Abstract
Pathogenic and sexual development of the fungus Ustilago maydis, the causal agent of corn smut disease, is regulated by heterodimerization of two unrelated homeodomain proteins bE and bW, both encoded by the multi-allelic b mating-type locus. This complex can only be formed if the two proteins are derived from different alleles. The heterodimer is believed to function as a transcriptional regulator that binds to target sites upstream of developmentally regulated genes. We have synthesized a translational fusion in which bE is tethered to bW by a designed flexible kink region. U. maydis strains expressing this synthetic b-fusion become pathogenic for corn illustrating that the single-chain fusion substitutes for the active bE/bW heterodimer. Synthetic b-fusions in which bE and bW originate from the same allele as well as fusions deleted for the dimerization domains were shown to be active while both homeodomains were required for function. Such active fusion proteins are expected to be instrumental in the identification of pathogenicity genes.
The basidiomycete fungus Ustilago maydis is a phytopathogen causing the formation of large tumors on aerial parts of maize plants. In the infected tissue the fungus proliferates and produces a massive amount of black spores (1–4). U. maydis exists in two morphologically distinct forms: a nonpathogenic haploid phase that grows yeast-like and a filamentous dikaryon that is generated after fusion of two compatible haploid cells and that is pathogenic for corn. Formation of the dikaryon can be monitored on artificial media containing charcoal (5). For further proliferation, however, the dikaryon needs to enter the host plant.
Cell fusion, the dimorphic switch to filamentous growth, and pathogenic development are genetically controlled by two unlinked mating-type loci, the biallelic a locus and the multi-allelic b locus. A stable, infectious dikaryon can only be formed and maintained if the fusing haploid sporidia carry different alleles at both loci (6–10). The a locus codes for the components of a pheromone and pheromone-receptor-based cell recognition system. The a locus is responsible for the fusion event (11, 12) and for maintenance of the filamentous dikaryon through autocrine stimulation of the pheromone signaling cascade. The b locus is the master control locus for all further sexual and pathogenic development (8, 13, 14). The link between a and b controlled events is provided by Prf1, a high mobility group domain protein, that is activated through the pheromone signaling cascade and then provides for high levels of b gene expression (15).
The b locus codes for two homeodomain proteins bE and bW that are unrelated in sequence (16, 17) but similar in organization: a highly polymorphic N-terminal region (variable domain) is followed by a homeodomain and a conserved C terminus (see Fig. 1A). Recent experiments have shown that bE and bW must dimerize to trigger pathogenic development and that the variable domains are crucial for dimerization (18). Heterodimers can only be formed if bE and bW are derived from different alleles (i.e., in the dikaryon) but not when the proteins originate from a single allele, which explains why haploid cells are usually nonpathogenic. However, haploid strains, genetically engineered to express bE and bW proteins from different alleles, become pathogenic. The bE/bW heterodimer is thought to be the central transcription factor regulating genes involved in pathogenic development. However, until now, none of these genes have been identified. Several approaches to search for potential target genes of the bE/bW heterodimer have failed presumably because two distinct proteins are needed which have to dimerize in addition.
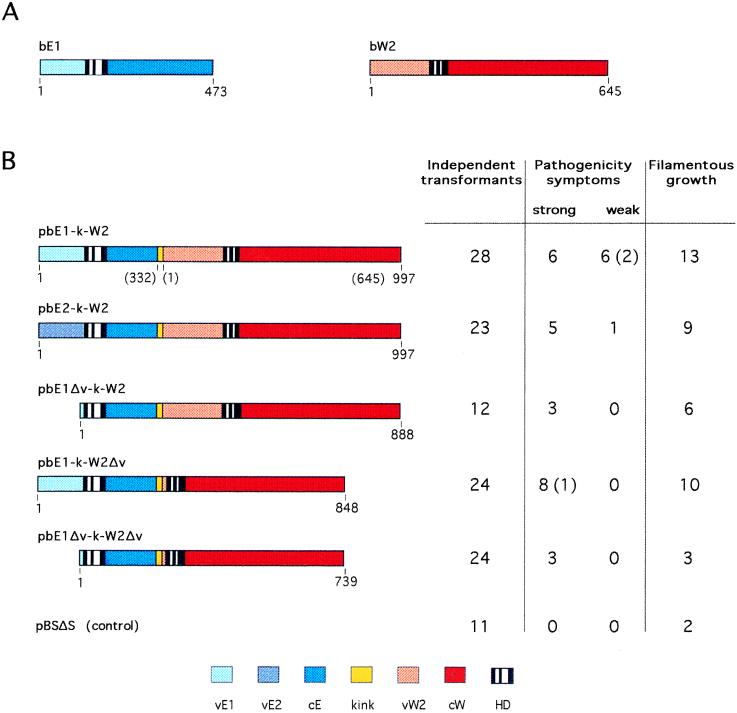
Figure 1.
Pathogenicity and filamentous growth induced by single-chain fusions of bE and bW. (A) Schematic organization of the two unrelated homeodomain proteins bE and bW encoded by the b mating-type locus of U. maydis. Variable domain, homeodomain, and constant region of bE and bW are shown in different colors as indicated below. (B) Different single-chain fusions of bE and bW were created by linking the two homeodomain proteins via a flexible kink region of 20 amino acids (shown in yellow). Plasmids encoding the translational fusion proteins shown are listed. The proteins are drawn to scale; numbers indicate the protein size in amino acids; numbers in parentheses indicate corresponding amino acid positions of the bE and bW proteins from which the portions were derived. Plasmids carrying the chimeric genes were transformed in the U. maydis Δb-strain RK2176; expression of the fusion constructs is driven by the constitutive TEF promoter. Independent transformants (numbers are listed on the left in the table) were scored for inducing pathogenicity symptoms on young maize seedlings. Strong symptoms indicate that transformants were able to induce tumor formation and anthocyanin synthesis; in the case of weak symptoms transformants caused anthocyanin synthesis only. The number of transformants that displayed filamentous growth as assayed on charcoal plates (listed on the right in the table) is indicated. Numbers in parenthesis indicate that these transformants caused pathogenicity symptoms but grew yeast-like on plates. All other pathogenic transformants showed filamentous growth.
In the present study we have synthesized single-chain fusions of the bE and bW polypeptides linked via a flexible kink region. The synthetic b-fusions were scored for activity after transformation in an U. maydis strain deleted for the b locus. Such transformants were able to induce tumors in corn, which indicates that the fusion construct substitutes for an active bE/bW heterodimer. In addition, we tested the contribution of the dimerization domains and the homeodomains to the biological activity of the single-chain fusion polypeptides.
MATERIALS AND METHODS
Plasmid Constructs.
pbE1-k-W2. To generate the bE1-k-W2 fusion, a 84-bp DNA fragment coding for the 20-amino acid kink region (GGSYPYDVPDYASLGGPSGG) including the hemagglutinin-tag (HA-tag, shown in boldface letters) (19) was amplified by PCR from plasmid pHA (A. Klippel, personal communication) with primers Pkink1 (5′-GCTCTAGACCCGGGGGCAGCTATCCTTATGACGTG-3′) and Pkink2 (5′-CGGGATCCATATGGCCTCCGGAAGGTCCTCCCAGGCT-3′). From this fragment a 74-bp XbaI–BamHI fragment was cloned in plasmid pBS KS(+) (Stratagene) in which the SmaI site was deleted to yield pBS-KINK. A 2408-bp NdeI–BamHI fragment from pTEFW2 containing an intronless copy of the bW2 gene was cloned into the corresponding sites of pBS-KINK to generate pBS-k-W2. In plasmid pTEFW2 a NotI/NdeI fragment with the promoter of the gene for the translation elongation factor 2 (TEF) (15) is fused to the intronless coding region of bW2 on an NdeI–NheI fragment in a pCM54 (20) derivative containing a NotI site. The bE1 gene was isolated as a 1274-bp NotI–SalI (blunt) fragment from plasmid pbcon (15) and was cloned into the NotI–XmaI (blunt) sites of pBS-k-W2 to yield pBSbE1-k-W2. This fragment contains the TEF promoter region (15) and a portion of the bE1 gene encoding amino acids 1–332, which have been shown to be functional in U. maydis (21). As a selection marker a 3.2-kb PvuII fragment from pCM54 carrying the hygromycin resistance gene under the control of the U. maydis hsp70 promoter (20, 22) was modified by the addition of NotI linkers and inserted in the NotI site to yield pbE1-k-W2.
pbE2-k-W2.
To construct the bE2-k-W2 fusion a 491-bp NdeI–XhoI fragment from pTRvE2 (18) that encompasses the entire variable domain and the first two helices of the homeodomain of bE2 was used to replace the corresponding fragment in pbE1-k-W2.
pbE1-k-W2Δv.
To delete the variable domain of bW2 a 334-bp DNA fragment from pBSbE1-k-W2 encompassing a deletion corresponding to amino acids 5–149 of bW2 was amplified by PCR using primers PΔvW2 (5′-GAAGATCTTTCCTACGAAGGAGCTCCCCTCAAG-3′) and PbW2256/250 (5′-CCAACACGCCGTATGATTTGC-3′). From this product a 244-bp BglII fragment was isolated and used to exchange the corresponding fragment in pbE1-k-W2 to yield pbE1-k-W2Δv.
pbE1Δv-k-W2, pbE1Δv-k-W2Δv.
The deletion of the variable domain of bE1 was accomplished by amplification of a 484-bp DNA fragment from pbE1-k-W2 that encompasses a deletion corresponding to amino acids 2–111 of bE1 with primers PΔvE1 (5′-GGCCGGTCACCATATGTGTCGAAATCTTTCGGAGGATCTTC-3′) and PbE1299/293 (5′-GGTTTAGTTTTGCGCGCTGGAT-3′). From this fragment a 165-bp BstEII–XhoI fragment was isolated and used to replace the corresponding fragments in pbE1-k-W2 and pbE1-k-W2Δv to generate pbE1Δv-k-W2 and pbE1Δv-k-W2Δv, respectively.
pbE1-k-W2ΔHD.
To generate the deletion of 10 amino acids of the bW2 homeodomain a 736-bp BglII-fragment from pUBW2Δ48–57 that contains a 30-bp deletion coding for amino acids 201–210 of bW2 (R. Schlesinger, R.K., and J.K., unpublished data) was used to replace the corresponding fragment in pbE1-k-W2.
pbE1ΔHD-k-W2, pbE1ΔHD-k-W2ΔHD.
The homeodomain of bE1 was deleted by exchanging a 392-bp MluNI–BssHII fragment from pUBE1Δ48–57 that contains a 30-bp deletion coding for amino acids 172–181 of bE1 (R. Schlesinger, R.K., and J.K., unpublished data) in pbE1-k-W2 and pbE1-k-W2ΔHD, respectively.
pBSΔS.
pBSΔS is a pBS KS(+) derivative in which the SmaI site has been deleted. The hygromycin resistance gene was inserted as a NotI fragment (see above) into the NotI site. This plasmid was used as a control.
For integrative transformation all plasmids containing the fusion constructs were linearized with SspI and ectopically integrated in the genome.
U. maydis Procedures.
Strain RK2176 (a2Δb) is a haploid U. maydis strain deleted for the bE2 and bW2 genes (15). The strain was propagated in liquid yeast extract peptone sucrose (YEPS) medium (20) at 28°C. Yeast-like growth was distinguished from the mycelial phenotype (Fuz+) on potato dextrose (PD) plates containing 1.5% potato dextrose (Difco), 2% agar, and 1% activated charcoal (5). Transformation was performed as described (17, 18).
Pathogenicity Tests.
To assay for pathogenicity cultures of independent transformants containing the fusion constructs were injected with a syringe into 1-week-old corn seedlings of the variety Early Golden Bantam. For each transformant eight plants were infected. Plants were propagated in a 14-h light (at 28°C) and 10-h dark (at 18°C) cycle. Pathogenicity symptoms such as tumor formation (Tum+) and anthocyanin biosynthesis (Ac+) were recorded 6–10 days after inoculation. When none of the eight plants infected by a given strain displayed an Ac+ or Tum+ phenotype, this was recorded as nonpathogenic. Strong symptoms indicate that at least one of the eight plants infected developed tumors and some of the others displayed an Ac+ phenotype. In the experiments recorded here at least four of the plants infected by a given strain developed tumors. Weak symptoms indicate that none of the infected plants developed tumors but at least two developed an Ac+ phenotype.
RESULTS
Single-Chain Fusions of bE and bW Homeodomain Proteins Are Active in U. maydis.
To circumvent the need for heterodimer formation of the b proteins and to facilitate the application of in vitro approaches we constructed a single-chain fusion bE1-k-W2. In this fusion two homeodomain proteins bE1 and bW2 (Fig. 1A) were linked to each other in a head-to-tail arrangement via a 20-amino acid tether segment (Fig. 1B). The flexible kink region was designed to include an hemagglutinin-tag flanked by stretches of small amino acids commonly found between domains existing in nature (23). The coding region of bE1-k-W2 was fused to the TEF1 promoter (15) which drives strong constitutive expression in U. maydis. After transformation into the haploid strain RK2176 lacking the b locus (15), hygromycin-resistant transformants, which must contain ectopic integrations of the transforming DNA, were selected and assayed for filamentous growth and for pathogenicity on young maize seedlings. About 20% of all independent transformants tested were able to induce tumors, and an additional 20% displayed weak pathogenicity symptoms (Fig. 1). This indicates that the synthetic single-chain fusion bE1-k-W2 is fully functional in vivo. Because activity depends on the integration of the entire b coding region plus the region encoding the hygromycin resistance, it is likely that the nonpathogenic transformants have integrated truncated fragments of the transforming DNA. For some transformants this was confirmed by PCR (data not shown). In addition, the DNA may have become integrated at sites where it is not expressed due to position effects.
There are three possibilities to rationalize the activity of this fusion protein: (i) an intramolecular interaction could occur between the variable domains of the bE and bW, (ii) intermolecular interaction could involve parts of the variable domains of bE and bW of two fusion protein molecules, and (iii) the fusion protein could be active without contacts between the dimerization domains. To discriminate between these possibilities we constructed a second fusion bE2-k-W2 in which both proteins were derived from the same allele (Fig. 1B). In nature bE2 and bW2 cannot dimerize and are therefore inactive (17, 18). Interestingly, the bE2-k-W2 fusion also promoted pathogenic development when introduced into a haploid strain deleted for the b locus (Fig. 1B). Thus the requirement for recognition of a compatible partner through contacts in the variable domains has been bypassed in the synthetic fusion molecules.
Role of the Dimerization Domains.
To investigate whether the variable domains of the b proteins are required at all in the fusion protein we constructed single-chain b-fusions in which the variable domains of bE1, bW2, or both protein moieties were deleted in their entirety (see bE1Δv-k-W2, bE1-k-W2Δv, and bE1Δv-k-W2Δv in Fig. 1B). The deletion ended 10 and 14 amino acids before the homeodomain motifs in bE and bW, respectively. All three synthetic fusion constructs were able to elicit tumor formation after transformation in a haploid b-null strain (Fig. 1). Notably, the tumors induced by such strains were significantly larger than tumors induced by strains expressing the full-length bE1-k-W2 fusion (data not shown). This supports the view that the variable domains govern primarily the dimerization process. In addition, they may effect the activity of the heterodimer by regulating the efficiency of dimerization. Once the two proteins are tethered as in the single-chain fusions the variable domains become dispensable for sexual and pathogenic development.
Role of the Homeodomains in the bE-k-W Fusion.
Next we established whether in the bE1-k-W2 single-chain fusion both homeodomains are necessary to confer activity. We deleted 10 amino acids of helix 3 of the homeodomain motifs in bE1 (bE1ΔHD-k-W2), bW2 (bE1-k-W2ΔHD), or both (bE1ΔHD-k-W2ΔHD). The deletion encompasses amino acids from position 48–57 (WFINARRRSG in bE1 and WFQNRRNRKG in bW2) according to the standard nomenclature of the eukaryotic homeodomain motif (24). The introduction of respective deletions into wild-type bE and bW genes had already been shown to interfere with b-dependent development (R. Schlesinger, R.K., and J.K., unpublished data). As can be seen from Table 1 all three types of deletions in the single-chain fusions rendered transformants nonpathogenic, indicating a crucial role for both homeodomains in triggering pathogenicity. This could imply that both homeodomains are directly involved in specific DNA binding; alternatively, one of the homeodomains could fulfill a more structural role in enabling another functional domain to adopt a critical conformation.
Table 1.
Homeodomain deletions in the bE1-k-W2 fusion
| Plasmid | Independent transformants in U. maydis (a2Δb) | Pathogenicity symptoms*
|
Filamentous growth† | |
|---|---|---|---|---|
| Strong | Weak | |||
| pbE1ΔHD-k-W2 | 20 | 0 | 0 | 3 |
| pbE1-k-W2ΔHD | 23 | 0 | 0 | 3 |
| pE1ΔHD-k-W2ΔHD | 15 | 0 | 0 | 2 |
Strong and weak pathogenicity symptoms scored as tumor formation and anthocyanin synthesis, respectively, were assayed as described.
Filamentous growth was assayed by spotting cultures of the respective transformant on charcoal plates (see Material and Methods).
Pathogenicity and Filamentous Growth.
Heterokaryons resulting from cell fusion of two compatible haploid strains are usually able to induce tumors in corn plants (Tum+ phenotype) and display filamentous growth on charcoal plates (Fuz+ phenotype) (4). In contrast, diploid strains carrying different b alleles but identical a alleles are pathogenic but grow yeast-like (3). The latter holds true also for haploid strains with the one a allele and a chimeric b locus consisting of the bE1 and bW2 gene (25). From recent experiments it is known that filamentous growth requires high expression of the b genes (15). This is accomplished through an activated pheromone signaling cascade and the activation of Prf1 (ref. 15; also see the Introduction). Therefore, transformants expressing biologically active single-chain fusion b proteins in the haploid strain RK2176 were expected either to display a Fuz−/Tum+ or a Fuz+/Tum+ phenotype, depending on the level of transgene expression. In transformants the expression level can be influenced by the copy number of the integrated plasmid and by position effects. The majority of those transformants that were pathogenic displayed the Tum+/Fuz+ phenotype, indicating that in most cases the expression driven by the constitutive TEF1 promoter is sufficient to initiate filamentous growth (Fig. 1). Some transformants showed a Fuz−/Tum+ phenotype; in these cases the expression of the transformed gene is presumably too low to cause filamentous growth. Transformants expressing b-fusion proteins with no biological activity were expected to have a Tum−/Fuz− phenotype, which was observed for the majority of these transformants (Fig. 1). However, in all transformations we obtained nonpathogenic transformants that grew filamentous on plates (Fig. 1 and Table 1). In particular, this class of transformants was also obtained with the control plasmid pBSΔS which does not contain a b gene. This effect is most likely caused by instability of strain RK2176; even in untransformed cells of this strain Fuz+ colonies appeared spontaneously (data not shown). Thus, the only reliable test for functionality of the chimeric b genes in U. maydis is the assay for pathogenicity of respective transformants.
DISCUSSION
The dimerization of the unrelated homeodomain proteins bE and bW via their variable domain is the key regulatory event for sexual and pathogenic development in U. maydis. Here we present data showing that a single-chain fusion of the two proteins can substitute for the natural heterodimer. Such linked dimers have already been described for single-chain antigen-binding proteins consisting of an antibody variable light chain sequence tethered to a variable heavy chain sequence (26), for enzyme subunits from human superoxide dismutase (27), and for gene V protein from bacteriophage f1 that binds cooperatively to single-stranded nucleic acids (28). These linked dimers could all be synthesized in Escherichia coli and showed nearly normal enzymatic activity and/or specificity.
The variable domains of the bE and bW proteins have been shown to serve as dimerization domains (18). It was unclear, however, whether these domains play additional roles essential for biological activity of the complex. One attractive possibility was that in the heterodimer the variable domains could induce the correct spacing of the two homeodomains as has been described for various homo- or heterodimeric eukaryotic transcription factors (29, 30). Since in U. maydis both dimerization domains in the synthetic b-fusion polypeptide can be deleted without affecting biological activity, we consider it unlikely that these domains have additional functions besides dimerization. This situation may be analogous to the one found for the homeodomain proteins MATa1 and MATα2 in yeast. In the MATa1/MATα2 heterodimer the coiled–coil dimerization motifs are separated from the homeodomains by a relative long flexible spacer, suggesting minor effects of the coiled–coil motifs on the orientation of the homeodomains (31). Furthermore, in the synthetic bE-k-W fusion proteins described in this paper the length of the tether segment linking the bE with the bW homeodomain can be of variable length (333 amino acids in bE1-k-W2; 183 amino acids in bE1-k-W2Δv and bE1Δv-k-W2Δv). Two fusion constructs with linkers of 157 and 132 amino acids, respectively, were also functional (A. Grandel and T.R., unpublished data). Thus, it is obvious that no defined distance between the two homeodomains is needed. This is in line with the assertion that this region does not have a structural role in homeodomain spacing. It will be interesting, however, to analyze whether further length reductions of the spacer between the two homeodomains are tolerated.
In the b-fusion polypeptide both homeodomains are needed for function, and this is also true for the native bE1 and bW2 protein (R. Schlesinger, R.K., and J.K., unpublished data). Multi-allelic homeodomain proteins have been shown to control development also in other basidiomycetes fungi like Coprinus cinereus and Schizophyllum commune. In these two fungi the molecular analysis of the multi-allelic mating-type loci has revealed that they contain several gene pairs encoding homeodomain proteins (see ref. 32). All these gene pairs act independently of each other, and the homeodomain proteins specified by one allele are unable to interact. In the dikaryon the formation of one active heterodimer is sufficient for the initiation of sexual development. As in U. maydis, heterodimerization of two homeodomain proteins occurs via their N-terminal variable domains (33, 34). In contrast to U. maydis, however, it has been shown for C. cinereus and S. commune that in the heterodimeric complex only one of the homeodomains is required for biological activity (35, 36).
Interestingly, in C. cinereus rare dominant mutations resulting in constitutive promotion of sexual development without mating have been isolated (37). One of these mutations was analyzed molecularly and was shown to have created a chimeric homeobox gene through an illegitimate recombination event (38). In the resulting fusion protein the N-terminal 387 amino acids of one homeodomain protein (corresponding to the variable domain, homeodomain, and part of the constant region of bW in U. maydis) were linked to the C-terminal 394 amino acids of the other (corresponding to the constant region of bE). This fusion contains only a single homeodomain that is the one critical for function also in the heterodimer (36). Furthermore, the chimeric gene fusion in C. cinereus still triggers sexual development when the single residual N-terminal dimerization domain is deleted (33), which is comparable to the bE1Δv-k-W2Δv fusion in U. maydis.
In the spontaneously derived chimeric homeodomain gene of C. cinereus functional domains of two distinct proteins are newly combined while other domains of the progenitor genes are deleted. Interestingly, attempts to engineer biologically active synthetic fusions of the respective two homeodomain genes were not very successful: from six different fusion proteins generated only one was found that displayed a very weak activity while the others were inactive (ref. 36; R. N. Asante-Owusu and L. A. Casselton, personal communication). Thus, in C. cinereus the positioning of the fusion point appears crucial for function. In U. maydis the bE an bW proteins were linked through a flexible kink region. It is quite likely that this flexible linker bypasses the requirement for a defined fusion point between the two proteins, and as long as no functional domains are deleted, the fusion protein remains biologically active.
The single-chain bE-k-W fusion proteins are expected to provide invaluable tools for the in vivo and in vitro search of b-target sequences and should enable us to gain new insights into pathways regulating pathogenic and sexual development in the large group of basidiomycete fungi. Once such genes have been isolated the synthetic b-fusions appear ideally suited to map functional domains and to determine whether these have a positive or negative effect on transcription.
Acknowledgments
We thank A. Jamnischek for plasmid constructions, A. Travers for comments on the manuscript, and K. H. Braun and J. Görl for providing plants for pathogenicity tests. This work was supported by grants from the Deutsche Forschungsgemeinschaft through SFB 190 and through the Leibniz program.
References
- 1.Christensen, J. J. (1963) Am. Phytopath. Soc. Monogr. 2.
- 2.Banuett F, Herskowitz I. Adv Plant Pathol. 1988;6:427–455. [Google Scholar]
- 3.Banuett F. Trends Genet. 1992;8:174–180. doi: 10.1016/0168-9525(92)90220-x. [DOI] [PubMed] [Google Scholar]
- 4.Banuett F. Annu Rev Genet. 1995;29:179–208. doi: 10.1146/annurev.ge.29.120195.001143. [DOI] [PubMed] [Google Scholar]
- 5.Holliday R. In: Handbook of Genetics. King R C, editor. Vol. 1. New York: Plenum; 1974. pp. 575–595. [Google Scholar]
- 6.Rowell J B, DeVay J E. Phytopathology. 1954;44:356–362. [Google Scholar]
- 7.Rowell J B. Phytopathology. 1955;45:370–374. [Google Scholar]
- 8.Holliday R. Genet Res. 1961;2:201–230. [Google Scholar]
- 9.Puhalla J E. Genet Res. 1970;16:229–232. [Google Scholar]
- 10.Day P R, Anagnostakis S L, Puhalla J E. Proc Natl Acad Sci USA. 1971;68:533–535. doi: 10.1073/pnas.68.3.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bölker M, Urban M, Kahmann R. Cell. 1992;68:441–450. doi: 10.1016/0092-8674(92)90182-c. [DOI] [PubMed] [Google Scholar]
- 12.Spellig T, Bölker M, Lottspeich F, Frank R W, Kahmann R. EMBO J. 1994;13:1620–1627. doi: 10.1002/j.1460-2075.1994.tb06425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puhalla J E. Genetics. 1968;60:461–474. doi: 10.1093/genetics/60.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banuett F, Herskowitz I. Proc Natl Acad Sci USA. 1989;86:5878–5882. doi: 10.1073/pnas.86.15.5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartmann H A, Kahmann R, Bölker M. EMBO J. 1986;15:1632–1641. [PMC free article] [PubMed] [Google Scholar]
- 16.Schulz B, Banuett F, Dahl M, Schlesinger R, Schäfer W, Martin T,, Herskowitz I, Kahmann R. Cell. 1990;60:295–306. doi: 10.1016/0092-8674(90)90744-y. [DOI] [PubMed] [Google Scholar]
- 17.Gillissen B, Bergemann J, Sandmann C, Schroeer B, Bölker M, Kahmann R. Cell. 1992;68:1–20. doi: 10.1016/0092-8674(92)90141-x. [DOI] [PubMed] [Google Scholar]
- 18.Kämper J, Reichmann M, Romeis T, Bölker M, Kahmann R. Cell. 1995;81:73–83. doi: 10.1016/0092-8674(95)90372-0. [DOI] [PubMed] [Google Scholar]
- 19.Wilson I A, Niman H L, Houghten R A, Cherenson A R, Connolly M L, Lerner R A. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- 20.Tsukuda T, Carleton S, Fotheringham S, Holloman W K. Mol Cell Biol. 1988;8:3703–3709. doi: 10.1128/mcb.8.9.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kahmann R, Romeis T, Bölker M, Kämper J. Curr Opin Genet Dev. 1995;5:559–564. doi: 10.1016/0959-437x(95)80023-9. [DOI] [PubMed] [Google Scholar]
- 22.Wang K J, Holden D W, Leong S A. Proc Natl Acad Sci USA. 1988;85:865–869. doi: 10.1073/pnas.85.3.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Argos P. J Biol Chem. 1990;211:943–958. doi: 10.1016/0022-2836(90)90085-Z. [DOI] [PubMed] [Google Scholar]
- 24.Bürglin T R. In: Guidebook to the Homeobox Genes. Duboule D, editor. Oxford: Oxford Univ. Press; 1994. pp. 27–71. [Google Scholar]
- 25.Bölker M, Genin S, Lehmler C, Kahmann R. Can J Bot. 1994;73:320–325. [Google Scholar]
- 26.Bird R E, Hardman K D, Jacobson J W, Johnson S, Kaufman B M, Lee S M, Lee T, Pope S H, Riordan G S, Whitlow M. Science. 1988;242:423–426. doi: 10.1126/science.3140379. [DOI] [PubMed] [Google Scholar]
- 27.Hallewell R A, Laria I, Tabtizi A, Carlin G, Getzoff E D, Tainer J A, Cousens L S, Mullenbach G T. J Biol Chem. 1989;264:5260–5268. [PubMed] [Google Scholar]
- 28.Liang H, Sandberg W S, Terwilliger T C. Proc Natl Acad Sci USA. 1993;90:7010–7014. doi: 10.1073/pnas.90.15.7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sessa G, Morelli G, Ruberti I. EMBO J. 1993;12:3507–3517. doi: 10.1002/j.1460-2075.1993.tb06025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beckmann H, Kadesch T. Genes Dev. 1991;5:1057–1066. doi: 10.1101/gad.5.6.1057. [DOI] [PubMed] [Google Scholar]
- 31.Ho C-Y, Adamson J G, Hodges R S, Smith M. EMBO J. 1994;13:1403–1413. doi: 10.1002/j.1460-2075.1994.tb06394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Casselton L A, Kües U. In: The Mycota. Wessels J G H, Meinhardt F, editors. Vol. 1. Berlin: Springer; 1994. pp. 307–321. [Google Scholar]
- 33.Banham A H, Asante-Owusu R N, Göttgens B, Thompson S A J, Kingsnorth C S, Mellor E J C, Casselton L A. Plant Cell. 1995;7:773–783. doi: 10.1105/tpc.7.6.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Magae Y, Novotny C, Ullrich R. Biochem Biophys Res Commun. 1995;211:1071–1076. doi: 10.1006/bbrc.1995.1920. [DOI] [PubMed] [Google Scholar]
- 35.Luo Y, Ullrich R C, Novotny C P. Mol Gen Genet. 1994;244:318–324. doi: 10.1007/BF00285460. [DOI] [PubMed] [Google Scholar]
- 36.Asante-Owusu R N, Banham A H, Bönhert H U, Mellor E J C, Casselton L A. Gene. 1996;172:25–31. doi: 10.1016/0378-1119(96)00177-1. [DOI] [PubMed] [Google Scholar]
- 37.Day P R. Genet Res. 1963;4:55–65. [Google Scholar]
- 38.Kües U, Göttgens B, Stratmann R, Richardson W V J, O’Shea S F, Casselton L A. EMBO J. 1994;13:4054–4059. doi: 10.1002/j.1460-2075.1994.tb06722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]