Scholar]
- Smith AM, Zeeman SC, Smith SM (2005) Starch degradation. Annu Rev Plant Biol 56 73–97 [DOI] [PubMed] [Google Scholar]
- Smith SM, Fulton DC, Chia T, Thorneycroft D, Chapple A, Dunstan H, Hylton C, Zeeman SC, Smith AM (2004) Diurnal changes in the transcriptome encoding enzymes of starch metabolism provide evidence for both transcriptional and posttranscriptional regulation of starch metabolism in Arabidopsis leaves. Plant Physiol 136 2687–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolov LN, Dominguez-Solis JR, Allary AL, Buchanan BB, Luan S (2006) A redox-regulated chloroplast protein phosphatase binds to starch diurnally and functions in its accumulation. Proc Natl Acad Sci USA 103 9732–9737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparla F, Costa A, Schiavo FL, Pupillo P, Trost P (2006) Redox regulation of a novel plastid-targeted β-amylase of Arabidopsis. Plant Physiol 141 840–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steup M, Robenek H, Melkonian M (1983) In vitro degradation of starch granules isolated from spinach chloroplasts. Planta 158 428–436 [DOI] [PubMed] [Google Scholar]
- Stitt M, Lilley RMcC, Gerhardt R, Heldt HW (1989) Metabolite levels in specific cells and subcellular compartments in plant leaves. Methods Enzymol 174 518–552 [Google Scholar]
- Takeda Y, Hizukuri S (1981) Re-examination of the action of sweet-potato beta-amylase on phosphorylated 1,4-α-D-glucan. Carbohydr Res 89 174–178 [Google Scholar]
- Tetlow IJ, Morell MK, Emes MJ (2004) Recent developments in understanding the regulation of starch metabolism in higher plants. J Exp Bot 55 2131–2145 [DOI] [PubMed] [Google Scholar]
- Waffenschmidt S, Jaenicke L (1987) Assay of reducing sugars in the nanomole range with 2,2′-bicinchoninate. Anal Biochem 165 337–340 [DOI] [PubMed] [Google Scholar]
- Wattebled F, Dong Y, Dumez S, Delvalle D, Planchot V, Berbezy P, Vyas D, Colonna P, Chatterjee M, Ball S, et al (2005) Mutants of Arabidopsis lacking a chloroplastic isoamylase accumulate phytoglycogen and an abnormal form of amylopectin. Plant Physiol 138 184–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worby CA, Gentry MS, Dixon JE (2006) Laforin: a dual specificity phosphatase that dephosphorylates complex carbohydrates. J Biol Chem 281 30412–30418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano R, Nakamura M, Yoneyama T, Nishida I (2005) Starch related α-glucan/water dikinase is involved in the cold induced development of freezing tolerance in Arabidopsis. Plant Physiol 138 837–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TS, Kofler H, Häusler RE, Hille D, Flügge UI, Zeeman SC, Smith AM, Kossmann J, Lloyd J, Ritte G, et al (2001) The Arabidopsis sex1 mutant is defective in the R1 protein, a general regulator of starch degradation in plants, and not in the chloroplast hexose transporter. Plant Cell 13 1907–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TS, Zeeman SC, Thorneycroft D, Fulton DC, Dunstan H, Lue WL, Hegemann B, Tung SY, Umemoto T, Chapple A, et al (2005) α-Amylase is not required for breakdown of transitory starch in Arabidopsis leaves. J Biol Chem 280 9773–9779 [DOI] [PubMed] [Google Scholar]
- Zeeman S, Smith SM, Smith AM (2007) The diurnal metabolism of leaf starch. Biochem J 401 13–28 [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Thorneycroft D, Schupp N, Chapple A, Weck M, Dunstan H, Haldimann P, Bechtold N, Smith AM, Smith SM (2004) Plastidial α-glucan phosphorylase is not required for starch degradation in Arabidopsis leaves but has a role in the tolerance of abiotic stress. Plant Physiol 135 849–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.