Abstract
To isolate new peptide signal molecules involved in regulating developmental processes in hydra, a novel screening project was developed. Peptides extracted from the tissue of Hydra magnipapillata were systematically purified to homogeneity using HPLC. A fraction of each purified peptide was examined by differential display–PCR for its ability to affect gene expression in hydra. Another fraction was used to determine the tentative structure using an amino acid sequence analyzer and/or a mass spectrometer. Based on the results, peptides of potential interest were selected for chemical synthesis, followed by confirmation of the identity of the synthetic with the native peptides using HPLC. Using this approach, 286 peptides have been isolated, tentative amino acid sequences have been determined for 95 of them, and 19 synthetic peptides identical to native ones were produced. The 19 synthetic peptides were active in a variety of biological tests. For example, Hym-54 stimulated muscle contraction in adult polyps of hydra and sea anemone, Anthopleura fuscoviridis, and induced metamorphosis of planula, the larval stage, into polyps in a marine hydrozoan species, Hydractinia serrata. Another peptide, Hym-33H, inhibited nerve cell differentiation in hydra and induced tissue contraction in planula of Hydractinia serrata. The evidence obtained so far suggests that hydra contains a large number (>350) of peptide signal molecules involved in regulating developmental or other processes in cnidaria. These peptides can be isolated and their functions examined systematically with the new approach developed in this study.
Peptide signal molecules (<50 amino acid residues) occur widely in animals, and many play important regulatory roles in a variety of physiological processes. For example, they serve as hormones in endocrine systems, as neurotransmitters or neuromodulators in nervous systems, and as activating factors for lymphocytes in immune systems. In contrast, only a few peptides that play regulatory roles in developmental processes are known. For example, the hypothalamic PACAP peptides containing 27 or 38 amino acid residues stimulate proliferation of primordial germ cells in mice (1). The hexapeptide metamorphosin A induces metamorphosis of the planula into a polyp in the marine hydrozoan Hydractinia echinata (2, 3).
Most of the signaling molecules regulating development are proteins (>50 amino acid residues) (4–6). Why have so few peptide signal molecules involved in developmental regulation been uncovered? It could be that peptides rarely play signaling roles in development. Alternatively, there could be a large number that do, but most remain undiscovered. Assuming the latter explanation to be correct, we have started a novel screening project to systematically isolate peptide signal molecules from a freshwater cnidarian species, Hydra magnipapillata.
In previous studies, signaling molecules were usually isolated one at a time. The process was often time-consuming and laborious since it required many steps of fractionation involving a large number of biological assays at each step. In this study, a drastically simplified approach has been developed; this approach consists of the following five steps. (i) Peptides extracted from hydra tissue were purified to homogeneity in a systematic manner using HPLC in the absence of any biological assays. (ii) The quantity of each isolated peptide (1–10 nmol) was insufficient for conventional biological assays. To select the peptides of potential interest, hydra were treated with a peptide, poly(A) RNA isolated from the treated and untreated control animals, and used in the differential display (DD)-PCR assay. Peptides capable of affecting gene expression were identified by differences in the DD-PCR band patterns between treated and control animals. (iii) Another fraction of this peptide was used to determine its amino acid sequence using an automated amino acid sequence analyzer and/or a tandem mass spectrometer. (iv) Those peptides which affected gene expression, and for which a sequence was obtained, were selected for chemical synthesis. The identity of the synthetic product with the native peptide was confirmed using HPLC. (v) The synthetic peptides produced were then subjected to a series of biological tests to examine their functions.
Using this approach, a large number of peptides were isolated from hydra tissue and characterized to varying degrees, depending on individual peptides. In this report, we describe the procedures developed for the project and present 19 peptides whose final structures were established. We also describe the biological activities of two families of peptides among the 19 that play regulatory roles in cnidarian development.
MATERIALS AND METHODS
Peptide Chemistry.
About 500 g of frozen tissue of Hydra magnipapillata was homogenized in 2 liters of acetone, and the homogenate was centrifuged at 16,000 × g for 30 min. The precipitate was homogenized once more in 5 volumes of a 3% acetic acid solution and centrifuged again. The first and second supernatants were combined and reduced in volume by rotary evaporation, a 1/10 volume of 1 M HCl was added, and the mixture was centrifuged. The clear supernatant obtained was applied to two large cartridges of reverse-phase chromatography (Analytichem Mega Bod Elut, Varian). After the cartridges were washed with 10% methanol, peptides were eluted with 60% methanol, and the eluates were rotary evaporated to dryness. Peptides were dissolved in a small volume of 0.1% trifluoroacetic acid, pH 2.2, and then subjected to a standard HPLC fractionation procedure for purification of individual peptides to homogeneity (see Fig. 1). Structural determination of isolated peptides was carried out by the automated Edman degradation method and/or tandem mass spectrometry. Chemical synthesis of peptides was carried out with the automated solid-phase procedure. Finally, the identity of the synthetic peptide with the native peptide was confirmed by co-chromatography using HPLC.
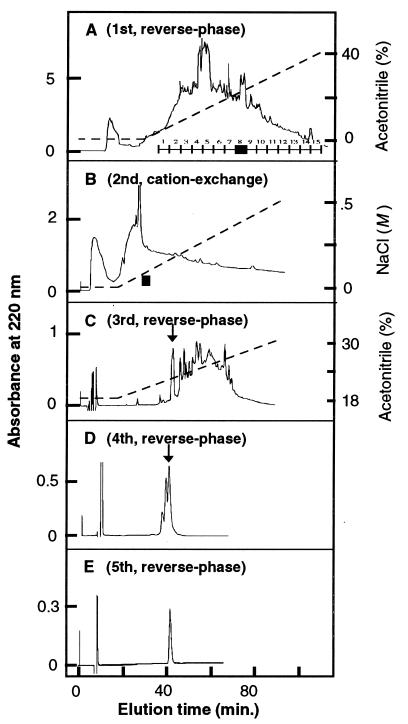
Figure 1.
Chromatographic profiles for the five steps of HPLC fractionation used for the isolation of the peptide Hym-54.
DD-PCR.
The procedure of Liang and Pardee (7) was used to examine the effect of peptides on gene expression in hydra. About 1/4 to 1/2 of a purified peptide was used to produce a peptide solution with a roughly estimated concentration of 10−9–10−7 M. This solution was used to treat 20 polyps, half of which were harvested after 4 h, and the rest after 20 h. To aid the penetration of a peptide into the tissue, dimethyl sulfoxide (DMSO) was added in some cases to a final concentration of 2% during the first hour of peptide treatment (8, 9). When DMSO was added, the controls were also treated with DMSO. Thereafter, animals were washed and reincubated in a peptide solution without DMSO. After treatment, poly(A) RNA was extracted from treated and untreated hydra, and subjected to DD-PCR analysis using an anchored primer, d(T12AG), and an arbitrarily selected deca-deoxynucleotide 5′ end primer. The DD-PCR products were separated by gel electrophoresis, and the band patterns of control and peptide-treated hydra were compared. Differences were interpreted as effects of the peptide on gene expression. To confirm that a synthetic peptide elicited the same response, each was similarly tested using a concentration range of 10−9–10−5 M.
Biological Assays.
All biological assays were carried out at 18°C using a concentration range of 10−8–10−5 M for a synthetic peptide.
Bud detachment from parental hydra.
A stream of culture medium was directed at individual buds with a Pasteur pipette to dislodge and remove any mature buds loosely attached to adults in a group of hydra. These animals were incubated in culture medium containing a peptide for 1 h and then transferred to a new dish containing fresh culture medium without the peptide. Thereafter, the group of animals was again subjected to the stream flow as described above to remove buds whose attachment had been loosened by the peptide treatment.
Rates of cell division and differentiation in hydra.
These were measured using standard procedures (10, 11). Groups of 10 young polyps were treated with a range of concentrations of a peptide for 48 h. The polyps were also pulse-labeled with BrdUrd by incubating them in 0.1 mM BrdUrd for 1 h at the beginning of the peptide treatment. At 48 h, each group of animals was macerated into a suspension of single cells, and the labeling index was determined for four cell types using immunocytochemistry with an anti-BrdUrd antibody (Becton Dickinson). The four types of cells were epithelial cells, interstitial cells (single and pairs of large interstitial cells), dividing nematoblasts (nests of four interstitial cells), and nerve cells.
Metamorphosis of Hydractinia planula larva.
Using the procedure described by Leitz et al. (2), groups of 10–20 planula of Hydractinia serrata were treated with a range of peptide concentrations in artificial seawater for 24 h. The treated groups of larva were washed twice and maintained in artificial seawater without peptide for 2 additional days. They were examined for tissue contraction at 1 and 24 h after the beginning of treatment as well as for metamorphosis (tentacle and stolon formation) at 3 days.
Muscle contraction in sea anemone.
This test was carried out as described by McFarlane et al. (12) using longitudinal parietal muscles of the sea anemone Anthopleura fuscoviridis. Individual muscle pieces were mounted in a chamber (2 ml) filled with artificial seawater. Different concentrations of a peptide, each dissolved in artificial seawater, were injected into the chamber, and tension changes produced in the tissue were recorded.
RESULTS
Peptide Isolation and Structure Determination.
Using one of the peptides, Hym-54, as an example, the procedure for isolation and structure determination of a peptide was as follows. Fig. 1 shows the five steps of HPLC used to isolate the peptide. The first step of reverse-phase HPLC was used to separate the peptide extract into 15 groups, each containing 2–3 fractions (Fig. 1A). Group 8 eluting at approximately 22% acetonitrile was subjected to the second step of cation-exchange HPLC (Fig. 1B). A pool of three fractions (indicated by the bar above the abscissa) eluting at approximately 0.15 M NaCl was then subjected to three successive steps of reverse-phase chromatography (Fig. 1 C–E), resulting in the isolation of a single sharp peak of one peptide, Hym-54.
About 1/10 of the Hym-54 peptide was examined with a fast atom bombardment mass spectrometer and found to consist of a single homogeneous substance with a molecular mass (unprotonated) of 759.3 Da (data not shown). About 1/2 of the peptide was subjected to DD-PCR and shown to a have a strong effect on gene expression in hydra; this effect will be described below. Finally, another 1/10 of the peptide was used to determine the tentative amino acid sequence, which was found to be Gly-Pro-Met-Thr-Gly-Leu-Trp. The calculated molecular weight for this structure was 760.3 if the C terminus was free, and 759.3 if amidated. To determine which was correct, both forms were synthesized, and each was cochromatographed with the native peptide using reverse-phase or cation-exchange HPLC. The amidated version eluted at exactly the same position as the native peptide, but the other version did not (data not shown). Thus, it was concluded that Hym-54 has the structure Gly-Pro-Met-Thr-Gly-Leu-Trp-NH2.
Characteristics of the Purified Peptides.
A total of 286 peptides were purified to homogeneity using the procedure similar to that shown in Fig. 1. Of these peptides, 108 were tested by DD-PCR, and 35 (32%) were found affect gene expression in hydra. Amino acid sequences were determined tentatively for 95 peptides (93 by automated Edman degradation and 2 by the tandem mass spectrometric method). Of the 95 peptides, 19, presented in Table 1, have been completely characterized as described for Hym-54 above. For each of them, a synthetic peptide was produced whose structure was confirmed to be identical to the native peptide by HPLC. All 19 are active in the DD-PCR assay, placing them in the class of putative signaling molecules.
Table 1.
Final structure of 19 peptides
| Family | Peptide | Structure |
|---|---|---|
| LWamide | Hym-53 | NPYPGLW-NH2 |
| Hym-54 | GPMTGLW-NH2 | |
| Hym-248 | EPLPIGLW-NH2 | |
| Hym-249 | KPIPGLW-NH2 | |
| Hym-331 | GPPPGLW-NH2 | |
| Hym-38* | GPPhPGLW-NH2 | |
| PW | Hym-33H | AALPW |
| Hym-35 | EPSAAIPW | |
| Hym-37 | SPGLPW | |
| Hym-310 | DPSALPW | |
| Hym-6† | Ac-ANTNPVILA | |
| Hym-31 | VSWQL | |
| Hym-36 | YLVGL | |
| Hym-99 | WPWQ | |
| Hym-126 | WKRLTLFG | |
| Hym-301 | KPPRRCYLNGYCSP-NH2 | |
| Hym-311 | FW | |
| Hym-323 | KWVQGKPTGEVKQIKF | |
| Hym-346 | AGEDVSHELEEKEKALANHS |
hP, hydroxyproline.
Structure initially determined by tandem mass spectroscopy.
Of the 19 peptides, 10 belong to two groups of structurally related peptides. The LWamide family has six members that share the same C-terminal structure of Gly-Leu-Trp-NH2. A peptide having the same terminal structure, metamorphosin A (pGlu-Gln-Pro-Gly-Leu-Trp-NH2), was previously isolated from the sea anemone Anthopleura elegantissima by Leitz et al. (2). At the N terminus, a proline residue is present in the second position (X-Pro) or in the second and third positions (X-Pro-Pro) in each of the six peptides, whereas a pyro-glutamyl residue is present in metamorphosin A. Both types of N-terminal structures confer resistance to amino-peptidase digestion (13). The second group (PW family) has four members that share the common sequence of Leu (or Ile)-Pro-Trp at the C terminus, and X-Pro at the N terminus (except for Hym-33H).
Three other peptides in Table 1 are related to peptides previously described. Hoffmeister (14) recently reported two peptides that stimulate foot regeneration in hydra. One of them, “pedibin,” is identical to Hym-346 (Table 1) in structure except for an additional glutamine residue at the C terminus in pedibin. Hym-311 was previously isolated from the echiuroid worm Urechis unicinctus on the basis of an inhibitory effect on the body wall muscle of this organism (15). Hym-323 may be related to head-activator (pGlu-Pro-Pro-Gly-Gly-Ser-Lys-Val-Ile-Leu-Phe) isolated by Schaller and Bodenmüller (16). These two peptides share identical amino acid residues in 5 of 10 positions in the C-terminal region of the peptide.
Tentative amino acid sequences were also determined for the other 76 peptides (data not shown). Tentative sequences of an additional 32 peptides indicated that 12 were duplicates of peptides already identified, and that 20 (17%) as judged by a homology search were fragments of degraded proteins.
Effects on Gene Expression of Peptides of the LWamide and PW Families.
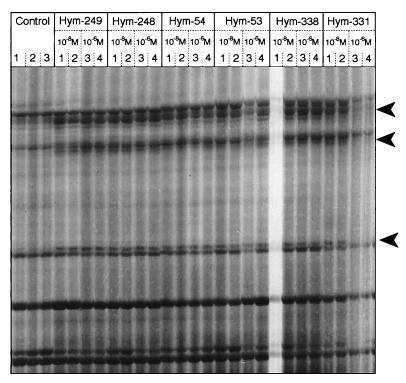
To determine if all members of the LW family, or the PW family, had a similar effect on gene expression, the patterns of gene expression induced by each member of a family were compared. For the six LWamide peptides, these patterns are shown in Fig. 2. The first three lanes in Fig. 2 show the products of three independent DD-PCR reactions for the control group of hydra. Approximately 20 major and 50 minor bands are visible in the same positions in the three lanes, suggesting a high reproducibility of the PCR reaction. All the other lanes represent the reverse transcription–PCR products for hydra treated with one of the six LWamide members at 10−8 or 10−6 M in duplicate. The three major bands marked by arrowheads in Fig. 2 are present in all the lanes for the six LWamide peptides. The only exceptions are lane 1 for Hym-338 and lanes 3 and 4 for Hym-331, where the three additional bands are not clearly visible due to sample loss. The major bands were greatly reduced in intensity in the three control lanes. This observation suggests that expression of the three genes represented by the three bands is specifically up-regulated in hydra treated with any of the six LWamide peptides in an identical manner.
Figure 2.
DD-PCR analysis for changes in gene expression produced in hydra by the six LWamide peptides. The peptide treatment was carried out for 4 h in the presence of 2% DMSO. Controls were treated with 2% DMSO for 4 h. Arrowheads indicate bands present at higher intensities in lanes for peptide-treated animals than in lanes for control animals. The primers used for reverse transcription–PCR were 5′-d(T12AG)-3′ and 5′-d(GACCGCTTGT)-3′.
Similar tests were also carried out using eight different primer combinations for reverse transcription–PCR amplification. Specific band changes common to all of the six LWamide peptides were produced by 3 combinations, whereas the remaining five did not produce any detectable band changes for any peptides. These observations suggest that all members of the LWamide family produce the same gene expression changes in the treated animals and, therefore, suggest that they perform the same function(s) in hydra. A similar assay was also carried out for the four members of the PW family using seven primer combinations. Two produced common band changes for all members, but the remaining five did not (data not shown), suggesting that all members within the PW family also perform similar function(s). The band changes stimulated by the PW peptides did not overlap with those caused by the LW peptides, indicating that the two families affect different biological activities in hydra.
Biological Activity of the LWamide Family.
Three different assays indicate that the members of this family have two different functions. One involves metamorphosis, and the other involves muscle contraction.
Metamorphosis in Hydractinia planula.
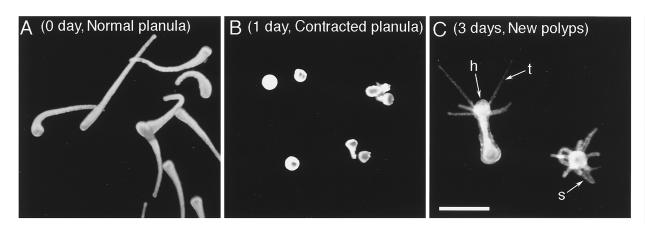
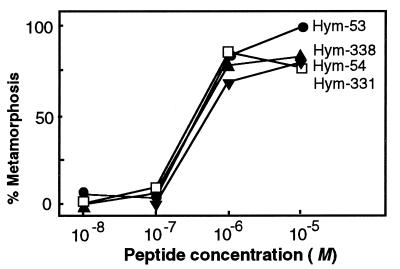
Metamorphosin A (2) and two other LWamide peptides (17) were previously found to induce metamorphosis of the planula, the larval stage, into polyps for the species Hydractinia echinata. The six LWamide peptides isolated in this study exhibited a similar activity in Hydractinia serrata. The planula of this species has a long and slender form (Fig. 3A). When treated with Hym-54, they contracted into a spherical form in 1 day (Fig. 3B) and then metamorphosed into polyps having a hypostome and tentacles at one end, and stolons at the other in 2–3 days (Fig. 3C). The dose–response curves for Hym-54 and three other members tested were very similar to one another as shown in Fig. 4. Metamorphosis was barely detectable at 10−8 and 10−7 M but occurred with higher frequency at 10−6 and 10−5 M. Although the dose–response was not examined, two other members of the family (Hym-248 and Hym-249) were also active in inducing metamorphosis.
Figure 3.
Metamorphosis in Hydractinia serrata induced by Hym-54 (10−6 M). (A) Normal planula larva. (B) Contracted planula 1 day after the beginning of treatment. (C) Metamorphosed polyps exhibiting tentacles (t), a hypostome (h), and stolons (s) at 3 days. (Bar = 100 mm.)
Figure 4.
Dose–response curves for induction of metamorphosis in Hydractinia serrata planula by four LWamide peptides.
Bud detachment in hydra.
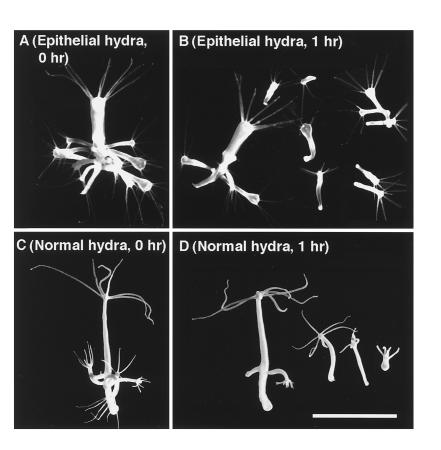
In hydra, a closely related species without a larval stage, the LWamide peptides caused a specific muscle contraction. Epithelial hydra, which are animals devoid of the interstitial cell lineage, do not undergo muscle contraction, nor do their mature buds detach (18). As shown in Fig. 5A, they accumulate a large number of buds. Treatment of that animal with Hym-54 resulted in the release of most of these buds within 1 h (Fig. 5B). A similar effect was also observed in normal hydra (Fig. 5 C and D). This effect was studied in more detail for two family members, Hym-54 and Hym-248. They induced bud detachment in normal and epithelial hydra in a concentration-dependent manner. Both were inactive at 10−7 M but caused detachment of 1/4 to 1/2 of the buds at 10−6 M, and about 3/4 at 10−5 M (data not shown). In this assay, some buds were observed to detach within a few minutes after the start of treatment. This relatively rapid effect suggests that these two peptides may stimulate the muscle processes whose contraction is required for bud detachment.
Figure 5.
Bud detachment induced by Hym-54. (A) A parental polyp of epithelial hydra carrying 11 mature buds. (B) The same polyp after 1 h of treatment with Hym-54 (10−5 M). Only three mature buds remain attached to the parent. (C) A parental polyp of normal hydra. (D) The same polyp 1 h after treatment with Hym-54 (10−5 M). (Bar = 1 mm.)
Muscle contraction in a sea anemone.
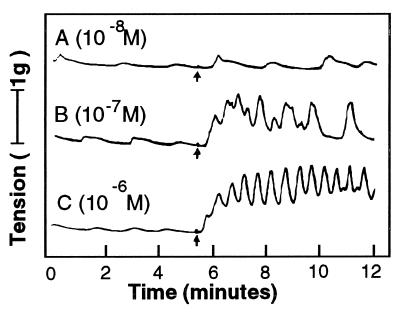
The effect of the LWamide family on muscle contraction was more directly observed in a second organism. When applied to the longitudinal parietal muscle isolated from the sea anemone A. fuscoviridis Hym-54 produced increases in rhythmic tension in a concentration-dependent manner starting at 10−8 M (Fig. 6). Similar results were also obtained with all the other LWamide family members, including metamorphosin A (data not shown).
Figure 6.
Typical examples of rhythmic tension increases produced by treatment with Hym-54 in the longitudinal parietal muscle of the sea anemone A. fuscoviridis. Arrows indicate the time of peptide addition.
Biological Activity of the PW Family.
As described above for the LWamide family, this family also exhibited different types of effects on hydra polyps and Hydractinia planula.
Inhibition of nerve cell differentiation.
To examine the effect of a peptide on cell division and differentiation, animals were treated with the peptide and pulse-labeled with BrdUrd, and labeling indices were measured 2 days later (10, 11). As summarized in Table 2, Hym-33H had no effect on the rates of cell division of epithelial cells of the two tissue layers, nor on interstitial cells, which give rise to several classes of differentiation products. Similarly, it had no effect on the dividing nematoblasts, which are early differentiation intermediates in the nematocyte pathways. In contrast, Hym-33H at concentrations of 10−6 M and 10−5 M caused a significant reduction in nerve cell differentiation. These decreases took place throughout the animal as similar reductions were observed in the upper, middle, and lower body regions (data not shown). Another family member, Hym-310, caused similar effects (data not shown). These results indicate that both peptides interfere with nerve cell differentiaton.
Table 2.
Effect of Hym-33H on nerve cell differentiation
| Labeling index*
| |||||
|---|---|---|---|---|---|
| Peptide | Concentration, M | Epithelial cells | Interstitial cells | Nematoblasts | Nerve cells |
| Hym-33H | 10−7 | 0.20 ± 0.01 | 0.50 ± 0.01 | 0.61 ± 0.04 | 0.17 ± 0.04 |
| 10−6 | 0.21 ± 0.01 | 0.51 ± 0.03 | 0.72 ± 0.07 | 0.13 ± 0.01† | |
| 10−5 | 0.20 ± 0.02 | 0.49 ± 0.01 | 0.68 ± 0.04 | 0.11 ± 0.01† | |
| Control | 0.21 ± 0.02 | 0.49 ± 0.02 | 0.65 ± 0.02 | 0.22 ± 0.03 | |
Values are means ± SD.
Statistically significantly different from the control at the 95% level.
Contraction of Hydractinia planula.
Hym-33H and Hym-37 were found to have strong effects on Hydractinia serrata planula, but in a very different manner from the LWamide peptides described above. They induced a rapid and strong contraction of planula tissue within 1 h after treatment began (instead of 1 day as for LWamide peptides) at 10−6 or 10−5 M (data not shown). Within 20 h, however, the effect was reversed, and the planula returned to the normal, or nearly normal, extended form even in the presence of the peptide. No metamorphosis occurred in the presence or absence of the peptide in the following days.
DISCUSSION
Peptide Signal Molecules in Hydra.
Evidence obtained in this study suggests that hydra tissue contains a large number of peptides that affect gene expression, and thus, are putative signaling molecules. The majority (218 of 286) of the peptides isolated in this study were derived from groups 8 and 9, which account for approximately 17% of the total area of the peptide elution profile in Fig. 1A. These numbers suggest that about 1300 peptides are present in the entire peptide extract. Of these, about 20% (≈250-300) will be breakdown products of other proteins. DD-PCR analysis showed that 32% (35 of 108) of the peptides examined were active in altering gene expression in hydra, suggesting that there may be ≈350 putative signal peptides in hydra tissue.
However, this is a conservative estimate since only a few primer combinations (≈2–6) were used for the routine DD-PCR analysis. Therefore, only a small fraction of all the transcripts were used to examine the effect of the peptide on gene expression. Had more transcripts been screened by using more primer pairs, most likely a higher fraction of the peptides would have been shown to affect gene expression. In addition, this study examined only relatively abundant peptides isolated in the range of 1–10 nmol, and hence, other peptides that are less abundant may have been missed. Therefore, hydra may have significantly more peptide signal molecules than the present estimate of 350.
The LWamide Family.
The six members of the LWamide family contain seven or eight amino acid residues, and are characterized by a Gly-Leu-Trp-NH2 sequence at the C terminus. Peptides of this size often arise as part of the proteolytic processing of a precursor protein. That also appears to be the case for the LWamide family. Genes encoding a precursor protein containing amino acid sequences corresponding to peptides of this family have been isolated from hydra (I. Leviev, M. Williamson, and C.J.P. Grimmelikhvijzen, personal communication) as well as from two other cnidarian species (17, 19). Leviev and coworkers have shown that the six LWamide peptides described here are among the eight members of that family encoded by the gene in hydra (I. Leviev, M. Williamson, and C.J.P. Grimmelikhvijzen, personal communication.
The six members of this family produced identical changes in gene expression in hydra (Fig. 2), suggesting that they perform the same functions in hydra. Evidence obtained suggests that this family plays two different roles, depending on the developmental stage or the species examined. In one role, the peptides affect morphogenesis. In Hydractinia, the six peptides examined here as well as metamorphosin A and two other LWamide peptides examined previously (2, 17) all induce the larval stage, the planula, to metamorphose into a polyp.
Although hydra develop directly from embryos into the adult form of polyps without a larval stage, there may be similarities in the development of both hydra and Hydractinia. After gastrulation, the hydra embryo has an organization resembling a Hydractinia planula: an outer ectodermal epithelial layer surrounding an interior mass of poorly organized endodermal cells and a small numbers of interstitial cells (20, 21). The embryo remains in this state for many days. Then, without obvious external stimuli, a series of changes occur which rapidly transform the embryo into an adult polyp form, including conversion of the unorganized interior cell mass into the endoderm, differentiation of nerve cells and nematocytes from the interstitial cells, and the formation of the head and foot (21). This final stage of embryonic development in hydra, which appears to resemble metamorphosis in Hydractinia, could be controlled by members of the LWamide family.
The other role of the LWamide family is stimulation of muscle contraction. All members of the family produce rhythmic tension increases in an isolated longitudinal muscle preparation in sea anemone (Fig. 6). The dramatic effect of two peptides (Hym-54 and Hym-248) on bud detachment in epithelial hydra (Fig. 5) is apparently also due to stimulation of muscle contraction. A unique type of muscle process, termed the “sphincter muscle,” is formed in the basal disk during a late stage of bud development and is thought to play an important role in bud detachment (R. Campbell, personal communication). The rapid detachment of mature buds induced by the LWamide peptides in epithelial hydra suggests that these peptides normally stimulate the rhythmic contraction of the sphincter muscle during bud detachment. These peptides, which are normally localized in nerve cells (3), are likely to be absent in epithelial hydra, which lack nerve cells. Thus, parental epithelial hydra often carry a large number of buds (Fig. 5A), which rapidly detach upon the peptide treatment (Fig. 5B).
Thus, the LWamide peptides appear to function as neurotransmitters, or neuromodulators, to directly induce contraction of specific muscle types in adult animals in hydra and sea anemone. Whether or not they also have this function in adult polyps of Hydractinia remains to be examined.
The PW Family.
Each of the four members of this family consists of five to eight amino acid residues, with Leu (or Ile)-Pro-Trp at the C terminus. The four members produced identical changes in gene expression in DD-PCR analysis, suggesting that they perform the same, or similar, functions in vivo. As with the LWamide family, the PW family may also play two different roles, depending on the developmental stage or species examined.
In Hydractinia larva, Hym-33H induced a rapid tissue contraction, although unlike the LWamide peptides, it did not induce subsequent metamorphosis. This observation suggests that the PW family plays an as yet unidentified role in these larvae.
In adult hydra polyps, Hym-33H exhibited a strong inhibitory effect on the differentiation of nerve cells from interstitial cells. Treatment with Hym-33H for 2 days reduced the number of newly formed nerve cells by about half throughout the animal. The peptide could act by blocking commitment of stem cells among the interstitial cells to nerve cell differentiation, or by interfering with the traverse of the nerve cell differentiation pathway. However, long-term treatment does not reduce the number of nerve cells in an adult polyp. These two apparently contradictory effects can be explained in the context of the tissue dynamics of the adult hydra. As part of these dynamics, the nerve cell populations are in a steady state of production and loss of nerve cells (22). Such a steady state is most likely governed by homeostatic mechanisms, which would include both inhibitors and stimulators of nerve cell differentiation. Hym-33H and other PW peptides could act as inhibitors in such mechanisms.
A point that remains unclear in these studies is the effective concentration of a peptide in either the LW or PW peptide families that elicits the observed effects on gene expression and biological activity. Because the peptides are applied outside the animals and most likely enter the tissue through the septate junctions that connect the epithelial cells of a layer, the actual concentration in the interstitial spaces in the two layers is unknown. More than likely, it is lower than the concentration in the external medium. In addition, the rate of breakdown that could also result in a lower concentration of the introduced peptide is unknown.
Conclusion.
The approach developed in this study provides a powerful means for isolating and characterizing peptide signal molecules of hydra. Further, the effects of a peptide on the expression of genes such as Cnox-2, KS1, and Cnash will provide more refined assays for examining its role in patterning processes or position-dependent differentiation (23–25). Novel peptides isolated in this project are likely to provide a new and effective means for exploring the mechanisms underlying developmental processes in cnidaria, and most likely in other animals as well.
Acknowledgments
We thank Mr. A. Takada (Hokkaido University) for the gift of Hydractinia serrata, and Drs. T. Takao and Y. Shimonishi (Osaka University) for mass spectrometric analyses. This work was supported in part by grants from the Ministry of Science, Education and Culture of Japan to O.K. (Grant 07454230), T.F. (Grant 06640803), and T.S. (Grants 06454689 and 072832210); the Sumitomo Foundation to T.S.; the Suntory Institute for Bioorganic Research to Y.M; Deutsche Forschungsgemeinschaft to C.N.D. and T.C.G.B. (Grant 446JAP-113/41/0); and the National Institutes of Health to H.R.B. (Grants HD 08086 and HD 27173).
Footnotes
Abbreviations: DD, differential display; DMSO, dimethyl sulfoxide.
References
- 1.Pesce M, Canipari R, Ferri G, Siracusa G, De Felici M. Development (Cambridge, UK) 1996;122:215–221. doi: 10.1242/dev.122.1.215. [DOI] [PubMed] [Google Scholar]
- 2.Leitz T, Morand K, Mann M. Dev Biol. 1994;163:440–446. doi: 10.1006/dbio.1994.1160. [DOI] [PubMed] [Google Scholar]
- 3.Leitz T, Lay M. Roux’s Arch Dev Biol. 1995;204:276–279. doi: 10.1007/BF00208495. [DOI] [PubMed] [Google Scholar]
- 4.Wall N A, Hogan B L. Curr Opin Genet Dev. 1994;4(4):517–522. doi: 10.1016/0959-437x(94)90066-c. [DOI] [PubMed] [Google Scholar]
- 5.Ingham P W. Curr Opin Genet Dev. 1995;5(4):492–498. doi: 10.1016/0959-437x(95)90054-k. [DOI] [PubMed] [Google Scholar]
- 6.Artavanis-Tsakonas S, Matsuno K, Fortini M. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- 7.Liang P, Pardee A B. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 8.Fraser S E, Green C R, Bode H R, Gilula N B. Science. 1987;237:49–55. doi: 10.1126/science.3037697. [DOI] [PubMed] [Google Scholar]
- 9.Zhang X, Sarras M P. Development (Cambridge, UK) 1994;120:425–432. doi: 10.1242/dev.120.2.425. [DOI] [PubMed] [Google Scholar]
- 10.Plickert G, Kroiher M. Development (Cambridge, UK) 1988;103:791–794. doi: 10.1242/dev.103.4.791. [DOI] [PubMed] [Google Scholar]
- 11.Bode H R, Gee L W, Chow M A. Dev Biol. 1990;139:321–243. doi: 10.1016/0012-1606(90)90292-q. [DOI] [PubMed] [Google Scholar]
- 12.McFarlane I D, Anderson P A V, Grimmelikhuijzen C J P. J Exp Biol. 1991;156:419–431. doi: 10.1242/jeb.156.1.419. [DOI] [PubMed] [Google Scholar]
- 13.Carstensen K, Rinehart K L, McFarlane I D, Grimmelikhuijzen C J P. Peptides (Tarrytown, NY) 1992;13:851–857. doi: 10.1016/0196-9781(92)90040-a. [DOI] [PubMed] [Google Scholar]
- 14.Hoffmeister S A H. Development (Cambridge, UK) 1996;122:1941–1948. doi: 10.1242/dev.122.6.1941. [DOI] [PubMed] [Google Scholar]
- 15.Ikeda T, Kubota I, Miki W, Nose T, Takao T, Shimonishi Y, Muneoka Y. In: Peptide Chemistry. Yanaihara N, editor. Leiden, The Netherlands: ESCOM; 1993. pp. 583–585. [Google Scholar]
- 16.Schaller H C, Bodenmüller H. Proc Natl Acad Sci USA. 1981;78:7000–7004. doi: 10.1073/pnas.78.11.7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gajewski M, Leitz T, Schlossherr J, Plickert G. Roux’s Arch Dev Biol. 1996;205:232–242. doi: 10.1007/BF00365801. [DOI] [PubMed] [Google Scholar]
- 18.Campbell R D. J Cell Sci. 1976;21:1–13. doi: 10.1242/jcs.21.1.1. [DOI] [PubMed] [Google Scholar]
- 19.Leviev I, Grimmelikhuijzen C J P. Proc Natl Acad Sci USA. 1995;92:11647–11651. doi: 10.1073/pnas.92.25.11647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van de Vyver G. In: Developmental and Cellular Biology of Coelenterates. Tardent P, Tardent R, editors. New York: Elsevier/North–Holland; 1980. pp. 109–120. [Google Scholar]
- 21.Tardent P. In: Morphogenese der Tiere. Seidel F, editor. Jena, Germany: Fischer; 1978. pp. 71–391. [Google Scholar]
- 22.Bode H R. Trends Genet. 1992;8:279–284. doi: 10.1016/0168-9525(92)90254-2. [DOI] [PubMed] [Google Scholar]
- 23.Shenk M A, Bode H R, Steele R E. Development (Cambridge, UK) 1993;117:657–667. doi: 10.1242/dev.117.2.657. [DOI] [PubMed] [Google Scholar]
- 24.Weiziger R, Salgado L M, David C N, Bosch T C G. Development (Cambridge, UK) 1994;120:2511–2517. doi: 10.1242/dev.120.9.2511. [DOI] [PubMed] [Google Scholar]
- 25.Grens A, Mason E, Marsh J L, Bode H R. Development (Cambridge, UK) 1995;121:4027–4035. doi: 10.1242/dev.121.12.4027. [DOI] [PubMed] [Google Scholar]