Abstract
Terminal differentiation is associated with repression in the expression of the proliferation potential proteins (P2P) subset of heterogeneous nuclear ribonucleoprotein (hnRNP) proteins. We report here the cloning and characterization of a 5173-bp P2P-related (P2P-R) cDNA that contains a 4214-bp open reading frame. Probes to this cDNA detect a single 8-kb mRNA in multiple murine tissues and in proliferating 3T3T cells, but not in terminally differentiated 3T3T adipocytes. Evidence that this cDNA can encode peptides with domains for hnRNP association was established by showing that such peptides are recognized by two monoclonal antibodies known to detect core hnRNP proteins, and by showing that the C130 monoclonal antibody, produced against a cDNA-derived fusion protein, also selectively detects native P2P hnRNP proteins. In addition, P2P-R cDNA-derived fusion proteins bind single-stranded nucleic acids, and a P2P-R cDNA-derived antisense oligonucleotide selectively represses P2P expression. Because terminal differentiation is associated with modulation in Rb1 function, we assayed if products of this cDNA might interact with Rb1. Evidence that the P2P-R cDNA encodes a protein domain that binds Rb1 was established using a glutathione S-transferase fusion protein to selectively precipitate Rb1 from cellular extracts. Data also show that this binding is reduced by competition with the adenovirus E1a protein, indicating that binding occurs through the “pocket” domain of Rb1. These results establish that the P2P-R cDNA encodes protein domains involved in both hnRNP association and Rb1 binding and complement recent reports that localize Rb1 to sites of RNA processing in the nucleus.
Differentiation in many cell lineages has been established to be a multistep process. This is perhaps best illustrated by analysis of the differentiation of 3T3T mesenchymal stem cells into adipocytes (1). Undifferentiated 3T3T cells first arrest their proliferation in the G1 phase of the cell cycle at a distinct state before differentiation. Associated with this process, expression of the PPARγ2 lineage specific transcription factor is induced (2). Thereafter, the C/EBP family of transcription factors are expressed followed by induction of a series of adipocyte differentiation genes that include 422, glyceraldehyde-3-phosphate dehydrogenase, lipoprotein lipase, and adipsin (3–5). The resultant adipocytes are nonterminally differentiated, because they can be induced to reinitiate proliferation and reenter the cell cycle. Adipocytes at the nonterminal state of differentiation can, however, be induced to terminally differentiate by exposure to aproliferin and thereby irreversibly lose their growth factor responsiveness (6). When terminal adipocyte differentiation occurs, a marked repression in the expression of proliferation potential proteins (P2P) is evident (7). Repression in the expression of P2Ps also has been shown to be associated with the terminal differentiation of normal human keratinocytes (7) and with the senescence of human cells (8).
P2Ps comprise a group of highly basic 35- to 40-kDa nuclear proteins that can bind to RNA and are associated with heterogeneous nuclear ribonucleoprotein (hnRNP) particles as determined by sucrose gradient sedimentation of nuclear components (7). Antibodies prepared against core hnRNPs recognize P2Ps, and two-dimensional gel electrophoresis established that P2Ps are members of the A/B class of hnRNP proteins, which are involved in RNA processing (7, 9).
Terminal differentiation also has recently been demonstrated to require the expression of the tumor suppressor protein Rb1 (10). In studies using myoblasts derived from normal animals that express Rb1 and myoblasts from transgenic animals that lack Rb1, it was established that cells lacking Rb1 cannot terminally differentiate. Instead, they are blocked at a state of nonterminal differentiation. These observations suggest that a function of the Rb1 tumor suppressor gene product may involve the control of terminal differentiation. Additionally, the Wilms tumor suppressor gene product WT1 also is involved in the terminal differentiation of renal blastema cells during neonatal development (11). Both the Rb1 and WT1 proteins also have been localized in the nucleus to sites of RNA processing (12, 13).
We therefore designed studies to clone and characterize P2P-related (P2P-R) cDNAs and now describe the P2P-R cDNA with the ability to encode domains for both Rb1 binding and hnRNP association. The P2P-R mRNA is expressed in a variety of murine tissues and in growing, undifferentiated murine 3T3T cells. However, expression of P2P-R mRNA specifically is repressed during the terminal phase of adipocyte differentiation.
MATERIALS AND METHODS
Cell Lines and Cell Culture Methods.
The BALB/c 3T3T mesenchymal stem cell line has been described previously in detail (14). Growing monolayer cultures of these cells were maintained at 37°C in 5% CO2 in DMEM (Sigma) supplemented with 10% bovine calf serum (HyClone). In some studies, undifferentiated 3T3T cells were treated with P2P-R cDNA-derived antisense (5′-CAGCAGGAGCTGTGTT-3′) or sense (5′-CTACTAAGCCATCGGC-3′) oligonucleotides at 50 μg/ml for various times to determine if P2P expression could be selectively repressed. Oligonucleotides were prepared by both St. Jude Children’s Research Hospital (Memphis, TN) and Biosynthesis (Lewisville, TX). Quiescent undifferentiated 3T3T cells were prepared by culture in DMEM containing 0.5% bovine calf serum for 3–4 days at low cell densities—i.e., 1 × 104 cells per cm2.
The human hematopoietic stem cell line K-562 also has been well characterized (15). These cells were grown as suspension cultures in RPMI 1640 medium supplemented with 10% bovine calf serum.
Cell Differentiation.
The procedure to induce 3T3T cells to undergo differentiation into adipocytes previously has been described (16). It is possible to obtain highly enriched populations of cells at various adipocyte differentiation states by using well documented culture conditions and reagents (1, 14, 16). These methods were used to prepare cell populations for the current studies.
Preparation of Cellular Lysates.
Cellular lysates were prepared as described by Kaelin et al. (17). Growing murine BALB/c 3T3T and human K562 cells were washed twice with ice-cold PBS (8 g of NaCl 0.2 g of kcl, 1.15 g of Na2HPO4·7H2O, and 0.2 g of KH2PO4 per liter of H2O, pH 7.4) and lysed for 30 min at 4°C in ice-cold EBC buffer (50 mM Tris, pH 8.0/120 mM NaCl/0.5% Nonidet P-40/200 mM sodium orthovanadate) containing 10 μg/ml each of the protease inhibitors aprotinin, leupeptin, and phenylmethylsulfonyl fluoride (Sigma). The lysates were cleared of cellular debris by centrifugation at 14,000 × g for 15 min at 4°C.
P2P-R cDNA Cloning and Sequencing.
To clone P2P-R sequences, an oligo(dT) random-primed λgt11 murine 3T3 fibroblast cDNA expression library (CLONTECH) was screened using standard procedures (18) with monoclonal antibodies AC88 or FA12. The AC88 antibody, generated against heat shock protein 90 (hsp90), cross-reacts with the P2P proteins and previously has been described (19). FA12 also recognizes P2Ps and was prepared against core hnRNP proteins (20). Clones positive for both AC88 and FA12 were identified and isolated by multiple rounds of plaque purification. The resulting P2P-R cDNAs were subcloned for dideoxynucleotide sequencing of both strands (21).
Additional 5′ P2P-R sequences were cloned using the rapid amplification of cDNA ends (5′ RACE) method (22). For RACE, gene-specific oligonucleotides were used to prime first-strand cDNA synthesis from murine 3T3T total RNA using the cDNA cycle kit (Invitrogen), and 5′-RACE was performed using a variety of primer sets. Amplified products were characterized by size analysis and cloned, and their DNA sequences were determined. Throughout this sequencing procedure periodic searches of the DNA databases using the BLAST programs were performed for related sequences. As we were completing the sequencing of the 5′ end of the P2P-R cDNA one significant homology was discovered. A human cDNA, designated RBQ1 (23), was found to have extensive homology to the 5′ region of the murine P2P-R cDNA. Therefore, primers for the 5′-most domains of RBQ-1 also were used in characterizing the P2P-R cDNA using reverse transcription-PCR.
RNA Isolation and Northern Analysis.
Total cellular RNA was isolated from growing cells, quiescent undifferentiated cells, cells at the nonterminal differentiation state, and terminally differentiated cells. Total cellular RNA (20 μg) from each sample was denatured and fractionated on formaldehyde/1.2% agarose gels and transferred to nitrocellulose filters. Hybridizations were carried out overnight at 42°C using random-primed 32P-labeled P2P-R cDNA probes. After hybridization the filters were washed and autoradiographed with intensifying screens at −70°C. Tissue-specific expression of the P2P-R mRNA was determined using a mouse multiple tissue Northern blot (CLONTECH) according to the manufacturer’s protocol.
Fusion Protein Expression for Monoclonal Antibody Production.
P2P-R cDNAs were cloned into the pET5a, -b, and -c vectors using standard procedures. Individual clones containing the cDNAs in all six possible reading frames were used for subsequent analysis. Expression was achieved using the procedure described by Studier et al. (24). For each of the cDNAs only one reading frame, which corresponded to the largest open reading frame (ORF), resulted in expression of a fusion protein antigenically related to P2Ps. These fusion proteins then were used to produce an anti-P2P-R specific monoclonal antibody, C130, at the University of Tennessee, Memphis, Molecular Resource Center Hybridoma Laboratory. The bacterial expression system, consisting of the pET5 series of expression vectors, the bacteriophage CE6, and Escherichia coli strain HMS174, was a generous gift from F. W. Studier.
Expression of P2P–GST and 6×-His–E1a Fusion Proteins.
P2P-R cDNA sequences coding for P2P-R peptides were generated using RT-PCR and ligated into the pGEX-KG vector to generate the following glutathione S-transferase (GST) fusion proteins (specific amino acid residues are given parenthetically): GST–P2P-(1–332), GST–P2P-(494–688), GST–P2P-(753–909), GST–P2P-(918–1095), and GST–P2P-(1221–1384). Expression and purification of the GST fusion proteins were performed as described (17, 25).
An E1a vector used to express the E1a protein as a 6×-His fusion protein was the generous gift of Margaret Quilan, University of Tennessee, Memphis. Expression and purification of this fusion protein was carried out using the His-Bind Kit and following the manufacturer’s protocol (Novagen).
For analysis of bound bacterial GST- or 6×-His proteins, specimens were boiled in 1× SDS sample buffer and analyzed by SDS/polyacrylamide gel electrophoresis, and then the proteins were visualized by staining with Coomassie blue.
pRb1-Binding Assay and Immunoprecipitation.
GST–P2P fusion proteins were expressed and recovered on glutathione-Sepharose beads as described above. Whole-cell lysates of K562 cells (1 × 107 cells per sample) were rocked with the beads for 1 h at 4°C and then washed five times with NETN buffer (17). The beads then were boiled in 1 × SDS loading buffer and the proteins were separated on SDS/polyacrylamide gels and transferred to nitrocellulose membranes. Competition experiments were performed by adding an excess of the 6×-His–E1a fusion protein to the cellular lysates before the addition of the GST–P2P fusion proteins. The Rb1 protein was visualized by immunoblotting using anti-Rb1 antibodies IF8 or C15 (Santa Cruz Biotechnology). These antibodies also were used to immunoprecipitate native Rb1 from the cellular lysates to serve as a positive control, following the manufacturer’s protocol.
RESULTS
Cloning and Characterization of a P2P-Related cDNA.
To clone a P2P-related cDNA, a 3T3 cDNA λgt11 library (CLONTECH) was screened using the AC88 monoclonal antibody, which detects both P2Ps and hsp90 (7, 19). AC88-positive clones were rescreened with the monoclonal antibody FA12, which was raised against core hnRNP proteins and previously has been shown to react with the P2Ps (7, 20). Two independent clones, designated clone A (1398 bp) and clone B (1943 bp), were found to be recognized by both antibodies. Nucleotide sequencing of the cDNAs showed that the 3′-most region of clone A and the 5′-most region of clone B were 100% homologous over a 863-bp region, suggesting that these were overlapping clones derived from a single RNA species. The overlapping clones were joined through a unique HindIII restriction endonuclease site in the overlapping region to generate 2478-bp cDNA clone. This includes a 1658-bp ORF and 820 bp of 3′ untranslated sequence.
Additional screens of the cDNA library using this cDNA as the probe failed to give new clones with any additional 5′ cDNA sequence. Therefore, the cDNA clone was extended toward the 5′ end using RACE methods whereby gene-specific oligonucleotides were used to prime first-strand cDNA synthesis from murine 3T3T total RNA, and 5′-RACE was performed. Amplified products were cloned, and their DNA sequences were determined. This extended the 5′ sequence by 1015 bp, and a GST–P2P fusion protein derived from this region was found to bind Rb1, as described below.
Throughout the sequencing procedure, periodic searches of the DNA databases using BLAST programs also were performed to search for related sequences, especially those encoding Rb1-binding domains. One significant homology to the 5′ region of the P2P-R cDNA was found with a human cDNA, designated RBQ1 (23). Primers to RBQ1 therefore were used to further extend the P2P-R cDNA sequence using RT-PCR methods to give a 5173-bp P2P-R cDNA (Fig. 1).
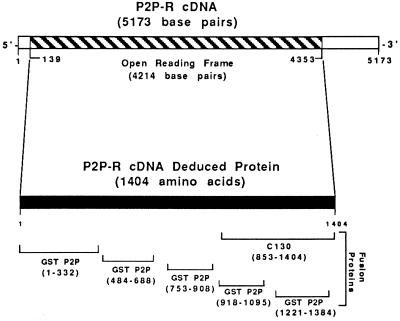
Figure 1.
Model for the P2P-R cDNA and its deduced protein. The 5173-bp P2P-R cDNA contains a 4214-bp ORF. Domains of the deduced 1404 amino acid protein expressed as fusion proteins are also shown. These include five GST–P2P fusion proteins and one β-galactosidase fusion protein designated C130. The amino acid residues of each fusion protein are given parenthetically.
Analysis of this cDNA reveals a single long ORF. The presence of two in-frame stop codons near the 5′ end and several in-frame stop codons at the 3′ end suggest that the cDNA contains the entire coding region of the gene. This ORF has the potential to code for a 1404 amino acid protein having a predicted molecular mass of 156.9 kDa (Fig. 2). This highly basic protein (pI, 9.6) has multiple potential nuclear localization signals, which agrees with our previous findings that P2Ps represent a subset of nuclear hnRNP proteins (7). In addition, computer analysis of the P2P-R cDNA-derived ORF shows a unique cysteine-rich domain near the amino terminus (amino acids 61–101), which closely resembles the consensus sequence of the “ring” class of Zn2+ finger domains (26) and another domain near the amino terminus (amino acids 79–97) that has been implicated in cell growth control—i.e., the cell division sequence motif (27).
Figure 2.
P2P-R cDNA-deduced protein consisting of 1404 amino acids. A hnRNP-associated domain is encoded by amino acids 853-1404, and Rb1-binding domain is encoded by amino acids 753–908. Potential nuclear localization signals are present between amino acids 717 and 1323 (underlined), and a cysteine-rich domain resembling a “ring” zinc finger is also present from amino acid 61 to amino acid 101 (boxed). The cell division sequence motif from amino acid 79 to amino acid 97 (bold) is also shown.
P2P-R mRNA Expression in Multiple Tissues and Repression by Terminal Adipocyte Differentiation.
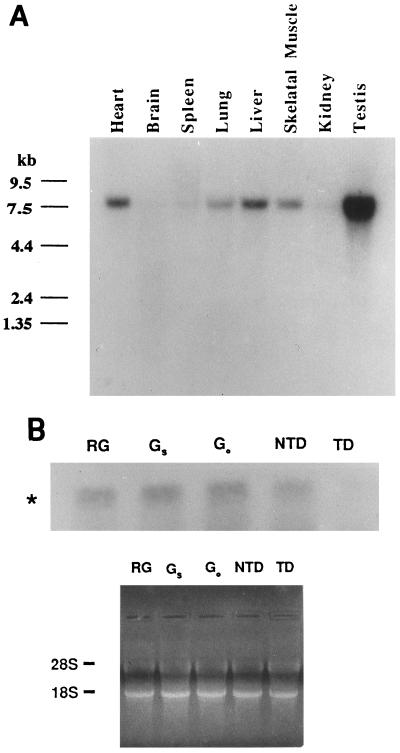
To establish the tissue distribution and the size of the P2P-R mRNA, a mouse multiple tissue Northern blot was probed with a P2P-R cDNA probe. A single 8-kb mRNA was found in all tissues examined. Very low, but detectable, levels of P2P-R mRNA were found in kidney, brain, and spleen, while moderate levels of P2P-R mRNA were found in heart, lung, liver, and skeletal muscle. The highest levels of P2P-R mRNA expression were detected in testis (Fig. 3A). The use of probes to different 3′ and 5′ P2P-R cDNA domains detected the same 8-kb RNA by Northern blotting (data not shown).
Figure 3.
Tissue distribution of the P2P-R mRNA and its specific repression by terminal adipocyte differentiation. (A) A murine multiple tissue Northern blot (CLONTECH) was analyzed using 32P-labeled random-primed P2P-R cDNA probes under high-stringency conditions. Size markers in kilobases (kb) are shown on the left. (B) Total cellular RNA (20 μg) isolated from growing undifferentiated 3T3T cells (RG), quiescent serum-starved undifferentiated 3T3T cells (Gs), quiescent predifferentiation arrested 3T3T cells (GO/GD), nonterminally differentiated 3T3T adipocytes (NTD), and terminally differentiated 3T3T adipocytes (TD) were hybridized with 32P-labeled random-primed P2P-R cDNA probes under high-stringency conditions. The 8-kb P2P-R mRNA (∗) is shown. A photograph of the ethidium bromide-stained gel before nucleic acid transfer to the nitrocellulose membrane is shown to indicate equivalent amounts of RNA in each lane.
To determine if terminal adipocyte differentiation has an effect on P2P-R mRNA expression, total RNA was isolated from rapidly growing 3T3T cells, quiescent serum-starved undifferentiated 3T3T cells, quiescent predifferentiated 3T3T cells, nonterminally differentiated 3T3T adipocytes, and terminally differentiated 3T3T adipocytes. Northern analysis was used to compare P2P-R mRNA levels in cells at these states. Fig. 3B shows that the 8-kb P2P-R mRNA is expressed in all specimens except those derived from cells at the terminal stage of adipocyte differentiation, where its expression is markedly repressed. This result agrees with our previous findings that P2P protein expression is repressed when murine 3T3T mesenchymal stem cells and normal human keratinocytes irreversibly lose their proliferative potential in association with terminal differentiation (7) or senescence (8).
Evidence That the P2P-R cDNA Encodes Peptides with hnRNP Characteristics.
The first evidence that P2P-R cDNA-encoded peptides are related to hnRNPs was established by the fact that the cDNA was isolated by screening an expression library with two monoclonal antibodies that detect core hnRNP proteins. These antibodies, which recognized different hnRNP epitopes and also react with native P2Ps, include AC88 and FA12.
To further establish a connection between the cloned cDNA and core hnRNP proteins, we generated the C130 monoclonal antibody. The carboxyl-terminal portion of the P2P-R cDNA ORF (base pairs 2695–4353) was subcloned into the pET5 series expression vectors. In this system, the cDNA was placed proximal to the bacteriophage T7 gene 10 translation initiation site such that individual plasmids were isolated containing the cDNA in each of the six reading frames in phase with the gene 10 protein product. Only one reading frame, which corresponds to the 3′ end of the large ORF (Fig. 1), resulted in expression of fusion protein antigenically related to P2Ps. The fusion protein was electroeluted from preparative gels and used to produce a P2P-R-specific monoclonal antibody at the University of Tennessee, Memphis, Molecular Resource Center Hybridoma Laboratory.
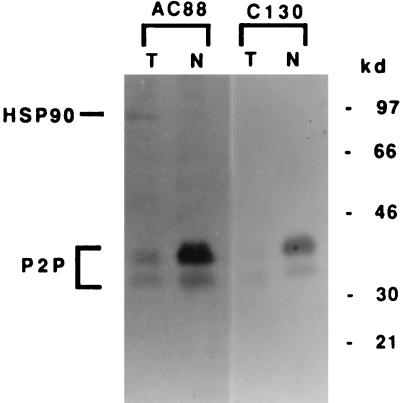
One hybridoma so generated was reactive against the purified fusion protein. The antibody, termed C130, then was used to probe 3T3T nuclear and total cell extracts by Western blot analysis. Fig. 4 shows that the C130 monoclonal antibody specifically detects native P2P proteins in a manner similar to the pattern seen with AC88. However, C130 and AC88 recognize separate epitopes, because C130 detects only P2Ps, whereas AC88 shows cross-reactivity to hsp90. These results show that P2P-R cDNA-derived peptides contain at least three hnRNP-associated epitopes detected by the monoclonal antibodies AC88, FA12, and C130.
Figure 4.
Monoclonal antibody C130 derived from a P2P-R cDNA fusion protein detects native P2P. Rapidly growing undifferentiated 3T3T total (T) cellular extracts (100 μg per lane) or nuclear (N) extracts (40 μg per lane) were separated on SDS/10% polyacrylamide gels and transferred to nitrocellulose membranes. Blots were probed with monoclonal antibody AC88 to detect native P2P proteins or with the P2P-R cDNA-derived monoclonal antibody C130. The locations of P2P proteins and hsp90 are indicated. Size standards are shown in kilodaltons.
We next evaluated if P2P-R fusions proteins could bind single-stranded nucleic acids as do hnRNPs. Of the five fusion proteins tested (Fig. 1), only two showed single-stranded DNA binding. Of these, GST–P2P-(1221–1384) showed the highest affinity. More specifically, significant single-stranded DNA binding to this fusion protein was detected after sequential buffer washes with 0.1 M NaCl, 0.1 M NaCl + 1 mg/ml heparin, and 0.2 M NaCl. This GST–P2P-(1221–1384) fusion protein was derived from the 3′ end of P2P-R in the same region that encoded the hnRNP antigen detected by the C130 monoclonal antibody.
Studies also were performed to determine the effect of a P2P-R cDNA-derived antisense oligonucleotide on the expression of native P2Ps. The results show that when undifferentiated 3T3T cells were treated with 50 μg/ml P2P-R antisense the expression of the 35- to 40-kDa P2P hnRNPs was repressed by 83%. This was determined by densitometric analysis of Western blots stained with the AC88 antibody, which detects both P2P hnRNPs and hsp90. In contrast, treatment of cells with a P2P sense oligonucleotide repressed P2P hnRNP expression by ≤6% and neither P2P-R antisense nor sense reagents had any effect on hsp90 expression.
The P2P-R cDNA Encodes a Rb1-Binding Peptide.
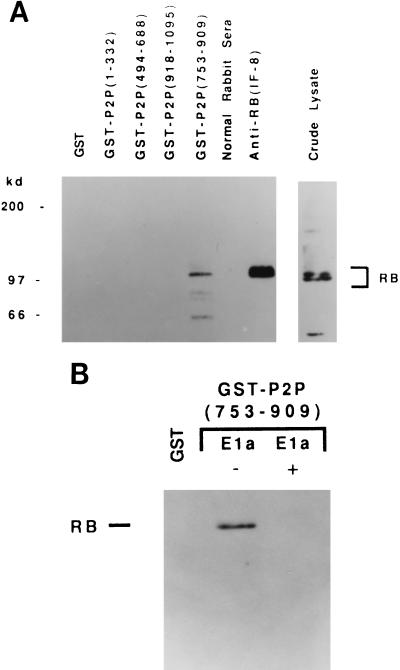
Because of the recent report showing that Rb1 is required for muscle cell terminal differentiation (10) and our data showing that P2P expression is modulated during terminal adipocyte differentiation, we performed studies to determine if P2P-R cDNA products might interact with Rb1. To accomplish this, GST–P2P fusion proteins were produced to different P2P-R cDNA domains. Cellular lysates were prepared from human K-562 hematopoietic stem cells that contain abundant Rb1 protein, and these lysates then were precipitated with each of the four GST–P2P fusion proteins—i.e., GST–P2P-(1–332), -(484–688), -(753–908), and -(918–1095) as illustrated in Fig. 1. The lysates also were precipitated with GST protein alone as a negative control in these experiments. Fig. 5A demonstrates that one fusion protein, GST–P2P-(753–909), specifically precipitates a protein that is detected by the anti-Rb1 antibody IF8.
Figure 5.
GST–P2P-(753–909) specifically binds Rb1 through the “pocket” domain. (A) Aliquots of K562 total cell lysate (1 × 107 cells per sample) were incubated with the GST leader sequence, or with GST–P2P-(1–332), GST–P2P-(494–688), GST–P2P-(918–1095), or GST–P2P-(753–909). As precipitation controls, aliquots of the K562 lysate were immunoprecipitated with anti-Rb1 antibody IF8 or normal rabbit serum. Bound proteins were separated by electrophoresis in an SDS/7% polyacrylamide gel and transferred to nitrocellulose. An aliquot of the K-562 crude lysate was included as a positive control for Western blot analysis. Proteins were visualized by probing with anti-Rb1 antibodies IF-8 or C15. (B) Binding of GST–P2P-(753–909) to Rb1 is blocked by competition with E1a protein. Aliquots of a K562 lysate were incubated as above with GST or GST–P2P-(752–909) in the presence (+E1a) or absence (−E1a) of purified adenovirus E1a protein. Bound proteins were separated by electrophoresis in an SDS/7% polyacrylamide gel and transferred to nitrocellulose. Proteins were visualized by probing with anti-Rb1 antibodies IF8 or C15.
Most proteins that associate with Rb1 bind to a region of Rb1 that has been termed the “pocket” domain (28). To determine if the interaction between Rb1 and GST–P2P-(753–909) occurs through the Rb1 pocket domain, competition experiments were conducted using purified viral E1a protein. E1a is known to bind specifically to the Rb1 pocket domain and to inhibit cellular proteins from binding to this region (29). Fig. 5B shows that the interaction between the GST–P2P-(753–909) fusion protein and Rb1 is blocked by the addition of purified E1a protein. This inhibition is specific for the E1a protein, because the addition of another protein, dihydrofolate reductase, did not block the interaction of Rb1 and the GST–P2P fusion protein (data not shown). Therefore, GST–P2P-(753–909) binds specifically to Rb1 via an interaction with the Rb1 pocket domain.
DISCUSSION
Evidence that the irreversible loss of proliferative potential is associated with repression in the expression of hnRNP-associated proteins that are involved in RNA processing was published by our research group in 1989 (7). We specifically demonstrated that the terminal differentiation of 3T3T adipocytes correlates with a markedly decreased expression of a set of proteins designated P2Ps. P2Ps were shown to have a pI of greater than 9.0, to range in size from 35 to 40 kDa, and to localize to nuclear hnRNP particles as determined using sucrose gradient sedimentation methods. Additional studies established that P2Ps are recognized by the FA12 monoclonal antibody, which detects purified core hnRNP proteins. The results of two-dimensional gel electrophoresis further established that P2Ps are type A/B hnRNP proteins. P2Ps also share an epitope in common with hsp90 as determined by use of the AC88 monoclonal antibody even though P2Ps are not heat shock proteins. Subsequently, the terminal differentiation of human keratinocytes also was shown to be associated with a marked decrease in P2P expression (7), and P2P expression was shown to markedly decrease in association with the senescence of normal human cells (8).
This paper describes the cloning and characterization of the P2P-related cDNA, P2P-R. The results of this effort define a 5173-bp cDNA containing a 4214-bp ORF encoding a highly basic (pI, 9.6) 156.9-kDa protein. Probes to the P2P-R cDNA detect a single 8-kb mRNA in murine kidney, liver, testes, lung, and other tissues and in growing 3T3T mesenchymal stem cells. In contrast, P2P-R mRNA expression is markedly decreased when 3T3T cells undergo the terminal step in the process of adipocyte differentiation. However, P2P-R mRNA expression is not repressed in nonterminally differentiated adipocytes, suggesting that regulation of P2P-R expression is associated specifically with terminal differentiation. This correlates with our previous findings that native P2P expression also is selectively repressed by terminal differentiation (7).
This paper also establishes that the P2P-R cDNA encodes domains for hnRNP association. We first showed that the P2P-R cDNA product contains three distinct epitopes in common with hnRNP core proteins. These include those epitopes recognized by AC88, FA12, and C130 monoclonal antibodies. GST–P2P-(1221–1384) fusion proteins also bind single-stranded nucleic acid, as do hnRNPs. The data showing that treatment of cells with a P2P-R cDNA-derived antisense oligonucleotide represses native P2P expression suggest a close relationship of P2P-R cDNA products with hnRNPs. This relationship could result from several possibilities. We favor the possibility that the P2P-R cDNA directly encodes 35- to 40-kDa P2Ps by a precursor–product relationship. However, P2P-R mRNA might share nucleotide sequence homology in the region that is recognized by the antisense oligonucleotide with another mRNA that encodes 35- to 40-kDa P2Ps. It is also possible that P2P-R cDNA could encode a protein that regulates the expression of native 35- to 40-kDa P2Ps such that when P2P-R antisense represses P2P-R protein levels, down-regulation of the 35- to 40-kDa P2P proteins results.
Studies next were performed to determine if Rb1 might interact with P2P-R cDNA products. This possibility was suggested by data showing that Rb1 is involved in terminal differentiation and in other growth-control mechanisms. Evidence that the P2P-R cDNA encodes a Rb1-binding protein was obtained by analysis of the Rb1 binding characteristic of GST–P2P fusion proteins. GST–P2P-(753–909) was specifically shown to bind Rb1. The fact that Rb1 binding to this fusion protein is specifically blocked by competition with E1a suggest that the binding occurs to the Rb1 pocket domain (28, 29). These data are compatible with data concerning the RBQ1 cDNA, which was selected on the basis of its ability to bind Rb1 and the fact that the RBQ1 cDNA shows significant homology to the 5′ portion of the P2P-R cDNA (23).
The deduced P2P-R cDNA product contains additional interesting domains. The first of these is a cell division sequence motif that has been proposed to be characteristic of proteins involved in the regulation of cell division (27). Examples of proteins that contain this motif include cdc25, c-myc, and several viral proteins, including E1a, E7, and simian virus 40 large T antigen. The presence of the cell division sequence motif in the P2P-R cDNA product is compatible with the evidence showing that P2Ps are involved in regulating a cell’s proliferative potential. The 5′ portion of the P2P-R cDNA also encodes a cysteine-rich region that is related to “ring” zinc fingers (30). Zinc finger domains are thought to define protein conformation characteristics that are involved in nucleic acid binding and protein–protein interactions. These attributes are compatible with the fact the P2Ps and P2P-R products bind to single-stranded DNA and that P2Ps associate with hnRNPs (31).
These data together suggest that the P2P-R cDNA can encode protein domains that are important in growth control and that can be modulated by differentiation. The fact that P2P-R cDNA products can bind Rb1 is highly significant, especially because it recently has been shown that both the Rb1 and WT1 tumor suppressor proteins localize to sites of RNA processing as do P2Ps (12, 13). Additionally, the ability of Rb1 to bind to nuclear matrix proteins (12, 13), such as p84 (12) and lamin A (32), is of interest, because hnRNP particles are also known to be associated with the nuclear matrix (33).
On the basis of these results, we propose that the ability of Rb1 and perhaps other tumor suppressor proteins to affect cotranscriptional or post-transcriptional regulation of RNA expression may be mediated through interactions with P2P-R gene products. If this proves to be true, these P2P-R gene products would be critically important in many biological and pathological processes, including growth control, differentiation, tumor suppression, and carcinogenesis.
Acknowledgments
We acknowledge the technical assistance of Robin Cox and the professional contributions of a series of associates, including Drs. D. Toft, P. Minoo, L. Solomon, and W. Sullivan, who helped with various aspects of this difficult cloning project. This research was funded by the Muirhead Chair of Excellence to R.E.S. and by the Gerwin Pathology Cancer Research Endowment.
Note Added in Proof.
While this work was under review, a highly homologous cDNA sequence was deposited in the GenBank database (accession no. U28789U28789). This cDNA was stated to encode peptides capable of binding the tumor suppressor protein p53.
Footnotes
Abbreviations: P2P, proliferation potential proteins; hnRNP, heterogeneous nuclear ribonucleoprotein; P2P-R, proliferation potential protein related; GST, glutathione S-transferase; RACE, rapid amplification of cDNA ends; ORF, open reading frame; hsp90, heat shock protein 90.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. U83913U83913).
References
- 1.Scott R E, Hoerl B J, Wille J J, Jr, Florine D L, Krawisz B R, Hun K. J Cell Biol. 1982;94:400–405. doi: 10.1083/jcb.94.2.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tontonoz P, Erding H, Spiegelman B M. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 3.Smyth M J, Sparks R L, Wharton W. J Cell Sci. 1993;106:1–9. doi: 10.1242/jcs.106.1.1. [DOI] [PubMed] [Google Scholar]
- 4.Smas C M, Sul H S. Biochem J. 1995;309:697–710. doi: 10.1042/bj3090697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McKnight S L. In: Transcriptional Regulation. McKnight S L, Yamamoto K R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. pp. 771–795. [Google Scholar]
- 6.Wier M L, Scott R E. Am J Pathol. 1986;125:546–554. [PMC free article] [PubMed] [Google Scholar]
- 7.Minoo P, Sullivan W, Solomon L R, Martin T E, Toft D O, Scott R E. J Cell Biol. 1989;109:1937–1946. doi: 10.1083/jcb.109.5.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Witte M M, Scott R E. Mol Cell Diff. 1993;1:185–195. [Google Scholar]
- 9.Dreyfuss G, Matunis M J, Pinol-Roma S, Burd C G. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 10.Schneider J W, Gu W, Zhu L, Mahdavi J, Nadal-Ginard B. Science. 1994;264:1467–1471. doi: 10.1126/science.8197461. [DOI] [PubMed] [Google Scholar]
- 11.Haber D A, Duckler A J. New Biol. 1992;4:97–106. [PubMed] [Google Scholar]
- 12.Durfee T, Mancini M A, Jones D, Elledge S J, Lee W-H. J Cell Biol. 1994;127:609–622. doi: 10.1083/jcb.127.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsson S H, Charlieu J-P, Miyagawa K, Endelkamp D, Rassoulzadegan M, Ross A, Cuzin F, van Heyningen V, Hastie N D. Cell. 1995;81:391–401. doi: 10.1016/0092-8674(95)90392-5. [DOI] [PubMed] [Google Scholar]
- 14.Krawisz B R, Scott R E. J Cell Biol. 1982;94:394–399. doi: 10.1083/jcb.94.2.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hay R, Macy M, Chen T R, McClintock P, Reid Y, editors. American Type Culture Collection Catalogue of Cell Lines and Hybridomas. 6th Ed. Rockville, MD: ATCC Press; 1988. p. 134. [Google Scholar]
- 16.Wang H, Sturtevant D, Scott R E. In: Cell Biology: A Laboratory Handbook. Celis J E, editor. I. San Diego: Academic; 1994. pp. 193–198. [Google Scholar]
- 17.Kaelin W G, Pallas D C, DeCaprio J A, Kaye F J, Livingston D M. Cell. 1991;64:521–532. doi: 10.1016/0092-8674(91)90236-r. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T, editors. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. pp. 12.16–12.24. [Google Scholar]
- 19.Toft D O, Sullivan W P, Smith D F, Beito T G, Krco C J. In: Steroid and Steroid Hormone Action. Spelsberg T C, Kumar R, editors. Boston: Nijhoff; 1987. pp. 25–39. [Google Scholar]
- 20.Leser G P, Escara-Wilke J, Martin T E. J Biol Chem. 1984;259:1827–1833. [PubMed] [Google Scholar]
- 21.Sanger F, Coulson A P, Barrell B G, Smith A J M, Roe A. J Mol Biol. 1980;143:161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- 22.Frohman M A. In: PCR Primer: A Laboratory Manual. Dieffenbach C W, Dveksler G A, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1995. pp. 381–409. [Google Scholar]
- 23.Sakai Y, Saijo M, Coelho K, Kishino T, Niikawa N, Taya Y. Genomics. 1995;30:98–101. doi: 10.1006/geno.1995.0017. [DOI] [PubMed] [Google Scholar]
- 24.Studier F W, Mofatt B A. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 25.Guan K, Dixon J E. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 26.Freemont P S, Hanson I M, Trowsdale J. Cell. 1991;64:483–484. doi: 10.1016/0092-8674(91)90229-r. [DOI] [PubMed] [Google Scholar]
- 27.Figge J, Smith T F. Nature (London) 1988;334:109. doi: 10.1038/334109a0. [DOI] [PubMed] [Google Scholar]
- 28.Fattaey A R, Helin K, Dembski M S, Dyson N, Harlow E, Vuocolo G A, Hanobik M G, Haskell K M, Oliff A, Defeo-Jones D, Jones R E. Oncogene. 1993;8:3149–3156. [PubMed] [Google Scholar]
- 29.Paggi M G, Martelli F, Fancuilli M, Felsani A, Sciacchitano S, Varmi M, Bruno T, Carapella C M, Floridi A. Cancer Res. 1994;54:1098–1104. [PubMed] [Google Scholar]
- 30.Berg J M, Shi Y. Science. 1996;271:1081–1085. doi: 10.1126/science.271.5252.1081. [DOI] [PubMed] [Google Scholar]
- 31.Minoo P, Martin T E, Riehl R M. Biochem Biophys Res Commun. 1991;176:747–755. doi: 10.1016/s0006-291x(05)80248-7. [DOI] [PubMed] [Google Scholar]
- 32.Ozaki T, Saijo M, Murakami K, Enomoto H, Taya Y, Sakiyama S. Oncogene. 1994;9:2649–2653. [PubMed] [Google Scholar]
- 33.He D, Martin T, Penman S. Proc Natl Acad Sci USA. 1991;88:7469–7473. doi: 10.1073/pnas.88.17.7469. [DOI] [PMC free article] [PubMed] [Google Scholar]