Abstract
Biochemical and biological activities of Myc oncoproteins are highly dependent upon their association with another basic region helix–loop–helix/leucine zipper (bHLH/LZ) protein, Max. Our previous observation that the DNA-binding/dimerization region of Max is absolutely conserved throughout vertebrate evolution provided the basis for a yeast two-hybrid interaction screen that led to the isolation of the Drosophila Myc (dMyc1) protein. Structural conservation in regions of known functional significance is consistent with the ability of dMyc1 to interact with vertebrate Max, to transactivate gene expression in yeast cells, and to cooperate with activated H-RAS to effect the malignant transformation of primary mammalian cells. The ability of P-element-mediated ectopic expression of dmyc1 to reverse a subset of the phenotypic alterations associated with the diminutive mutation suggests that diminutive may correspond to dmyc1. This finding, along with the localization of dmyc1 expression to zones of high proliferative activity in the embryo, implicates dMyc1 as an integral regulator of Drosophila growth and development.
Over the past decade, extensive efforts directed toward functional analysis of the Myc family of nuclear oncoproteins have pointed to their pivotal roles in the regulation of cellular proliferation, differentiation, and apoptosis (for reviews see refs. 1 and 2). In modulating these processes, Myc acts in part as a sequence-specific transcription factor in the form of a heterodimer with another basic region/helix–loop–helix/leucine zipper (bHLH/LZ) protein, Max (3–8). Max also can serve an antithetical role in the Myc pathway through its ability to heterodimerize with an expanding family of repressive bHLH/LZ proteins, whose members include Mad1, -3, and -4 and Mxi1 (9–11). These Mad/Max and Mxi/Max complexes antagonize Myc-induced transactivation and transformation by binding to the Myc/Max consensus sites and by tethering the Sin3 adapter protein, which serves to recruit active repressors affecting RNA polymerase II activity and chromatin structure (refs. 9 and 11–14, and L. Alland and R.A.D., unpublished observations).
Much of our understanding of the biological actions of vertebrate Myc family proteins (c-, N-, and L-Myc) has come from analysis of their oncogenic actions and developmental properties. All three myc family genes can cooperate with a mutant H-RAS gene to transform early-passage rat embryo fibroblasts (REF) and can generate tumors when overexpressed in transgenic mice (for review see ref. 15). The use of dominant-negative mutant forms of Myc has revealed that members of the Myc family function through common genetic pathways to transform cells and are linked to components governing progression through the G1 phase of the cell cycle (16). A role for the Myc family in normal development is supported by its dynamic pattern of stage- and cell type-specific expression (for review see ref. 17). During development, myc family gene expression is highest during embryonic stages and is down-regulated as mature organ systems become growth-arrested and terminally differentiated (18). A detailed analysis of c-, N-, and L-myc gene expression in midgestation coupled with gain-of-function studies performed in transgenic mice has supported the view that c-Myc plays a role in cellular proliferation, whereas N- and L-Myc are more closely linked to processes of differentiation (for reviews see refs. 15 and 17). Although L-Myc appears to be dispensable for gross morphological development (19), the loss of N- or c-Myc function is associated with midgestational embryonic lethality (refs. 20–23 and H.-W. Lee and R.A.D., unpublished observations). Mice devoid of N-Myc exhibit a marked delay in development as well as a decrease in size and diminished cellularity of organs that normally express abundant levels of N-myc mRNA during development (20–22). Homozygous null c-myc embryos exhibit a marked reduction in embryo size and a generalized delay in the early development of multiple organs (ref. 23 and H.-W. Lee and R.A.D., unpublished observations).
The above insights on Myc function in cancer and development have been complemented recently by several mechanistic and genetic clues as to how Myc may effect its activities on the molecular level through transactivation of specific gene targets and interaction with associated proteins. For example, Myc’s direct regulation of genes, such as those encoding ornithine decarboxylase (24) and the phosphatase cdc25A (25), suggests a role in DNA synthesis and G1 progression, respectively. Additionally, Myc has been shown to associate with the Rb-related protein p107 (26), the RNA polymerase II-associated TATA box-binding protein (27), and the general transcription factor YY1 (28), interactions consistent with a role in growth control and transcriptional regulation. Despite these advances, the limited repertoire of Myc targets on the gene and protein levels and the limited view of how Myc functions downstream of mitogenic and differentiative signals have hampered the forging of a clear genetic link between Myc and specific physiological pathways of growth, differentiation, and death.
Among the attempts to overcome this hurdle include those that have characterized Myc superfamily homologues in organisms that are more amenable to genetic analysis and experimental manipulation. Such studies in the lower vertebrates Xenopus laevis (29–33) and Brachydanio rerio (zebrafish) (34) as well as the invertebrate Asterias vulgaris (sea star) (35) have demonstrated striking structural and functional conservation over large evolutionary distances for many of the Myc superfamily members. In particular, the Max DNA binding/dimerization domain was found to be absolutely conserved from teleosts to man (34), leading us and others to speculate that intense structural constraints may be imposed on Max by its requirement for interaction with many different Myc superfamily proteins (34, 36). This observation provided the basis for the successful yeast two-hybrid interaction screening strategy described here that resulted in the isolation of a homologue of Myc in Drosophila melanogaster.
MATERIALS AND METHODS
Two-Hybrid Screens and Structural Analysis of Interacting Proteins.
For two-hybrid screening (37), mouse MaxΔ9 was fused in frame to LexA in the vector pBTM116 as reported previously (38) and tested for lack of autonomous activation of the yeast L40 strain reporter genes. The Drosophila embryonic MATCHMAKER cDNA library was obtained from CLONTECH and screened as described (13). Small-scale transformations to test for specificity and qualitative β-galactosidase filter assays were performed as described (38, 39). Nucleotide sequencing of two-hybrid isolates that showed interaction specificity for the bait was performed by automated and chain-terminating methods and analyzed by the Genetics Computer Group Software Analysis Package (40) and by the blast program. A 700-bp region between the GAL4 transactivation domain (GAD) and the dmyc1 open reading frame (ORF) in one of the two-hybrid isolates appears to be unrelated to the dmyc1 mRNA on the basis of several findings, including its lack of hybridization to cosmids bearing the dmyc1 gene, the failure to amplify an equivalent fusion transcript in reverse transcriptase-PCR assays using primers directed toward this interposed segment and dmyc1, and the inability of these sequences to detect the 7-kb dmyc1 transcript in Northern blot studies (not shown).
dmyc1 bHLH/LZ sequences from the two-hybrid isolate were used as a probe to identify a 2.4-kb cDNA from an early embryonic library subcloned in the plasmid NB40 (41); the sequence of this cDNA was submitted to GenBank under accession number (U81384U81384). The alcohol dehydrogenase (ADH)-driven dmyc1 cassette was prepared by excising a HindIII vector fragment from the pGAD-GH construct (CLONTECH) and replacing it with the 2.4-kb dmyc1 cDNA bearing the complete ORF.
For coupled in vitro transcription/translation reactions, a plasmid carrying the putative full-length dmyc1 ORF cloned into pNB40 (41) was linearized with NotI at the 3′ end of the gene, and transcribed in vitro using SP6 RNA polymerase (42). An aliquot of RNA was translated in rabbit reticulocyte lysate (Amersham) in the presence of [35S]methionine, and the translation products were visualized by autoradiography after fractionation by SDS/PAGE (7.5% acrylamide).
REF Cooperation Assays and Expression Constructs.
Early-passage cultures of REFs were prepared and cotransfected by the calcium phosphate method as described (16). Plates were seeded with 0.8 × 106 early-passage REFs the day before transfection. Each primary transfection plate received 2 μg of each of the listed expression constructs and 30 μg of genomic carrier DNA. The total number of foci was counted on six 10-cm2 plates that were split from two 10-cm2 primary transfected plates.
The dmyc1 expression construct contained a 2.4-kb cDNA fragment, encoding the complete dmyc1 ORF, subcloned into the pVNic vector (13) in the sense orientation relative to the Moloney murine leukemia virus long terminal repeats. The mouse c-myc expression construct used was pKO-myc, in which transcription of exons 2 and 3 of the c-myc gene is driven by a simian virus 40 promoter/enhancer, and pT24-ras contained the mutant H-Ras (Val-12) oncogene (16).
In Situ Hybridizations.
Whole-mount in situ hybridization to Drosophila embryos was carried out according to Tautz and Pfeifle (43) with modifications suggested by N. Hawkins (personal communication). Digoxigenin-labeled DNA probes were prepared by random priming; digoxigenin-labeled RNA probes were prepared by in vitro transcription. RNA antisense and DNA probes revealed identical patterns of dmyc1 expression (RNA antisense probes not shown); the sense RNA probe produced a low level of homogeneous background staining (not shown). In situ hybridization to polytene chromosomes from the wild-type stock Oregon R (described in ref. 44) was carried out using the pGAD-dmyc1 plasmid, containing DNA sequences encoding the entire dmyc1 ORF, labeled with biotinylated dUTP. An identical hybridization to region 3D4–7 was obtained using a probe prepared from a 1.1-kb PCR-generated fragment encompassing the bHLH/LZ and 3′ untranslated sequences of the dmyc1 cDNA.
PCR Amplification of Yeast Artificial Chromosomes (YACs).
YACs employed in the PCR amplifications were gifts of I. Duncan and P. Kiefel and included (YAC 1) DYR07–58, 320-kb insert with 3C1–2 to C5–7; (YAC 2) DY430, 150-kb insert with 3C1 to C3; (YAC 3) DYR22–11, 180-kb insert with 3A3–4 to C2–4; (YAC 4) DYN13–61, 300-kb insert with 3B5–6 to C1–2 and 67D; (YAC 5) DYR01–60, 150-kb insert with 3C4–5 to C10–12; (YAC 6) DYN11–88, 220-kb insert with 3D1–2 to E2–3; (YAC 7) DYN01–51, 150-kb insert with 3D1 to D4; and (YAC 8) DYE03–14, 260-kb with insert 3D4 to D7. YACs 1, 3, 4, 5, and 6 are described in ref. 45, and YACs 2, 7, and 8 are described in ref. 46. PCR primers, containing synthetic EcoRI sites, were as follows: forward primer 5′-GGAATTCGATACGATCGAGAAGCGCAATC-3′ and reverse primer 5′-CCGAATTCACTAACCGAGCGCGATTCGTTC-3′.
P-Element-Mediated Transformation.
The dmyc1 cDNA was subcloned into the pUAST vector (47) and introduced into the fly genome by conventional P-element transformation (48). An insert on the third chromosome was used in studies of phenotypic rescue. dmyc1 was expressed under the control of a GAL4 enhancer trap line inserted in the scabrous locus, scaGAL4, that was a kind gift of N. Baker (Albert Einstein College of Medicine, Bronx, NY). The diminutive (dm) mutant line was obtained from K. Matthews and the Bloomington stock center.
RESULTS AND DISCUSSION
Drosophila Myc (dMyc1) Can Interact with Mammalian Max, Transactivate Gene Expression, and Transform Primary Mammalian Cells.
Full-length mouse MaxΔ9 protein fused in-frame to bacterial LexA (38) was used as a bait to isolate interacting proteins encoded by an embryonic Drosophila melanogaster library subcloned into a GAD-containing vector. Of approximately 2.2 × 107 transformants screened, 20 yeast clones were identified that exhibited both growth in histidine-free media and activity in a β-galactosidase filter assay, suggesting an interaction between the two fusion proteins leading to the reconstitution of a functional transcriptional activator (37). A subset of these 20 library-encoded cDNAs hybridized to degenerate oligomers directed toward nucleotide sequences that were highly conserved in vertebrate myc family genes (i.e., those that encoded the basic region or helix-I structures) (data not shown). Nucleic acid sequence analysis of several clones from this subset demonstrated the presence of ORFs bearing substantial homology to the bHLH/LZ region of known c-Myc proteins (see Fig. 1C below).
Figure 1.
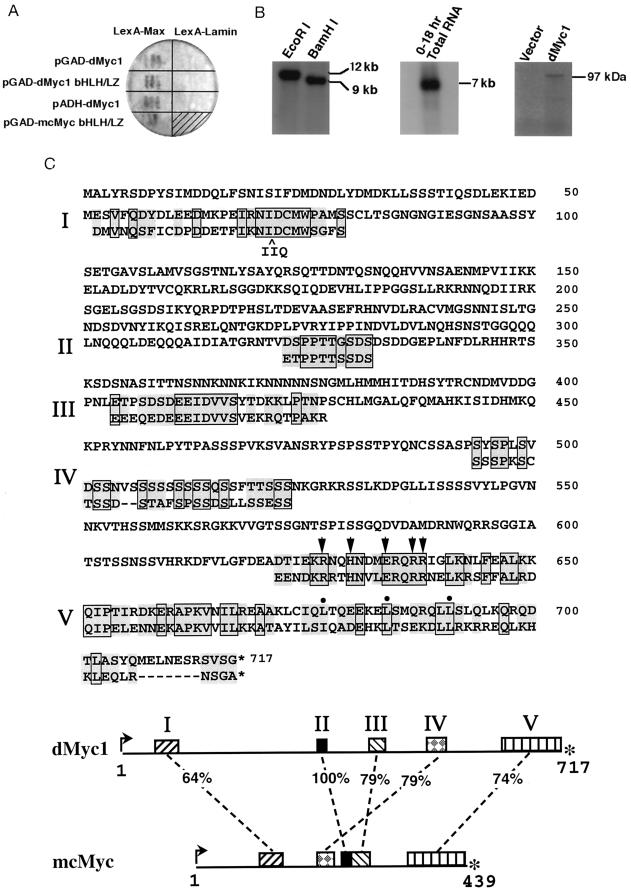
(A) Specificity of interaction between dMyc1 and mammalian Max. Yeast cells were simultaneously transformed with a plasmid encoding either dMyc1 or mouse c-Myc and a plasmid encoding either LexA-Max or LexA-lamin (generous gift of R. Sternglanz, SUNY at Stonybrook). The pGAD-dmyc1 two-hybrid isolate contained a 700-bp segment of non-dmyc1 sequences between the GAD sequences and the complete dmyc1 ORF. The pGAD-dMyc1[bHLH/LZ] two-hybrid isolate contained a 1.1-kb cDNA insert consisting of ORF sequences encompassing the dMyc1 bHLH/LZ region. The ADH-dmyc1 construct contained the dmyc1 cDNA encoding the complete ORF under the direct control of the yeast ADH promoter. The pGAD-mc-myc construct contained the mouse c-Myc bHLH/LZ region (38). Transformants were assayed by qualitative β-galactosidase filter assays as described (38, 39). (B) The dmyc1 gene, transcript, and gene product. (Left) Southern blot analysis of dmyc1 using a 280-bp BamHI/BglII dmyc1 ORF fragment on restriction enzyme-digested Drosophila genomic DNA identifies a unique genomic sequence. (Center) Northern blot analysis of 0- to 18-h embryonic total RNA using the same probe detects an abundant 7-kb transcript. (Right) Visualization of SDS/PAGE-fractionated products generated by in vitro coupled transcription/translation reactions programmed with either empty NB40 vector (41) or NB40 containing the dmyc1 cDNA. The latter generates a single product that migrates slower than that predicted from the dmyc1 ORF, perhaps due to post-translational modification or to the extremely charged nature of the protein. (C) Predicted dMyc1 protein and regions of sequence conservation with mouse c-Myc. dMyc1 and mouse c-Myc (54) protein alignment highlighting regions of strongest homology (segments of similarity are numbered I–V). Alignments were done with the Genetics Computer Group sequence analysis software package and by visual fit as described (34). Pairs of residues that are boxed and shaded are identical, and those that are shaded only are highly similar. Conserved basic region residues that have been shown to be critical for sequence-specific DNA binding (51, 52) are marked by arrowheads, and hydrophobic repeat residues of the leucine zipper are marked by filled circles. (Bottom) Locations in the dMyc1 and mcMyc proteins of homology regions I–V and their percent similarity. Intervening regions are poorly homologous. Interestingly, the split between regions II and III in dMyc1 occurs at the exon2/3 splice junction of mouse c-myc.
Specificity of interaction for two of the overlapping candidate Drosophila Myc (termed dMyc1) fusion proteins was verified by means of small-scale yeast transformations (39) in which their introduction along with plasmids encoding either LexA-Max or LexA-lamin baits yielded strong β-galactosidase activity in GAD-dmyc1 + LexA-max and GAD-dmyc1bHLH/LZ + LexA-max cotransformants only (Fig. 1A). As judged qualitatively by reactivity in the LacZ filter assay, activation of the integrated β-galactosidase reporter brought about by the interaction between dMyc1 and Max appeared comparable to that induced by the mouse c-Myc and Max interaction. Interestingly, one of the GAD-dmyc1 fusion clones contained a 700-bp segment between the sequences encoding the GAL4 transactivation domain and the ATG-initiated ORF of dmyc1; this unrelated 700-bp segment likely arose as a result of a cloning artifact (see Materials and Methods). The ability of this GAD-dmyc1 fusion clone to transactivate the β-galactosidase gene, despite the presence of translational terminators in the intervening 700-bp segment, suggested that the transactivation function provided by this two-hybrid library plasmid may stem exclusively from the dMyc1 protein itself. To test this possibility, the dmyc1 ORF was placed directly under the control of the yeast ADH promoter and cointroduced with LexA-max as above. ADH-dmyc1 + LexA-max cotransformants exhibited robust growth in histidine-free media (not shown) and a strong LacZ+ phenotype (Fig. 1A). Together, these findings are consistent with the view that, similar to its mammalian counterpart (49), dMyc1 can associate with Max in vivo and transactivate gene expression when placed in a promoter context.
Radiolabeled dmyc1 sequences from the GAD-dmyc1 plasmid were used to probe Southern blots of Drosophila genomic DNA, identifying a single-copy gene, and Northern blots of total embryonic RNA, detecting an abundant transcript of approximately 7 kb (Fig. 1B). Similar probes were used to screen a Drosophila early embryonic cDNA library, resulting in the isolation of cDNA clones containing an ATG-initiated ORF that was identical to that of the original two-hybrid dmyc1 clone shown to possess endogenous transactivation potential. This ORF comprises 2151 bp encoding a putative protein of 717 amino acids (Fig. 1C) with a predicted molecular mass of 79 kDa; when subjected to in vitro coupled transcription/translation, the dmyc1 cDNA generated a protein of 97 kDa (Fig. 1B). Database analyses of the predicted dMyc1 protein showed interspersed stretches of homology between the Drosophila and vertebrate c-Myc proteins (Fig. 1C, only mouse c-Myc comparison shown), similar to what has been reported previously for Drosophila homologues of other mammalian protooncogenes and growth factors (reviewed in ref. 50). Structures known to be essential for vertebrate Myc function are strikingly well conserved, particularly the bHLH/LZ domain, which bears 74% similarity and 37% identity at the amino acid level to mouse c-Myc (Fig. 1C, V). The relatedness in this region is further evidenced by the absolute conservation of specific key residues, such as those in the basic region previously shown to be critical for sequence-specific DNA binding (refs. 51 and 52; marked by arrows in Fig. 1C) and those in the leucine zipper essential for establishing the heptad repeat (reviewed in ref. 17; marked by filled circles in Fig. 1C). Moreover, like its vertebrate homologues, dMyc1 bears the signature feature of placement of the bHLH/LZ at the extreme carboxylterminus. Another conserved structure is the “myc homology region 2” (Fig. 1C, I), which is invariant in all previously reported Myc-family proteins and is essential for both oncogenic activity (reviewed in ref. 17) and transcriptional repression function (53).
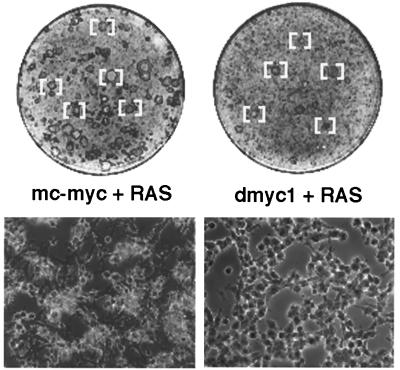
To determine the degree of functional relatedness between invertebrate and vertebrate Myc proteins, a dmyc1 expression construct was tested for its ability to cooperate with activated human RAS to effect the malignant transformation of early-passage REFs (55). In independent transfection experiments, dmyc1 proved to be quite effective, in comparison to the highly potent mouse c-myc expression construct, at generating foci in the REF monolayer (Table 1 and Fig. 2). In particular, the level of cooperation activity exhibited by dmyc1 appeared comparable to or slightly better than that observed for the mammalian N- and L-myc genes in this assay (16). Although arising after the same latency period, dmyc1/RAS-generated foci exhibited a range in the morphology that was, on average, slightly less transformed than that of mouse c-myc/RAS-generated foci. In addition, fewer of dmyc1/RAS-generated foci were capable of establishing permanent cell lines (data not shown). This apparent variance in the degree of oncogenic activity is similar to that we have documented previously for the zebrafish c-myc homologue (34) and may reflect differences in protein stability or affinity for associated factors or binding sites. Notwithstanding the variance in oncogenic activity, our results imply that dMyc1 can bind to mammalian Max and recognize and presumably regulate mammalian Myc-responsive gene targets required for cellular transformation. As such, many of the biochemical and biological properties of Myc appear to be remarkably well conserved over a phylogenetic distance of at least 500 million years.
Table 1.
Oncogenic activity of dMyc1 in comparison to mouse c-Myc
| Transfected DNA | Total foci count
|
|
|---|---|---|
| Exp. 1 | Exp. 2 | |
| mc-myc + RAS | 648 | 642 |
| dmyc1 + RAS | 122 | 42 |
| Empty vector + RAS | 7 | 7 |
The total number of transformed foci on six plates was counted on day 8 or day 11 posttransfection, respectively, for independent experiments 1 and 2.
Figure 2.
Gross and microscopic appearance of transformed foci generated in REF experiments presented in Table 1. (Upper) A subset of mc-myc/RAS- or dmyc1/RAS-transformed foci [bracketed] as seen in a Wright–Giemsa-stained monolayer on day 11 post-transfection (experiment 2 from Table 1). All foci were confirmed by microscopic inspection. (Lower) Microscopic appearance of a representative transformed focus under phase contrast photographed on a Zeiss axioscope at 130× magnification.
dmyc1 Is Expressed in the Proliferating Endoderm and Mesoderm.
To gain insight into the role of c-Myc in invertebrate development, its cellular expression was analyzed in a panel of Drosophila embryos by whole mount in situ hybridization (43) using digoxigenin-labeled probes derived from the dmyc1 bHLH/LZ region. A low level of maternally derived dmyc1 RNA was observed throughout early embryos before cellular blastoderm formation and appeared to be particularly concentrated in the pole plasm (Fig. 3A, embryo a). Zygotic expression was detected first during the cellular blastoderm stage in the endodermal anlagen of the anterior and posterior midgut at the two poles of the embryo (Fig. 3A, embryo b). Soon after, at the onset of gastrulation, expression was apparent in the mesoderm as it formed the invaginating ventral furrow (Fig. 3A, embryo c). During germ-band extension, dmyc1 expression continued in both the anterior and posterior midgut, as well as in the mesoderm (Fig. 3A, embryo d). At the end of germ-band retraction, expression remained detectable in the fusing midgut and in tissues that appeared to be developing somatic musculature (Fig. 3A, embryo e). Expression levels declined during subsequent stages of embryogenesis (not shown). Together, this pattern of expression is consistent with a role for dMyc1 in the proliferative phase of the development of the midgut and the somatic mesoderm.
Figure 3.
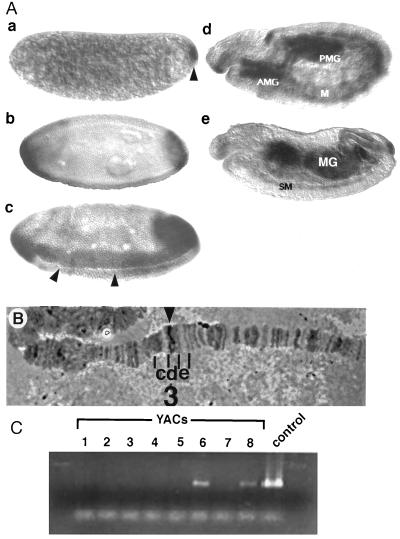
(A) Spatial pattern of dmyc1 transcript distribution during embryogenesis. Side views of whole-mount embryos with anterior to the left and dorsal up × 80. Embryo a, cleavage stage. dmyc1 RNA is concentrated in the pole plasm at the posterior pole (arrowhead); a low level of uniform expression is detected throughout the rest of the embryo. Embryo b, cellular blastoderm stage before the onset of gastrulation. dmyc1 exhibits two caps of expression at the poles of the embryo. The initiation of expression along the ventral surface also can be detected. Embryo c, gastrulation stage. Expression is seen in the anlagen of the anterior and posterior midgut at the poles and in the mesoderm flanking the invaginating ventral furrow (arrowheads). Embryo d, germ-band-extension. dmyc1 RNA is detected in the invaginated anterior midgut (AMG) and posterior midgut (PMG), and in tissues that appear to be the somatic mesoderm (M). Embryo e, after germ-band retraction. Expression persists in the developing somatic musculature (SM) and in the AMG and PMG as they fuse to form the midgut (MG). (B) Cytogenetic localization of the dmyc1 locus. A dmyc1 cDNA probe identifies a unique hybridization signal at 3D4–7 on Drosophila polytene chromosomes (arrowhead). Polytene segments 3 c, d, and e are indicated. (C) Confirmation of chromosomal localization by detection of dmyc1 in YACs bearing various segments of the Drosophila X chromosome. Primers directed to sequences that amplify the bHLH/LZ region of dmyc1 were used in a PCR screen of a series of 3CD-containing YACs. The 300-bp PCR amplification product was detected only in YAC 6 (containing regions 3D1–2 to 3E2–3) and YAC 8 (containing regions 3D4–3D7), and in the dmyc1 cDNA positive control.
Colocalization of dmyc1 and dm Loci and Phenotypic Rescue of the Bristle Defect Through Genetic Complementation.
As a first step in using genetic analysis to identify mutants that may result from dMyc1 gain- or loss-of-function, we determined the cytological location of the gene. In situ hybridization to Drosophila polytene chromosomes localized dmyc1 to bands 3D4–7 of the euchromatic region of the X chromosome (Fig. 3B). The assignment of dmyc1 to this interval was confirmed by PCR analysis of YACs that have been independently mapped to the region (45, 46) using oligonucleotides capable of specifically amplifying the sequences encoding the dMyc1 bHLH/LZ region (see Materials and Methods). Only YACs containing the 3D4–3D7 interval yielded the expected 300-bp amplification product (Fig. 3C).
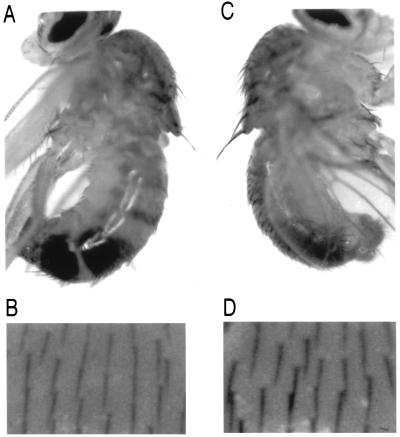
Several known mutations map to the region to which we have localized dmyc1 (56). The overt small and slender bristles and body of one such mutant, dm (57, 58), provided the opportunity to assess the ability of a dmyc1 transgene to achieve phenotypic rescue in vivo. Using the binary system developed by Brand and Perrimon (47), dmyc1 expression was placed under the control of GAL4 transactivation activity. Flies were generated carrying both an upstream activation sequence (UAS)-driven dmyc1 cDNA and a GAL4 enhancer-trap insert in the scabrous (sca) (59) locus. The sca locus was selected because mutations therein affect the number and spacing of bristles on the adult. Moreover, among the cells that express sca are ones thought to be adult sensory organ precursor cells (60). dm mutant flies carrying sca-GAL4/UAS-dmyc1 demonstrated a marked increase in the size of the micro- and macrochaetae of the adult cuticle (Fig. 4A and B, dm mutant; C and D, rescued flies). In separate experiments, we have expressed dmyc1 under the control of the Hsp70 promoter in the plasmid CaSpeR-Hsp70 (61) and observed a similar rescue of bristle size of the dm mutant, as well as an increase in body size. The ability of dmyc1 to rescue these components of the dm mutant phenotype, together with the mapping of dmyc1 and dm to the same chromosomal interval, provides support for the notion that abnormalities associated with the dm mutation arise from an alteration in the locus encoding dmyc1.
Figure 4.
Phenotypic rescue of the dm mutant phenotype by dmyc1 expression. Genotypes: (A and B) Fm6, dm1/Y; +/+; p[w+, UAS-dmyc1/+. (C and D) Fm6, dm1/Y; scaGAL4/+; p[w+, UAS-dmyc1/+. A and C show side views of adult flies × 20. Note in particular the difference in size of the scutellar macrochaetae extending from the dorsal surface of the thorax. Note also the size difference of the abdominal bristles. B and D show images of fields of microchaetae present on the dorsal surface of the thorax × 125. Note the larger size of bristles in D. For C and D, bristles produced by flies of the genotype Fm6, dm1/Y; scaGAL4/+; p[w+, UAS-dmyc1/+ are similar in size to those produced by wild-type flies (data not shown).
The identification of a Myc family member in Drosophila melanogaster and the demonstration of conservation of its biochemical and biological properties indicate that lessons learned in the fly regarding Myc function should be readily extrapolated to vertebrate systems. In the immediate future, dMyc1 will allow for the genetic analysis of functional interactions among conserved members of the Myc superfamily. In addition, the dmyc1 reagent will allow for the determination of the cell biological basis for the dm mutation that first was described 60 years ago. Ultimately, the power of the Drosophila model system (62) will permit one to place the dMyc network at a nexus where mitogenic and differentiative signals are integrated in the execution of a normal developmental program.
Acknowledgments
We thank R. Sternglanz for generously providing advice and reagents for two-hybrid screens, I. Duncan and P. Kiefel for providing yeast strains carrying YACs, K. Matthews and the Bloomington stock center for the dm mutant, N. Baker for the scaGAL4 enhancer trap line, N. Hawkins for advice regarding in situ hybridization, and L. Chin for her help in preparation of the manuscript. This work was supported by grants from the National Institutes of Health and an award from the Irma T. Hirschl Fund to R.A.D., grants from the National Institutes of Health and the American Cancer Society to D.S., and grants from the National Institutes of Health to L.S.
Note Added in Proof.
A similar study (63) was published recently and confirmed our hypothesis that phenotypic alterations associated with dm result from mutations in dmyc1.
Footnotes
Abbreviations: ADH, alcohol dehydrogenase; dmyc, Drosophila myc; dm, diminutive; GAD, GAL4 transactivation domain; ORF, open reading frame; bHLH/LZ, basic region helix–loop–helix/leucine zipper; REF, rat embryo fibroblast; YAC, yeast artificial chromosome. Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. U81384U81384).
References
- 1.Blackwood E M, Kretzner L, Eisenman R N. Curr Opin Genet Dev. 1992;2:227–235. doi: 10.1016/s0959-437x(05)80278-3. [DOI] [PubMed] [Google Scholar]
- 2.Evan G I, Littlewood T D. Curr Opin Genet Dev. 1993;3:44–49. doi: 10.1016/s0959-437x(05)80339-9. [DOI] [PubMed] [Google Scholar]
- 3.Kato G J, Barrett J, Villa-Garcia M, Dang C V. Mol Cell Biol. 1990;10:5914–5920. doi: 10.1128/mcb.10.11.5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kretzner L, Blackwood E M, Eisenman R N. Nature (London) 1992;359:426–429. doi: 10.1038/359426a0. [DOI] [PubMed] [Google Scholar]
- 5.Prendergast G C, Lawe D, Ziff E B. Cell. 1991;65:395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- 6.Blackwood E M, Eisenman R N. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 7.Amati B, Littlewood T D, Evan G I, Land H. EMBO J. 1993;12:5083–5087. doi: 10.1002/j.1460-2075.1993.tb06202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amati B, Brooks M W, Levy M, Littlewood T D, Evan G I, Land H. Cell. 1993;72:233–245. doi: 10.1016/0092-8674(93)90663-b. [DOI] [PubMed] [Google Scholar]
- 9.Ayer D E, Kretzner L, Eisenman R N. Cell. 1993;72:211–222. doi: 10.1016/0092-8674(93)90661-9. [DOI] [PubMed] [Google Scholar]
- 10.Zervos A S, Gyuris J, Brent R. Cell. 1993;72:223–232. doi: 10.1016/0092-8674(93)90662-a. [DOI] [PubMed] [Google Scholar]
- 11.Hurlin P J, Queva C, Koskinen P J, Steingrimsson E, Ayer D E, Copeland N G, Jenkins N A, Eisenman R N. EMBO J. 1995;14:5646–5659. doi: 10.1002/j.1460-2075.1995.tb00252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koskinen P J, Ayer D E, Eisenman R N. Cell Growth Diff. 1995;6:623–629. [PubMed] [Google Scholar]
- 13.Schreiber-Agus N, Chin L, Chen K, Torres R, Rao G, Guida P, Skoultchi A I, DePinho R A. Cell. 1995;80:777–786. doi: 10.1016/0092-8674(95)90356-9. [DOI] [PubMed] [Google Scholar]
- 14.Ayer D E, Lawrence Q A, Eisenman R N. Cell. 1995;80:767–776. doi: 10.1016/0092-8674(95)90355-0. [DOI] [PubMed] [Google Scholar]
- 15.Morgenbesser S D, DePinho R A. Semin Cancer Biol. 1994;5:21–36. [PubMed] [Google Scholar]
- 16.Mukherjee B, Morgenbesser S D, DePinho R A. Genes Dev. 1992;6:1480–1492. doi: 10.1101/gad.6.8.1480. [DOI] [PubMed] [Google Scholar]
- 17.DePinho R A, Schreiber-Agus N, Alt F W. Adv Cancer Res. 1991;57:1–46. doi: 10.1016/s0065-230x(08)60994-x. [DOI] [PubMed] [Google Scholar]
- 18.Zimmerman K A, Yancopoulos G D, Collum R G, Smith R K, Kohl N E, Denis A, Nau M M, Witte O N, Toran-Allerand D, Gee C E, Minna J D, Alt F W. Nature (London) 1986;319:780–783. doi: 10.1038/319780a0. [DOI] [PubMed] [Google Scholar]
- 19.Hatton K S, Mahon K, Chin L, Chiu F-C, Lee H-W, Peng D, Morgenbesser S D, Horner J, DePinho R A. Mol Cell Biol. 1996;16:1794–1804. doi: 10.1128/mcb.16.4.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charron J, Malynn B, Fisher P, Stewart V, Jeannotte L, Goff S P, Robertson L, Alt F W. Genes Dev. 1992;6:2248–2257. doi: 10.1101/gad.6.12a.2248. [DOI] [PubMed] [Google Scholar]
- 21.Sawai S A, Shimono K, Yakamatsu Y, Palmes C, Hanaoka K, Kondoh H. Development (Cambridge, UK) 1993;117:1445–1455. doi: 10.1242/dev.117.4.1445. [DOI] [PubMed] [Google Scholar]
- 22.Stanton B R, Perkins A, Tessarollo L, Sassoon D, Parada L. Genes Dev. 1992;6:2235–2247. doi: 10.1101/gad.6.12a.2235. [DOI] [PubMed] [Google Scholar]
- 23.Davis A C, Wims M, Spotts G D, Hann S, Bradley A. Genes Dev. 1993;7:671–682. doi: 10.1101/gad.7.4.671. [DOI] [PubMed] [Google Scholar]
- 24.Bello-Fernandez C, Packham G, Cleveland J L. Proc Natl Acad Sci USA. 1993;90:7804–7808. doi: 10.1073/pnas.90.16.7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galaktionov K, Chen X, Beach D. Nature (London) 1996;382:511–517. doi: 10.1038/382511a0. [DOI] [PubMed] [Google Scholar]
- 26.Gu W, Bhatia K, Magrath I T, Dang C V, Dalla-Favera R. Science. 1994;264:251–254. doi: 10.1126/science.8146655. [DOI] [PubMed] [Google Scholar]
- 27.Hateboer G, Timmers H T, Rustgi A K, Billaud M, van’t Veer L J, Bernards R. Proc Natl Acad Sci USA. 1993;90:8489–8493. doi: 10.1073/pnas.90.18.8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shrivastava A, Saleque S, Kalpana G V, Artandi S, Goff S P, Calame K. Science. 1993;262:1889–1892. doi: 10.1126/science.8266081. [DOI] [PubMed] [Google Scholar]
- 29.Schreiber-Agus N, Torres R, Horner J, Lau A, Jamrich M, DePinho R A. Mol Cell Biol. 1993;13:2456–2468. doi: 10.1128/mcb.13.4.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vize P D, Vaughan A, Krieg P. Development (Cambridge, UK) 1990;110:885–896. doi: 10.1242/dev.110.3.885. [DOI] [PubMed] [Google Scholar]
- 31.Vriz S, Taylor M, Mechali M. EMBO J. 1989;8:4091–4097. doi: 10.1002/j.1460-2075.1989.tb08593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Principaud G, Spohr G. Nucleic Acids Res. 1991;19:3081–3088. doi: 10.1093/nar/19.11.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.King M W, Blackwood E M, Eisenman R N. Cell Growth Diff. 1993;4:85–92. [PubMed] [Google Scholar]
- 34.Schreiber-Agus N, Horner J, Torres R, Chiu F-C, DePinho R A. Mol Cell Biol. 1993;13:2765–2775. doi: 10.1128/mcb.13.5.2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walker C W, Boom J D, Marsh A G. Oncogene. 1992;7:2007–2012. [PubMed] [Google Scholar]
- 36.Atchley W R, Fitch W M. Proc Natl Acad Sci USA. 1995;92:10217–10221. doi: 10.1073/pnas.92.22.10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fields S, Song O-K. Nature (London) 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 38.Schreiber-Agus N, Chin L, Chen K, Torres R, Thomson C, Sacchettini J C, DePinho R A. Oncogene. 1994;9:3167–3177. [PubMed] [Google Scholar]
- 39.Vojtek A B, Hollenberg S M, Cooper J A. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 40.Devereux J, Haeberli P, Smithies O. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown N H, Kafatos F C. J Mol Biol. 1988;203:425–437. doi: 10.1016/0022-2836(88)90010-1. [DOI] [PubMed] [Google Scholar]
- 42.Melton D A, Krieg P A, Rebagliati M R, Maniatis T, Zinn K, Green M R. Nucleic Acids Res. 1984;12:7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tautz D, Pfeifle C. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- 44.Ashburner M. Drosophila, A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 45.Cai H, Kiefel P, Yee J, Duncan I. Genetics. 1994;136:1385–1399. doi: 10.1093/genetics/136.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ajioka J W, Smoller D A, Jones R W, Carulli J P, Vellek A E, Garza D, Link A J, Duncan I W, Hartl D L. Chromosoma. 1991;100:495–509. doi: 10.1007/BF00352200. [DOI] [PubMed] [Google Scholar]
- 47.Brand A, Perrimon N. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 48.Rubin G M, Spradling A C. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 49.Amati B, Dalton S, Brooks M W, Littlewood T D, Evan G I, Land H. Nature (London) 1992;359:423–426. doi: 10.1038/359423a0. [DOI] [PubMed] [Google Scholar]
- 50.Hoffmann F M. Curr Top Microbiol Immunol. 1989;147:1–29. doi: 10.1007/978-3-642-74697-0_1. [DOI] [PubMed] [Google Scholar]
- 51.Ferre-D’Amare A R, Prendergast G C, Ziff E B, Burley S K. Nature (London) 1993;363:38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- 52.Fisher D E, Parent L A, Sharp P A. Cell. 1993;72:467–476. doi: 10.1016/0092-8674(93)90122-7. [DOI] [PubMed] [Google Scholar]
- 53.Li L H, Nerlov C, Prendergast G, MacGregor D, Ziff E B. EMBO J. 1994;13:4070–4079. doi: 10.1002/j.1460-2075.1994.tb06724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stanton L W, Fahrlander P D, Tesser P M, Marcu K B. Nature (London) 1984;310:423–425. doi: 10.1038/310423a0. [DOI] [PubMed] [Google Scholar]
- 55.Land H, Parada L F, Weinberg R A. Nature (London) 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- 56.Lindsley D L, Zimm G G. The Genome of Drosophila Melanogaster. San Diego: Academic; 1992. [Google Scholar]
- 57.Bridges C B. Drosophila Inf Serv. 1935;3:5–19. [Google Scholar]
- 58.Schalet A P. Mutat Res. 1986;163:115–144. [Google Scholar]
- 59.Baker N E, Yu S, Han D. Curr Biol. 1996;6:1290–1301. doi: 10.1016/s0960-9822(02)70715-x. [DOI] [PubMed] [Google Scholar]
- 60.Mlodzek M, Baker N E, Rubin G M. Genes Dev. 1990;4:1848–1861. doi: 10.1101/gad.4.11.1848. [DOI] [PubMed] [Google Scholar]
- 61.Bang A G, Posakony J W. Genes Dev. 1992;6:1752–1769. doi: 10.1101/gad.6.9.1752. [DOI] [PubMed] [Google Scholar]
- 62.Rubin G M. Science. 1988;243:1453–1459. doi: 10.1126/science.3131880. [DOI] [PubMed] [Google Scholar]
- 63.Gallant P, Shiio Y, Cheng P F, Parkhurst S M, Gisenman R N. Science. 1996;274:1523–1527. doi: 10.1126/science.274.5292.1523. [DOI] [PubMed] [Google Scholar]