Abstract
Uncontrolled protein aggregation is a constant challenge in all compartments of living organisms. The failure of a peptide or protein to remain soluble often results in pathology. So far, more than 40 human diseases have been associated with the formation of extracellular fibrillar aggregates—known as amyloid fibrils—or structurally related intracellular deposits. It is well known that molecular chaperones and elaborate quality control mechanisms exist in the cell to counteract aggregation. However, an increasing number of reports during the past few years indicate that proteins have also evolved structural and sequence-based strategies to prevent aggregation. This review describes these strategies and the selection pressures that exist on protein sequences to combat their uncontrolled aggregation. We will describe the different types of mechanism evolved by proteins that adopt different conformational states including normally folded proteins, intrinsically disordered polypeptide chains, elastomeric systems and multimodular proteins.
Keywords: evolutionary pressure, cross-β, misfolding, negative design, natural selection
Introduction
The deposition of insoluble fibrillar protein aggregates occurs in more than 40 pathological conditions in humans (Chiti & Dobson, 2006). These fibrils, which are generally known as amyloid fibrils when their deposition occurs extracellularly, are typically 7–13 nm wide and are stabilized by an extensive β-sheet structure in which the β-strands are perpendicular to the fibril axis (Sunde & Blake, 1997). For many of these diseases, evidence exists that either the fibrils or their precursor protein oligomers have a clinically important role in pathogenesis.
Until almost ten years ago, the ability to form amyloid fibrils was thought to be a property of only a few polypeptide chains, such as those associated with disease. However in 1998, Dobson and co-workers showed that even a protein with no link to protein deposition diseases—the src homology domain of bovine phosphatidylinositol 3-kinase—has, at acidic pH, the ability to aggregate into fibrils that are structurally indistinguishable from those associated with human disease (Guijarro et al, 1998). This observation occurred almost by chance, as the investigators had intended to explore the structure and dynamics of a soluble, partly unfolded state of the protein. After that first observation, there have been many reports of the formation of amyloid-like fibrils by non-disease proteins under appropriate conditions (Stefani & Dobson, 2003; Uversky & Fink, 2004), resulting in the conclusion that amyloid fibril formation is a generic property of polypeptide chains, rather than a peculiar characteristic of a few unfortunate sequences (Dobson, 2003).
The increasing awareness that most proteins, if not all, have the potential to convert into amyloid-like fibrils has increased the number of studies aimed at elucidating the mechanisms of protein aggregation. It has also led to the search for potentially interesting new nanomolecular materials that could be exploited by industry, research and medicine (Hamada et al, 2004; Rajagopal & Schneider, 2004). However, these have not been the only consequences. If proteins have a generic ability to aggregate into amyloid-like fibrils, then it is likely that living systems have evolved mechanisms to prevent the formation of these detrimental species, so that any protein can remain functional in its native state. Molecular chaperones and quality control systems, such as the unfolded protein response, endoplasmic reticulum-associated degradation and autophagy, certainly serve this purpose (reviewed by Bukau et al, 2006; Young et al, 2004). However, several reports indicate that even proteins themselves have acquired sequence and structural adaptations that enable them not only to escape undesired general protein aggregation, but also to avoid the specific formation of amyloid-like fibrils. This review aims to detail these intrinsic strategies of protection against amyloid-like aggregation.
Folding is a primary strategy to prevent aggregation
The propensity of a protein to aggregate decreases abruptly when it is folded into a stable globular structure (Dobson, 2003; Kelly, 1998; Uversky & Fink, 2004). The existence of strong selection pressure on the conformational stability of the native state, at least for amino-acid residues not directly involved in catalysis or binding, is well established (Monsellier & Bedouelle, 2006; Sanchez et al, 2006; Xia & Levitt, 2004). In addition, for some familial forms of protein deposition disease, there is clear evidence that destabilization of the folded structure of the amyloid precursor protein is the primary mechanism through which natural mutations mediate their pathogenicity (Canet et al, 2002; Hammarstrom et al, 2002; Raffen et al, 1999). It has been shown experimentally that Top7—an α/β single-domain protein designed de novo with no evolutionary history—folds through a multiplicity of phases with stable non-native structures that co-exist at equilibrium with the native state (Watters et al, 2007). Fragments of this protein were also shown to be folded stably in isolation. This suggests that folding cooperatively into a unique globular fold through a smooth energy landscape, as is observed for natural proteins, is not inherent to the chemical properties of protein molecules, but instead is the result of natural selection to escape aggregation (Watters et al, 2007).
Sanchez and colleagues studied a data set of 488 point mutations within globular domains for which the effects on conformational stability had been measured in vitro (Sanchez et al, 2006). Interestingly, they showed that mutations that are predicted by specific algorithms to decrease the propensity of local regions of the sequence to form amyloid structures or to aggregate more generally, cause a larger destabilization of the folded state than the mutations that increase such a propensity. The former mutations are selected against by evolution more than the latter, as inferred from the distributions of the corresponding residues at the position of mutation in a family of homologous sequences (Sanchez et al, 2006). This shows that local regions with an intrinsically high propensity to form misfolded aggregates are generally useful for reaching a stable folded state, and for this reason might be positively selected. Folding into a compact conformation therefore seems to be the main selective pressure against misfolding, and is also an effective means by which local aggregating regions of the sequence are sequestered (Sanchez et al, 2006). These data confirm the existence of a trade-off between the requirement for sufficient stability and the necessity to avoid aggregation (Bastolla et al, 2004; Bastolla & Demetrius, 2005).
Negative design to control the assembly of folded proteins
Although folding into a stable cooperative structure is an effective way to escape aggregation, the fact remains that even fully folded proteins can maintain a significant, albeit small, propensity to aggregate. Mechanisms of aggregation have been described for some proteins in which the native or quasi-native state assembles initially—with no need to unfold—into oligomers in which the constituent protein molecules retain their native topology and only later undergo a structural reorganization to form amyloid-like protofibrils or fibrils (Bouchard et al, 2000; Pedersen et al, 2004; Plakoutsi et al, 2005; Soldi et al, 2005). For some proteins it has been reported that even mature fibrils retain a native-like conformation under some conditions (Bousset et al, 2002; Laurine et al, 2004).
By analysing the three-dimensional structures of approximately 75 proteins that are representative of the various known all-β folds, Richardson and Richardson noticed that such proteins have evolved structural adaptations to protect their peripheral β-strands—that is, the external strands of a native β-sheet (Richardson & Richardson, 2002). Peripheral β-strands have only one adjacent strand in the same sheet and can therefore interact with the peripheral β-strands of other protein molecules. They are generally protected by a combination of structural strategies, including covering them with a loop or a helix, the formation of a continuous sheet to yield a β-barrel, the use of a short and very twisted peripheral strand, and the use of other features that create considerable distortions of the β-structure, such as inward-pointing charges, prolines, β-bulges, glycine-promoted bends and twists (Richardson & Richardson, 2002). By means of these negative design strategies, a folded all-β protein can protect the few aggregation-promoting motifs that remain after folding.
Conservation of proline and glycine residues
The alignment of protein sequences from the fibronectin type III (fnIII) superfamily showed that three proline residues—Pro 5, Pro 25 and Pro 64—are highly conserved (Steward et al, 2002). When these proline residues were mutated to alanine in two representative members of the superfamily, the resulting six variants were found to have conformational stabilities and folding rates that were not significantly lower than those of the corresponding wild-type protein domains. However, double-module protein constructs containing the ninth and tenth fnIII domains of human fibronectin were shown to aggregate when one of these three proline residues was substituted by alanine in the tenth domain. By contrast, the corresponding constructs with no substitutions or with mutations involving a non-conserved proline were found to be soluble. As proline residues have structural constraints that make it difficult to adapt them into a β-sheet structure, it was concluded that the three conserved prolines have been maintained during evolution owing to their ability to inhibit aggregation (Steward et al, 2002). The three conserved prolines are located in the domain boundaries in the multimodular fnIII proteins, suggesting that this protective effect is carried out both during biosynthesis, when the domains are fully or partly unfolded, and after folding.
Similarly, it was found that several glycine residues are highly conserved in the acylphosphatase-like structural family (Parrini et al, 2005). Four glycine-to-alanine artificially constructed variants—G19A, G45A, G53A and G69A—of one representative member, human muscle acylphosphatase (AcP), were enzymatically active and had conformational stabilities similar to those of other mutants with non-conserved residues. However, they were found to aggregate more markedly than the wild-type protein under both native and denaturing conditions. Owing to their small size and lack of a β-carbon atom, glycine residues have a high level of conformational flexibility and a high entropic cost associated with their secondary structure formation. It was therefore concluded that the high level of conservation of these four glycine residues in this structural family arises from the need to have inhibitors of aggregation at strategic positions that act at both the unfolded and folded levels (Parrini et al, 2005). It will be interesting to assess whether the use of strategically positioned proline and glycine residues as ‘guardians' of protein solubility is a generic strategy used by other proteins.
The use of gatekeeper residues to control aggregation
The conservation of strategically positioned residues with a low propensity to form β-sheet structures is not the only adaptation of proteins to escape aggregation. By using an experimentally validated algorithm that is able to identify segments, within a sequence, with a high propensity to promote the formation of β-structured aggregates (TANGO), Rousseau and co-workers have surveyed the proteomes of Escherichia coli and Homo sapiens (Rousseau et al, 2006). In both organisms, it was found that the positions flanking aggregating stretches are enriched with residues such as proline, lysine, arginine, glutamate and aspartate. Prolines are β-breakers, as mentioned above. The other four residues are at the bottom of the aggregation propensity scales devised by various investigators, mainly owing to their very low hydrophobicity, charge and a β-sheet propensity that is not particularly high. A striking 90% of the 26,000 aggregating sequence segments found in the E. coli proteome have at least one of these five residues at the first position on either side of the segment.
Such residues, called ‘gatekeepers' by the authors that have described them, are not generally conserved in evolutionarily related proteins because each type of residue can exert its role at slightly different positions or on the other side of the segment, and can be replaced by one of the other four residues. Furthermore, aggregating sequences seem to be located at different positions along the sequence, even in related proteins. Therefore, the use of such residues at the flanking positions of aggregating sequences is not as restrictive as the conservation of specific residues, such as the proline and glycine residues described in the previous paragraph. This type of evolutionary pressure, other examples of which will be presented in the following paragraphs, can allow a certain freedom in the search for evolutionarily new sequences and the adaptation of old sequences to environmental changes.
Limiting β-propensity, hydrophobicity and low net charge
A pattern of alternating polar (p) and non-polar (n) residues is, in principle, highly favourable for the formation of a β-strand in an amphiphilic β-sheet, as p and n residues would all point to the solvent and hydrophobic sides, respectively. Such a pattern seems to be the most represented in protein sequence databases when the analysis is restricted to regions that adopt an exposed β-strand conformation, which confirms its ability to fit in, and provide stability to amphiphilic β-sheets (West & Hecht, 1995; Mandel-Gutfreund & Gregoret, 2002). However, a pattern of this type is also highly favourable for the formation of amyloid-like aggregates, as tested experimentally (West et al, 1999). Broome and Hecht have analysed the frequency of patterns of alternating p and n residues in more than 250,000 proteins derived from the OWL database (Broome & Hecht, 2000). The analysis showed that sequence segments containing alternating p and n residues, for example, pnpnp, are the least represented among all possible patterns with the same ratio of p and n residues, for example, pnnpp, pnppn. Such patterns are therefore generally disfavoured by evolution; those that are present are buried in cooperatively folded units, particularly as β-strands in amphiphilic β-sheets, providing further support to the concept, described above, that burying a potentially amyloidogenic sequence segment through the folding process is a primary strategy to annihilate, or markedly reduce, its aggregation potential.
Other authors have carried out similar analyses focusing on stretches of consecutive hydrophobic residues, considering either the whole sequence (Schwartz et al, 2001) or just the hydrophobic core of soluble proteins (Patki et al, 2006). In all these studies, long stretches of consecutive hydrophobic residues, typically more than five residues, were found to be represented significantly less than would be predicted if the residues were selected independently. This global under-representation indicates the existence of a negative selection pressure against such stretches, which would otherwise promote aggregation. Indeed, using a lattice-based computer simulation, it was shown that for a given number of hydrophobic residues, the propensity to aggregate increases as the hydrophobic residues became concentrated in fewer continuous sub-sequences (Istrail et al, 1999).
The aggregation propensity of an unfolded polypeptide chain is also inversely correlated with its overall net charge (Chiti & Dobson, 2006). The natural mutations that are associated with familial forms of protein deposition diseases and that decrease the net charge of the protein are much more numerous than those that increase the net charge (Chiti et al, 2002). This points to the possibility that charge is a crucial factor controlling the aggregation propensity of a polypeptide chain and that an evolutionary pressure exists to render the net charge of a protein sufficiently high. This is particularly true for intrinsically disordered proteins (see below).
By using the TANGO algorithm to analyse the proteome of E. coli, it was observed that aggregation-promoting regions do occur but that the vast majority have a low aggregation propensity (Rousseau et al, 2006). Using a related algorithm that also defines the propensity to form β-structured aggregates (Pawar et al, 2005), we studied the aggregation propensity of a peptide that encompasses the first 29 residues of horse heart apomyoglobin, apoMb1–29. This model peptide can form amyloid-like fibrils in vitro (Picotti et al, 2007). By studying the aggregation kinetics of scrambled variants of apoMb1–29—variants having the same amino-acid composition as the wild-type peptide but a reshuffled sequence—we found that the clustering of the residues with the highest propensity to aggregate within a narrow sequence segment increases the aggregation rate of this peptide (Monsellier et al, 2007). In addition, by comparing wild-type apoMb1–29 with homologous segments of globins, we showed that stretches of consecutive residues with a high propensity to aggregate are under-represented in this superfamily (Monsellier et al, 2007). This suggests that a negative selection pressure exists to maintain the aggregation propensity of aggregation-prone regions below a certain threshold.
How intrinsically disordered proteins remain soluble
If folding protects against aggregation, the question arises as to what mechanisms maintain the solubility of intrinsically disordered proteins. Uversky compared 102 natively unfolded proteins with 275 folded proteins selected from the SWISS-PROT and TrEMBL databases for having no disulphide bonds, no interactions with ligands and a length between 50 and 200 residues (Uversky, 2002). The two groups of proteins are well separated when shown in a graph of their mean net charge and mean hydrophobicity on the x and y axes, respectively. In addition to a higher net charge and lower hydrophobicity, intrinsically disordered proteins also have a lower number of aggregating sequences, as determined using the TANGO algorithm (Linding et al, 2004), and a higher proline content (Tompa, 2002). Therefore, intrinsically disordered proteins use, to a greater extent, the ‘classical' strategies used by folded proteins to avoid amyloid aggregation.
Elastomeric proteins assemble but do not form amyloids
Inhibition of aggregation is not the only necessity for proteins. Elastomeric proteins have clearly evolved to aggregate into polymeric structures, but they do not form the highly organized β-structure that is the characterizing and stabilizing element of amyloid fibrils. For example, the sequence of tropoelastin—the precursor of elastin—consists of crosslinking domains alternating with hydrophobic domains (Vrhovski & Weiss, 1998). In the relaxed state, the crosslinking domains from different tropoelastin molecules are covalently bonded, whereas the hydrophobic domains are aggregated through hydrophobic interactions. These intermolecular interactions are largely disrupted under a stretching force, whereas the crosslinking domains ensure that elastin polymers do not break apart under the same force (Rauscher et al, 2006). This aggregated but non-amyloid-like state is essential for the protein to be functional.
How can elastomeric proteins polymerize without forming stable, highly organized amyloid-like fibrils? A comparison of elastomeric proteins with those that form amyloid-like fibrils either physiologically or artificially shows that the elastomeric proteins are characterized by a higher combined content of proline and glycine residues (Rauscher et al, 2006). Elastomeric and amyloidogenic proteins are well separated when shown in a graph of their glycine and proline contents on the x and y axes, respectively. In elastomeric proteins, the relatively high hydrophobicity favours aggregation, but the high glycine and proline content prevents the conversion of the aggregates into a stable β-structure—a process that would compromise the responsiveness of the polymers under a deforming stimulus and interfere with the overall function of these proteins.
Low sequence identity in multimodular proteins
The increasing evidence that protein and peptide molecules aggregate into amyloid fibrils by forming a parallel in-register alignment of their sequence segments suggests that sequence diversity is an important tool to prevent the co-aggregation of similar proteins. In multimodular proteins, the significant sequence identity shared by the constituent domains and their high local concentration potentially favour aggregation. To understand how two homologous sequences avoid co-aggregation, Wright and colleagues studied the in vitro aggregation of several double-module constructs in which the 27th immunoglobulin domain from human cardiac titin, TI27, was covalently coupled to another domain with a sequence identity ranging from 0 to 100% (Wright et al, 2005). It was found that the efficiency of co-aggregation increases with sequence identity, and no co-aggregation was observed for domains with less than 30% sequence identity. In the same study, it was shown that in the immunoglobulin and fnIII superfamilies, only 30% and 10% of adjacent domain pairs share more than 30% and 40% sequence identity, respectively, whereas these proportions are significantly higher for pairs of non-adjacent domains (Wright et al, 2005). Therefore, the maintenance of a low sequence identity between adjacent domains is an important evolutionary pressure that aims to inhibit co-aggregation between distinct domains within multimodular proteins.
Protein solubility and expression level are correlated
As protein aggregation is a multimolecular process, its rate is highly dependent on protein concentration. Interestingly, the sequence of a protein seems to have been finely regulated in the course of evolution so that its aggregation propensity is just below the limit of solubility in vivo (Tartaglia et al, 2007). Indeed, the aggregation rates of 11 human proteins measured in vitro are inversely correlated with their expression levels in vivo, as estimated from measurements of the cellular mRNA concentrations (Tartaglia et al, 2007). This implies that solubility of a protein is precisely adapted to counteract the possible aggregation induced by high expression levels in vivo. Proteins typically expressed at high levels have a sequence with a low aggregation propensity, whereas polypeptide chains expressed in tiny amounts have a lower overall solubility (Tartaglia et al, 2007). The existence of diverse and complementary mechanisms to protect proteins from amyloid-like aggregation probably allow the fine regulation of their sequences, and maintain the delicate equilibrium between folding, aggregation and expression levels.
Conclusions
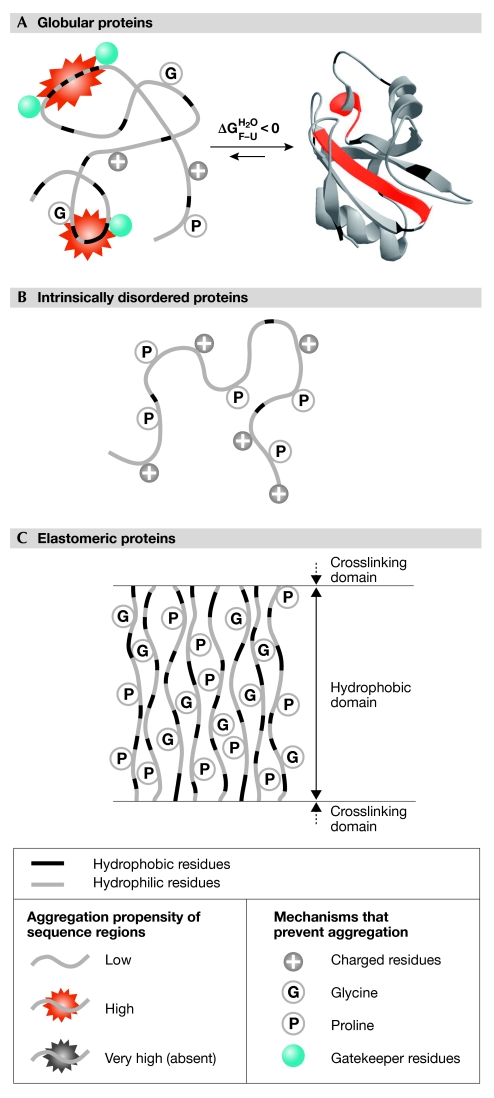
Amyloid fibrils and structurally related intracellular deposits are stabilized by the cross-β motif and extensive hydrophobic interactions, and grow when the repulsive electrostatic interactions between the constituent protein molecules are not too severe. The molecules involved contribute to the various β-strands with a limited portion of their sequence, and the resulting strands are generally parallel and in register. The precursor oligomers that are likely to cause cell dysfunction more effectively than mature fibrils, such as the spherical or chain-like protofibrils, also have many of these structural characteristics (Chiti & Dobson, 2006). It is fascinating to see how the adaptations that proteins use to escape aggregation are aimed at counteracting these structural characteristics. This review cites examples for many of these features, which are also shown schematically in Fig 1. Importantly, a protein generally uses several of these strategies at different sites along the sequence.
Figure 1.
Sequence and structural adaptations evolved by proteins to counteract amyloid aggregation. (A) Globular proteins have evolved to fold cooperatively into a compact structure, to minimize clusters of consecutive hydrophobic residues and patterns of alternating polar and non-polar residues, and to have a sufficiently high net charge and conserved glycine and proline residues (particularly in loops). Aggregation-promoting regions are present, but they do not have a dramatically high aggregation propensity and are flanked by gatekeeper residues. (B) Intrinsically disordered proteins have few aggregation-promoting regions, a high fraction of proline residues, a high net charge and a low content of hydrophobic residues. (C) Elastomeric proteins, such as elastin, aggregate in a non-amyloid form. To this end, they have evolved high overall hydrophobicity, but also a high proportion of combined glycine and proline residues. Additional adaptations (not shown) involve the protection of peripheral β-strands in all-β globular proteins, and sequence divergence in adjacent domains of multimodular proteins (see text for details). Individual proteins use more than one adaptation described here.
Elucidating such adaptations is important not only for understanding how protein sequences have evolved, but also for identifying those strategies that nature has designed to protect proteins from the uncontrolled formation of amyloid-like fibrils and their precursors. Indeed, the cellular machineries, as well as the various individual macromolecules, operating inside any living organism are very efficient. This nature-derived knowledge is extremely important as it can provide inspiration to rationally control aggregation when it is not desired, such as in pathology and biotechnology, as well as to promote it in a controlled manner when it is desired, as in the construction of new materials of biotechnological interest.

Elodie Monsellier

Fabrizio Chiti
References
- Bastolla U, Demetrius L (2005) Stability constraints and protein evolution: the role of chain length, composition and disulfide bonds. Protein Eng Des Sel 18: 405–415 [DOI] [PubMed] [Google Scholar]
- Bastolla U, Moya A, Viguera E, van Ham RC (2004) Genomic determinants of protein folding thermodynamics in prokaryotic organisms. J Mol Biol 343: 1451–1466 [DOI] [PubMed] [Google Scholar]
- Bouchard M, Zurdo J, Nettleton EJ, Dobson CM, Robinson CV (2000) Formation of insulin amyloid fibrils followed by FTIR simultaneously with CD and electron microscopy. Protein Sci 9: 1960–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousset L, Thomson NH, Radford SE, Melki R (2002) The yeast prion Ure2p retains its native α-helical conformation upon assembly into protein fibrils in vitro. EMBO J 21: 2903–2911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broome BM, Hecht MH (2000) Nature disfavors sequences of alternating polar and non-polar amino acids: implications for amyloidogenesis. J Mol Biol 296: 961–968 [DOI] [PubMed] [Google Scholar]
- Bukau B, Weissman J, Horwich A (2006) Molecular chaperones and protein quality control. Cell 125: 443–451 [DOI] [PubMed] [Google Scholar]
- Canet D, Last AM, Tito P, Sunde M, Spencer A, Archer DB, Redfield C, Robinson CV, Dobson CM (2002) Local cooperativity in the unfolding of an amyloidogenic variant of human lysozyme. Nat Struct Biol 9: 308–315 [DOI] [PubMed] [Google Scholar]
- Chiti F, Dobson CM (2006) Protein misfolding, functional amyloid and human disease. Annu Rev Biochem 75: 333–366 [DOI] [PubMed] [Google Scholar]
- Chiti F, Calamai M, Taddei N, Stefani M, Ramponi G, Dobson CM (2002) Studies of the aggregation of mutant proteins in vitro provide insights into the genetics of amyloid diseases. Proc Natl Acad Sci USA 99: 16419–16426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson CM (2003) Protein folding and misfolding. Nature 426: 884–890 [DOI] [PubMed] [Google Scholar]
- Guijarro JI, Sunde M, Jones JA, Campbell ID, Dobson CM (1998) Amyloid fibril formation by an SH3 domain. Proc Natl Acad Sci USA 95: 4224–4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada D, Yanagihara I, Tsumoto K (2004) Engineering amyloidogenicity towards the development of nanofibrillar materials. Trends Biotechnol 22: 93–97 [DOI] [PubMed] [Google Scholar]
- Hammarstrom P, Jiang X, Hurshman AR, Powers ET, Kelly JW (2002) Sequence-dependent denaturation energetics: a major determinant in amyloid disease diversity. Proc Natl Acad Sci USA 99: 16427–16432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Istrail S, Schwartz R, King J (1999) Lattice simulations of aggregation funnels for protein folding. J Comput Biol 6: 143–162 [DOI] [PubMed] [Google Scholar]
- Kelly JW (1998) The alternative conformations of amyloidogenic proteins and their multi-step assembly pathways. Curr Opin Struct Biol 8: 101–106 [DOI] [PubMed] [Google Scholar]
- Laurine E et al. (2004) Lithostathine quadruple-helical filaments form proteinase K-resistant deposits in Creutzfeldt–Jakob disease. J Biol Chem 278: 51770–51778 [DOI] [PubMed] [Google Scholar]
- Linding R, Schymkowitz J, Rousseau F, Diella F, Serrano L (2004) A comparative study of the relationship between protein structure and β-aggregation in globular and intrinsically disordered proteins. J Mol Biol 342: 345–353 [DOI] [PubMed] [Google Scholar]
- Mandel-Gutfreund Y, Gregoret LM (2002) On the significance of alternating patterns of polar and non-polar residues in β-strands. J Mol Biol 323: 453–461 [DOI] [PubMed] [Google Scholar]
- Monsellier E, Bedouelle H (2006) Improving the stability of an antibody variable fragment by a combination of knowledge-based approaches: validation and mechanisms. J Mol Biol 362: 580–593 [DOI] [PubMed] [Google Scholar]
- Monsellier E, Ramazzotti M, Polverino de Laureto P, Tartaglia GG, Taddei N, Fontana A, Vendruscolo M, Chiti F (2007) The distribution of residues in a polypeptide sequence is a determinant of aggregation optimized by evolution. Biophys J (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrini C, Taddei N, Ramazzotti M, Degl'Innocenti D, Ramponi G, Dobson CM, Chiti F (2005) Glycine residues appear to be evolutionarily conserved for their ability to inhibit aggregation. Structure 13: 1143–1151 [DOI] [PubMed] [Google Scholar]
- Patki AU, Hausrath AC, Cordes MH (2006) High polar content of long buried blocks of sequence in protein domains suggests selection against amyloidogenic non-polar sequences. J Mol Biol 362: 800–809 [DOI] [PubMed] [Google Scholar]
- Pawar AP, Dubay KF, Zurdo J, Chiti F, Vendruscolo M, Dobson CM (2005) Prediction of ‘aggregation-prone' and ‘aggregation-susceptible' regions in proteins associated with neurodegenerative diseases. J Mol Biol 350: 379–392 [DOI] [PubMed] [Google Scholar]
- Pedersen JS, Christensen G, Otzen DE (2004) Modulation of S6 fibrillation by unfolding rates and gatekeeper residues. J Mol Biol 341: 575–588 [DOI] [PubMed] [Google Scholar]
- Picotti P, De Franceschi G, Frare E, Spolaore B, Zambonin M, Chiti F, Polverino de Laureto P, Fontana A (2007) Amyloid fibril formation and disaggregation of fragment 1–29 of apomyoglobin: insights into the effect of pH on protein fibrillogenesis. J Mol Biol 347: 1237–1245 [DOI] [PubMed] [Google Scholar]
- Plakoutsi G, Bemporad F, Calamai M, Taddei N, Dobson CM, Chiti F (2005) Evidence for a mechanism of amyloid formation involving molecular reorganisation within native-like precursor aggregates. J Mol Biol 351: 910–922 [DOI] [PubMed] [Google Scholar]
- Raffen R, Dieckman LJ, Szpunar M, Wunschl C, Pokkuluri PR, Dave P, Wilkins Stevens P, Cai X, Schiffer M, Stevens FJ (1999) Physicochemical consequences of amino acid variations that contribute to fibril formation by immunoglobulin light chains. Protein Sci 8: 509–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal K, Schneider JP (2004) Self-assembling peptides and proteins for nanotechnological applications. Curr Opin Struct Biol 8: 480–486 [DOI] [PubMed] [Google Scholar]
- Rauscher S, Baud S, Miao M, Keeley FW, Pomes R (2006) Proline and glycine control protein self-organization into elastomeric or amyloid fibrils. Structure 14: 1667–1676 [DOI] [PubMed] [Google Scholar]
- Richardson JS, Richardson DC (2002) Natural β-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc Natl Acad Sci USA 99: 2754–2759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau F, Serrano L, Schymkowitz JW (2006) How evolutionary pressure against protein aggregation shaped chaperone specificity. J Mol Biol 55: 1037–1047 [DOI] [PubMed] [Google Scholar]
- Sanchez IE, Tejero J, Gomez-Moreno C, Medina M, Serrano L (2006) Point mutations in protein globular domains: contributions from function, stability and misfolding. J Mol Biol 363: 422–432 [DOI] [PubMed] [Google Scholar]
- Schwartz R, Istrail S, King J (2001) Frequencies of amino acid strings in globular protein sequences indicate suppression of blocks of consecutive hydrophobic residues. Protein Sci 10: 1023–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldi G, Bemporad F, Torrassa S, Relini A, Ramazzotti M, Taddei N, Chiti F (2005) Amyloid formation of a protein in the absence of initial unfolding and destabilization of the native state. Biophys J 89: 4234–4244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani M, Dobson CM (2003) Protein aggregation and aggregate toxicity: new insights into protein folding, misfolding diseases and biological evolution. J Mol Med 81: 678–699 [DOI] [PubMed] [Google Scholar]
- Steward A, Adhya S, Clarke J (2002) Sequence conservation in Ig-like domains: the role of highly conserved proline residues in the fibronectin type III superfamily. J Mol Biol 318: 935–940 [DOI] [PubMed] [Google Scholar]
- Sunde M, Blake C (1997) The structure of amyloid fibrils by electron microscopy and X-ray diffraction. Adv Protein Chem 50: 123–159 [DOI] [PubMed] [Google Scholar]
- Tartaglia GG, Pechmann S, Dobson CM, Vendruscolo M (2007) Life on the edge: a link between gene expression levels and aggregation rates of human proteins. Trends Biochem Sci 32: 204–206 [DOI] [PubMed] [Google Scholar]
- Tompa P (2002) Intrinsically unstructured proteins. Trends Biochem Sci 27: 527–533 [DOI] [PubMed] [Google Scholar]
- Uversky VN (2002) Natively unfolded proteins: a point where biology waits for physics. Protein Sci 11: 739–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN, Fink AL (2004) Conformational constraints for amyloid fibrillation: the importance of being unfolded. Biochim Biophys Acta 1698: 131–153 [DOI] [PubMed] [Google Scholar]
- Vrhovski B, Weiss AS (1998) Biochemistry of tropoelastin. Eur J Biochem 258: 1–18 [DOI] [PubMed] [Google Scholar]
- Watters AL, Deka P, Corrent C, Callender D, Varani G, Sosnick T, Baker D (2007) The highly cooperative folding of small naturally occuring proteins is likely the result of natural selection. Cell 128: 613–624 [DOI] [PubMed] [Google Scholar]
- West MW, Hecht MH (1995) Binary patterning of polar and nonpolar amino acids in the sequences and structures of native proteins. Protein Sci 4: 2032–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West MW, Wang W, Patterson J, Mancias JD, Beasley JR, Hecht MH (1999) De novo amyloid proteins from designed combinatorial libraries. Proc Natl Acad Sci USA 8: 11211–11216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright CF, Teichmann SA, Clarke J, Dobson CM (2005) The importance of sequence diversity in the aggregation and evolution of proteins. Nature 438: 878–881 [DOI] [PubMed] [Google Scholar]
- Xia Y, Levitt M (2004) Simulating protein evolution in sequence and structure space. Curr Opin Struct Biol 14: 202–207 [DOI] [PubMed] [Google Scholar]
- Young JC, Agashe VR, Siegers K, Hartl FU (2004) Pathways of chaperone-mediated protein folding in the cytosol. Nat Rev Mol Cell Biol 5: 781–791 [DOI] [PubMed] [Google Scholar]